Abstract
The lacrimal functional unit (LFU) is defined by the 2007 International Dry Eye WorkShop as ‘an integrated system comprising the lacrimal glands, ocular surface (cornea, conjunctiva and meibomian glands) and lids, and the sensory and motor nerves that connect them’. The LFU maintains a healthy ocular surface primarily through a properly functioning tear film that provides protection, lubrication, and an environment for corneal epithelial cell renewal. LFU cells express thousands of proteins. Over two hundred new LFU proteins have been discovered in the last decade. Lacritin is a new LFU-specific growth factor in human tears that flows through ducts to target corneal epithelial cells on the ocular surface. When applied topically in rabbits, lacritin appears to increase the volume of basal tear secretion. Lacritin is one of only a handful of tear proteins preliminarily reported to be downregulated in blepharitis and in two dry eye syndromes. Computational analysis predicts an ordered C-terminal domain that binds the corneal epithelial cell surface proteoglycan syndecan-1 (SDC1) and is required for lacritin’s low nanomolar mitogenic activity. The lacritin binding site on the N-terminus of SDC1 is exposed by heparanase. Heparanase is constitutively expressed by the corneal epithelium and appears to be a normal constituent of tears. Binding triggers rapid signaling to downstream NFAT and mTOR. A wealth of other new proteins, originally designated as hypothetical when first identified by genomic sequencing, are expressed by the human LFU including: ALS2CL, ARHGEF19, KIAA1109, PLXNA1, POLG, WIPI1 and ZMIZ2. Their demonstrated or implied roles in human genetic disease or basic cellular functions are fuel for new investigation. Addressing topical areas in ocular surface physiology with new LFU proteins may reveal interesting new biological mechanisms and help get to the heart of ocular surface dysfunction.
Keywords: dry eye, cornea, lacrimal, meibomian, LACRT, LFU
1. Introduction
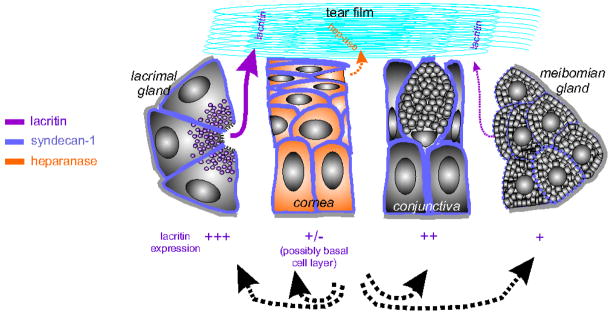
The lacrimal functional unit (LFU; Fig. 1) is ‘an integrated system comprising the lacrimal glands, ocular surface (cornea, conjunctiva and meibomian glands) and lids, and the sensory and motor nerves that connect them’ (2007 International Dry Eye WorkShop; reviewed by Rolando & Zierhut, 2001, and Stern et al., 2004). Corneal mechanonociceptors, polymodal nociceptors and cold receptors send afferent impulses to the CNS. Reflex efferent innervation stimulates secretion by main and accessory lacrimal glands. It also likely stimulates secretion by conjunctival goblet cells and the Meibomian gland (Stern et al., 2004). Stimulated or basal protein and lipid secretions flow onto the ocular surface to form the biophysically complex tear film. An estimated 15% of the tear film is expended per minute increasing to as much as 500% upon irritation (Jordan & Baum, 1980). Trans epithelial water transport also contributes to the tear film and replaces water loss from evaporation (Levin & Verkman, 2004). This constant state of sensory and secretory flux - likely involving thousands of soluble agonists, surface receptors and signaling mediators - is responsible for maintaining the health of the ocular surface epithelium.
Figure 1.
Schematic depiction of key cellular components of the lacrimal functional unit (LFU), and expression of prosecretory mitogen lacritin, cell surface proteoglycan syndecan-1 (to which lacritin binds) and heparanase (necessary for lacriting binding of syndecan-1). The LFU consists of lacrimal gland acinar cells, cornea epithelial cells, the goblet cell-enriched conjunctiva, the lipid and protein releasing meibomian gland from the lid margin, and (not shown) sensory and motor nerves. Corneal afferent fibers are sensory for mechanical, chemical and thermal reduction stimuli from mechanonociceptor, polymodal nociceptor and cold receptors. Impulses pass to the CNS. Efferents (mainly parasympathetic) help coordinate ocular surface wetting. They are motor to main and accessory lacrimal gland secretion. They are also thought to promote goblet cell and meibomian gland secretion (Stern et al., 2004).
Much of the cellular and molecular LFU machinery is well-known, but many gaps in the understanding of LFU proteins remain. Largely unaddressed are contributions of alternative splicing, post-translational modifications, upstream regulators and heterocomplex formation. Also, little is known about transient molar amounts of LFU proteins in tissues and how these doses affect other transient agonists and antagonists. At a proteomic level, the complexity of new proteins and their interactions with others is challenging. The human corneal BodyMap and NEIBank human keratocornea databases separately list over 2000 distinct mRNAs, and over 200 are listed in the human conjunctival BodyMap database, each coding for distinct LFU proteins. Similarly, the human tear proteome is more complex than previously appreciated with over 400 proteins apparently constituting the tear proteome (de Souza et al., 2006; Laurie et al., 2008). Preliminary studies suggest that only 4 – 5% may be downregulated in dry eye or dry eye-related syndromes, one of which is lacritin (Koo et al., 2005; Green-Church et al., 2007; Kitagawa et al., 2007).
This review of new proteins of the human LFU proteome begins with lacritin (Sanghi et al., 2001; Ma et al., 2008), a 12.3 kDa secreted glycoprotein in tears whose normal expression is largely restricted to the LFU. Lacritin is mitogenic for subconfluent human corneal epithelial cells (Wang et al., 2006), and promotes rat lacrimal acinar (Sanghi et al., 2001) and human corneal epithelial cell secretion. Although a tear protein, when applied topically in rabbits lacritin appears to increase the volume of basal tear secretion. Lacritin also appears to be protective against the inflammatory cytokines interferon-γ and TNF (Wang & Laurie, unpublished). Orthologs of the human LACRT gene are represented in more than thirteen different mammalian genomes and may be absent from lower vertebrates.
1.1 Lacritin structural features
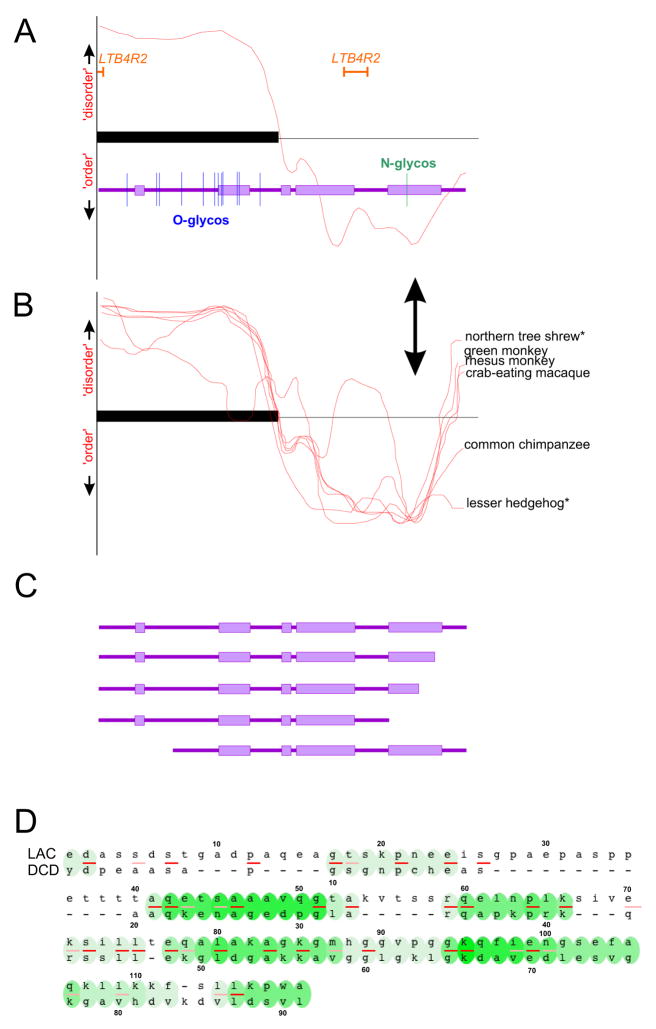
Considering lacritin’s widespread expression in the LFU and its presence in tears, it is surprising that lacritin was not discovered earlier. Lacritin’s protein backbone has a predicted isolectric point (pI) of 5, and eleven to twelve predicted O-glycosylation sites (Fig. 2; NetOGlyc 3.1 above threshold prediction) that support some FASTA homology with the mucin-like region of fibulin-2 and glycosaminoglycan binding region of neuroglycan C (Sanghi et al., 2001). Predicted O-glycosylation sites are concentrated in the N-terminal half of human lacritin (Fig. 2A) and orthologs (not shown). Unglycosylated recombinant human lacritin generated in E. coli is mitogenic (Sanghi et al., 2001; Wang et al., 2006). This activity is unaffected by deletion of up to 65 N-terminal amino acids (Wang, McKown, Laurie, unpublished). Expression of N-65 lacritin (lacking 65 amino acids from the N-terminus) in mammalian cells would exclude all predicted O-glycosylation sites, and yet O-glycosylation of the N-terminal half appears to be conserved in all lacritins, although possibly at different levels.
