Abstract
Predicting the behavior of living organisms is an enormous challenge given their vast complexity. Efforts to model biological systems require large datasets generated by physical binding experiments and perturbation studies. Genetic perturbations have proven important and are greatly facilitated by the advent of comprehensive mutant libraries in model organisms. Small-molecule chemical perturbagens provide a complementary approach, especially for systems that lack mutant libraries, and can easily probe the function of essential genes. Though single chemical or genetic perturbations provide crucial information associating individual components (for example, genes, proteins or small molecules) with pathways or phenotypes, functional relationships between pathways and modules of components are most effectively obtained from combined perturbation experiments. Here we review the current state of and discuss some future directions for ‘combination chemical genetics’, the systematic application of multiple chemical or mixed chemical and genetic perturbations, both to gain insight into biological systems and to facilitate medical discoveries.
An important challenge facing the life sciences is to quantitatively describe the bewildering complexity of living organisms1, both to appreciate the elegance of nature and to make medically relevant predictions. The scope of this complexity is vast. Even the function of a single mammalian cell typically involves coordinated activities among over 20,000 genes, 100,000 proteins2, and thousands of small-molecule lipids, carbohydrates and metabolites, each of which may be expressed at differing levels over time. These components interact in physical complexes and functional modules that operate at many levels of organization1. The systems biology approach to understanding such processes involves constructing large-scale models of cellular function, using networks of metabolic and signaling pathways extended and buttressed by incorporating interaction data obtained from physical binding experiments3.
Systems biology models are refined by exposing a system to experimental perturbations and assessing responses for consistency with model predictions3. A striking example of this approach was the construction of a detailed model of the pathways controlling sea urchin larval gut development that could predict specific abnormal phenotypes (for example, gut duplication) upon mutation4. Genetic perturbations are attractive because they make it possible to manipulate individual macromolecular components with little ambiguity about which target has been directly affected. Moreover, the advent of comprehensive mutant libraries for several model organisms permits the large-scale investigation of phenotypic sensitivities across the entire genome, which can be extended through the use of complementary DNA and RNA interference libraries5,6. However, genetic perturbations are labor intensive, cannot resolve protein functions that differ between contexts, and generate secondary effects by modulating the target protein's own abundance. Because chemical perturbagens avoid many of these concerns, genetic screens can be complemented by ‘chemical genetics’ studies that use organic molecules as perturbers7.
Although single perturbations are effective at determining which components in a system are essential for a phenotype, functional connections between components are best identified either by direct interaction data or through combination effects8. For example, paired genetic mutations can distinguish whether two nonessential genes have serial or parallel functionalities9,10, and analyses across larger sets of perturbations can resolve the system into functional modules and pathways11,12. With the arrival of genome-scale mutant libraries, comprehensive combination genetic experiments are now being undertaken in model organisms such as yeast8, nematodes13 and bacteria14. This approach has also been extended to chemical-gene interactions15-17 and purely chemical combinations18,19, thus marking the advent of a new area of investigation: combination chemical genetics.
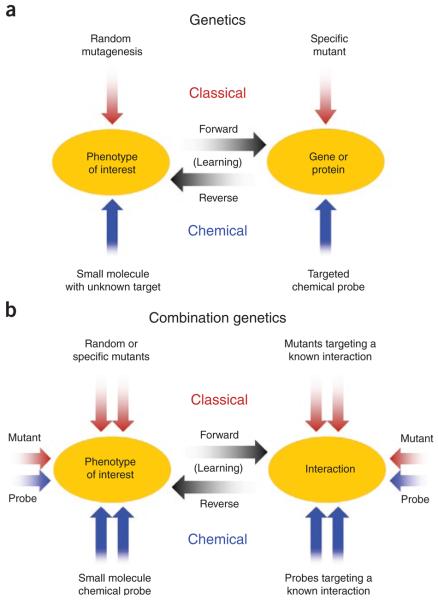
Combination chemical genetics (CCG) can be defined as the systematic testing of multiple perturbations involving chemical agents and can include purely chemical combinations or mixed chemical and genetic perturbations. Classical and chemical genetics are generally divided into ‘forward’ screens, in which uncharacterized perturbers are tested against a chosen phenotype to identify genes affecting that phenotype, and ‘reverse’ studies, in which a specific gene or protein is modulated and multiple phenotypes are monitored to determine the effects of that target7,20. Studies involving combined perturbations can be similarly classified (Fig. 1) with the mechanistic focus shifted from individual targets to interactions between them. Here we present an overview of this emerging field and discuss uses of CCG for both scientific understanding and medical discoveries.
Figure 1.
Combined perturber studies in the context of forward and reverse genetics. (a) In classical and chemical genetics, “forward” screens use many uncharacterized perturbers and a known phenotype to discover genes or proteins that affect that phenotype, and “reverse” studies test many phenotypes using a perturbagen with a known target to determine which phenotypes are affected by that target. In both cases, the questions under investigation center on the function of individual genes or proteins. (b) For combination chemical genetics, the focus of investigations shifts from individual targets to interactions between them or conditional target dependencies, and the perturbations are applied as combinations. Here forward screens use combinations of many perturbagens to discover interactions, and reverse studies involve modulating a known interaction with a set of probes targeting its components to determine which of many tested phenotypes are affected by that interaction. Figure adapted from ref. 20.
Chemical genetics
Research in chemical genetics developed over time as a field derived from classical genetics, and most of the methods and terminology used reflect that history. Genetic knockouts have their counterparts in chemical ‘knockdowns’, and studies can be designed to be either forward or reverse depending on the direction of learning that underlies their motivation7,20. Forward studies involve testing many chemical probes against one or a few phenotypes in order to identify active agents, and reverse studies perform multiple phenotype measurements on a few related chemicals to characterize their function. In both cases, chemical agents can be analyzed across a panel of phenotypic assays to identify either broad activity or selectivity between the phenotypes. The essential tools for chemical genetics include large, diverse libraries of chemicals with both known and unknown biological targets21 (Table 1). These can comprise approved drugs22 and mechanistically characterized chemical probes23, which are often informed by chemical-protein associations from the literature24.
Table 1.
