Summary
We have identified a highly conserved RNA motif that occurs upstream of genes involved in S-adenosyl-l-methionine (SAM) recycling in many Gram-positive and Gram-negative species of bacteria. The phylogenetic distribution and the conserved structural features of representatives of this motif are indicative of riboswitch function. Riboswitches are widespread metabolite-sensing gene control elements that are typically found in the 5′ untranslated regions (UTRs) of bacterial mRNAs. We experimentally verified that examples of this RNA motif specifically recognize S-adenosylhomocysteine (SAH) in protein-free in vitro assays, and confirmed that these RNAs strongly discriminate against SAM and other closely related analogs. A representative SAH motif was found to activate expression of a downstream gene in vivo when the metabolite is bound. These observations confirm that SAH motif RNAs are distinct ligand-binding aptamers for a riboswitch class that selectively binds SAH and controls genes essential for recycling expended SAM coenzymes.
Introduction
RNA aptamers are highly structured polynucleotides that form selective binding pockets for specific ligands (Gold et al., 1995; Osborne and Ellington, 1997). Engineered aptamers can be made to exhibit high affinity and selectivity for their targets, and can have considerable potential for applications in basic research, biotechnology and therapeutics (Breaker, 2004; Bunka and Stockley, 2006). RNA aptamers also can exist naturally in the 5′ UTRs of many bacterial messenger RNAs, where they serve as the sensor domain of gene control elements called riboswitches (Mandal and Breaker, 2004a; Soukup and Soukup, 2004; Winkler and Breaker, 2005). Like their engineered counterparts, natural aptamers can bind their targets with high affinity and specificity, which is required for RNA aptamers that are used by cells to precisely regulate the expression of critical metabolic genes. However, natural aptamers are typically much larger (ranging between 34 and 200 nucleotides) than engineered aptamers (Breaker, 2006), which presumably allows natural aptamers to exhibit the high level of performance required to compete with gene control factors made of protein.
Each riboswitch usually harnesses metabolite binding by its aptamer domain to bring about structural changes in an adjoining expression platform. These structural changes establish the level of protein production from the open reading frame (ORF) or from multiple ORFs located downstream. However, folding changes are not always brought about by metabolite binding. For example, one riboswitch class uses the target metabolite as a coenzyme for a self-cleaving ribozyme whose action represses expression of the downstream ORF (Winkler et al., 2004; Cochrane et al., 2007). Regardless of the mechanism used, riboswitches often control genes whose protein products are involved in the synthesis, degradation, or transport of the metabolite or precursors of the metabolite whose cellular concentration is sensed by the aptamer domain.
Riboswitch aptamers must form precision binding pockets for their target metabolites using only the four common nucleotides, unless modified bases or supporting protein factors are involved. This places strong selective pressure on each aptamer to maintain certain structural characteristics throughout evolution, although some ligands such as SAM (Corbino et al., 2005; Fuchs et al., 2006) and preQ1 (Weinberg et al., 2007) are detected by multiple riboswitch classes with entirely different aptamer structures. The representatives of riboswitch aptamer classes identified from even distantly related organisms exhibit considerable conservation in sequence and secondary structure. This conservation, coupled with the identity of the ligand bound by the aptamer, forms the basis for the organization of riboswitches into distinct classes.
The conserved features of riboswitch aptamers have been exploited by computer-aided search algorithms to expand the number of representatives of known riboswitch classes (e.g.Rodionov et al., 2002; Rodionov et al., 2003a; Nahvi et al., 2004; Rodionov et al., 2004;) and to discover previously unknown riboswitch classes (e.g.Rodionov et al., 2003b; Barrick et al., 2004; Corbino et al., 2005; Weinberg et al., 2007). In contrast, expression platforms vary widely in sequence and structure, even within the same riboswitch class. For example, TPP riboswitches integrate a consensus aptamer with various expression platform architectures to control transcription termination and translation initiation in bacteria (Winkler et al., 2002; Mironov et al., 2002; Rentmeister et al., 2007), and RNA splicing in eukaryotes (Sudarsan et al., 2003a; Kubodera et al., 2003; Bocobza et al., 2007; Cheah et al., 2007; Wachter et al., 2007).
As noted above, riboswitches with aptamers from different structural classes can recognize the same target metabolite. Four riboswitch classes have been discovered that specifically recognize the coenzyme SAM (or AdoMet), and these have been termed SAM-I (Epshtein et al., 2003; McDaniel et al., 2003; Winkler et al., 2003), SAM-II (Corbino et al., 2005, Lim et al., 2006b), SAM-III or SMK (Fuchs et al., 2006) and SAM-IV (Weinberg et al., 2007; unpublished data). Among these classes, SAM-I riboswitches have the most widespread phylogenetic distribution known, with examples found in many bacterial groups (Rodionov et al., 2004). SAM-II riboswitches are largely found in Gram-negative proteobacteria and bacteroidetes (Corbino et al., 2005), while SAM-III riboswitches are reported only in Gram-positive lactic acid bacteria (Fuchs et al., 2006). The aptamers of all four classes preferentially bind SAM over SAH (or AdoHcy), which is a byproduct of SAM-dependent methyl group transfer (Takusagawa et al., 1998). For example, representatives of the SAM-I and SAM-II aptamers discriminate ∼80-fold and more than 1000-fold against SAH, respectively (Lim et al., 2006b). This level of discrimination is striking considering the compounds differ by only a single methyl group.
In the current study, we describe the discovery of a bacterial RNA element with highly conserved primary sequence and secondary structure that is always found upstream of genes responsible for converting SAH to l-methionine. The RNA element represents a distinct class of aptamers that selectively binds SAH and discriminates at least 1000-fold against SAM, which is in stark contrast to the molecular recognition characteristics of the four known SAM aptamers. In the ahcY mRNA of the bacterium Pseudomonas syringae, the structural changes brought about by metabolite binding to the SAH aptamer activates expression of the downstream gene in vivo, thus demonstrating that the SAH aptamer forms the sensor component of a distinct class of SAH-responsive riboswitches.
Results
Identification of a Conserved RNA Motif Associated with SAH Recycling
In an effort to identify additional riboswitch classes and other structured regulatory RNA motifs, systematic searches of the intergenic regions (IGRs) of gene families in numerous bacterial classes were carried out (Weinberg et al., 2007) using the CMfinder comparative genomics pipeline (Yao et al., 2007). Numerous RNA motifs were identified in bacteria using this search strategy, and several have the characteristics expected for RNAs that function as metabolite-sensing riboswitches.
One candidate riboswitch class was termed the SAH motif because the genes most commonly associated with representatives of this RNA are S-adenosylhomocysteine hydrolase (ahcY, COG0499), cobalamin-dependent methionine synthase (metH, COG1410) and methylenetetrahydrofolate reductase (metF, COG0685) (Figure 1). The genomic contexts of SAH motif representatives, and their high degree of conservation, are consistent with a role in sensing SAH and activating the expression of genes whose products are required for SAH recycling to resynthesize SAM. The SAH byproduct of SAM-dependent methylation reactions (Takusagawa et al., 1998) binds a number of SAM-dependent methyltransferases with similar or even better affinities compared to SAM, and therefore functions a potent inhibitor of these enzymes (Ueland, 1982). It is likely that, during times of high methytransferase activity, cells could accumulate toxic levels of SAH unless the compound is converted into less toxic products. Thus, rapid sensing of SAH and activation of genes for recycling of SAH to reform SAM would be advantageous.
Figure 1. Typical SAM Metabolic Cycle in Eubacteria.