Figure 2.
Structural and functional features of lacritin. (A) Comparison of lacritin PONDR (Predictor Of Naturally Disordered Regions; red line) analysis to a linear model of lacritin (purple) with boxes indicating PSIPRED-predicted alpha helices. Lacritin’s N-terminal and C-terminal halves are respectively predicted to be disordered and ordered. Disordered implies absence of fixed tertiary structure. The ordered half contains more predicted α-helices. Vertical lines are sites of predicted glycosylation. Most are N-terminal. An IPR003982 Leukotriene B4 type 2 receptor (LTB4R2) domain is predicted by InterPro. The cytokine TNFSF8 also contains an LTB4R2 domain. Shown is human lacritin without signal peptide. (B) Similar PONDR analysis of six lacritin orthologues, all without signal peptide. (* lacritin ortholog length is shorter; tracing was scaled to match the others). (C) The C-terminal α-helix in the ordered half contains mitogenic and syndecan-1 binding domain(s), as determined by deletion analysis (Wang et al., 2006). (D) Near-Optimal Global Alignment (Smoot et al., 2004) of human lacritin and dermcidin. Level of green indicates robustness of alignment. Red and pink underline respectively indicates identity and similarity.
These and other properties in some non-human primates appear to differ. Predicted molecular weights, pI and O-glycosylation site values for secreted lacritin in confirmed or predicted orthologs include the primates: chimpanzee (12.6 kDa, 5.2, 13), orangutan (12.4 kDa, 5, 13), rhesus monkey (12.2 kDa, 5, 14) and mouse lemur (12.3 kDa, 4.9, 9); and non-primates: house cat (10.7 kDa, 5, 14), common shrew (12.9 kDa, 10.2, 4), northern tree shrew (11.9 kDa, 5.4, 20), lesser hedgehog (10.43 kDa, 6.6, 17), little brown bat (9.3 kDa, 5, 10), and nine-banded armadillo (10 kDa, 10.1, 10). Human lacritin’s single predicted N-linked glycoslyation site towards the C-terminus (Fig. 2A) is conserved in chimpanzee and orangutan, but not in the others. Estimated molecular weights and isoelectic points for several other orthologues were provided in Ma et al. (2008). Mouse and rat lacritin orthologs are currently not listed (Ensembl release version 49), as is the case for abundant human tear protein proline rich 4 (PRR4). We noted previously that human genes displaying preferential lacrimal gland expression in humans do not do so in mouse (Ozylidirim et al., 2005). Possibly these human and rat or mouse LFU differences (Ozyildirim et al., 2005) are habitat-related and linked to absence of a human Harderian gland. If so, our dependence on other animals for disease models and preclinical studies may require more informed interpretations, perhaps from parallel use of human LFU cell lines.
An InterPro IPR003982 Leukotriene B4 type 2 receptor domain extending into the signal peptide has been assigned to human (Ma et al., 2008) and chimp lacritin, but not to other orthologues. Other proteins with an IPR003982 domain vary widely in function and include a transcription factor, an NFκB complex inhibitor, a cytokine, a receptor, a phosphatase inhibitor and a furin-dependent secreted protein. Deletion mutants appear to rule out a functional role for this domain (Wang, McKown, Laurie, unpublished).
PONDR (Predictor of Naturally Disordered Regions) analyses of human lacritin (Fig. 2A) and orthologues (Fig. 2B) suggest that the N-and C-terminal halves are respectively disordered and ordered. This is in keeping with more α-helical structure in the C-terminal half (Fig. 2A), as confirmed by circular dichroism using synthetic peptides (Wang et al., 2006) and recombinant lacritins (McKown, unpublished). The disordered domain may contribute to secreted lacritin’s anomalously slow migration in SDS-PAGE (observe 18 kDa vs expected 12.3 kDa for non-glycosylated) as is the case for XPA (Iakoucheva et al., 2001). Also slowing migration may be its uneven distribution of charge residues (Garfin, 1990), ie. N-71 is highly acidic (pI 10.7) and α-helix-rich, whereas C-59 is basic (pI 3.8).
Progressive deletion analysis (Fig. 2C; Wang et al., 2006) localized lacritin’s mitogenic and syndecan-1 binding activities to the α-helical region (NGSEFAQKLL) between C-terminal amino acids 99 and 109 of secreted lacritin. Helical wheel predicts that NGSEFAQKLL and flanking residues form a conserved amphipathic α-helix. The hydrophobic face of amphipathic α-helices is capable of mediating low nanomolar protein-protein interactions. An amphipathic α-helix in parathyroid hormone is responsible for ligating parathyroid hormone receptor 1 (Pioszak and Xue, 2008), a G-protein coupled receptor (GPCR). A similar mechanism mediates the interaction of exendin-4 with the GPCR glucagon-like peptide-1 receptor (Runge et al., 2008), and binding of small ankyrin 1 of the sarcoplasmic reticulum with obscurin (Borzok et al., 2007). At micromolar concentrations, some amphipathic α-helices insert in membranes. This is the approach by which RGS2 (regulator of G-protein signaling2) targets the plasma membrane to inhibit the M1 muscarinic receptor (Gu et al., 2007) and by which antimicrobial peptides are effective (Hawrani et al., 2008). Actual molar levels of lacritin in human tears are currently unknown, but one preliminary estimate (McKown, unpublished) puts it in the nanomolar range. In vitro dose response studies with human recombinant lacritin reveal a biphasic low nanomolar dose optimum for signaling and mitogenesis. This is in keeping with a co-receptor or receptor-binding mechanism (see below 1.3 Lacritin cell targeting).
Dermcidin (DCD) was proposed to be homologous to lacritin (Porter et al., 2003), but is not listed as a homologue by Homologene. Dermcidin’s structure is not known. Although the best alignment shown was between signal peptides (Porter et al., 2003), several other regions are similar (Fig. 2D; alignment without signal peptide), and the functional and structural parallels are intriguing. Dermcidin has been reported to be mitogenic (Porter et al., 2003). Like lacritin dermcidin displays PONDR-predicted N-terminal half disorder and C-terminal half order, as well as aberrant mobility in SDS PAGE. Both appear to be relatively unstable in solution, a condition possibly attributable to the disordered half (Majczak et al., 2007). Dermcidin is proteolytically processed to generate N- and C-terminal peptides respectively with neural cell survival and skin antimicrobial activities, a process that has neither been demonstrated nor found necessary for lacritin mitogenic and prosecretory activities. Interestingly, LACRT and DCD genes are closely adjacent on human chromosome 12q13 and have been reported to be co-amplified in some metastatic breast cancers (Porter et al., 2003).