Perturbation sets for combination chemical genetics
| Chemical sets | Number of probes | Number of targets | Notes |
|---|---|---|---|
| Existing drugs | 1,357 | 324–1,048 | Drugs approved in the United States22 |
| Drugs and probes | 4,765 | 4,447 | DrugBank23 (www.drugbank.ca/, on 2008.08.18) |
| Bioactives | 37,349 | 6,128 | PubChem actives100 (http://pubchem.ncbi.nlm.nih.gov/) |
| Literature | 128,120 | 1,320 | WOMBAT24,100 (version 2006.1) |
| Registered chemicals | >3.7 × 107 | – | CAS101 (http://www.cas.org/, on 2008.08.18) |
| Genetic libraries | Number of probes | % of genome | Notes |
| Bacteria, E. coli | ~3,900 | 93% | KO77, ORF78 |
| Bacteria, S. aureus | ∼2,600 | 95% | ORF79 |
| Bacteria, Brucella | ∼3,000 | 90% | ORF80 |
| Bacteria, Campylobacter | ∼1,600 | 98% | ORF81 |
| Bacteria, F. tularensis | ∼1,500 | 96% | ORF82 |
| Fungi, S. cerevisiae | ∼6,100 | 98% | KO15, OX89, ORF102 |
| Fungi, S. pombe | ∼5,000 | 95% | KO (http://www.bioneer.com/), ORF83 |
| Fungi, C. albicans | ∼2,800 | 45% | KO84 |
| Fungi, C. glabrata | ∼500 | 10% | KO85 |
| Worm, C. elegans | ∼11,000 | 50% | RNAi87 |
| Fly, Drosophila | ∼13,000 | 95% | RNAi88, ORF (http://www.fruitfly.org/) |
| Vertebrate, Mus musculus | ∼11,000 | 50% | RNAi6, shRNA5 |
| Vertebrate, Homo sapiens | ∼22,000 | 90% | RNAi6, ORF103 |
KO, knockout; OX, overexpression; ORF, open reading frame; RNAi/shRNA, RNA interference.
Chemical perturbations provide information that is distinct from and complementary to the information provided by genetic mutations, given the differences between how they modulate protein functions7. The advantages of chemical perturbations are that they (i) can target a single domain of a multidomain protein, (ii) allow precise temporal control that is critical for rapidly acting processes, (iii) can target orthologous or paralogous proteins, enabling comparisons between species or redundant functions, and (iv) do not directly alter the concentrations of a targeted protein, thus avoiding indirect effects on multiprotein complexes. Small molecules also lend themselves more readily to combination interventions, making them especially useful for integrating systems and chemical biology9,19.
The main challenges with using small molecules are that they are generally pleiotropic—they have multiple dose-dependent molecular targets that are often not fully characterized, which leads to unexpected activities. Some small molecules also have additional liabilities that can hamper their use, such as in vitro aggregation, poor solubility, difficulty traversing biological membranes and reactive or toxic functionalities. Moreover, compounds are subject to metabolic modifications in vivo that can substantially alter their activities and toxicities. Such liabilities can be minimized in many cases, but suitable precautions need to be taken; for example, multiple structurally distinct probes sharing a known target can be used to distinguish off-target from on-target effects. Finally, despite the impressive size of chemical databases (Table 1), the known targets of bioactive libraries still cover only a small fraction of the proteome22. This deficiency is due to the difficulty of finding biological targets, constraints such as cellular compartmentalization or varying protein levels that make some targets inaccessible, and the limited structural diversity of libraries generated by combinatorial chemistry25.
Forward chemical genetic approaches have recently yielded medically relevant and biologically informative insights. For example, comparing activity profiles across ∼70,000 compounds revealed that modulating the activity of mitochondrial voltage-dependent anion channels causes the selective death of RAS-transformed tumor cells26, and subsequent mechanistic studies27 showed that three such compounds achieve this selectivity through a new, non-apoptotic cell death process. Similarly, gene expression profiles in cancer cell lines identified genetic markers for acute myelogenous leukemia that were then used for a chemical genetic screen across a diverse set of 1,739 compounds, to find 8 drug candidates that induced favorable cell differentiation signatures28. As a final example, a cell-based screen of small-molecule libraries identified chemicals that induce stem cell self renewal through modulating specific combinations of targets29.
Reverse chemical genetics studies have been similarly revealing. For example, a number of annotated compounds were screened for their ability to prevent the enigmatic cell death process that occurs in olfactory sensory neurons, among the few types of neurons that turn over in vivo30. The study found that jun kinase (JNK) inhibitors could prevent this cell death by means of a new functional role for the target, and it validated this new signaling function for JNK both in vitro and in vivo. As another example, after discovering inhibitors of the yeast transcription factor component Hap3p in forward screens using immobilized small-molecule arrays, the new inhibitor haptamide B was mechanistically characterized using whole-genome transcriptional profiling of wild-type and knockout yeast cells31. Finally, a compendium of expression profiles was used to functionally characterize small-molecule treatments32, which demonstrates that transcriptional profiling is one of the most effective tools for reverse chemical genetics.
Experiments that test many compounds against a comparable number of phenotypes can be used to integrate both forward and reverse approaches in a single study. For example, 100 diverse drugs and research probes were tested in cancer cells across ∼100 phenotypes (for 11 cancerrelevant proteins, each with ∼10 microscopic measurements of stains or fluorescent antibodies tested at 12 concentrations of each drug)33, and the resulting dataset was used both for assigning targets to previously uncharacterized drugs and for measuring the functional responses to drugs with remarkable detail. Also in this category is the Connectivity Map project34, which is systematically collecting whole-genome expression profiles across multiple tumor cell lines for a large number of chemical agents, with both a forward goal of identifying chemicals with desirable selectivity across a multigene expression phenotype and a reverse objective of mechanistically characterizing drug responses.
Combined perturbers
Combined perturbation studies are inherently more complex than those based on single agents. In particular, the interaction needs to be compared to the individual single-agent effects in order to determine whether there is “synergy,” where the agents cooperate toward a phenotype, or “antagonism,” where they impede each other's activity. Systematic combination experiments also require quantitative models that represent the expected combination effects, against which synergy and antagonism can be measured.