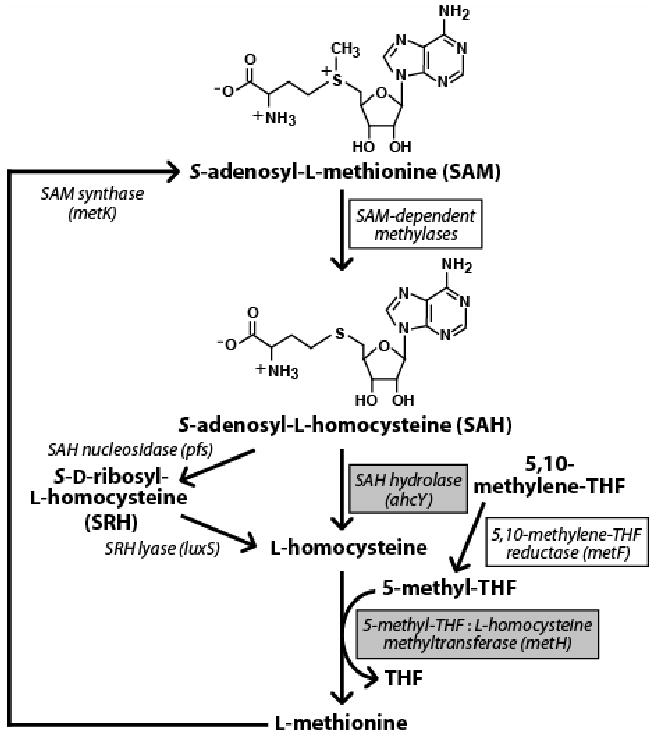
Names of genes from Escherichia coli (strain K-12) or Pseudomonas syringae are given in parentheses following each enzyme. Shaded boxes identify ORFs that are located immediately downstream of an SAH element, while open boxes identify ORFs that sometimes reside downstream of (and possibly in the same operon as) the SAH hydrolase gene ahcY when an SAH element is present. THF is tetrahydrofolate.
There are two major routes in eubacteria for SAH catabolism that both convert SAH to homocysteine (Figure 1). In many bacterial species, SAH nucleosidase (EC 3.2.2.9) catalyzes cleavage of the N-glycosidic bond of SAH to produce adenine and S-ribosylhomocysteine (Della Ragione et al., 1985). The latter compound can be subsequently hydrolyzed by the gene product of luxS to form homocysteine (Schauder et al., 2001). The alternative route involves the ahcY gene that is commonly associated with SAH motif representatives. SAH hydrolase, the product of the ahcY gene, catalyzes the breakdown of the thioether bond of SAH to produce adenosine and homocysteine (Shimizu et al., 1984). The gene product of metH subsequently methylates homocysteine to l-methionine using 5-methyltetrahydrofolic acid (5-methylTHF) as the methyl donor (Old et al., 1988), which is generated by the gene product of metF from 5,10-methylTHF (Sheppard, 1999). SAM synthase, encoded by the metK gene, regenerates SAM from methionine and ATP to complete the cycle (Tabor and Tabor, 1984). It is interesting to note that the metK gene resides in close proximity upstream of the SAH RNA motif in some species of β-proteobacteria and γ-proteobacteria, and might be in the same operon. No homologs of the luxS gene product (COG1854) were identified in genomes where the conserved RNA element was found, so it is possible that in these organisms the catabolism of SAH proceeds exclusively by the route involving SAH hydrolase.
In total, 95 occurrences of the SAH RNA motif conforming to a revised structural model were found, and 68 of these matches are non-duplicative (see Figure S1 and text in the Supplemental Data available with this article online). All examples of the SAH RNA motif identified occur adjacent to SAH recycling genes (Figure S2) except for those identified from environmental sequences or from incompletely-sequenced genomes where the gene identities adjacent to the RNA motif were not available. In most instances, SAH RNA representatives are located upstream of what appear to be operons that, almost without exception, code for SAH hydrolase (ahcY), methylenetetrahydrofolate reductase (metF) and occasionally methionine synthase (metH). In some species of Burkhoderia, Dechloromonas, and Ralstonia, the operon includes a predicted membrane protein (COG1950) of unknown function. Other genes appearing rarely in SAH RNA-associated operons include those that encode an rRNA methylase (COG1189), a regulator of competence-specific genes (tfoX, COG3070) and an enzyme involved in guanosine polyphosphate metabolism (spoT, COG0317).
A consensus sequence and structural model of the RNA motif (Figure 2A) was constructed based on the 68 non-duplicative sequences in the alignment (see Experimental Procedures for details). The consensus RNA motif is composed of a central core of highly-conserved nucleotides flanked by base-paired regions designated P1 and P2, wherein P2 is capped by a variable-sequence loop (L2). The RNA also forms a pseudoknot (P4) between the central core and highly-conserved nucleotides at the 3′ terminus of the RNA. A hairpin (P3 and L3) occurs in some representatives between the P1 and P4 stems. The proposed secondary structure elements are all supported by covariation.
Figure 2. The SAH Element is a Highly Conserved RNA Motif Found in Many Gram-positive and Gram-negative Bacteria.
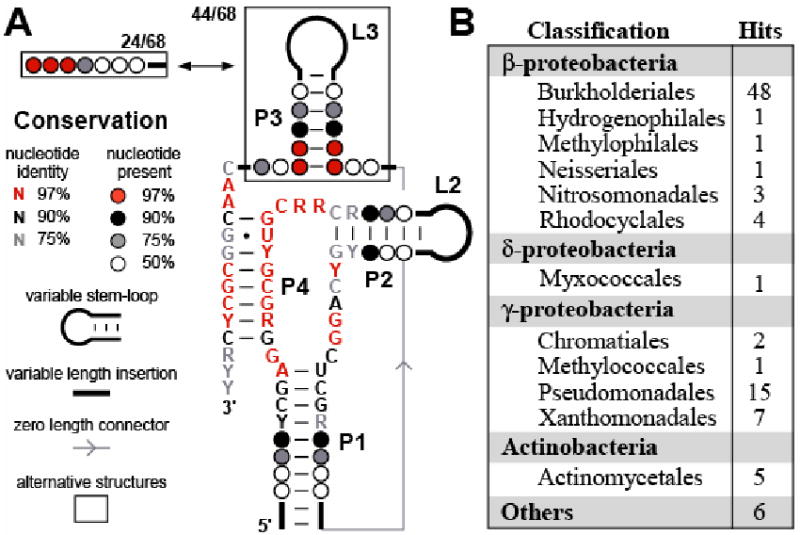
(A) Consensus nucleotide sequence and secondary structure model of the conserved RNA element located in the 5′ UTRs of genes for SAH recycling. The consensus sequence and structural model were determined by examining 68 representatives of the motif (Figure S1) excluding duplicates identified from near identical genomic DNAs (see Experimental Procedures). P1 through P4 identify predicted base-paired regions, and the numbers of non-duplicative representatives that carry or exclude the optional P3 stem are indicated.
(B) Phylogenetic distribution of SAH elements identified by bioinformatics searching (Experimental Procedures) and verified by genomic context, including duplicative hits of the same sequence in closely related bacterial strains. “Others” category refers to hits found in environmental sequences.
In addition to β-proteobacteria and γ-proteobacteria, examples of RNAs matching this consensus sequence and structure were also found in a species of δ-proteobacteria and in some Gram-positive Actinobacteria (Figure 2B). Most organisms have one occurrence of the RNA element, except for Acidovorax sp., Dechloromonas aromatica, Azoarcus sp., Polaromonas naphthalenivorans, Polaromonas sp., Rhodoferax ferrireducens, and Rubrivivax gelatinosus, where two incidents were identified in front of both the predicted ahcY and metH coding sequences that are located in different regions of the genome.
Selective binding of SAH by the 68 metH RNA
The structural features and genomic arrangements of SAH RNAs strongly suggest that they serve as aptamers for riboswitches that sense SAH. We therefore set out to experimentally determine whether examples of the RNA motif could specifically recognize metabolites in a protein-free in vitro system and function as a gene control element, which are two defining characteristics of riboswitches. We chose to examine a 68-nucleotide RNA encompassing the SAH RNA identified upstream of the metH gene in Dechloromonas aromatica (designated 68 metH, Figure 3A). The D. aromatica 68 metH RNA is nearly the shortest sequence that closely corresponds to the consensus sequence and structure for this motif. Its reduced length is due in part to the absence of the optional P3 stem (Figure 2A).