1.2 Lacritin expression and regulation
Lacritin expression is mainly restricted to the LFU (Fig. 1) and in tissue sections is detected most strongly in lacrimal acinar cell secretory granules (Sanghi et al., 2001) from which it is released after carbachol stimulation (Nakajima et al., 2007). In the monkey LFU, quantitative PCR suggests the following decreasing levels of tissue expression: lacrimal gland>conjunctiva>meibomian gland>cornea (Nakajima et al., 2007). Recent mass spec studies have detected lacritin in human meibomian gland secretions (Tsai et al., 2005), in human tears (Zhou et al., 2006; Koo et al., 2005) and deposited on contact lenses (Green-Church & Nichols, 2008). Its eye-specific transcriptional regulation appears to exceed that of α-crystallin, rhodopsin and keratocan. Lacritin is also expressed in salivary gland (staining of some ductal cells [Sanghi et al., 2001], in saliva [Ramachandran et al., 2006]) and thyroid (Sanghi et al., 2001) glands, and in lung bronchoalveolar lavage (Human Proteinpedia: HuPA_00022). No information is available on mechanisms of transcriptional regulation. Lacritin mRNAs are the sixth most common behind lysozyme, proline rich 4, lipocalin-1, lactotransferrin and proline rich 1 in NEIBank’s human lacrimal EST database (Ozylidirim et al., 2005) from mixed female (51 yrs) and male (75 yrs) normal lacrimal gland.
Aceview documents lacritin-b, lacritin-c (Ma et al., 2008) and lacritin–d splice variants largely from NEIBank lacrimal transcripts (Ozylidirim et al., 2005). The most common form is lacritin-a. Currently 80 accessions are reported for lacritin-a versus one accession each for lacritin–b and –d; and eight accessions for –c. Lacritin-a is the representative RefSeq transcript. The LACRT gene is composed of five exons. Lacritin-b mRNA encodes a predicted secreted glycoprotein (MW 11.1 kDa, pI 5.3, 12 predicted O-linked sites) deficient in the sequence SIVEKSILTE from an abbreviated exon 4. This was not detected in an early RT-PCR that in retrospect was capable of resolving lacritin-b (but not lacritin-c or -d; Sanghi et al., 2001). Amplification was performed from human lacrimal and submandibular RNA each pooled from three females (66, 69, 71 yrs) and one male (38 yr). Further variant-specific PCR and antibody-based assays are warranted. Lacritin-c mRNA encodes a predicted secreted glycoprotein (MW 10.7 kDa, pI 4.6, 13 predicted O-linked sites) lacking sequence from exons 4 and 5 and instead displaying a novel C-terminus from intron 3. The lacritin-d prediction is not as strong. Its putative mRNA lacks exon 1 and part of exon 2, and codes for a non-secreted, possibly nuclear protein (MW 8.7, pI 7.3) with a potential PEST sequence for rapid turnover. The incidence of lacritin splice variants in tears of normal versus dry eye populations has not been studied but could be fruitful for both lacritin-b and -c are predicted to be inactive.
1.3 Lacritin cell targeting
Screening of seventeen different cell types suggests that lacritin may target an unusually narrow selection of epithelia, and not lymphoblastic, fibroblastic or glioma cells (Wang et al., 2006). Lacritin appears to promote constitutive tear secretion by cultured rat lacrimal acinar cells (Sanghi et al., 2001), and both secretion (Wang & Laurie, unpublished) and mitogenesis of human corneal epithelial cells (Wang et al., 2006). The lacrimal acinar cell response implies an autocrine or paracrine stimulatory mechanism since lacritin is itself a stimulated secretory product of acinar cells.
A mechanism of targeting has been partially elucidated by recombinant protein engineering and receptor screening strategies (Fig. 2B, 3A; Wang et al., 2006; Ma et al., 2006). The lacritin C-terminal amphipathic α-helix targets the cell surface heparan sulfate proteoglycan syndecan-1 (SDC1), but not syndecan-2 or -4. Syndecan-1 is a common cell surface heparan sulfate proteoglycan that surrounds all human corneal and conjunctival epithelia cells (Fig. 1; Heimann et al., 2001). Deletion analysis narrowed the lacritin binding site to the first 51 N-terminal amino acids of SDC1. Binding requires heparanase (HPSE) cleavage of the three heparan sulfate chains attached in this region (Fig. 3; Ma et al., 2006). This previously undescribed mechanism may at first glance seem counterintuitive to the health of the ocular surface since corneal stromal release of heparanase by infiltrating lymphocytes is a component of the inflammatory response (reviewed by Nasser, 2008). However, heparanase is constitutively expressed throughout the normal corneal epithelium (Fig. 1; Berk et al., 2004), and may be an important element in normal epithelial physiology. It also appears to be a normal constituent of human tears (Ma and Laurie, unpublished). In skin, heparanase is involved in the migration of stem cell progeny (Zcharia et al., 2005), and antisense knockown of heparanase in Xenopus embryos promotes cell death, a phenotype that can be rescued by co-transfection of a heparanase construct (Bertolesi et al., 2008). Salivary branching morphogenesis is dependent on heparanase cleavage of heparan sulfate to release FGF10 (Patel et al., 2007). Secretion of heparanase that has been processed from its latent 65 kDa to active 58 kDa heterodimeric form can be stimulated by UTP (Shafat et al., 2006). UTP is in late clinical trials for the treatment of dry eye (Tauber et al., 2004) via a mechanism thought to involve the production of mucins (Murakami et al., 2003). These observations suggest a potential linkage between lacritin, UTP and heparanase in ocular surface physiology.
Figure 3.
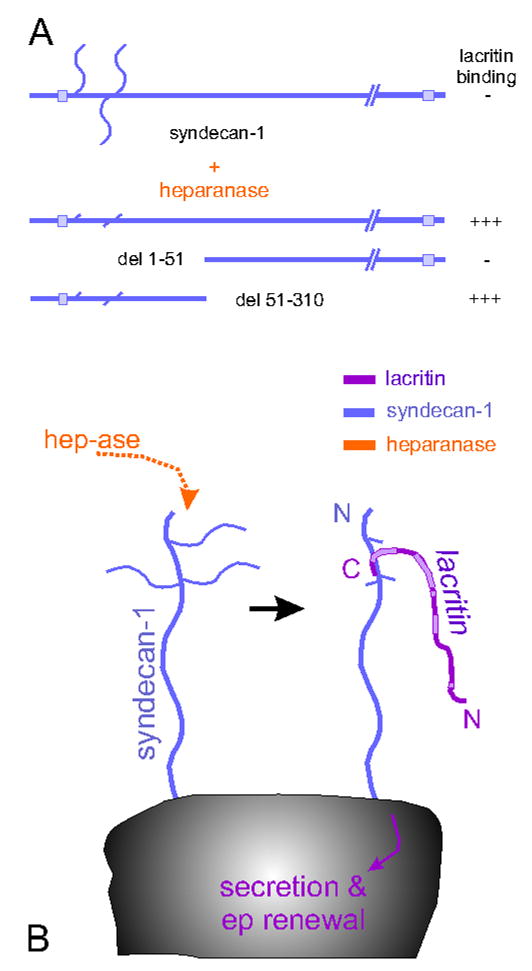
Off/on switch for lacritin function. (A) Partial deletion analysis reveals that lacritin binds an N-terminal region in the core protein of syndecan-1 after heparanase (hep-ase) treatment. (B) Heparanase cleaves heparan sulfate chains to expose the binding site.
1.4 Lacritin cell signaling
Lacritin binding of syndecan-1 sets up a cascade of intracellular prosecretory and promitogenic signaling that has been partially worked out using pharmacological inhibitors and siRNA transfections (Fig. 4; Wang et al., 2006). Interestingly, proximal signaling is inhibited by pertussis toxin. Pertussis toxin blocks the ADP-ribosylation of the G proteins Gαi and Gαo. This suggests that lacritin signaling is generated via a G-protein coupled receptor (GPCR) or G-protein-dependent ion channel that develops affinity for lacritin when lacritin is immobilized on syndecan-1. Lacritin signaling proceeds from Gαi and Gαo to the rapid dephosphorylation of protein kinase Cα (PKCα by an unidentified phosphatase. This targets PKCα to the perinuclear Golgi region where it complexes and activates phospholipase D1 (PLD1) and phospholipase Cγ2 (PLCγ2). Activation is not detected after knockdown of PKCα by siRNA.