Historically, interactions between genes have been described as epistatic relationships deriving from statistical concepts35. In these models, the fitness of a double mutant in the absence of a genetic interaction is expected to be the product of the individual fitness measurements of the corresponding single mutants. Another definition of epistasis derives from the work of Bateson and is typically used to describe situations in which the activity of one gene masks effects at another locus, allowing inferences about the order of gene action. Classical examples of Bateson-type epistasis analysis include studies of signaling pathways that control the yeast cell cycle, nematode pheromone responses and sex determination in Drosophila melanogaster35. More recently, defining the nature of genetic interactions has been expanded36,37 and modeled in the context of metabolic10 or other cellular networks38.
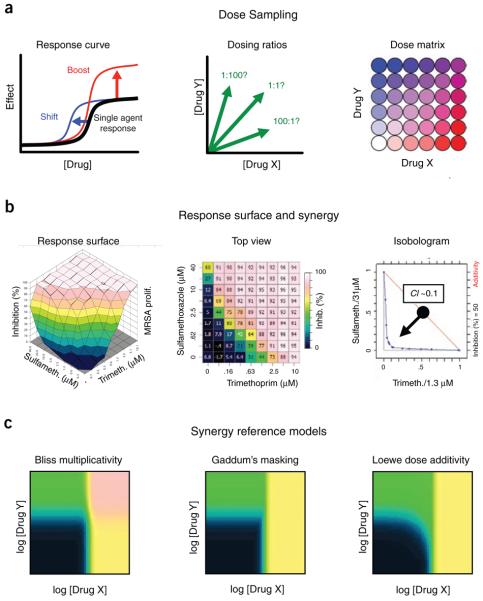
When working with chemicals, the effects of varying the concentrations need to be considered39 (Fig. 2). Chemical agents generally transition from inactive to a fixed effect level as the dose increases, and the rate of transition depends on the mechanistic interaction between the perturber and its target (for example, whether the chemical is a competitive or allosteric inhibitor). Introducing a second agent can cause synergy or antagonism either by boosting the high-dose response to a different effect level or by shifting the transition to a higher or lower concentration. Many chemical perturbers also affect multiple targets, in some cases at distinct doses, leading to several transition levels in their response curves and further complicating any combination analysis. Chemical responses can be measured for drug-gene interactions using serially diluted dose series experiments with and without the genetic perturbation. For chemical combinations, however, the optimal dosing ratio between the agents is unknown, so it is useful either to test the combination as a fixed doseratio series, where component drugs are mixed at a high concentration and the mixture is diluted serially, or to use a factorial “dose matrix” testing all pairs of serially diluted single-agent doses. The results from a dose matrix experiment can be visualized as a three-dimensional response surface, or from the top using color to represent the different effect levels. By focusing on a chosen effect contour with an “isobologram,” it is possible to visualize how much dose shifting has occurred at that level owing to synergy or antagonism between the agents.
Figure 2.
Measuring synergy for chemical combinations. (a) Continuous perturbations with sigmoidal response curves can cooperate either to boost the high-dose effect to new levels or to shift the effective concentration to lower doses, and the optimal dosing ratio is usually not known. A factorial dose matrix design can capture all of these possibilities. (b) The resulting interaction can be analyzed using the full three-dimensional response surface or using an isobologram to measure linear dose shifting at a chosen effect level via a combination index CI. For this example, we show a strongly synergistic antibiotic combination that targets folate metabolism enzymes19. (c) Synergy reference models will differ depending on the null-effect assumption. Bliss independence (multiplicative epistasis) or Gaddum's non-interaction (Bateson masking) are generally used to analyze genetic epistasis, and Loewe dose additivity is most widely used for drug combinations. The multiplicative model produces stronger effects than either of the single agents at high combined doses, whereas masking simply follows the strongest single agent at corresponding doses. In dose-additive combinations, the agents cooperate in the same way as increasing the dose of a single drug. All three models can be adapted for analyzing pairs of agonists and generalized for three or more agents.
Determining synergy for chemical combinations is also more complicated39. The genetic interaction models have their counterparts (Fig. 2) in Bliss independence (multiplicativity) and Gaddum's non-interaction (masking). For medical applications, however, the most relevant reference is Loewe additivity, which is the dose-additive expectation for a drug combined with itself, usually compared to observed synergies via a combination index40. Although all three reference models are indistinguishable when one of the perturbations has no effect alone, there has been considerable disagreement concerning which null-interaction model is most generally applicable to biological contexts19,37,39. For now, it seems that genetic interactions in proliferation assays find mostly multiplicative effects37, whereas chemical combinations are dominated by masking responses19. Ultimately, comparisons between genetic and chemical effects should be made using a single null-effect reference that is mechanistically appropriate to a common measured endpoint.
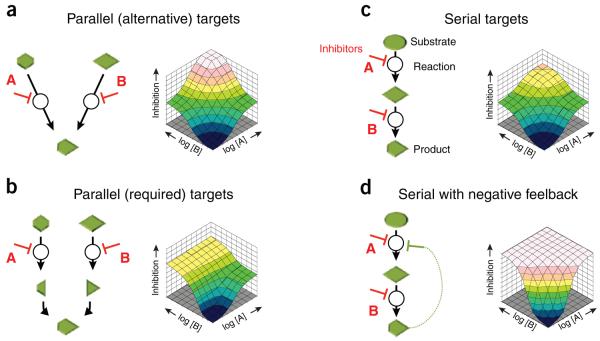
Going beyond simply measuring synergy or antagonism, dose matrix responses to chemical combinations can also provide detailed mechanistic information. Although modeling combination effects for theoretical networks of reactions has historically proven difficult41, largely because the combination index at a chosen effect level can be very sensitive to the assumed kinetic parameters in each reaction, more recent efforts focused on the dynamic responses of multiply inhibited simulated networks across different effect levels have yielded more stable predictions19,42. When performed in dose matrices, such simulations produce a variety of synergistic and antagonistic responses whose surface shapes contain information on the topological connectivity between inhibited targets19 (Fig. 3). Experimental dose matrices can be morphologically compared to responses from many simulations like these, each of which represents a mechanistic hypothesis about how the targets are connected in the network under study, to determine which hypotheses are most consistent with the data.