Figure 3. The SAH Element from the metH 5′-UTR of Dechloromonas aromatica Undergoes Structural Changes When Selectively Binding SAH.
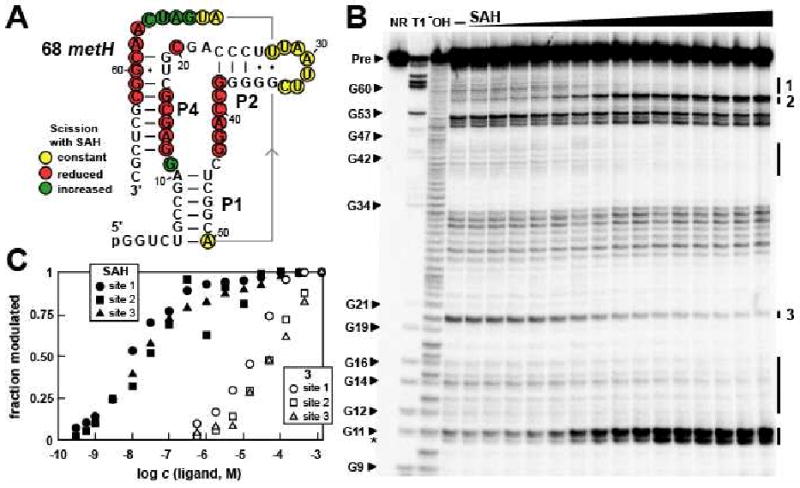
(A) Sequence and secondary structure model of the 68 metH RNA, which contains the conserved SAH aptamer. Sites of spontaneous RNA cleavage during in-line probing of 5′ 32P-labeled RNAs were identified from the data presented in B. Two guanosyl residues not present in the natural RNAs were added to the 5′ end of the construct to facilitate in vitro transcription by T7 RNA polymerase. Nucleotides in regions 1 through 8 and 63 through 68 could not be visualized on the gel depicted in B, and therefore strand scission was not quantitated for these residues (see Experimental Procedures).
(B) Spontaneous cleavage patterns resulting from in-line probing of the 68 metH RNA in the absence (–) and presence of SAH at concentrations ranging from 100 pM to 300 μM. NR, T1, and −OH lanes identify precursor RNAs without treatment, partially digested with RNase T1, or partially digested at elevated pH, respectively. Some bands generated by RNase T1 digestion (cleavage after G residues) are identified (arrowheads), and Pre identifies the uncleaved precursor RNA bands. Regions of modulation of spontaneous RNA cleavage are identified with vertical bars, and sites that were quantitated to estimate binding affinity are labeled 1 through 3. The asterisk identifies a product band that corresponds to cleavage after G11, but where the 2′, 3′-cyclic phosphate intermediate has been hydrolyzed to yield a mixture of 2′- and 3′-phosphate products that exhibit slightly faster mobility during PAGE.
(C) Plot depicts the dependence of the 68 metH RNA spontaneous cleavage at the sites designated in B on the concentration (c) of SAH and compound 3. Apparent KD values are estimated for each compound by using a best fit curve for the collective data and assuming a two-state binding model. The concentration of ligand needed to induce half-maximal structural modulation reflects the apparent KD.
An in-line probing assay (Soukup and Breaker, 1999) was used to corroborate the predicted secondary structure (Figure 3A), and to establish that 68 metH RNA directly binds SAH. This assay exploits the fact that spontaneous cleavage of RNA occurs faster at internucleotide linkages that are unstructured, and therefore can reveal structural features of RNAs and the changes they undergo when binding ligands. 5′ 32P-labeled 68 metH RNA was incubated in reactions containing progressively higher concentrations of SAH (see Experimental Procedures for details) and the resulting spontaneous cleavage products were separated by polyacrylamide gel electrophoresis (Figure 3B). In the absence of any ligand, higher band intensities were observed in regions of the RNA predicted to form L2, the internal loop separating P1 and P2, and the 3′ terminal portion of the RNA. These observations suggest that P1 and P2 are formed, but that the vast majority of the highly conserved nucleotides in the core of the RNA and in the 3′ tail (including the nucleotides of the P4 pseudoknot) are less structured in the absence of ligand.
In the presence of increasing concentrations of SAH, progressive changes in band intensities were observed at several locations throughout the RNA, confirming that SAH causes changes in RNA folding, as expected if the RNA serves as an aptamer for this metabolite. Moreover, the structural changes occur mostly in the phylogenetically conserved regions (Figure 2A, Figure 3A), as is typically observed for many other riboswitch aptamers. The most noteworthy changes in response to SAH binding are the stabilization of pseudoknot P4, and reduction of flexibility in other conserved nucleotides of the core. This suggests that the highly-conserved nucleotides are instrumental in forming the binding pocket for SAH.
The apparent dissociation constant KD of 68 metH RNA for SAH was estimated to be ∼20 nM by quantifying the extent of spontaneous cleavage at several nucleotide positions over a range of SAH concentrations (Figure 3B, C). This value for apparent KD is typical of riboswitch aptamers, which have been found to range from ∼200 μM (Winkler et al., 2004) to ∼200 pM (Welz and Breaker, 2007; Sudarsan et al., 2006). Similar SAH-mediated shape changes revealed by in-line probing assays were obtained with a 122-nucleotide SAH aptamer construct derived from the ahcY gene of P. syringae. Although 122 ahcY RNA carries the optional P3 stem (Figure S3A), the RNA undergoes key structural changes in the core of the aptamer on ligand binding (Figure S3B), and the apparent KD value is similar to that measured for 68 metH RNA.
It is important to note that the KD values determined for riboswitch aptamers are useful for comparing the binding affinities of various ligands to establish molecular recognition properties, but the absolute KD values might be different in cells. Furthermore, even if the KD values of riboswitch aptamers measured in vitro matched those inside cells, it is not assured that they will be tuned to the concentrations of metabolites. Recent findings (Wickiser et al., 2005a; 2005b; Gilbert et al, 2006) have revealed that some riboswitches are kinetically rather than thermodynamically driven. In other words, sometimes it is not the KD of the aptamer that establishes the metabolite concentration needed to modulate riboswitch function, but rather the rate of metabolite-aptamer association and the speed of transcription or translation initiation that is most important.
For the SAH aptamer to function as the sensor for an SAH-responsive riboswitch, it must be able to discriminate against biologically relevant compounds that are structurally similar to SAH. Perhaps most importantly, the aptamer must discriminate against SAM, which differs from SAH only by a single methyl group and the associated positive charge at the sulfur center. Using in-line probing and a commercial source of SAM, we found that the 68 metH RNA, as well as other SAH aptamers, exhibited apparent KD values that were no more than 10-fold higher than the apparent KD values established for SAH (data not shown). However, SAM spontaneously degrades to produce substantial levels of SAH contamination, and even freshly prepared commercial samples of SAM contain ∼10% SAH (Sigma-Aldrich Technical Service and data not shown). Thus, a greater level of discrimination against SAM by the 68 metH RNA might be obscured by the presence of SAH in SAM samples.
To obtain a more accurate estimate of the ability of SAH aptamers to discriminate against SAM, we conducted in-line probing using a sulfone analog of SAH that has been shown previously to closely mimic the binding properties of SAM with a SAM-I class riboswitch aptamer (Lim et al., 2006b). Specifically, the sulfone analog of SAH lacks the methyl group on sulfur that is present in SAM, but carries two electronegative oxygen atoms that withdraw electron density from sulfur to increase the relative positive charge of this atom similar to that of the natural coenzyme. In-line probing results (Figure 3C) reveal that the sulfone analog is bound by the 68 metH RNA with an apparent KD that is approximately three orders of magnitude poorer than SAH.