Figure 4.
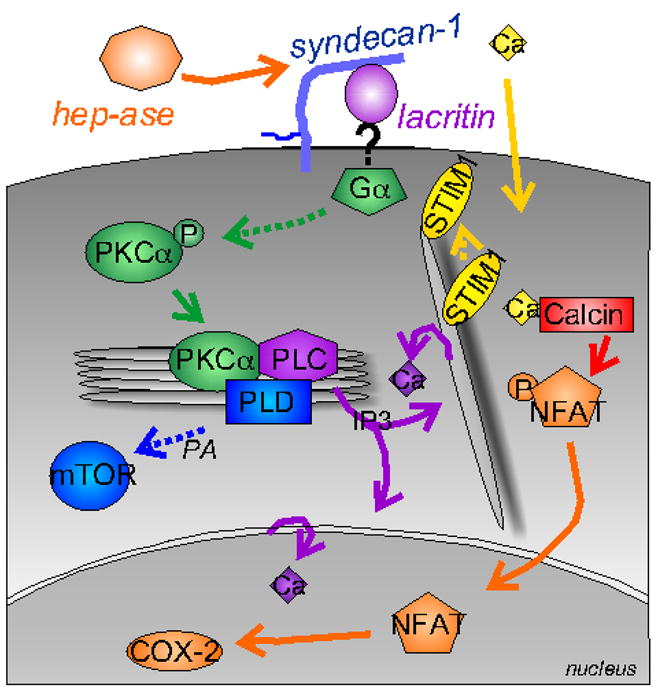
Lacritin signaling. Lacritin binding of syndecan-1 after the heparanase off/on switch triggers signaling through Gαi or Gαo/PKCα-PLC/Ca2+/calcineurin/NFATC1 and Gαi or Gαo/PKCα-PLC/PLD/mTOR signaling (Ma et al., 2006). Signaling proceeds in a biphasic dose-dependent manner requiring Ca2+ sensor STIM1.
PLD1 is a key mediator of cell secretion and proliferation by catalyzing the generation of phosphatidic acid to promote secretory granule release from the Golgi apparatus (Chen et al., 1997). Phosphatidic acid is important in regulated insulin secretion from pancreatic β-cells (Hughes et al., 2004). It also activates mTOR. mTOR is a serine/threonine kinase regulating multiple cellular processes including protein translation, cell proliferation and survival (Bhaskar & Hay, 2007). Cells depleted of mTOR (or PKCα by siRNA are unresponsive to lacritin in mitogenic assays (Wang et al., 2006).
Activation of PLCγ2 triggers a separate chain of events. Activated PLCγ2 catalyzes the hydrolysis of phosphatidylinositol biphosphate to generate inositol triphosphate (IP3) and diacylglycerol (DAG), likely in the perinuclear Golgi region. IP3 then binds IP3 receptors on the endoplasmic reticulum to drive Ca2+ mobilization from endoplasmic reticulum into the cytoplasm and then Ca2+ entry into the cell via the Ca2+ sensor STIM1. Prolonged Ca2+ entry activates the cyclosporin A-sensitive serine/threonine phosphatase calcineurin to in turn dephosphorylate the cytoplasmic form of transcription factor nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 - also known as NFATC1. Despite its name, NFATs are widely expressed in epithelia and other cell types (Horsley et al., 2008). Dephosphorylated NFATC1 translocates into the nucleus where it functions as a transcription factor in an obligatory cooperative manner with several other transcription factors to regulate growth and secretion (Heit et al., 2006). Cells depleted of NFATC1 by siRNA are unresponsive to lacritin in mitogenic assays (Wang et al., 2006). Thus lacritin signaling involves Gαi and Gαo/PKCα/PLCγ2/Ca2+/calcineurin/NFATC1 and Gαi and Gαo/PKCα/PLD1/mTOR.
The two diverging downstream pathways may interact (Wang et al., 2006), but are far from being completely understood. Deciphering cell signaling is at the heart of new drug design and understanding how drugs work. Although cyclosporin A suppresses T cell function to inhibit the inflammatory component of dry eye, a potential overlooked adverse effect is blockage of calcineurin/NFAT signaling in ocular surface epithelium. Many epithelial cells are absolutely dependent on calcineurin/NFAT signaling, ie. pancreatic β-cells (proliferation, differentiation and insulin production; Heit et al., 2006; Gurda et al., 2008) and lung epithelium (expression of aqaporin-1 and -5 and lysozyme; Dave et al., 2006).
1.5 Lacritin function: preliminary preclinical testing
In a small preclinical study, recombinant human lacritin was topically applied to rabbit eyes and basal tear flow measured (Samudre et al., submitted). A tear volume increase of 40% over vehicle control emerged, despite variability inherent in the Schirmer strip basal tear volume assay and problems with recombinant protein stability. Tear composition was normal with no significant changes noted in tear sodium, potassium, pH or protein concentration. Efficacy was apparent at the first time point (60 min) and was most significant at the last time point (240 min). Topical application of lacritin was also well tolerated by ocular tissues as indicated by negative Rose Bengal and lissamine green staining. Additional testing in normal and dry eye rodent models, and in non-human primates is needed to confirm these preliminary results. How might lacritin be prosecretory on the eye? The mechanism is unknown, but new data suggests that lacritin is a secretogogue for the ocular surface mucin MUC16 in corneal epithelial cell culture (Wang & Laurie, unpublished). MUC16 and other ocular mucins have short O-linked oligosaccharide chains, at least in human, rabbit and dog. Mucins together may form a continuous hydrophilic barrier on the ocular surface. Shedding of MUC16 leaves MUC1, MUC4 and other elements of the glycocalyx to help shield the cell surface. However, experimental knockdown of MUC16 (Blalock et al., 2007), dysregulation of MUC16 glycation (Sumiyoshi et al., 2008), or shedding of MUC16 (Blalock et al., 2008) appears to partially expose the corneal epithelial surface to bacteria and disrupt the normal tear film.
1.6 Lacritin and disease
Although it is still not clear what proteins make up the human tear proteome, over 400 have been reported. Preliminary studies involving small trials suggest that only 4 – 5% of these may be dysregulated in ocular surface disease, of which lacritin is the only one apparently capable of promoting tear production. Most have utilized 2-D SDS PAGE to compare quantities of proteins from normal tears versus tears from dry eye-related conditions. An estimate is generated from densitometry of silver-stained protein dots or by fluourescent measurement of cye3/cye5-stained protein dots, and protein identity of extracted dots is determined by mass spectrometric protein sequencing. In tears from patients with blepharitis, lacritin was downregulated ‘about 50%’ (Koo et al., 2005). Blepharitis is a common inflammation of the eyelid, associated with evaporative dry eye. 27 normals and 19 patients participated. A smaller study (Green-Church et al., 2007) comparing tears from 10 normals and 10 contact lens-related dry eye patients reported that ‘lacritin showed a ~7-fold downregulation’. The same group recently reported that lacritin is one of six tear proteins non-specifically absorbed to four different contact lenses (Green-Church & Nichols, 2008). LC-MS/MS suggests that lacritin may also be downregulated in Sjögren’s syndrome tears (Kitagawa et al., 2007). This follows an earlier SELDI study by the same group indicating differences in peak heights of unidentified proteins from normal versus Sjögren’s syndrome tears (Tomosugi et al., 2005). Large trials and quantitative capture assays are needed to transform lacritin tear data into molar levels that have physiological implications. Attention should also be paid to cell targeting (ie. levels of tear heparanase, disease-associated shedding of syndecan-1) and even cell signaling components.