Figure 3.
The response shape in dose matrix experiments depends on target connectivity. In simulations of multiply inhibited metabolic networks19, the response surface morphology depended strongly on how the inhibited targets were connected in the network. Here we show four representative target connectivities, where substrates (green symbols) are metabolized by reactions (black arrows), and the reaction enzymes (white circles) are modulated by inhibitors (red markers). The resulting response surfaces from dynamic simulations are shown to the right of each such pathway. (a,b) Inhibitor pairs with parallel targets produced either saturated (a) or masking (b) effects in combination, depending on whether the targets affected independent alternatives or codependent ingredients of the final reaction. (c,d) Inhibiting serial targets along a pathway yielded multiplicative effects (c) for partially effective single agents, but serial targets in pathways regulated by negative feedback (d) produced strong dose shifting like that seen in our antibacterial example (Fig. 2b). Each of these cases represents a mechanistic hypothesis relating response shape to target connectivity, generated by the pathway simulations.
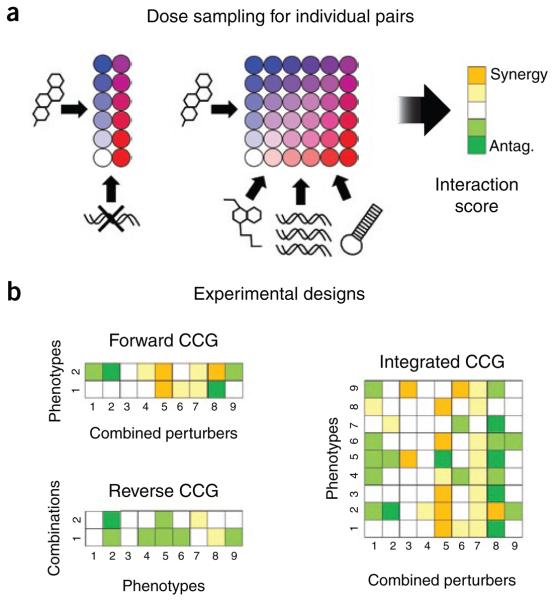
The experimental designs for CCG studies bear a direct resemblance to those used for classical and chemical genetics (Fig. 4). Tests in each phenotype become either paired-dose series or full-dose matrices, depending on whether the perturbers can be continuously dosed, and in each case the combination data are used to derive a score quantifying synergy or antagonism between the perturbers. By analogy with chemical genetics, forward CCG studies involve testing multiple combinations against one or a few phenotypes in order to identify synergistic interactions or conditional sensitivities, and reverse studies perform multiple phenotype measurements on combinations aimed at a few related interactions to characterize their function. Both approaches can be integrated by collecting profiles across many combinations for many phenotypes, thus enabling large-scale profile comparisons in either direction.
Figure 4.
Designing combination chemical genetics experiments. (a) The dose sampling possible for individual pairs depends on whether the perturbagens are discrete (for example, knockouts) or continuous (for example, chemicals, overexpression or RNAi). When the perturbers have known cellular targets, interactions can be described in terms of those targets. (b) Following the practices of chemical genetics, CCG involves many such experiments, either testing multiple combinations against a few phenotypes to discover synergistic interactions (forward) or testing a few combinations against many phenotypes to characterize the function of an interaction (reverse). These approaches can be integrated by collecting profiles across many combinations for a comparable number of phenotypes, allowing profiles in either direction to be compared for similarity or selective effects.
Chemogenomics
The most advanced area of combined chemical genetics involves the joint application of chemical and genetic perturbations. The budding yeast Saccharomyces cerevisiae has dominated this field, owing to the comprehensive genetic tools that have been developed. These resources include a systematic gene deletion set, where precise start-to-stop mutant strains were constructed for ∼6,000 genes (homozygous for inessential and heterozygous for essential genes)15. The strains were constructed with molecular tags to permit individual mutant sensitivities to be extracted from genome-wide competitive growth in a single culture43. Mutant libraries are being developed for a number of other model organisms, and the construction of large RNA interference (RNAi) probe sets and open reading frame (ORF) libraries (Table 1) has opened up still more organisms for systematic genetic testing. Using these tools, “chemogenomic” chemical sensitivity screens can be carried out that can test large sets of cells with mutated, silenced or overexpressed genes against panels of chemical perturbations.
Early large-scale efforts in CCG focused on discovering interactions between chemical and genetic perturbations in bacteria, beginning with statistical epistasis between random mutagenesis and chemical stresses44. The introduction of large-scale yeast mutant libraries, however, caused a rapid transition to chemogenomic screening, which had both the forward and reverse aspects of integrated chemical genetics. For example, a screen of 74 drugs and chemical probes against a panel of ∼3,000 heterozygous yeast mutants identified sensitive strains as likely targets for most of the chemicals17. Another study that tested ten drugs at multiple doses against a comprehensive panel of yeast mutants identified multiple genes that conferred sensitivity to each of the drugs16. Mutant sensitivities involving haploinsufficient essential genes were used to identify direct targets of the drug, but those from homozygous deletions discovered gene products that act indirectly on the drug's targets16. When such drug sensitivity profiles are compared with double-mutant fitness profiles involving a drug target's pathway45, the degree of similarity can provide further information on the drug's mechanism of action. As an example of reverse CCG among these studies, chemogenomic screening with homozygous strains produced insights on the machinery of RNA processing16. Chemogenomic screens in yeast cells have also been used to discover a new antineoplastic mechanism that induces mitotic arrest46 and to identify potential toxicities related to the off-target effects of psychoactive drugs47.
Chemogenomic screening in other organisms has also advanced, often with a more direct focus on drug discovery applications. For example, screening natural compounds using a Candida albicans mutant set identified fungal-specific inhibitors of mRNA polyadenylation48, thereby opening a new mechanistic class for potential antifungal therapies. In human cells, combining RNAi perturbation with small molecules49 offers insights into the activity of chemical combinations on both normal and abnormal disease-relevant cell types. For example, small interfering RNA screens with commonly used antineoplastics in tumor cell lines have identified potential targets for cotherapeutic cancer treatment50. Whole-genome expression signatures have also been used as a phenotype to identify a beneficial interaction between the mTOR inhibitor rapamycin and the glucocorticoid receptor as a potential therapy for lymphoid malignancies51.
