Abstract
Genetic imprinting, found in flowering plants and placental mammals, uses DNA methylation to yield gene expression that is dependent on the parent of origin1. DNA methyltransferase 3a (Dnmt3a) and its regulatory factor, DNA methyltransferase 3-like protein (Dnmt3L), are both required for the de novo DNA methylation of imprinted genes in mammalian germ cells. Dnmt3L interacts specifically with unmethylated lysine 4 of histone H3 through its amino-terminal PHD (plant homeodomain)-like domain2. Here we show, with the use of crystallography, that the carboxy-terminal domain of human Dnmt3L interacts with the catalytic domain of Dnmt3a, demonstrating that Dnmt3L has dual functions of binding the unmethylated histone tail and activating DNA methyltransferase. The complexed C-terminal domains of Dnmt3a and Dnmt3L showed further dimerization through Dnmt3a–Dnmt3a interaction, forming a tetrameric complex with two active sites. Substitution of key non-catalytic residues at the Dnmt3a–Dnmt3L interface or the Dnmt3a–Dnmt3a interface eliminated enzymatic activity. Molecular modelling of a DNA–Dnmt3a dimer indicated that the two active sites are separated by about one DNA helical turn. The C-terminal domain of Dnmt3a oligomerizes on DNA to form a nucleoprotein filament. A periodicity in the activity of Dnmt3a on long DNA revealed a correlation of methylated CpG sites at distances of eight to ten base pairs, indicating that oligomerization leads Dnmt3a to methylate DNA in a periodic pattern. A similar periodicity is observed for the frequency of CpG sites in the differentially methylated regions of 12 maternally imprinted mouse genes. These results suggest a basis for the recognition and methylation of differentially methylated regions in imprinted genes, involving the detection of both nucleosome modification and CpG spacing.
In both flowering plants and placental mammals, DNA methylation has a central role in imprinting, but in neither case is it clear how imprinted genes are targeted for methylation. Imprinted genes in mammals are often associated with differentially methylated regions (DMRs)3, which show DNA methylation patterns that depend on the parent of origin. How the imprinting machinery recognizes DMRs is unknown. The Dnmt3 family includes three members: two de novo CpG methyltransferases, namely Dnmt3a and Dnmt3b (ref. 4), and an enzymatically inactive paralogue, Dnmt3L, that functions as a regulatory factor in germ cells5. Inactivating both Dnmt3a and Dnmt3b abolishes de novo methylation in mouse embryos4. Although Dnmt3b conditional germline knockout animals and their offspring show no apparent phenotype, the phenotype of a corresponding Dnmt3a conditional knockout6 is indistinguishable from that of Dnmt3L knockout mice5,7 with altered sex-specific de novo methylation of DNA sequences in male and female germ cells. These results indicate that Dnmt3a and Dnmt3L are both required for the methylation of most imprinted loci in germ cells.
We undertook structural and biochemical studies of a homogeneous complex of Dnmt3L and Dnmt3a2, generated by a co-expression and co-purification system (Supplementary Fig. 1). Dnmt3a2 is a shorter isoform of Dnmt3a that is the predominant form in embryonic stem cells and embryonal carcinoma cells and can also be detected in testis, ovary, thymus and spleen8. For crystallography, we focused on a stable complex of the C-terminal domains from both proteins (Dnmt3a-C and Dnmt3L-C) that retains substantial methyltransferase activity (Supplementary Fig. 1)9. The structure of the C-terminal complex was determined to a resolution of 2.9 Å in the presence of cofactor product S-adenosyl-l-homocysteine (AdoHcy) (Supplementary Table 1).