2.1 Other new LFU proteins - originally hypothetical proteins
Over two hundred new human LFU proteins have been discovered over the past decade, most from unbiased sequencing of thousands of human lacrimal gland cDNAs for NEIBank’s human lacrimal gland EST database (Ozyildirim et al., 2005). Here we define new proteins as those that were originally designated as hypothetical indicating a protein predicted from informatic analysis of the human genome. Of these, only the cytoskeletal protein myosin heavy chain 14 (MYH14) overlaps with the de Souza et al. (2006) tear proteome. Dermokine (DMKN) is extracellular but not apparently in tears. The remainder appear to be intracellular. We previously noted that more hypotheticals per total number of sequenced transcripts are concentrated in human lacrimal gland than in any other human organ (Ozyildirim et al., 2005). Although this term is slowly disappearing from GenBank with gradual protein characterization, we have the original human lacrimal gland EST data. Table 1 documents all past and current LFU hypothetical proteins integrated from NCBI Protein, BodyMap and NEIBank databases. Their current RefSeq protein designation, and known or predicted functions are indicated. Many are highly conserved (Ensembl). Some are disease genes or associated with disease genes (OMIM). Others may be important in LFU cell biology and androgen or estrogen responsiveness. We briefly highlight several in the following sections.
TABLE 1.
Other new (formerly hypothetical) proteins of the LFU
| Gene Symbol | Protein |
|---|---|
| (i) Apoptosis/Autophagy | |
| FAM82C | family with sequence similarity 82, member C3,6 |
| RNF144B | ring finger 144B1,6 |
| WIPI1 | WD repeat dom., phosphoinositide interact. 11,6 |
| ZC3H12A | Zn finger CCCH-type containing 12A1,6 |
| (ii) ATPase/Transferase/Other Enzyme | |
| ANKRD36B | ankyrin repeat domain 36B1,7 |
| ATP13A3 | ATPase type 13A31,6 |
| C14orf172 | chromosome 14 open reading frame 1721,6 |
| C18orf25 | chromosome 18 open reading frame 251,6 |
| CARS2 | cysteinyl-tRNA synth. 2, mitochon. (putative)3,10 |
| CCDC46 | coiled-coil domain containing 461,6 |
| CCDC57 | coiled-coil domain containing 571,6 |
| CHD1L | chromodom. helicase DNA bp 1-like1,9 |
| COX18 | COX18 cytochr. c oxidase assem. hom.1,10 |
| DALRD3 | DALR anticodon binding domain containing 31,6 |
| EME2 | essential meiotic endonuclease 1 homolog 23,6 |
| FLJ20628 | hypothetical protein FLJ206281,6 |
| HECTD3 | HECT domain containing 31,6 |
| LOC100134-860 | hypothetical protein LOC1001348603,9 |
| MANSC1 | MANSC domain containing 11,6 |
| N4BP2L2 | NEDD4 binding protein 2-like 21,6 |
| PCMTD1 | prot.-L-isoasp. O-methyltransf. dom. contain. 13,6 |
| POLG | polymerase (DNA directed), gamma3,8,9 |
| TARS2 | threonyl-tRNA synthetase 2, mitochond. (put.) 3,6 |
| UPRT | uracil phosphoribosyltransf. (FUR1) homol. 3,6 |
| ZDHHC21 | zinc finger, DHHC-type containing 213,6 |
| (iii) Calcium | |
| C9orf140 | chromosome 9 open reading frame 1402,6 |
| (iv) Cell Adhesion/Motility/Structure | |
| CEP192 | centrosomal protein 192kDa1,6 |
| CGNL1 | cingulin-like 11,6 |
| FNBP1L | formin binding protein 1-like5,7 |
| MPZL3 | myelin protein zero-like 33,7 |
| MYH14 | myosin, heavy chain 145,6 |
| (v) Cell Growth | |
| C2orf29 | chromosome 2 open reading frame 292,6 |
| LYAR | Ly1 antibody reactive homolog (mouse)3,10 |
| (vi) Endosomal/Lysosomal | |
| ALS2CL | ALS2 C-terminal like1,6 |
| CHMP7 | CHMP family, member 73,6 |
| KIAA0528 | KIAA05282,6 |
| NECAP2 | NECAP endocytosis associated 23,8,9 |
| STX17 | syntaxin 171,6 |
| TTC13 | tetratricopeptide repeat domain 131,6 |
| UFM1 | ubiquitin-fold modifier 12,6 |
| VPS37A | vacuolar protein sorting 37 homolog A 3,6 |
| ZFYVE27 | zinc finger, FYVE domain containing 271,6 |
| (vii) Extracellular | |
| DMKN | dermokine3,6 |
| FAM20B | family with sequence similarity 20, member B2,6 |
| (viii) Immune | |
| KIAA1109 | KIAA11091,6 |
| (ix) Lipid/Cholesterol | |
| IAH1 | soamyl acetate-hydrolyz. esterase 1 homol. 3,6 |
| LASS4 | LAG1 homolog, ceramide synthase 43,6 |
| MTMR10 | myotubularin related protein 101,6 |
| PLCXD1 | PI-spec. phospholipase C, X dom. contain. 11,6 |
| (x) Other/Unknown | |
| ARGLU1 | arginine and glutamate rich 11,6 |
| C1orf58 | chromosome 1 open reading frame 581,6 |
| C1orf80 | chromosome 1 open reading frame 801,6 |
| C1orf109 | chromosome 1 open reading frame 1092,6 |
| C1orf163 | chromosome 1 open reading frame 1632,6 |
| C4orf18 | chromosome 4 open reading frame 182,6 |
| C4orf28 | chromosome 4 open reading frame 282,6 |
| C5orf24 | chromosome 5 open reading frame 242,6 |
| C5orf28 | chromosome 5 open reading frame 282,6 |
| C5orf30 | chromosome 5 open reading frame 302,10 |
| C6orf70 | chromosome 6 open reading frame 701,6 |
| C6orf106 | chromosome 6 open reading frame 1061,6 |
| C7orf47 | chromosome 7 open reading frame 472,10 |
| C8orf42 | chromosome 8 open reading frame 421,6 |
| C9orf41 | chromosome 9 open reading frame 412,6 |
| C10orf104 | chromosome 10 open reading frame 1041,6 |
| C11orf48 | chromosome 11 open reading frame 481,10 |
| C11orf56 | chromosome 11 open reading frame 561,6 |
| C12orf30 | chromosome 12 open reading frame 301,6 |
| C12orf35 | chromosome 12 open reading frame 351,6 |
| C12orf44 | chromosome 12 open reading frame 441,6 |
| C14orf159 | chromosome 14 open reading frame 1591,8 |
| C16orf58 | chromosome 16 open reading frame 582,6 |
| C17orf45 | chromosome 17 open reading frame 452,6 |
| C17orf56 | chromosome 17 open reading frame 562,10 |
| C19orf42 | chromosome 19 open reading frame 421,10 |
| CAMSAP1 | calmodulin regulated spectrin-assoc. prot. 13,8,9 |
| CCDC49 | coiled-coil domain containing 493,6 |
| CCDC94 | coiled-coil domain containing 943,6 |
| CCDC109A | coiled-coil domain containing 109A3,6 |
| CCDC117 | coiled-coil domain containing 1173,6 |
| CCNYL1 | cyclin Y-like 13,6 |
| CXorf39 | chromosome X open reading frame 392,6 |
| FAM46A | family with sequence similarity 46, member A1,6 |
| FAM46C | family with sequence similarity 46, member C1,6 |
| FAM65A | family with sequence similarity 65, member A1,6 |
| FAM76B | family with sequence similarity 76, member B1,6 |
| FAM83E | family with sequence similarity 83, member E1,6 |
| FAM96A | family with sequence similarity 96, member A1,10 |
| FAM107B | family with sequence similarity 107, member B2,6 |
| FAM114A1 | family with sequence similarity 114, mem. A12,6 |
| FAM128B | family with sequence simil. 128, member B1,10 |
| FAM133B | family with sequence simil. 133, member B1,10 |
| FAM116A | family with sequence simil. 116, member A2,10 |
| FCHO2 | FCH domain only 23,6 |
| FLJ14154 | hypothetical protein FLJ141541,10 |
| FLJ20254 | FLJ202541,6 |
| FLJ23584 | hypothetical FLJ235844,9 |