The KD value of the 68 metH aptamer for SAM also was assessed by using equilibrium dialysis to compare the functions of the SAH aptamer with a previously-reported SAM-II aptamer termed 156 metA (Corbino et al., 2005). Either 68 metH RNA or 156 metA RNA (10 μM) were added to one chamber of an equilibrium dialysis apparatus and 100 nM of [3H]SAM (Figure 4A) was added to the other chamber. The chambers were separated by a permeable membrane with a molecular weight cut-off of 5000 Daltons. Since the methyl group at the sulfur center of [3H]SAM is radiolabeled, contaminating SAH is not radioactive and therefore does not affect the equilibrium distribution of radioactivity between the two chambers.
Figure 4. The 68 metH RNA Discriminates against SAM by Several Orders of Magnitude.
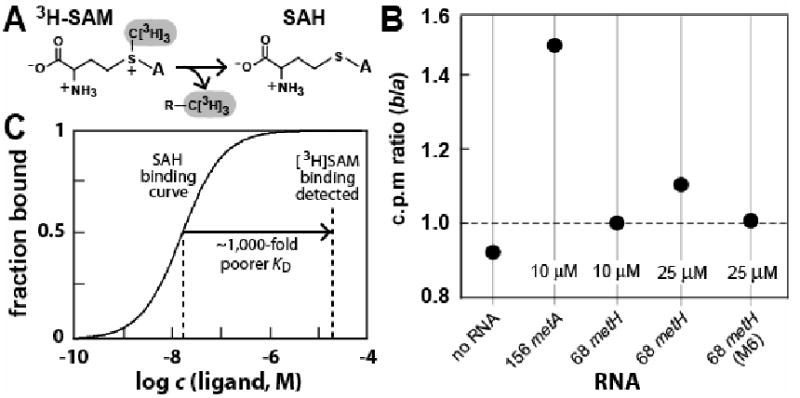
(A) Breakdown of radioactive [3H]SAM by demethylation yields non-radioactive SAH.
(B) Equilibrium dialysis was used to compare SAM binding affinity of a known SAM aptamer (156 metA) (Corbino et al., 2005) with 68 metH RNA. Chambers a and b of each equilibrium dialysis system are separated by a permeable membrane with a molecular weight cut-off (MWCO) of 5000 Daltons. [3H]SAM (100 nM) or RNA as indicated are added to chamber a and b, respectively. The apparent KD of the SAM-II aptamer 156 metA for SAM is ∼1 μM (Corbino et al., 2005), therefore the shift in tritium to chamber b should be maximal under the conditions used. Since 68 metH RNA is present in excess, the effect of non-radioactive SAH (from spontaneous demethylation of [3H]SAM) on tritium distribution between the chambers should be negligible. Data shown are representative of several repeats.
(C) Comparison of the expected curve for binding of SAH with 68 metH RNA and the concentration of 68 metH RNA used for equilibrium dialysis. The observation that 68 metH RNA does not maximally shift [3H]SAM when the RNA is present at 25 μM suggests that the apparent KD for SAM ∼1,000-fold poorer or worse. M6 carries two mutations (G12U and A13U) that are equivalent in location and nucleotide identity to construct M6 depicted in Figure 6A.
156 metA RNA shifts the distribution of [3H]SAM to favor the chamber containing the SAM-II aptamer by ∼1.5-fold, compared to no shift when the 68 metH RNA is present under identical assay conditions (Figure 4B). The 156 metA RNA has an apparent KD of ∼1 μM for SAM as determined by in-line probing (Corbino et al., 2005). Therefore, the KD value of 68 metH for SAM must be higher than 1 μM. Furthermore, the absence of a shift in radioactivity equivalent to that observed for the SAM-II riboswitch when 25 μM 68 metH RNA is used indicates that its KD for SAM is likely to be no better than 25 μM. This suggests that 68 metH RNA recognizes SAH with an affinity that is likely to be greater than 1000-fold better than for SAM (Figure 4C). This discrimination between SAH and SAM should be sufficient to precisely control the expression of SAH recycling genes in response to changing SAH levels even if the riboswitch were exposed to concentrations of SAM that are equivalent to SAH, or even in modest excess.
The molecular recognition characteristics of SAH aptamers were assessed in greater detail by establishing the apparent KD values of 68 metH RNA for 15 analogs of SAH (Figure 5A). The carboxyl and amino groups of the amino acid, various positions on the sugar ring and the nucleobase, as well as the thioester linkage were modified. All analogs, except S-adenosylcysteine (SAC), exhibit decreases of at least 3 orders of magnitude in affinity. Interestingly, the shorter amino acid side chain of SAC caused only a modest loss of affinity, unlike that observed for SAM-sensing riboswitches (Lim et al., 2006b).
Figure 5. Molecular Recognition Characteristics of the 68 metH SAH Aptamer.
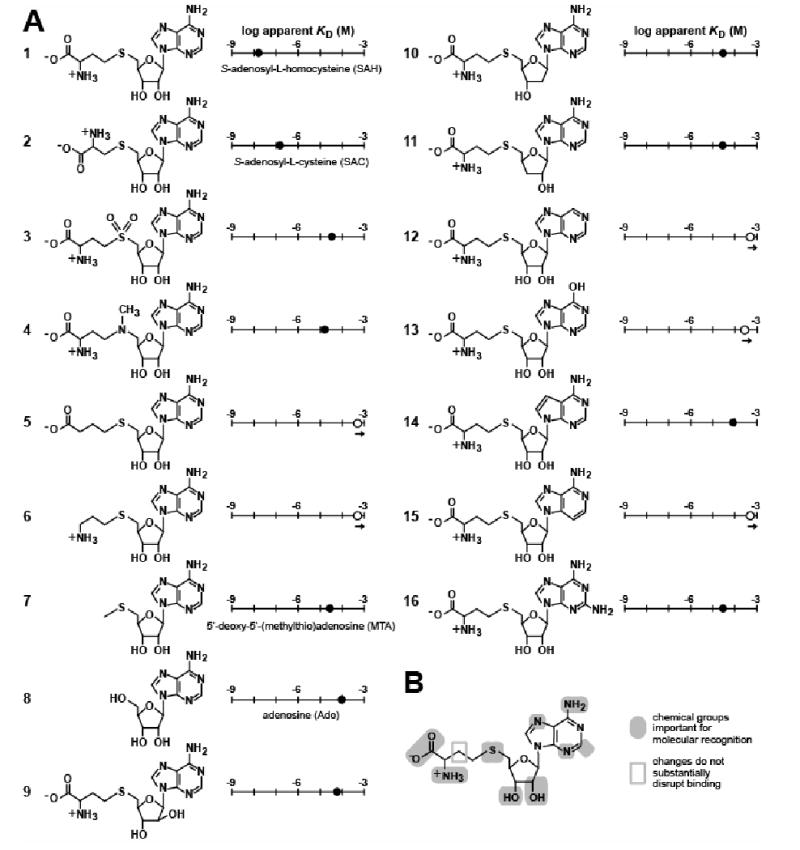
(A) Chemical structures and apparent KD values of compounds whose ligand-binding affinities were established by in-line probing with 68 metH RNA. Apparent KD values were estimated as described for SAH in Figure 3C.
(B) Summary of the known molecular recognition features of 68 metH RNA for SAH.