Both Dnmt3a-C and Dnmt3L-C have the classical fold characteristic for S-adenosyl-l-methionine (AdoMet)-dependent methyltransferases10, but AdoHcy was found only in Dnmt3a-C, not in Dnmt3L-C (Fig. 1a). This is consistent with Dnmt3a-C being the catalytic component of the complex, whereas Dnmt3L is inactive and unable to bind cofactor9. The overall complex is elongated (about 160 × 60 × 50 Å3) with a butterfly shape (Fig. 1a, b). The complex contains two monomers of Dnmt3a-C and two of Dnmt3L-C, forming a tetramer (Dnmt3L–Dnmt3a–Dnmt3a–Dnmt3L) with two Dnmt3L–Dnmt3a interfaces (about 906 Å2 interface area) and one Dnmt3a–Dnmt3a interface (about 944 Å2). The Dnmt3L–Dnmt3a interface of Dnmt3L also supports a Dnmt3L homodimer (Supplementary Fig. 2a, b). Dnmt3a2 might use the same interface to form a Dnmt3a2 homo-oligomer, as suggested by analytical size exclusion chromatography (a broad peak of about 500 kDa; Supplementary Fig. 2c, d). An F728A mutation of Dnmt3a2, which eliminates a hydrophobic interaction at the Dnmt3a–Dnmt3L interface, disrupted the Dnmt3a2 homo-oligomer to yield a roughly 150-kDa dimer (Supplementary Fig. 2c, d; the calculated mass of a Dnmt3a2 monomer is 78 kDa) and abolished methyltransferase activity (Fig. 1c; compare lanes 1 and 4). The equivalent mutant in Dnmt3L, F261A, lost its ability to form a homodimer (Supplementary Fig. 2a, b) and simultaneously its ability to stimulate wild-type Dnmt3a2 activity (Fig. 1c; compare lanes 1–3). At the Dnmt3a–Dnmt3a interface, an R881A mutation of Dnmt3a that eliminates a network of polar interactions (Fig. 1d) abolished the activity of Dnmt3a-C (ref. 11). These data indicate that both interfaces (Dnmt3a–Dnmt3L and Dnmt3a–Dnmt3a) are essential for catalysis. Dnmt3L might stabilize the conformation of the active-site loop of Dnmt3a (residues 704–725 before helix αD, containing the key nucleophile Cys 706), by means of interactions with the C-terminal portion of the active-site loop (Supplementary Fig. 3). These stabilizing interactions could explain the stimulation of Dnmt3a2 activity by Dnmt3L (Fig. 1c; lanes 1 and 2)9,12-14, as well as the linked loss of the Dnmt3a–Dnmt3L interface and of catalytic activity in Dnmt3a2 F728A (Fig. 1c, lanes 4–6).
Figure 1. Structure of the Dnmt3a–Dnmt3L (3a–3L) C-terminal domain complex.

a, Side view of the tetramer Dnmt3L (blue)–Dnmt3a (green) –Dnmt3a (green)–Dnmt3L (blue), with the bound AdoHcy shown as a stick model. The inset shows the two pairs of phenylalanine residues (F728 and F768 of Dnmt3a, and F261 and F301 of Dnmt3L) in the centre of the Dnmt3a–Dnmt3L interface. b, Top view showing the Dnmt3a–Dnmt3a interface with two pairs of salt bridges formed between R881 and D872 (enlarged). c, Activities of Dnmt3a2 and its point mutant F728A in the presence and absence of Dnmt3L. Error bars represent s.d. calculated from two independent experiments. d, A network of polar interactions between two Dnmt3a molecules (coloured blue and purple) involves R881–D872, D872–H869, H869–E851 and E851–R881.
Among the known active DNA methyltransferases, Dnmt3a and Dnmt3b have the smallest DNA-binding domain (absent from Dnmt3L; Supplementary Fig. 4). This domain includes about 50 residues in Dnmt3a/Dnmt3b in comparison with, for example, about 85 residues in the bacterial GCGC methyltransferase M.HhaI (ref. 15). However, dimerization by means of the Dnmt3a–Dnmt3a interface brings two active sites together and effectively doubles the DNA-binding surface. We superimposed the Dnmt3a structure on that of M.HhaI complexed with a short oligonucleotide15. This yielded a model of a Dnmt3a–DNA complex with a short DNA duplex bound to each active site (Fig. 2a). The two DNA segments can be connected easily to form a contiguous DNA, such that the two active sites are located in the major groove about 40 Å apart (Fig. 2b). This model indicates that dimeric Dnmt3a could methylate two CpGs separated by one helical turn in one binding event.