| FLJ31818 | hypothetical protein FLJ318181,6 |
| JAGN1 | jagunal homolog 1 (Drosophila)3,6 |
| KIAA0515 | KIAA05151,6 |
| KIAA0913 | KIAA09131,9 |
| KIAA1545 | KIAA1545 protein4,10 |
| KRCC1 | lysine-rich coiled-coil 13,10 |
| LMF2 | lipase maturation factor 23,6 |
| LOC26010 | viral DNA polymerase-transactivated protein 61,8 |
| LOC129293 | hypothetical protein LOC1292932,10 |
| LOC201229 | hypothetical protein LOC2012291,10 |
| LOC283951 | hypothetical protein LOC2839512,10 |
| LOC441150 | similar to RIKEN cDNA 2310039H083,10 |
| MGC3207 | hypothetical protein MGC32071,10 |
| MGC70857 | similar to RIKEN cDNA C030006K11 gene2,10 |
| NHLRC2 | NHL repeat containing 23,6 |
| NOL10 | nucleolar protein 101,6 |
| NBPF15 | neuroblastoma breakpoint family, member 152,10 |
| NPC-A-5 | NPC-A-51,10 |
| ODAM | odontogenic, ameloblast asssociated1,6 |
| PRR14 | proline rich 143,10 |
| PRRC1 | proline-rich coiled-coil 13,6 |
| RWDD4A | RWD domain containing 4A1,6 |
| SERTAD4 | SERTA domain containing 43,6 |
| SETD5 | SET domain containing 53,6 |
| SPG11 | spastic paraplegia 11 (autosomal recessive) 1,6 |
| SPTY2D1 | SPT2, Suppressor of Ty, domain containing 11,6 |
| STAG3L4 | stromal antigen 3-like 41,10 |
| SYCP2L | synaptonemal complex protein 2-like3,6 |
| TMCO4 | transmembrane and coiled-coil domains 43,6 |
| TMEM25 | transmembrane protein 253,6 |
| TMEM32 | transmembrane protein 323,6 |
| TMEM43 | transmembrane protein 433,6 |
| TMEM44 | transmembrane protein 441,6 |
| TMEM62 | transmembrane protein 621,6 |
| TMEM66 | transmembrane protein 663,6 |
| TMEM85 | transmembrane protein 853,6 |
| TMEM100 | transmembrane protein 1001,10 |
| TMEM109 | transmembrane protein 1093,6 |
| TMEM125 | transmembrane protein 1253,6 |
| TMEM127 | transmembrane protein 1273,6 |
| TMEM134 | transmembrane protein 1342,10 |
| TMEM168 | transmembrane protein 1681,6 |
| TMEM184A | transmembrane protein 184A1,6 |
| TTC17 | tetratricopeptide repeat domain 173,6 |
| TTC19 | tetratricopeptide repeat domain 195,6 |
| TTC21B | tetratricopeptide repeat domain 21B1,6 |
| TTC31 | tetratricopeptide repeat domain 313,6 |
| UHRF1BP1 | UHRF1 (ICBP90) binding protein 11,6 |
| WDR52 | WD repeat domain 523,6 |
| WDR60 | WD repeat domain 601,6 |
| (xi) Protein Production/Binding/Complex | |
| C4orf34 | chromosome 4 open reading frame 342,6 |
| CCDC130 | coiled-coil domain containing 1303,6 |
| EIF4B | eukaryotic translation initiation factor 4B 1,6 |
| FAM96A | FAM96A1,6 |
| FAM136A | family with sequence similarity 136, member A2,6 |
| NOC4L | nucleolar complex associated 4 homolog 3,6 |
| ORMDL1 | ORM1-like 13,6 |
| PHF15 | PHD finger protein 153,6 |
| (xii) Proteinase/Hydrolase/Lysase | |
| AMZ2 | archaelysin family metallopeptidase 21,6 |
| CMBL | carboxymethylenebutenolidase homolog1,6 |
| ECHDC2 | enoyl Coenzyme A hydratase dom. contain. 21,6 |
| FAM111A | family with sequence similarity 111, member A1,6 |
| FAM113B | family with sequence similarity 113, member B2,6 |
| FLJ35220 | hypothetical protein FLJ352202,10 |
| KIAA1434 | hypothetical protein KIAA14341,10 |
| NUDT16 | nudix-type motif 163,6 |
| SCRN2 | secernin 23,6 |
| SERHL | serine hydrolase-like1,6 |
| SPATA20 | spermatogenesis associated 203,6 |
| UFSP2 | UFM1-specific peptidase 23,10 |
| (xiii) Receptor/Channel/Transport | |
| GPRC5B | G prot.-coupled recept., fam. C, grp 5, mem. B5,6 |
| KCTD12 | K channel tetramerisation dom. containing 121,6 |
| PLXNA1 | plexin A11,6 |
| SHKBP1 | SH3KBP1 binding protein 13,6 |
| SLC25A46 | solute carrier family 25, member 462,6 |
| TMEM104 | transmembrane protein 1041,6 |
| (xiv) RNA Processing | |
| CCDC131 | coiled-coil domain containing 1311,6 |
| CNOT6L | CCR4-NOT transcripti. complex, subunit 6-like1,6 |
| CWF19L1 | CWF19-like 1, cell cycle control1,6 |
| INTS3 | integrator complex subunit 31,6 |
| INTS10 | integrator complex subunit 101,6 |
| MAGOHB | mago-nashi homolog B 3,6 |
| NOL12 | nucleolar protein 121,6 |
| NPIP | nuclear pore complex interacting protein1,6 |
| RBM35B | RNA binding motif protein 35B1,6 |
| SFRS2 | splicing factor, arginine/serine-rich 23,6 |
| ZMAT5 | zinc finger, matrin type 51,6 |
| (xv) Signal Transduction | |
| ARHGEF19 | Rho guanine nucleotide exchange factor 193,6 |
| C10orf81 | chromosome 10 open reading frame 812,6 |
| C19orf50 | chromosome 19 open reading frame 502,6 |
| DEF8 | differentially expressed in FDCP 8 homolog1,6 |
| FLJ10357 | hypothetical protein FLJ103571,6 |
| KIAA1975 | KIAA1975 protein similar to MRIP21,10 |
| LOC440456 | sim. plecks. homol. dom. fam. M memb. 14,6 |
| NOMO1 | NODAL modulator 15,6 |
| SH2D4A | SH2 domain containing 4A 3,6 |
| SPATA13 | spermatogenesis associated 133,6 |
| TBC1D17 | TBC1 domain family, member 173,6 |
| (xvi) Transcription/Nucleic Acid Binding | |
| AFTPH | aftiphilin1,5 |
| ANKZF1 | ankyrin repeat and Zn finger dom. containing 11,6 |
| APBB2 | amyloid beta prec. prot.-bind., fam. B, mem. 2)5,6 |
| ATPBD1B | ATP binding domain 1 family, member B1,9 |
| CREB3L2 | cAMP responsive element bind. prot. 3-like 21,6 |
| BSDC1 | BSD domain containing 13,6 |
| BTBD14B | BTB (POZ) domain containing 14B1,6 |
| C9orf86 | chromosome 9 open reading frame 861,6 |
| C9orf152 | chromosome 9 open reading frame 1521,6 |
| CCDC75 | coiled-coil domain containing 751,6 |
| CXXC5 | CXXC finger 53,6 |
| MED25 | mediator complex subunit 251,6 |
| MGC16385 | hypothetical protein MGC163851,6 |
| NSMCE4A | non-SMC element 4 homolog A 3,6 |
| RBPJ | recombin. signal bp for Ig kappa J region1,9 |
| STOX1 | storkhead box 13,6 |
| TCEAL4 | transcription elongation factor A (SII)-like 45,6 |
| ZCCHC6 | zinc finger, CCHC domain containing 61,6 |
| ZGPAT | zinc finger, CCCH-type with G patch domain1,6 |
| ZMAT2 | zinc finger, matrin type 23,6 |
| ZMIZ2 | zinc finger, MIZ-type containing 21,6 |
| ZNF280D | zinc finger protein 280D1,6 |
| ZNF517 | zinc finger protein 5171,6 |
| ZNF557 | zinc finger protein 5571,6 |
| ZNF618 | zinc finger protein 6181,6 |
| ZNF664 | zinc finger protein 6643,6 |
| ZNF700 | zinc finger protein 7003,6 |
| ZNF821 | zinc finger protein 8213,6 |
Validated1, predicted2, provisional3, model4 or reviewed5 RefSeq proteins from: (1) accession number search of all human lacrimal6 EST’s (Ozyilidirim et al., 2005) originally linked to proteins bioinformatically designated as hypothetical, and (2) keyword ‘eye and hypothetical and human’ search of the NCBI Protein database. The latter hits were then restricted to all sources associated with the human lacrimal functional unit: ‘cytoplasmic corneal stroma7’, ‘eye’ (then traced to lacrimal gland6 by accession number) and ‘eye anterior segment8’. Hits (‘hypothetical’) from keratoconus9 and BodyMap10 cornea were included. Most derive from the human lacrimal gland6.