As the number of genetic perturber sets expands (Table 1), chemogenomic approaches will provide ever larger sets of interaction data to aid with systems biology modeling efforts52. The analysis of chemogenomic screens presents a formidable challenge because they involve profiles with thousands of combinations, where each single agent might have multiple states of genetic perturbations (knockout, knockdown, overexpression) and many doses of chemical components. Such datasets will require both appropriate synergy analyses for each individual response matrix19 and global displays and statistics for determining mechanistic patterns between the interactions53.
Combinations of chemicals
The usefulness of synergistic drug combinations has long been appreciated9,39,54,55, especially in the therapeutic areas of cancer and infectious diseases. Multitarget synergies between drugs can be used to overcome resistance to one of the components, to make use of targets that are individually insufficient for therapy and to reduce side effects through dose sparing56. However, although there have been numerous individual studies of synergistic combinations39, it is only recently that systematic large-scale testing of chemical combinations has been pursued18,19,57. Owing to the required investment in concentration sampling, these efforts have focused on testing integrative phenotypes, such as growth, that are influenced by many cellular components, rather than monitoring many genes simultaneously.
The first systematic CCG efforts involving pure chemical combinations were aimed directly at drug discovery. Using a dedicated screening platform, an effort focused on cancer, inflammation and fungal assays identified a number of synergies as potential combination therapeutics57. All three assays yielded unexpected interactions involving drugs not normally administered toward the target therapy. Both the anticancer and the anti-inflammation synergy translated to animals57, and the latter has since been confirmed in a phase 2a clinical trial58. Drug discovery approaches of this kind must consider the possibility of synergistic toxicity and in vivo pharmacological interactions59 that can complicate delivery of the combination to targeted tissues or cause metabolic hepatotoxicity. Synergies in cell-based assays can also fail to translate when tested in animals or humans. To avoid such concerns, it is essential to carry out counterscreens and preclinical investigations, which can be guided by published toxicity or drug interaction data for the single agents, before considering any potential combination therapies for the clinic.
The use of chemical combination screens to extract mechanistic information is at an earlier stage. The first such screen tested all pairs of 21 antibiotic drugs in Escherichia coli bacterial proliferation at single combination dose points18. This study found a wide variety of combination effects between the drugs and was able to identify modules of drug pairs whose group interactions were purely synergistic or antagonistic, where each module comprised drugs targeting related cellular functions. Another chemical genetic screen in Candida glabrata yeast19, testing all pairs of ten antifungal drugs in six-by-six dose matrices, confirmed the relationship between response matrix morphology and target connectivity expected from dynamic pathway simulations19 (Fig. 3). Also in that study, an analysis of synergy profiles derived from an anticancer screen in HCT116 cells that tested all pairs of 90 drugs and research probes in six-by-six dose matrices found that probes with related targets had more similar synergy profiles than those with disparate functions19. These examples illustrate how CCG using chemical combinations, especially with varying doses, can reveal constraints on the topology of the underlying cellular network and assist with identifying unknown activities of chemical probes in the library. It is important to note that the relevance of combination effects from phenotypic experiments is limited to the cell systems under study and that mechanistic conclusions from in vitro synergies can fail to translate to other contexts (for example, due to cell type differences or in vivo pharmacological drug interactions). In any case, such studies are dependent on libraries of well-characterized chemical probes with known biological targets, and it is especially valuable to have several chemically distinct probes targeting the same protein when possible in order to separate on-target (consistent responses) from off-target (inconsistent response) effects.
Almost all of the past CCG screens involving combinations of chemicals have been essentially forward studies, aimed at uncovering unexpected synergies or interactions between drug targets. Reverse CCG studies are a more recent development but are increasingly undertaken, especially to elucidate the mechanisms of anticancer synergies. For example, whole-genome transcriptional profiling in prostate cancer cell lines of synergies between taxane microtubule binders and either estramustine60 or capecitabine61 identified genes associated with the synergistic response that were not present in the single agents' profiles. Another study testing the combination of the apoptosis inducer taurolidine with tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) used esophageal carcinoma cells and flow cytometry to characterize the synergistic responses in many apoptosis-related signaling proteins62. In yeast, chemogenomic screening performed over multiple doses of several drug combinations was used to map strain-specific interaction measurements onto the yeast genetic network and identify protein complexes that buffer the cell from the drug combination53. Given recent trends, we expect such studies to increase in frequency and complexity.
Discussion and future directions
Combination chemical genetics is an emerging field of research. Whether genetic and chemical perturbations are applied together or combinations are purely between chemicals, CCG allows the testing of interactions between cellular components to be studied in new contexts and with more detail than can be achieved with single agents or only genetic perturbations.
To make the best use of available resources, CCG researchers will need to agree on standards for the design, data collection and analysis of experiments, building on resources already established for chemical genetics. For example, the US National Institutes of Health (NIH) Molecular Library Initiative63 aims to create a publically available collection of biologically active compounds64 and experimental standards65. For CCG, it will be helpful to coordinate common probe libraries that can facilitate comparisons between experiments, and to agree on combination effect reference models for analyses across combination studies. Interaction data can be collected in existing public repositories (for example, BioGRID66), which will need to be equipped with cheminformatics tools to allow integration with the knowledge in chemical databases21.
Chemical genetic studies will benefit from continued improvement and extension of available chemical libraries. Efforts to catalog the druggable genome for existing chemicals22 have led to collections of biologically active compounds with known targets23,67. Ligands for protein targets can be identified using traditional affinity purification7, radiolabeling68 and live-cell target-labeling approaches69. Yeast three-hybrid techniques70 and the use of immobilized chemical or protein arrays71 can reveal direct drug-protein binding events, and haploinsufficiency fitness tests15 can associate small molecules with their target genes, which will be especially helpful toward characterizing the mechanisms of bioactive natural products72. Chemical genetics probe sets are also being extended by comparing response profiles across cell-based phenotypes33,67, by gene expression profiling28,34 and by analysis of drug activities and side effects73. Considerable progress has also been made with establishing libraries for specific target classes (for example, epidermal growth factor receptor kinases74). For modulating the many targets that are not covered by current probe sets, it remains critical to develop libraries with greater chemical diversity. Current efforts to this end involve assembling bioactive molecular fragments75, or using diversity-oriented synthesis76 to produce complex, natural product–like libraries.