The changes at the sulfur atom in the sulfone (compound 3) and the aza (compound 4) derivatives of SAH also decrease affinity by three orders of magnitude, indicating the importance of this position for ligand recognition. Compound 4 is largely uncharged under the conditions used for in-line probing (Lim et al., 2006b). Therefore, its weaker interaction with the RNA might result from steric interference due to the presence of the methyl group. The higher positive charge density of compound 3 at the sulfur atom (Lim et al., 2006b) might contribute to the slightly weaker binding of this compound compared to compound 4. In addition, the two set of lone pair electrons of the sulfur center in SAH might play a key role in ligand recognition of the RNA. The sulfur atom of SAH (and SAC) conceivably could make a productive interaction with the aptamer via charge transfer or van der Waals interactions. Finally, adenosine does interact weakly with the SAH aptamer (Kd ∼100 μM), but neither homocysteine nor methionine cause any modulation of the RNA structure at concentrations as high as 1 mM (data not shown).
These results have been used to create a model for molecular recognition of SAH by the 68 metH RNA (Figure 5B). This model indicates that essentially every functional group on the ligand serves as a critical molecular recognition determinant, as has been observed for several other riboswitch aptamer classes (e.g. Mandal et al., 2003; Sudarsan et al., 2003b; Mandal et al., 2004; Lim et al., 2006a; Lim et al., 2006b).
SAH Binding Activates Expression of a Reporter Gene
The sequence between the SAH element and the start codon of the ahcY mRNA from P. syringae is predicted to form an extra stem-loop structure where the two nucleotides at the 3′ end of the P4 stem alternatively could base pair with the ribosome binding site or Shine-Dalgarno (SD) sequence (Figure 6A). This latter interaction extends the extra stem-loop structure by two base pairs, and thus might function as a SD-sequestering stem. Therefore, we hypothesize that this RNA functions as an SAH-responsive riboswitch that activates gene expression on ligand binding, perhaps by exposing the SD sequence to facilitate ribosome interaction with the mRNA. Although a similar SAH-mediated rearrangement is predicted for the SAH aptamer from the D. aromatica metH gene, ligand binding to this RNA appears to control the formation of a transcription terminator stem (Figure S4).
Figure 6. An SAH Riboswitch Controls Reporter Gene Expression.
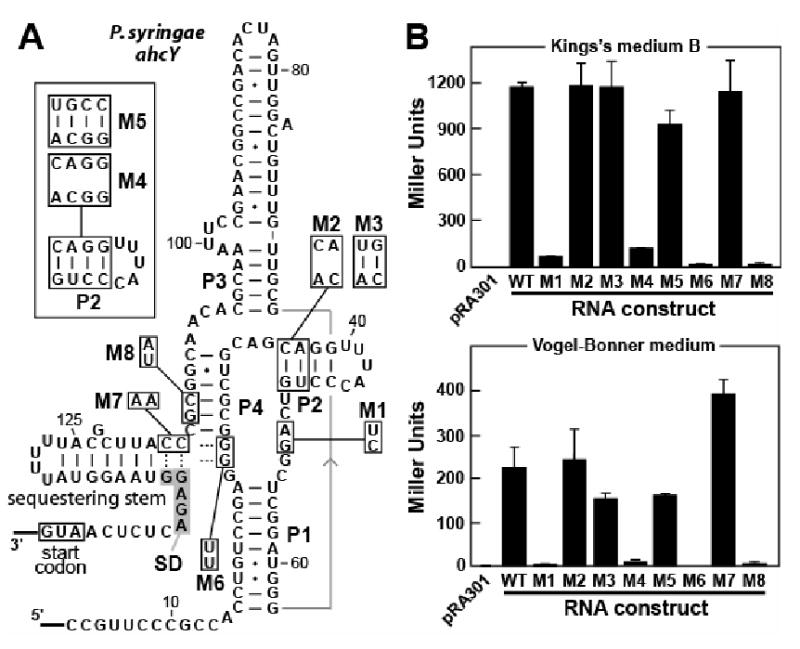
(A) Sequence and secondary structure model of the ahcY 5′-UTR of P. syringae and various mutations introduced for gene expression studies.
(B) Plots comparing expression levels for a β-galactosidase reporter gene (lacZ) fused downstream of wild-type (WT) and mutant ahcY leader sequences as defined in A. pRA301 denotes the reporter strain transformed by the original plasmid pRA301 without any insertion in front of lacZ. Different growth media used are defined in Experimental Procedures. Values are averages from three independent experiments and error bars represent standard deviation.
To test this hypothesis, an in vivo gene control assay was conducted wherein the IGR encompassing the 5′ UTR of the ahcY gene in P. syringae was fused immediately upstream of a lacZ coding sequence. This construct retained the start codon of the ahcY gene, which was fused in frame with the reporter gene ORF. The resulting translational fusion vector was transformed into P. syringae to produce the wild-type (WT) reporter strain (see Experimental Procedures). Similarly, a series of mutant reporter strains were constructed that are expected to disrupt or restore aptamer function (Figure 6A). Transformed P. syringae strains subsequently were grown in either King's medium B (rich) or Vogel-Bonner medium (minimal), and the reporter expression levels of the mutant constructs were compared to the results from the WT reporter construct (Figure 6B).
In rich medium, SAH concentrations are expected to be high because cells should be utilizing metabolic pathways that require the use of SAM as a cofactor. As expected, β-galactosidase reporter activity for the WT construct is elevated compared to construct M1 (Figure 6B, top) that carries mutations disrupting two conserved nucleotides in the junction between stems P1 and P2 or J1-2 (Figure 6A). This finding is consistent with our hypothesis that SAH binding should activate gene expression, and mutations that disrupt ligand binding will cause the riboswitch to default to an off state. Unfortunately, both M2 and M3 exhibit WT-like gene expression, despite the fact that destabilization of the P2 stem by the mutations carried by M2 was expected to disrupt riboswitch function. However, in-line probing (with and without SAH present) revealed that the M2 construct is grossly misfolded (data not shown), which likely causes the reporter construct to default to high gene expression. We examined additional mutations in P2 that yielded constructs that did not excessively misfold. As expected, mutants M4 (P2 disruption) and M5 (P2 restoration) exhibit a loss of gene expression and a subsequent restoration of gene expression, respectively. These findings again indicate that disruption of the aptamer causes the riboswitch to default to an off state for gene expression, and suggest that the M2 mutation indeed is abnormal in its misfolding and gene expression results.
Similarly, the effects of the mutations carried by constructs M6, M7 and M8 are consistent with a mechanism for gene control involving SD sequestration. These constructs each carry two mutations that reduce the stability of P4, and we observed a loss of SAH binding affinity for such constructs by using in-line probing (Figure S3C). Furthermore, constructs with disrupted SAH binding also would be expected to yield low β-galactosidase reporter activity. This result indeed is observed for M6 and M8, but M7 produces near WT expression (Figure 2A). Although the M7 mutations disrupt P4 formation, they also are predicted to disrupt the sequestering stem that overlaps the SD sequence. Therefore, the M7 riboswitch should no longer be capable of defaulting to an OFF state, and gene expression of the riboswitch-reporter fusion construct is expected to be high as is observed. A similar set of reporter gene assays was conducted with cells grown in a minimal medium (Vogel-Bonner medium). Although the same trends are observed with the various constructs grown in minimal medium (Figure 6B, bottom), the absolute values for reporter expression were higher in rich medium. This reduction in overall reporter expression in minimal medium might be due to a reduction in general metabolic activity and protein synthesis.
In-line probing was used to determine whether the competition between formation of the P4 stem and the SD-sequestering stem could be observed. In-line probing using longer constructs based on a 151 ahcY RNA, similar to that depicted in Figure 6A, reveal that the SD-sequestering stem indeed is disrupted in the WT construct when SAH is present (Figure S5). In contrast, the M6 version of this construct forms the SD-sequestering stem regardless of the presence or absence of SAH, whereas the M7 variant cannot form this stem. Thus, the mutually-exclusive formation of the full P4 stem and the full SD-sequestering stem might be a critical component of the expression platform for the SAH riboswitch from the P. syringae ahcY gene.