2.2 Other new LFU proteins: Risk factor locus for autoimmune disease
KIAA1109 is a large (556 kDa) integral membrane protein with at least twelve splice variants and a predicted nuclear location. Although highly conserved, no functional or structural information is available. The KIAA1109 gene is proximal to ADAD1 (adenosine deaminase domain containing 1), IL2 (interleukin 2) and IL21 (interleukin 21) genes. All three reside on a 480 kb section of human chromosome 4q27 in linkage disequilibrium for celiac disease, rheumatoid arthritis and type 1 diabetes - suggesting that this locus may be a general risk factor for autoimmune disease (Zhernakova et al., 2007). Secondary Sjögren’s syndrome, an autoimmune disease of the LFU, is associated with rheumatoid arthritis. Although IL2 and IL21 are the suspected disease genes, the KIAA1109 SNP (single nucleotide polymorphism) rs13119723 is more significantly linked to celiac disease than any other SNP outside of the HLA region. Further meta-analysis identified a strong association at SNP rs6822844 which is 24 kb 5′ of IL21 (van Heel et al., 2007).
2.3 Other new LFU proteins: Human genetic diseases
ALS2CL (ALS2 C-terminal like) is a conserved 108 kDa endosomal protein with eight MORN (membrane occupation and recognition nexus), one PH (pleckstrin homology) and one VPS9 (domain present in yeast vacuolar sorting protein 9 and other proteins) domains. Aceview predicts eighteen or more splice variants. Its C-terminal Rab5-GDP/GTP exchange (GEF) activity region (Hadano et al., 2004) is homologous with the larger Golgi protein ALS2 (amyotrophic lateral sclerosis 2) that also shares MORN and VPS9 domains. ALS2 is the disease gene for juvenile recessive amyotrophic lateral sclerosis. ALS2 is also associated with primary lateral sclerosis and hereditary spastic paraplegia (Chandran et al., 2007). All are neurological degenerative disorders of motor neurons. ALS2CL homodimers bind ALS2 oligomers on membranes to reduce ALS2-depedent enlargement of endosomes (Suzuki-Utsunomiya et al., 2007). Yang et al (1999) noted changes in the distribution of lacrimal acinar cell Rab5 in endosomal compartments with carbachol stimulation as part of a larger theme of aberrant endosomal sorting in development of LFU autoimmune disease. APBB2 (amyloid beta precursor protein-binding, family B, member 2) is a conserved 81 kDa nuclear or cytoplasmic protein with WW (two conserved tryptophan residues) and two PTP (phosphotyrosine binding) domains. WW domains bind proline and phosphoserine/phosphothreonine motifs. Aceview predicts seventeen splice variants.
APBB2 binds APP (amyloid beta precursor protein), whose mutated gene is associated with autosomal dominant early onset Alzheimer disease in individuals developing disease prior to 75 years old (Li et al., 2005).
CREB3L2 (cAMP responsive element binding protein 3-like 2) is a conserved 57.4 kDa integral membrane protein with a predicted nuclear location and single BRLZ (basic region leucine zipper), transmembrane and CC (coiled coil) domains. Aceview predicts nine splice variants. Translocation of CREB3L2 and FUS chromosome bands form a chimera associated with human fibrosarcomas (Mertens et al., 2005).
MYH14 (myosin, heavy chain 14) is a 229 kDa cytoskeletal protein with myosin, IQ (calmodulin-binding) and CC domains. Aceview predicts six splice variants. MYH14 gene point mutations in some individuals have been associated with progressive hearing loss (Donaudy et al., 2004). From NOD mouse studies and developmental expression of the transcription factor Hox11 (Tlx1) in pancreas, salivary gland, tongue and cochlea, Lonyai et al. (2008) recently proposed a Hox11-associated developmental link for the hearing loss observed in some Sjögren’s syndrome patients.
POLG (polymerase [DNA directed], gamma) is a conserved 139.6 kDa mitochondrial DNA polymerase. POLG contains one CC motif. Aceview predicts ten splice variants. Human missense mutations and deletions in the POLG gene are associated with several human genetic diseases including progressive external ophthalmoplegia or ophthalmoparesis (extraocular muscle weakness or paralysis; Van Goethem et al., 2002) and Alper’s syndrome (cerebral degenerative disease in children).
SPG11 (spastic paraplegia 11 [autosomal recessive]) is evolutionarily conserved and is predicted to be a large secreted protein of approximately 275 kDa without signal peptide. SPG11 contains one CC motif. Aceview predicts eighteen splice variants. Point mutations in the SPG11 gene are associated with neurodegenerative thinning of the corpus callosum leading to spastic paraplegia in children (Stevanin et al., 2007).
TMEM43 (transmembrane protein 43) is a conserved 44.9 kDa integral membrane protein likely of the endoplasmic reticulum and Golgi apparatus. TMEM43 contains four predicted transmembrane domains. Aceview predicts six splice variants. MIssense mutation of the TMEM43 gene causes arrhythmogenic right ventricular dysplasia (Merner et al., 2008).
ZFYVE27 (zinc finger, FYVE domain containing 27) is a conserved 46.4 kDa integral membrane protein with three transmembrane and one C-terminal FYVE (protein present in Fab1, YOTB, Vac1, and EEA1) domains. Aceview predicts 28 splice variants. Point mutation in the ZFYVE27 gene is associated with spastic paraplegia (Mannan et al., 2006). ZFYVE27 is an ERK phosphorylated regulator of Rab11 membrane trafficking (Shirane and Nakayama, 2006). Active Rab11 is an endosomal GTPase. Its major roles are in sorting within the endosomal pathway and in regulation of transcytosis. Changes in its activity in transcytosis might result in abnormalities in the flow of proteins into tears. Rab11 also appears to play a role in docking multivesicular bodies with autophagosomes (Fader et al., 2008). Autophagosomes and autophagy are mechanistically linked to cell survival or death mechanisms and innate defense.
2.4 Other new LFU proteins: Androgen or estrogen signaling
WIPI1 (WD repeat domain, phosphoinositide interacting 1) is a conserved 48.7 kDa cytoplasmic protein with two WD40 repeats suggesting a scaffolding function in protein-protein interactions. Aceview predicts eleven splice variants. In pull-down assays, WIPI1 binds androgen, estrogen and retinoic acid receptors. The LFU’s androgen-dependence has focused much attention on the contribution of androgen insufficiency to dry eye (reviewed by Sullivan, 2004). Retinoic acid is an LFU androgen antagonist (Ubels et al., 2002). Use of estrogen in postmenopausal hormone replacement therapy places women at increased risk of dry eye syndrome (Schaumberg et al., 2001). Thus WIPI1 potentially integrates three key elements of LFU physiology. WIPI1 appears to accumulate in trans-Golgi and endosomal membranes, and on LC3-positive autophagic membranes in a PI3K dependent manner (Proikas-Cezanne et al., 2007). Autophagy is a complicated cell survival mechanism that can be also utilized as a cell death pathway.