Figure 2. Model of Dnmt3a–Dnmt3L tetramer with DNA.

a, Surface representation of the Dnmt3a–Dnmt3L tetramer, with two short DNA molecules adopted by superimposition of the HhaI–DNA complex structure15 onto individual Dnmt3a-C. b, Two views of the Dnmt3a–Dnmt3L tetramer with one contiguous curved DNA molecule, approximately 25 base pairs in length, covering two active sites. c, Cooperative formation of large protein–DNA complexes (red arrow) after incubation of a 146-base-pair DNA with increasing concentrations of Dnmt3a-C or Dnmt3a-C–Dnmt3L-C complex. The lower band (black arrow) corresponds to one Dnmt3L-C bound to the DNA as a result of the presence of some free Dnmt3L-C in the mixture (Supplementary Fig. 5a).
Electrophoretic mobility-shift assays revealed cooperative multimerization on DNA of Dnmt3a-C alone or of the Dnmt3a-C–Dnmt3L-C complex, with each monomer of Dnmt3a-C binding to about 12 base pairs (Fig. 2c and Supplementary Fig. 5a). Gel-filtration experiments, using short oligonucleotides of different lengths, confirmed the oligomerization of Dnmt3a-C on DNA with one monomer bound for each roughly nine base pairs (Supplementary Fig. 5b). Oligonucleotides containing a single CpG site are substrates for the Dnmt3a–Dnmt3L C-terminal complex, but at least eight base pairs on each side of the CpG are required for substantial activity, which is consistent with a possible requirement for DNA contact by both Dnmt3a molecules (Supplementary Fig. 1d).
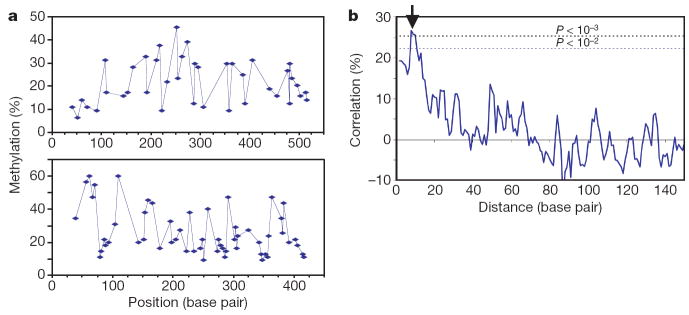
On longer DNAs, we tested the possibility that the Dnmt3a dimer, in one binding event, methylates two CpG sites separated by a helical turn. Two different DNA fragments were methylated in vitro by Dnmt3a-C. Methylation was analysed by bisulphite conversion, followed by cloning and sequencing, of 119 clones in total. At an overall methylation level of 22–26% there was a periodic fluctuation of the relative methylation at the various CpG sites (Fig. 3a and Supplementary Fig. 6). To determine whether there is a correlation between the methylation states of any two CpG sites at a given distance from one another, the autocorrelation of the methylation states was calculated for all pairs of CpGs in each individual clone. We observed a highly significant correlation of methylation status at distances of eight to ten base pairs between two CpG sites (Fig. 3b). These experiments were performed under conditions in which the DNA was saturated with the enzyme. As a result of its large interface with DNA, the enzyme oligomer or polymer cannot move along the DNA, in agreement to the observation that Dnmt3a-C methylates DNA in a non-processive manner16. The enzyme oligomer on the DNA presents the active sites in a regular spacing, which leads to a correlated methylation of CpG sites. In contrast, CpG sites positioned between the active sites are not readily available for methylation, which causes a correlation of absence of methylation that has the same period (Supplementary Fig. 6c).