Chemogenomics efforts are becoming increasingly complex and diverse with the introduction of expanded probe sets and higher content experimental platforms. The resources for genome-scale genetic perturbations in S. cerevisiae and other yeasts have grown dramatically, and the set of model organisms has expanded to include bacteria77-82, other yeasts83-85, invertebrates13,86-88 and select vertebrates5,6 (Table 1). Genome-wide overexpression libraries are being constructed in yeast that can be applied in conjunction with knockout experiments to provide a complete characterization of genetic perturbations89. Finally, the rapid adoption of RNAi technology promises a smooth transition from sequence to function in more complex cells and metazoans90, and flow cytometry readouts permit high-throughput analysis of changes in cell-cycle state or the expression of cytoplasmic and cell-surface markers.
Drug discovery will remain a major driving force for combination chemical genetics, and we expect to see expanded efforts involving forward, reverse and integrated CCG approaches toward this goal. One advantage of forward CCG is that it is a purely empirical approach, allowing new biological interactions to be revealed. Another potential advantage is suggested by a recent comparison of synthetic lethality in yeast (with deletion alleles) and nematodes (using double RNAi), which concluded that synthetic lethal interactions are not conserved91. This suggests that the kinds of interactions probed by CCG are likely to be organism and context specific, offering the possibility that combination therapies targeting such interactions may achieve higher levels of selectivity than single agents toward targeting infectious diseases or other context-dependent conditions such as cancer.
Considering theoretical simulations, an obvious extension of the current modeling is to simulate chemical combination effects at genome scales. This can be achieved by adapting the metabolic simulations10 previously used for genetic interaction predictions to allow partial inhibition rather than total knockouts for target genes. The response surfaces from such simulations could be compared to data from matched combination experiments, and inconsistencies could guide improvements in our understanding of the underlying biological network. Another promising approach is to use combination effects to infer topological models of the target network, using linear or nonlinear regression methods92 adapted for combination data constraints93. This approach provides a data-driven complement to the model-driven predictions from a priori reaction networks, and combining both methods should provide an effective strategy for refining predictive biological models.
An area with considerable promise involves high-order combinations of three or more perturbagens. Current CCG studies in yeast extend to third order (combinations of three agents), using designs that (i) test pairwise combinations against single mutants94 or chemogenomic profiles53, (ii) screen double mutants for sensitivity to drug treatments95 or varying conditions96 and (iii) investigate purely chemical combinations97. All three designs generally find synergies that pairwise interactions could not fully account for. Theoretically, high-order combinations should yield ever more selective control of complex systems42, and it should even be possible to use high-order testing to quantify the functional complexity of a biological system98. Extending chemical genetic studies to yet higher order combinations should provide constraints on the limits of medically useful synergy98 as well as mechanistic insights into biological networks99. Another dimension to explore is the effect on synergy due to nonsimultaneous drug application, both on heterogeneous and synchronized cell populations, which could reveal conditional dependencies on cellular state changes. Designing and analyzing high-order and phased combination experiments will be challenging, and it will be an area of considerable activity in coming years.
Combination chemical genetics brings together the traditions of genetic perturbation and synergistic drug discovery to enable the detailed study of network topology. This has been made possible by the assembly of large, diverse chemical libraries and comprehensive sets of genetic mutants or RNAi suppressors. This marriage of large-scale genomic approaches, synergy analysis and chemical genetic tools offers the promise of new insights into biology and a new avenue for drug discovery.
ACKNOWLEDGMENTS
B.R.S. is supported by a Beckman Young Investigator Award from the Arnold and Mabel Beckman Foundation and by the NIH (CA097061 and GM085081). C.N. and G.G. are supported by the Canadian Institutes of Health Research (CIHR MOP-81340 and CIHR MOP-84305, respectively).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturechemicalbiology/.
Reprints and permissions information is available online at http://npg.nature.com/ reprintsandpermissions/
References
- 1.Stelling J, Sauer U, Szallasi Z, Doyle FJ, III, Doyle J. Robustness of cellular functions. Cell. 2004;118:675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 3.Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 4.Davidson EH, et al. A genomic regulatory network for development. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 5.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 6.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 7.Stockwell BR. Exploring biology with small organic molecules. Nature. 2004;432:846–854. doi: 10.1038/nature03196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 9.Sharom JR, Bellows DS, Tyers M. From large networks to small molecules. Curr. Opin. Chem. Biol. 2004;8:81–90. doi: 10.1016/j.cbpa.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Segrè D, Deluna A, Church GM, Kishony R. Modular epistasis in yeast metabolism. Nat. Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 11.Wong SL, et al. Combining biological networks to predict genetic interactions. Proc. Natl. Acad. Sci. USA. 2004;101:15682–15687. doi: 10.1073/pnas.0406614101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang LV, et al. Motifs, themes and thematic maps of an integrated Saccharomyces cerevisiae interaction network. J. Biol. 2005;4:6. doi: 10.1186/jbiol23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 14.Butland G, et al. eSGA: E. coli synthetic genetic array analysis. Nat. Methods. 2008;5:789–795. doi: 10.1038/nmeth.1239. [DOI] [PubMed] [Google Scholar]
- 15.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 16.Giaever G, et al. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc. Natl. Acad. Sci. USA. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lum PY, et al. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell. 2004;116:121–137. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- 18.Yeh P, Tschumi AI, Kishony R. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 2006;38:489–494. doi: 10.1038/ng1755. [DOI] [PubMed] [Google Scholar]