To further establish SAH as the metabolite effector in vivo, reporter expression was measured for the WT strain growing in minimal medium supplemented with effectors that are known to increase cellular SAH levels (Figure 7). Adenosine-2′,3′-dialdehyde (an inhibitor of SAH hydrolase) and adenosine plus homocysteine were both reported previously to substantially increase cellular SAH levels (Hermes et al., 2004). Medium supplemented with either the SAH hydrolase inhibitor or the adenosine/homocysteine mixture increase expression of the WT riboswitch-reporter fusion construct by ∼10 to 20 fold (Figure 7A), and increasing adenosine-2′,3′-dialdehyde concentrations cause increasing gene expression with the WT but not the defective variants M1 and M7 (Figure 7B). The addition of methionine to the medium causes a similar increase in riboswitch-mediated expression. Excess methionine in growth medium leads to higher cellular SAM concentrations (Winkler et al., 2003), and it is possible that the cellular SAH concentration is increased as well.
Figure 7. Elevated SAH Levels Increase Expression of the Riboswitch-Reporter Fusion.
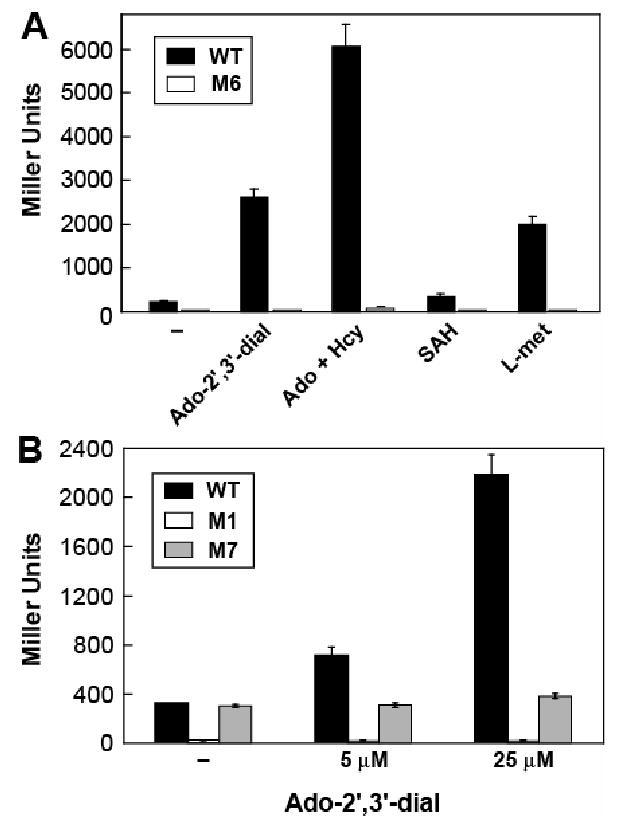
(A) Plotted are β-galactosidase expression levels of wild-type (WT) and mutant (M6) reporter strains (Figure 6A and Experimental Procedures) grown in Vogel-Bonner medium without supplementation (–), or supplemented with 25 μM of Ado-2′,3′-dial or 250 μM of the other compounds listed. Compound abbreviations: Ado-2′,3′-dial (adenosine-2′,3′-dialdehyde); Ado (adenosine); Hcy (homocysteine); L-met (L-methionine).
(B) WT, M1 and M7 constructs subjected to reporter assays as described in A with Ado-2′,3′-dial at indicated concentrations. Values are averages from three independent experiments and error bars represent standard deviation.
In contrast, the addition of 250 μM SAH to the medium only slightly increases reporter expression. SAH does not cross cell membranes as an intact molecule (Ueland, 1982), which explains why the addition of this compound has little effect on reporter expression levels. The up-regulation of reporter expression appears to be riboswitch dependent, since almost no gene expression was observed with an M6 riboswitch-reporter fusion construct carrying mutations that disrupt the formation of P4 and alter the core consensus sequence of the aptamer (Figure 6A).
Discussion
SAH is ubiquitous in living systems as the product of SAM-dependent methyl transfer reactions, but also functions as an inhibitor of these reactions when present in high concentrations (Ueland, 1982). Therefore, it is likely that many cells have a need to selectively sense SAH and dispose of the excess. In some organisms, SAH is sensed by a protein genetic factor (Rey et al., 2005), which regulates genes involved in sulfur metabolism. Our findings reveal that RNA molecules can serve in a similar capacity to sense SAH and control key genes required for its catabolism.
The identification of the SAH element provides the first example of a highly conserved and widely distributed bacterial RNA that selectively binds SAH. This RNA serves as the aptamer domain of a riboswitch class that always is located immediately upstream of ORFs whose predicted protein products are involved in SAH catabolism. This suggests that the vast majority of SAH riboswitches are likely to turn gene expression on in response to ligand binding (Figure S4). Indeed, direct ligand binding of the RNA element stabilizes certain secondary and tertiary structures, which might make accessible the SD sequence for the adjoining ORF to permit ribosome binding and subsequent translation.
Although there are several distinct classes of SAM-sensing riboswitches known that strongly discriminate against SAH binding (Lim et al., 2006b; Fuchs, 2006), the SAH riboswitch is the first class of RNAs identified that reverses this discrimination to favor SAH binding and reject SAM. As with other riboswitch classes, it is likely that high selectivity of ligand binding is required to prevent regulation of gene expression by other closely-related compounds. The strong molecular discrimination by SAH aptamers likely precludes other compounds such as SAM from unnecessarily activating SAH catabolic gene expression, which is required only when SAH accumulates to high levels. This discriminatory power of SAH aptamers is particularly important because SAM, the metabolic precursor of SAH, has a larger cellular pool than SAH under normal conditions (Ueland, 1982). Both the gene context of SAH riboswitches and their ability to discriminate against SAM by ∼3 orders of magnitude strongly suggest SAH is the metabolite sensed by the RNA element in vivo. Therefore, SAH riboswitches provide cells with a class of sensor and gene control element that permits cells to expend resources for SAH degradation only when SAH concentrations merit expression of the appropriate genes.
Like natural SAM aptamers, the SAH aptamer examined in greatest detail in this study uses essentially every functional group on the ligand for molecular recognition. One notable exception is the observation that SAH aptamers tolerate the removal of a methylene group from the amino acid portion of the ligand without substantial loss of affinity. S-adenosylcysteine (SAC; Figure 5A, compound 2) carries the same array of functional groups that are present in SAH, but some of these groups must be displaced by one or more angstroms in the binding pocket of the aptamer. A possible explanation for the tolerance of the shorter SAC analog is that the RNA aptamer might form two separate binding pockets wherein each interacts with one side of the ligand. This would allow for a certain level of adaptability that allows the binding site to recognize ligands of different lengths. Precedence for this type of riboswitch aptamer function has been reported for TPP-binding riboswitches that can bind the progressively shorter thiamin monophosphate and thiamin compounds, albeit with progressively poorer apparent KD values (Winkler et al., 2002). Atomic-resolution models of TPP riboswitch aptamers (Edwards and Ferré-D'Amaré, 2006; Serganov et al., 2006; Thore et al., 2006) reveal a bipartite ligand-binding pocket that can adjust their positions to bind the shorter thiamin monophosphate ligand with relatively high affinity.
The most highly conserved nucleotides in the SAH aptamer are largely located in P4 and in the junctions bridging P1, P2 and P4 (Figure 2A). These nucleotides are most likely involved in forming the binding pocket for SAH, as is evident by the substantial modulation of structure of these residues observed by in-line probing (Figure 3A, B). These nucleotides form a pocket that binds a ligand lacking charge at the sulfur center, and therefore they must form a structure that interacts with this atomic center in a way that is distinct from that of SAM-I aptamers. SAH aptamers reject both compounds tested in this study that carry modifications to the sulfur center (Figure 5A, compounds 3 and 4). The discrimination against compound 4 is particularly notable, as the nitrogen center replacing the sulfur of SAH is similarly uncharged. Therefore, it is possible that SAH riboswitches discriminate by steric exclusion of compounds like SAM, 3 and 4 that carry additional chemical moieties on the atom equivalent to the sulfur in SAH. However, productive interaction between the aptamer and the sulfur atom via charge transfer or van der Waals interactions has not been ruled out.