Figure 3. Periodicity of Dnmt3a-C activity on long DNA substrates.
a, Two DNA fragments (from λ phage (top; 520 base pairs, 40 CpGs) and the CpG island upstream of the human SUHW1 gene (bottom; 420 base pairs, 56 CpGs)) were used for methylation. A total of 119 clones (64 from λ phage, 55 from the human CpG island) were sequenced to determine their methylation pattern. b, Autocorrelation of CpG methylation. By averaging two substrates, the P values for the correlation factors observed for distances of eight to ten base pairs (arrow) are in the range (3–7) × 10−4. This periodicity is detectable for correlation of methylation of individual CpG sites as well as for correlation of the absence of methylation at two sites (Supplementary Fig. 6).
The similarity of defects observed in the Dnmt3a conditional germline knockout and the Dnmt3L-null mutants indicates that both Dnmt3a and Dnmt3L are required for the methylation of DMRs in imprinted genes5-7. We studied the distribution of CpG sites among 12 known maternally imprinted DMRs17 that are methylated in wild-type embryos and responsible for their germline targeting, including the three (Snrpn, Igf2r and Peg1) that were shown experimentally to be unmethylated in affected embryos6. The frequencies of the distances between CpG sites peak periodically, with an average interval of 9.5 base pairs (Fig. 4a; the λ DNA fragment and the CpG island used as methylation substrates in Fig. 3 do not contain such pattern). The periodic occurrence of CpG sites 9.5 base pairs apart on average (examples are shown in Fig. 4d) makes these DNA sequences an ideal substrate for the activity of two active sites in Dnmt3a, indicating that the CpGs on maternally imprinted DMRs could be methylated simultaneously by a Dnmt3a–Dnmt3L tetramer or oligomer (as shown in Fig. 2). The periodicity of the distribution of pairwise distances of CpGs was further analysed by calculating an autocorrelation function of the frequencies, which underscored the significance of the observation (Fig. 4b). As controls, ten CpG islands randomly taken from promoter regions of genes on human chromosome 21 (Fig. 4c and Supplementary Fig. 7) did not show any correlation in the positioning of CG sites.
Figure 4. Periodicity of CpG sites in maternally imprinted DMRs17.
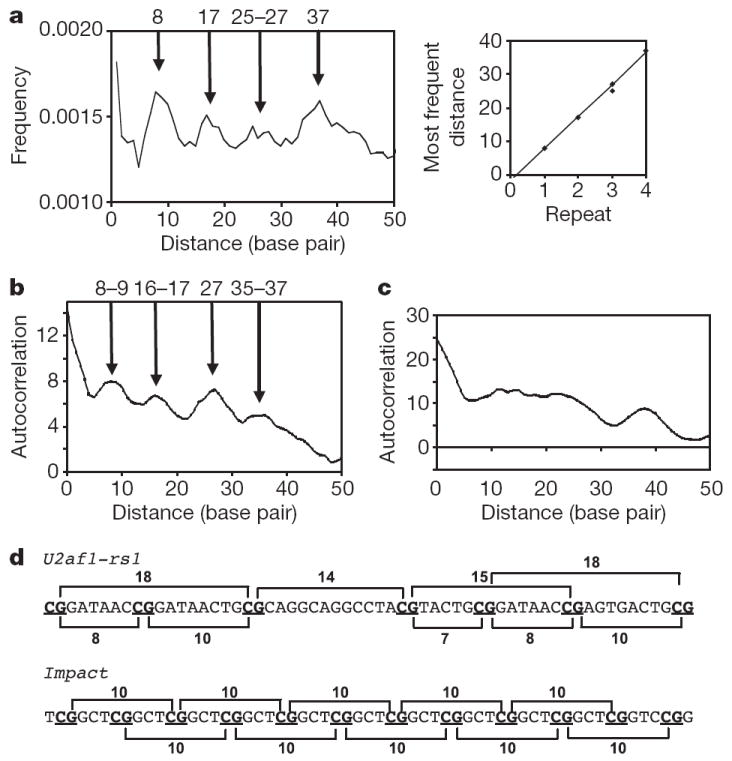
a, Frequencies of CpG pairs at a given distance with respect to all pairs, for 11 maternally imprinted DMRs. The slope of the graph in the inset is 9.5. b, The autocorrelation of peaks for 11 maternally imprinted DMRs. One sequence (Impact) was excluded from the plot because it contains repeats (see d), with each CpG site separated from the second subsequent one by ten base pairs; thus, it follows the same pattern seen by the other 11 DMRs, bringing the total number to 12 DMRs. c, Autocorrelation of peaks for ten CpG islands randomly selected from human chromosome 21. d, A region of DMR sequences of the U2af1-rs1 and Impact genes17.