- 19.Lehár J, et al. Chemical combination effects predict connectivity in biological systems. Mol. Syst. Biol. 2007;3:80. doi: 10.1038/msb4100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawasumi M, Nghiem P. Chemical genetics: elucidating biological systems with small-molecule compounds. J. Invest. Dermatol. 2007;127:1577–1584. doi: 10.1038/sj.jid.5700853. [DOI] [PubMed] [Google Scholar]
- 21.Oprea TI, Tropsha A, Faulon JL, Rintoul MD. Systems chemical biology. Nat. Chem. Biol. 2007;3:447–450. doi: 10.1038/nchembio0807-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 23.Wishart DS, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olah M, et al. In: Chemical Biology: from Small Molecules to Systems Biology and Drug Design. Schreiber SL, Kapoor T, Wess G, editors. Vol. 2. Wiley-VCH GmbH; Weinheim, Germany: 2007. pp. 760–779. [Google Scholar]
- 25.Feher M, Schmidt JM. Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. J. Chem. Inf. Comput. Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 26.Yagoda N, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem. Biol. 2008;15:234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stegmaier K, et al. Gene expression-based high-throughput screening(GE-HTS) and application to leukemia differentiation. Nat. Genet. 2004;36:257–263. doi: 10.1038/ng1305. [DOI] [PubMed] [Google Scholar]
- 29.Chen S, et al. Self-renewal of embryonic stem cells by a small molecule. Proc. Natl. Acad. Sci. USA. 2006;103:17266–17271. doi: 10.1073/pnas.0608156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gangadhar NM, Firestein SJ, Stockwell BR. A novel role for jun N-terminal kinase signaling in olfactory sensory neuronal death. Mol. Cell. Neurosci. 2008;38:518–525. doi: 10.1016/j.mcn.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koehler AN, Shamji AF, Schreiber SL. Discovery of an inhibitor of a transcription factor using small molecule microarrays and diversity-oriented synthesis. J. Am. Chem. Soc. 2003;125:8420–8421. doi: 10.1021/ja0352698. [DOI] [PubMed] [Google Scholar]
- 32.Hughes TR, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 33.Perlman ZE, et al. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 34.Lamb J, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 35.Avery L, Wasserman S. Ordering gene function: the interpretation of epistasis in regulatory hierarchies. Trends Genet. 1992;8:312–316. doi: 10.1016/0168-9525(92)90263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anastassiou D. Computational analysis of the synergy among multiple interacting genes. Mol. Syst. Biol. 2007;3:83. doi: 10.1038/msb4100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mani R, Onge RP, Hartman JL, IV, Giaever G, Roth FP. Defining genetic interaction. Proc. Natl. Acad. Sci. USA. 2008;105:3461–3466. doi: 10.1073/pnas.0712255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter GW, et al. Prediction of phenotype and gene expression for combinations of mutations. Mol. Syst. Biol. 2007;3:96. doi: 10.1038/msb4100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greco WR, Bravo G, Parsons JC. The search for synergy: a critical review from a response surface perspective. Pharmacol. Rev. 1995;47:331–385. [PubMed] [Google Scholar]
- 40.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 41.Jackson RC. Amphibolic drug combinations: the design of selective antimetabolite protocols based upon the kinetic properties of multienzyme systems. Cancer Res. 1993;53:3998–4003. [PubMed] [Google Scholar]
- 42.Araujo RP, Petricoin EF, Liotta LA. A mathematical model of combination therapy using the EGFR signaling network. Biosystems. 2005;80:57–69. doi: 10.1016/j.biosystems.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Costanzo M, Giaever G, Nislow C, Andrews B. Experimental approaches to identify genetic networks. Curr. Opin. Biotechnol. 2006;17:472–480. doi: 10.1016/j.copbio.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Elena SF, Lenski RE. Test of synergistic interactions among deleterious mutations in bacteria. Nature. 1997;390:395–398. doi: 10.1038/37108. [DOI] [PubMed] [Google Scholar]
- 45.Parsons AB, et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 46.Dorer RK, et al. A small-molecule inhibitor of Mps1 blocks the spindle-checkpoint response to a lack of tension on mitotic chromosomes. Curr. Biol. 2005;15:1070–1076. doi: 10.1016/j.cub.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 47.Ericson E, et al. Off-target effects of psychoactive drugs revealed by genome-wide assays in yeast. PLoS Genet. 2008;4:e1000151. doi: 10.1371/journal.pgen.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang B, et al. PAP inhibitor with in vivo efficacy identified by Candida albicans genetic profiling of natural products. Chem. Biol. 2008;15:363–374. doi: 10.1016/j.chembiol.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 49.Eggert US, et al. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. e379;2:2004. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat. Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 51.Wei G, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10:331–342. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 52.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoon S, et al. An integrated platform of genomic assays reveals small-molecule bio-activities. Nat. Chem. Biol. 2008;4:498–506. doi: 10.1038/nchembio.100. [DOI] [PubMed] [Google Scholar]
- 54.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat. Rev. Drug Discov. 2005;4:71–78. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- 55.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 56.Zimmermann GR, Lehár J, Keith CT. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov. Today. 2007;12:34–42. doi: 10.1016/j.drudis.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 57.Borisy AA, et al. Systematic discovery of multicomponent therapeutics. Proc. Natl. Acad. Sci. USA. 2003;100:7977–7982. doi: 10.1073/pnas.1337088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kvien TK, et al. Efficacy and safety of a novel synergistic drug candidate - CRx-102 - in hand osteoarthritis. Ann. Rheum. Dis. 2008;67:942–948. doi: 10.1136/ard.2007.074401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dresser GK, Spence JD, Bailey DG. Pharmacokinetic-pharmacodynamic consequences and clinical relevance of cytochrome P450 3A4 inhibition. Clin. Pharmacokinet. 2000;38:41–57. doi: 10.2165/00003088-200038010-00003. [DOI] [PubMed] [Google Scholar]
- 60.Li Y, et al. Gene expression profiling revealed novel molecular targets of docetaxel and estramustine combination treatment in prostate cancer cells. Mol. Cancer Ther. 2005;4:389–398. doi: 10.1158/1535-7163.MCT-04-0244. [DOI] [PubMed] [Google Scholar]