In addition to expanding the list of riboswitch classes, the identification of SAH aptamers could potentially lead to applications as highly selective SAH sensors. For example, SAH is a source of homocysteine, which can be used as a marker for cardiovascular disease (Nygård et al., 1999). However, creating precision engineered SAH aptamers by in vitro selection from randomized RNA sequences has proven to be difficult (Gebhardt et al., 2000). In vitro selection of aptamers that bind ligands carrying adenosyl moieties usually yields aptamers that largely recognize the adenosine or adenine moiety, with little recognition of the appendages that make them unique (Saran et al., 2003). For example, aptamers generated by in vitro selection for SAH are essentially adenosine/adenine aptamers, that discriminate against SAM by only ∼3 fold. The shortest example identified in sequenced bacterial genomes that conforms to the consensus of the SAH element is slightly more than 50 nucleotides in size (occurring upstream of ORF Daro_0186 in Dechloromonas aromatica, Figure S1). Coupled with nanomolar affinity for SAH and discrimination by several orders of magnitude against closely related analogs such as SAM, as well as readily available expression platforms, the SAH element possesses desired properties for in vitro and in vivo applications.
Experimental Procedures
Oligonucleotides, Chemicals, and Bacteria
Synthetic DNA oligonucleotides were purchased from the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University or from Sigma-Genosys, and were used without further treatment unless otherwise stated. [3H]SAM or S-(5′-adenosyl)-l-methionine-(methyl-3H) was purchased from Sigma-Aldrich, and [γ-32P]ATP was purchased from Amersham Biosciences. Compound 4 was the gift from Prof. G. Michael Blackburn at The University of Sheffield. All other chemicals were purchased from Sigma-Aldrich, except for SAH analogs 3, 5, 6, and 9-16, whose preparations were described elsewhere (Lim et al., 2006b). Pseudomonas syringae pv. tomato str. DC3000 was the gift of Prof. Gregory B. Martin at Cornell University.
RNA Preparation
The 68 metH and 122 ahcY RNAs were prepared by transcription in vitro using T7 RNA polymerase and the corresponding DNA templates. The DNA template for 68 metH was generated by extending a primer carrying the T7 promoter sequence hybridized to synthetic template DNA, using SuperScript II reverse transcriptase (Invitrogen) following the manufacturer's instructions. Synthetic template DNAs were purified by using denaturing (8 M urea) polyacrylamide gel electrophoresis (PAGE) prior to extension. The DNA templates for wild-type and mutant 122 ahcY RNAs were produced from PCR amplification using appropriate plasmids as described below. The RNA transcripts were purified by denaturing PAGE and eluted from gel fragments using a solution containing 10 mM Tris-HCl (pH 7.5 at 23°C), 200 mM NaCl and 1 mM EDTA. RNAs were concentrated by precipitation with ethanol, dephosphorylated with alkaline phosphatase (Roche Diagnostics) and 5′-radiolabeled using [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs) following the manufacturer's instructions. Radiolabeled RNAs were purified by denaturing PAGE and isolated as described above.
The wild-type and mutant 68 metH RNAs used for equilibrium dialysis were prepared and purified as described above, but without dephosphorylation and radiolabeling. The 156 metA RNA (Corbino et al., 2005) used for equilibrium dialysis was prepared as described above using DNA templates generated by whole-cell PCR of Agrobacterium tumefaciens strain GV2260.
In-line Probing Analysis
In-line probing assays were carried out essentially as previously described (Soukup and Breaker, 1999). 5′ 32P-labeled RNAs were incubated at room temperature for approximately 40 h in 10 μl volumes containing 50 mM Tris-HCl (pH 8.3 at 25°C), 20 mM MgCl2, 100 mM KCl, and included test compounds at the concentrations indicated. Spontaneous cleavage products were separated by denaturing 10% PAGE, visualized using a PhosphorImager (Molecular Dynamics), and quantified using SAFA v1.1 software (Das et al., 2005). The amount of RNA cleaved at each nucleotide position was obtained from SAFA analysis, which automatically determines reference nucleotide positions with invariant levels of spontaneous cleavage to correct for different amounts loaded in each lane. Individual band intensities at each modulated site were summed and normalized to obtain values for the fraction of RNA that was structurally modulated, assuming that there was no modulation in the absence of ligand and maximal modulation in the presence of the highest concentration of ligand. Values for the apparent dissociation constant (KD) were determined by fitting the plot of the fraction modulated (F), versus the ligand concentration, [L], at all three indicated modulation sites to the equation F = [L]/([L] + KD) using SigmaPlot 9 software (Systat Software, Inc.).
Equilibrium dialysis
Equilibrium dialysis was performed by delivering 20 μl samples to each side of a Dispo-Equilibrium Biodialyzer (The Nest Group, Inc., Southboro, MA, USA) wherein the separation between chambers is maintained by a dialysis membrane with a 5000 Dalton molecular weight cut-off. One chamber contained a buffer solution comprised of 100 mM Tris-HCl (pH 8.3 at 24°C), 20 mM MgCl2, 100 mM KCl, plus 100 nM [3H]SAM. The opposing chamber contained the same buffer solution plus either 156 metA or 68 metH RNAs at the concentrations indicated. After incubation for 11.5 h at 25°C, aliquots of 5 μl were withdrawn from both chambers and radioactivity was measured by a liquid scintillation counter.
In-vivo reporter gene expression assays
Nucleotides −367 to +6 relative to the predicted ahcY translation start site (GenBank accession: NC_004578.1; nucleotides 5,771,072 to 5,771,444) were amplified as a BamHI-HindIII fragment by whole-cell PCR of Pseudomonas syringae pv. tomato str. DC3000. The PCR product was cloned into the translational fusion vector pRA301 (Akakura and Winans, 2002) immediately upstream of the lacZ reporter gene. This reporter vector is stably maintained in pseudomonas since it harbors the origin of replication rep and stability region sta derived from the pseudomonas aeruginosa vector pVS1 (Itoh et al., 1984). Mutants M1-M8 were generated using the resulting plasmid as the template, appropriate mutagenic primers, and the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The integrity of all recombinant plasmids was confirmed by sequencing. Transformations of pRA301 variants into Pseudomonas syringae were accomplished by electroporation following a standard protocol (Ausubel et al., 1992). The correct transformants were identified by selecting for spectinomycin (75 μg ml-1) and rifampicin (50 μg ml-1) resistance.
To measure expression levels of the lacZ gene in wild-type (WT) and mutant Pseudomonas syringae reporter strains, cells were grown overnight with shaking at 28°C in either King's medium B (King et al., 1954) (20 g l-1 Proteose Peptone no. 3, 10 g l-1 glycerol, 1.5 g l-1 K2HPO4, 1.5 g l-1 MgSO4·7H2O, pH 7.2), 50 μg ml-1 rifampicin and 75 μg ml-1 spectinomycin, or Vogel-Bonner minimal medium (0.2 g l-1 MgSO4·7H2O, 2 g l-1 citric acid monohydrate, 10 g l-1 K2HPO4, 3.5 g l-1 NH4NaHPO4·4H2O, pH 7.0), 0.5 % w/v glucose, 50 μg ml-1 rifampicin and 75 μg ml-1 spectinomycin. The overnight cultures were diluted to A600 = 0.1 to 0.2 into the same medium, and supplemented with additional compounds as indicated. Cultures were incubated for an additional 4.5 h (King's medium B) or 6 h (Vogel-Bonner medium) before performing β-galactosidase assays following standard protocols (Miller, 1992). Miller units reported were averages of three repetitive experiments.