In contrast with the maternally imprinted DMRs, three paternally imprinted DMRs (H19, Dlk1–Gtl2 and Rasgrf1; Supplementary Fig. 8a) did not show such periodicity; only Rasgrf1 showed a weak periodic pattern similar to maternally imprinted DMRs. The three paternal DMRs showed different methylation levels in impaired spermatogenesis: first, the DMR of Rasgrf1 is normally methylated in both the Dnmt3a conditional mutant and Dnmt3L−/− as well as in wild-type males6, but unmethylated in Dnmt3L knockout animals in a different study18; second, the H19 DMR is unmethylated in both mutants6 but showed mosaic methylation in two different studies7,18; and third, the DMR at Dlk1–Gtl2 was methylated in Dnmt3L−/− animals but not in a Dnmt3a conditional mutant. It is possible that additional factors (such as RNA19) are involved in establishing paternal imprints at specific loci, including paternally imprinted retrotransposons (LINE-1 and IAP; Supplementary Fig. 8b). Dnmt3L and Dnmt3a are at present the only factors known to be required for establishing maternal imprints in germ cells. We conclude that the periodic arrangement of CpGs in maternally imprinted DMRs constitutes an environment that is favourable for methylation by the Dnmt3a–Dnmt3L tetramer, which is consistent with the tetramers having two active sites with similar spacing and might contribute to their preferential methylation in the female germ line. Comprehensive genome-wide studies will be required in the future to determine whether all CpG islands with a periodicity of eight to ten base pairs are maternally imprinted.
Finally, histone methylation has a function in epigenetic signalling in addition to DNA methylation. There have been reports of an inverse relationship between methylation of histone H3 lysine 4 (H3K4) and allele-specific DNA methylation at DMRs; that is, a lack of H3K4 methylation at the methylated allele and the presence of H3K4 methylation at the unmethylated allele20-23. The Dnmt3L–Dnmt3a complex structure presented here indicates a novel mechanism by which an absence of H3K4 methylation is recognized by the PHD (plant homeodomain)-like domain of Dnmt3L (ref. 2), whereas its C-terminal methyltransferase-like domain brings in the active DNA methyltransferase Dnmt3a to establish a heritable DNA methylation pattern. Hence, H3K4 methylation could protect unmethylated DMRs from DNA methylation by the Dnmt3a–Dnmt3L complex.
METHODS SUMMARY
Co-expression and co-purification of Dnmt3 proteins
Co-expression of full-length Dnm3a2 (residues 220–908 of Dnmt3a; National Center for Biotechnology Information (NCBI) accession number o88508) and Dnmt3L (NCBI accession number AAH83147) was achieved by engineering two expression cassettes in one plasmid. Co-expression of Dnmt3a-C (residues 623–908) and Dnmt3L-C (residues 160–386) was achieved by the sequential transformation of two plasmids (pXC528 and pXC510) into Escherichia coli strain BL21 (DE3). Dnmt3a2 or Dnmt3a-C contained an N-terminal His6 tag, and Dnmt3L or Dnmt3L-C was a glutathione S-transferase (GST) fusion protein. The protein complex was purified with the use of three-column chromatography (GSTrap HP column, Ni2+-chelating column and Superdex 75; Amersham-Pharmacia). The GST tag was cleaved by thrombin.