- 61.Li Y, et al. Gene expression profiling revealed novel mechanism of action of Taxotere and Furtulon in prostate cancer cells. BMC Cancer. 2005;5:7. doi: 10.1186/1471-2407-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daigeler A, et al. Synergistic apoptotic effects of taurolidine and TRAIL on squamous carcinoma cells of the esophagus. Int. J. Oncol. 2008;32:1205–1220. doi: 10.3892/ijo_32_6_1205. [DOI] [PubMed] [Google Scholar]
- 63.Austin CP, Brady LS, Insel TR, Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 64.Seiler KP, et al. ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36:D351–D359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tolliday N, et al. Small molecules, big players: the National Cancer Institute's Initiative for Chemical Genetics. Cancer Res. 2006;66:8935–8942. doi: 10.1158/0008-5472.CAN-06-2552. [DOI] [PubMed] [Google Scholar]
- 66.Stark C, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Root DE, Flaherty SP, Kelley BP, Stockwell BR. Biological mechanism profiling using an annotated compound library. Chem. Biol. 2003;10:881–892. doi: 10.1016/j.chembiol.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 68.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 69.Miller LW, Cornish VW. Selective chemical labeling of proteins in living cells. Curr. Opin. Chem. Biol. 2005;9:56–61. doi: 10.1016/j.cbpa.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 70.Lefurgy S, Cornish V. Finding Cinderella after the ball: a three-hybrid approach to drug target identification. Chem. Biol. 2004;11:151–153. doi: 10.1016/j.chembiol.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 71.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 72.Lopez A, Parsons AB, Nislow C, Giaever G, Boone C. Chemical-genetic approaches for exploring the mode of action of natural products. Prog. Drug Res. 2008;66:237, 239–71. doi: 10.1007/978-3-7643-8595-8_5. [DOI] [PubMed] [Google Scholar]
- 73.Campillos M, Kuhn M, Gavin AC, Jensen LJ, Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 74.Melnick JS, et al. An efficient rapid system for profiling the cellular activities of molecular libraries. Proc. Natl. Acad. Sci. USA. 2006;103:3153–3158. doi: 10.1073/pnas.0511292103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacDonald ML, et al. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat. Chem. Biol. 2006;2:329–337. doi: 10.1038/nchembio790. [DOI] [PubMed] [Google Scholar]
- 76.Schreiber SL. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science. 2000;287:1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 77.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 79.Brandner CJ, et al. The ORFeome of Staphylococcus aureus v 1.1. BMC Genomics. 2008;9:321. doi: 10.1186/1471-2164-9-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dricot A, et al. Generation of the Brucella melitensis ORFeome version 1.1. Genome Res. 2004;14:2201–2206. doi: 10.1101/gr.2456204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parrish JR, et al. High-throughput cloning of Campylobacter jejuni ORfs by in vivo recombination in Escherichia coli. J. Proteome Res. 2004;3:582–586. doi: 10.1021/pr0341134. [DOI] [PubMed] [Google Scholar]
- 82.Murthy T, et al. A full-genomic sequence-verified protein-coding gene collection for Francisella tularensis. PLoS ONE. 2007;2:e577. doi: 10.1371/journal.pone.0000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsuyama A, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 84.Xu D, et al. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog. 2007;3:e92. doi: 10.1371/journal.ppat.0030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoyer LL, Konopka J. Candida here, and Candida there, and Candida everywhere! Future Microbiol. 2008;3:271–273. doi: 10.2217/17460913.3.3.271. [DOI] [PubMed] [Google Scholar]
- 86.Boutros M, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 87.Reboul J, et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- 88.Foley E, O'Farrell PH. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2004;2:E203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jones GM, et al. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat. Methods. 2008;5:239–241. doi: 10.1038/nmeth.1181. [DOI] [PubMed] [Google Scholar]
- 90.Brasch MA, Hartley JL, Vidal M. ORFeome cloning and systems biology: standardized mass production of the parts from the parts-list. Genome Res. 2004;14:2001–2009. doi: 10.1101/gr.2769804. [DOI] [PubMed] [Google Scholar]
- 91.Tischler J, Lehner B, Fraser AG. Evolutionary plasticity of genetic interaction networks. Nat. Genet. 2008;40:390–391. doi: 10.1038/ng.114. [DOI] [PubMed] [Google Scholar]
- 92.Gardner TS, di Bernardo D, Lorenz D, Collins JJ. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–105. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- 93.Nelander S, et al. Models from experiments: combinatorial drug perturbations of cancer cells. Mol. Syst. Biol. 2008;4:216. doi: 10.1038/msb.2008.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haggarty SJ, Clemons PA, Schreiber SL. Chemical genomic profiling of biological networks using graph theory and combinations of small molecule perturbations. J. Am. Chem. Soc. 2003;125:10543–10545. doi: 10.1021/ja035413p. [DOI] [PubMed] [Google Scholar]
- 95.Onge RP, et al. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Musso G, et al. The extensive and condition-dependent nature of epistasis among whole-genome duplicates in yeast. Genome Res. 2008;18:1092–1099. doi: 10.1101/gr.076174.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee MS, et al. The novel combination of chlorpromazine and pentamidine exerts synergistic antiproliferative effects through dual mitotic action. Cancer Res. 2007;67:11359–11367. doi: 10.1158/0008-5472.CAN-07-2235. [DOI] [PubMed] [Google Scholar]
- 98.Lehár J, Krueger A, Zimmermann G, Borisy A. High-order combination effects and biological robustness. Mol. Syst. Biol. 2008;4:215. doi: 10.1038/msb.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Deutscher D, Meilijson I, Kupiec M, Ruppin E. Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nat. Genet. 2006;38:993–998. doi: 10.1038/ng1856. [DOI] [PubMed] [Google Scholar]
- 100.Southan C, Varkonyi P, Muresan S. Complementarity between public and commercial databases: new opportunities in medicinal chemistry informatics. Curr. Top. Med. Chem. 2007;7:1502–1508. doi: 10.2174/156802607782194761. [DOI] [PubMed] [Google Scholar]
- 101.Stobaugh RE. Chemical Abstracts Service Chemical Registry System. 11. Substance-related statistics: update and additions. J. Chem. Inf. Comput. Sci. 1988;28:180–187. doi: 10.1021/ci00060a003. [DOI] [PubMed] [Google Scholar]
- 102.Gelperin DM, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lamesch P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]