Supplementary Material
Acknowledgments
We thank Zasha Weinberg and Jeremy Gore for their contributions to bioinformatics analyses, Jeffrey Barrick for assistance with structure prediction, and Narasimhan Sudarsan for comments on the use of plasmids and bacterial strains, and SAH-sensing proteins. We are grateful to Cornell University Professors Gregory Martin, for his gift of Pseudomonas syringae pv. tomato str. DC3000, and Stephen Winans, for his gift of pRA301. We also thank University of Sheffield Prof. G. Michael Blackburn for the gift of compound 4. This work was supported by NIH (R33 DK07027 and GM 068819) grants to R.R.B. E.R.L. was supported by a training grant (T32GM007223) from the National Institute of General Medical Sciences. RNA research in the Breaker laboratory is also supported by the Howard Hughes Medical Institute.
Footnotes
Supplemental Data
Supplemental Data include four figures and can be found with this article online at…
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akakura R, Winans SC. Mutations in the occQ operator that decrease OccR-induced DNA bending do not cause constitutive promoter activity. Journal Of Biological Chemistry. 2002;277:15773–15780. doi: 10.1074/jbc.M200109200. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology. New York: John Wiley & Sons, Inc.; 1992. [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev. 2007;21:2874–2879. doi: 10.1101/gad.443907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. Natural and engineered nucleic acids as tools to explore biology. Nature. 2004;432:838–845. doi: 10.1038/nature03195. [DOI] [PubMed] [Google Scholar]
- Breaker RR. Riboswitches and the RNA World. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor Laboratory Press; Cold Spring Harbor Laboratory, NY: 2006. pp. 89–107. [Google Scholar]
- Bunka DH, Stockley PG. Aptamers come of age – at last. Nat Rev Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–U497. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA-a Publication of the RNA Society. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Ragione F, Porcelli M, Carteni-Farina M, Zappia V, Pegg AE. Escherichia coli S-adenosylhomocysteine/5′-methylthioadenosine nucleosidase. Purification, substrate specificity and mechanism of action. Biochem J. 1985;332:335–341. doi: 10.1042/bj2320335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TE, Ferré-D'Amaré AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RT, Grundy FJ, Henkin TM. The SMK box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- Gebhardt K, Shokraei A, Babaie E, Lindqvist BH. RNA aptamers to S-adenosylhomocysteine: kinetic properties, divalent cation dependency, and comparison with anti-S-adenosylhomocysteine antibody. Biochemistry. 2000;39:7255–7265. doi: 10.1021/bi000295t. [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J Mol Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- Hermes M, Osswald H, Mattar J, Kloor D. Influence of an altered methylation potential on mRNA methylation and gene expression in HepG2 cells. Experimental Cell Research. 2004;294:325–334. doi: 10.1016/j.yexcr.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Watson JM, Haas D, Leisinger T. Genetic and molecular characterization of the Pseudomonas plasmid pVS1. Plasmid. 1984;11:206–220. doi: 10.1016/0147-619x(84)90027-1. [DOI] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- Lim J, Grove BC, Roth A, Breaker RR. Characteristics of ligand recognition by a glmS self-cleaving ribozyme. Angew Chem Int Ed Engl. 2006a;45:6689–6693. doi: 10.1002/anie.200602534. [DOI] [PubMed] [Google Scholar]
- Lim J, Winkler WC, Nakamura S, Scott V, Breaker RR. Molecular-recognition characteristics of SAM-binding riboswitches. Angewandte Chemie-International Edition. 2006b;45:964–968. doi: 10.1002/anie.200503198. [DOI] [PubMed] [Google Scholar]
- Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004a;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004b;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- McDaniel BAM, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proceedings of the National Academy Of Sciences of the United States of America. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. A Short Course in Bacterial Genetics 72. New York: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygård O, Vollset SE, Refsum H, Brattström L, Ueland PM. Total homocysteine and cardiovascular disease. J Intern Med. 1999;246:425–454. doi: 10.1046/j.1365-2796.1999.00512.x. [DOI] [PubMed] [Google Scholar]
- Old IG, Hunter MG, Wilson DTR, Knight SM, Weatherston CA, Glass RE. Cloning and characterization of the genes for the two homocysteine transmethylases of Escherichia coli. Mol Gen Genet. 1988;211:78–87. doi: 10.1007/BF00338396. [DOI] [PubMed] [Google Scholar]
- Osborne SE, Ellington AD. Nucleic acid selection and the challenge of combinatorial chemistry. Chem Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- Rey DA, Nentwich SS, Koch DJ, Ruckert C, Puhler A, Tauch A, Kalinowski J. The McbR repressor modulated by the effector substance S-adenosylhomocysteine controls directly the transcription of a regulon involved in sulphur metabolism of Corynebacterium glutamicum ATCC 13032. Mol Microbiol. 2005;56:871–887. doi: 10.1111/j.1365-2958.2005.04586.x. [DOI] [PubMed] [Google Scholar]
- Rentmeister A, Mayer G, Kuhn N, Famulok M. Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm300. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of thiamin biosynthesis in prokaryotes. New genes and regulatory mechanisms. J Biol Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003a;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Regulation of lysine biosynthesis and transport genes in bacteria: yet another riboswitch? Nucleic Acids Res. 2003b;31:6748–6757. doi: 10.1093/nar/gkg900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 2004;32:3340–3353. doi: 10.1093/nar/gkh659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saran D, Frank J, Burke DH. The tyranny of adenosine recognition among RNA aptamers to coenzyme A. BCM Evol Biol. 2003;3:26. doi: 10.1186/1471-2148-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Molec Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard CA, Trimmer EE, Matthews RG. Purification and properties of NADH-dependent 5,10-methylenetetrahydrofolate reductase (MetF) from Escherichia coli. J Bact. 1999;181:718–725. doi: 10.1128/jb.181.3.718-725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Shiozaki S, Ohshiro T, Yamada H. Occurrence of S-adenosylhomocysteine hydrolase in prokaryote cells - characterization of the enzyme from Alcaligenes faecalis and role of the enzyme in the activated methyl cycle. Eur J Biochem. 1984;141:385–392. doi: 10.1111/j.1432-1033.1984.tb08203.x. [DOI] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA-a Publication of the RNA Society. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup JK, Soukup GA. Riboswitches exert genetic control through metabolite-induced conformational change. Curr Opin Struct Biol. 2004;14:344–349. doi: 10.1016/j.sbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003a;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003b;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- Tabor CW, Tabor H. Methionine adenosyltransferase (S-adenosylmethionine synthetase) and S-adenosylmethionine decarboxylase. Advances Enzymol. Related Areas Mol Biol. 1984;56:251–282. doi: 10.1002/9780470123027.ch4. [DOI] [PubMed] [Google Scholar]
- Takusagawa F, Fujioka M, Spies A, Schowen RL. S-adenosylmethionine (AdoMet)-dependent methyltransferases. In: Sinnott M, editor. Comprehensive Biological Catalysis. Academic Press; San Diego, CA: 1998. pp. 1–30. [Google Scholar]
- Thore S, Leibundgut M, Ban NN. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Ueland PM. Pharmacological and biochemical aspects of S-adenosylhomocysteine and S-adenosylhomocysteine hydrolase. Pharmacological Reviews. 1982;34:223–285. [PubMed] [Google Scholar]
- Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by alternative 3′ end processing of mRNAs. The Plant Cell. 2007 doi: 10.1105/tpc.107.053645. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm487. doi:10.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz R, Breaker RR. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA. 2007;13:573–582. doi: 10.1261/rna.407707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005a;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005b;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-binding ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Yao Z, Barrick JE, Weinberg Z, Neph S, Breaker RR, Tompa M, Ruzzo WL. A computational pipeline for high throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLoS Comput Biol. 2007;3:e126. doi: 10.1371/journal.pcbi.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