Crystallography
By combining the Se-anomalous diffraction data (Supplementary Table 1) and the molecular replacement with the use of the C-terminal domain of Dnmt3L homodimer as the initial search model, the structure of Dnmt3a-C–Dnmt3L-C complex was solved.
Sequence analyses
The analyses of the periodicity of CpG positioning and in Dnmt3a activity were performed with two in-house programs.
Supplementary Material
Acknowledgments
We thank J. R. Horton for assistance with X-ray diffraction data collection; Z. Yang for assistance in solving the structure; H. Sasaki and R. Hirasawa for providing DMR sequences; S. Devine and R. Mills for help with DMR sequence analysis; A. Pingoud for providing an R.EcoRV expression clone used for calibration of the EMSA experiments; R. M. Blumenthal for critical editing of the manuscript; and E. Bernstein and R. E. Collins for comments on the manuscript. This work was supported by grants from the National Institutes of Health to X.C. and grants from the Deutsche Forschungsgemeinschaft and BMBF (Biofuture programme) to A.J.
Footnotes
Author Information The X-ray structure of Dnmt3a–Dnmt3L C-terminal tetramer complex is deposited in the Protein Data Bank under ID code 2QRV. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Feil R, Berger F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007;23:192–199. doi: 10.1016/j.tig.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Ooi SKT, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferguson-Smith AC, Surani MA. Imprinting and the epigenetic asymmetry between parental genomes. Science. 2001;293:1086–1089. doi: 10.1126/science.1064020. [DOI] [PubMed] [Google Scholar]
- 4.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 5.Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 6.Kaneda M, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 7.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 8.Chen T, Ueda Y, Xie S, Li E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem. 2002;277:38746–38754. doi: 10.1074/jbc.M205312200. [DOI] [PubMed] [Google Scholar]
- 9.Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- 10.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gowher H, et al. Mutational analysis of the catalytic domain of the murine Dnmt3a DNA-(cytosine C5)-methyltransferase. J Mol Biol. 2006;357:928–941. doi: 10.1016/j.jmb.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 12.Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci USA. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 14.Kareta MS, Botello ZM, Ennis JJ, Chou C, Chedin F. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J Biol Chem. 2006;281:25893–25902. doi: 10.1074/jbc.M603140200. [DOI] [PubMed] [Google Scholar]
- 15.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 16.Gowher H, Jeltsch A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J Biol Chem. 2002;277:20409–20414. doi: 10.1074/jbc.M202148200. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi H, et al. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet. Genome Res. 2006;113:130–137. doi: 10.1159/000090824. [DOI] [PubMed] [Google Scholar]
- 18.Webster KE, et al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc Natl Acad Sci USA. 2005;102:4068–4073. doi: 10.1073/pnas.0500702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 20.Delaval K, et al. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J. 2007;26:720–729. doi: 10.1038/sj.emboj.7601513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fournier C, et al. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 2002;21:6560–6570. doi: 10.1093/emboj/cdf655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamasaki Y, et al. Neuron-specific relaxation of Igf2r imprinting is associated with neuron-specific histone modifications and lack of its antisense transcript Air. Hum Mol Genet. 2005;14:2511–2520. doi: 10.1093/hmg/ddi255. [DOI] [PubMed] [Google Scholar]
- 23.Vu TH, Li T, Hoffman AR. Promoter-restricted histone code, not the differentially methylated DNA regions or antisense transcripts, marks the imprinting status of IGF2R in human and mouse. Hum Mol Genet. 2004;13:2233–2245. doi: 10.1093/hmg/ddh244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.