ZMIZ2 is a conserved 96.5 kDa cyotplasmic or nuclear protein with one NLS (nuclear localization) domain that colocalizes with and binds the androgen receptor. Binding enhances androgen receptor transcription (Huang et al., 2005). Aceview predicts twelve splice variants.
2.5 Other new LFU proteins: Cell biological features
ARHGEF19 (Rho guanine nucleotide exhange factor [GEF] 19) is an 89.2 kDa conserved nuclear or cytoplasmic Rho activating GEF with RhoGEF, PH and SH3 (Src homology 3) domains. Aceview predicts 9 splice variants.
PLXNA1 (plexin A1) is a conserved 208.6 kDa plasma membrane protein with one Sema (semaphorin), two PSI (domain found in plexins, semaphorins and integrins) and three IPT (Ig-like, plexins, transcription factors) domains. Aceview predicts seven splice variants. PLXNA1 is a cell surface signaling receptor for its ligands Sema3A and Sema6D. Sema6D targeting of dendritic cells induces IL-12 production; and PLXNA1−/− dendritic cells poorly stimulate allogeneic and antigen-specific T-cells versus wild type cells. PLXNA1 thus appears to play a key role in T-cell stimulation (Takegahara et al., 2006). Although PLXNA1 has not apparently been examined in Sjögren’s syndrome, the disease is associated with invading dendritic cells and increased IL-12 (Manoussakis et al., 2007). PLXNA1 also is a negative regulator of epithelial and endothelial migration and axonal migration (Bachelder et al., 2003).
3.0 Conclusions
Exploring lacritin and the over two hundred new LFU proteins that have been discovered over the last decade together offer a unique opportunity to understand human ocular surface physiology at a level of complexity previously unimagined. Some will expand the frontiers of biology to benefit all fields. This back to basics molecular approach gets to the heart of ocular surface dysfunction.
Acknowledgments
GWL and RLM are supported by R01 EY013143 and EY018222, and in part by R42 EY015376 to PBW. This research was also supported by grant funding from Virginia’s Commonwealth Health Research Board (RLM).
Abreviations
- LACRT
lacritin
- SDC1
syndecan-1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- The definition and classification of dry eye disease: report of the Definition and Classification Subcommittee of the International Dry Eye WorkShop. Ocul Surf. 2007;2:75–92. doi: 10.1016/s1542-0124(12)70081-2. [No authors listed] [DOI] [PubMed] [Google Scholar]
- Bachelder RE, Lipscomb EA, Lin X, Wendt MA, Chadborn NH, Eickholt BJ, Mercurio AM. Competing autocrine pathways involving alternative neuropilin-1 ligands regulate chemotaxis of carcinoma cells. Cancer Res. 2003;63:5230–5233. [PubMed] [Google Scholar]
- Berk RS, Dong Z, Alousi S, Kosir MA, Wang Y, Vlodavsky I. Murine ocular heparanase expression before and during infection with Pseudomonas aeruginosa. Invest Ophthalmol Vis Sci. 2004;45:1182–1187. doi: 10.1167/iovs.03-0589. [DOI] [PubMed] [Google Scholar]
- Bertolesi GE, Michaiel G, McFarlane S. Two heparanase splicing variants with distinct properties are necessary in early Xenopus development. J Biol Chem. 2008;283:16004–16016. doi: 10.1074/jbc.M708525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar PT, Hay N. The two TORCs and Akt. Dev Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Blalock TD, Spurr-Michaud SJ, Tisdale AS, Heimer SR, Gilmore MS, Ramesh V, Gipson IK. Functions of MUC16 in corneal epithelial cells. Invest Ophthalmol Vis Sci. 2007;48:4509–4518. doi: 10.1167/iovs.07-0430. [DOI] [PubMed] [Google Scholar]
- Blalock TD, Spurr-Michaud SJ, Tisdale AS, Gipson IK. Release of membrane- associated mucins from ocular surface epithelial. Invest Ophthalmol Vis Sci. 2008;49:1864–1871. doi: 10.1167/iovs.07-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borzok MA, Catino DH, Nicholson JD, Kontrogianni-Konstantopoulos A, Bloch RJ. Mapping the binding site on small ankyrin 1 for obscurin. J Biol Chem. 2007;282:32384–32396. doi: 10.1074/jbc.M704089200. [DOI] [PubMed] [Google Scholar]
- Chandran J, Ding J, Cai H. Alsin and the molecular pathways of amyotrophic lateral sclerosis. Mol Neurobiol. 2007;36:224–231. doi: 10.1007/s12035-007-0034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Siddhanta A, Austin CD, Hammond SM, Sung TC, Frohman MA, Morris AJ, Shields D. Phospholipase D stimulates release of nascent secretory vesicles from the trans-Golgi network. J Cell Biol. 1997;138:495–504. doi: 10.1083/jcb.138.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davé V, Childs T, Xu Y, Ikegami M, Besnard V, Maeda Y, Wert SE, Neilson JR, Crabtree GR, Whitsett JA. Calcineurin/Nfat signaling is required for perinatal lung maturation and function. J Clin Invest. 2006;116:2597–2609. doi: 10.1172/JCI27331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza GA, Godoy LM, Mann M. Identification of 491 proteins in the tear fluid proteome reveals a large number of proteases and protease inhibitors. Genome Biol. 2006;7:R72. doi: 10.1186/gb-2006-7-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaudy F, Snoeckx R, Pfister M, Zenner HP, Blin N, Di Stazio M, Ferrara A, Lanzara C, Ficarella R, Declau F, Pusch CM, Nürnberg P, Melchionda S, Zelante L, Ballana E, Estivill X, Van Camp G, Gasparini P, Savoia A. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4) Am J Hum Genet. 2004;74:770–776. doi: 10.1086/383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader CM, Sánchez D, Furlán M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- Garfin DE. One-dimensional gel electrophoresis. Methods Enzymol. 1990;182:425–441. doi: 10.1016/0076-6879(90)82035-z. [DOI] [PubMed] [Google Scholar]
- Gu S, He J, Ho WT, Ramineni S, Thal DM, Natesh R, Tesmer JJ, Hepler JR, Heximer SP. Unique hydrophobic extension of the RGS2 amphipathic helix domain imparts increased plasma membrane binding and function relative to other RGS R4/B subfamily members. J Biol Chem. 2007;282:33064–33075. doi: 10.1074/jbc.M702685200. [DOI] [PubMed] [Google Scholar]
- Green-Church KB, Sessler R, Nichols JJ. Proteomic analyses in contact lens- related dry eye. Invest Ophthalmol Vis Sci. 2007;48 E-Abstract 5405. [Google Scholar]
- Green-Church KB, Nichols JJ. Mass spectrometry-based proteomic analyses of contact lens deposition. Mol Vis. 2008;14:291–297. [PMC free article] [PubMed] [Google Scholar]
- Gurda GT, Guo L, Lee SH, Molkentin JD, Williams JA. Cholecystokinin Activates Pancreatic Calcineurin-NFAT Signaling In Vitro and In Vivo. Mol Biol Cell. 2008;19:198–206. doi: 10.1091/mbc.E07-05-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadano S, Otomo A, Suzuki-Utsunomiya K, Kunita R, Yanagisawa Y, Showguchi- Miyata J, Mizumura H, Ikeda JE. ALS2CL, the novel protein highly homologous to the carboxy-terminal half of ALS2, binds to Rab5 and modulates endosome dynamics. FEBS Lett. 2004;575:64–70. doi: 10.1016/j.febslet.2004.07.092. [DOI] [PubMed] [Google Scholar]
- Hawrani A, Howe RA, Walsh TR, Dempsey CE. Origin of low mammalian cell toxicity in a class of highly active antimicrobial amphipathic helical peptides. J Biol Chem. 2008 Apr 23; doi: 10.1074/jbc.M709154200. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Heimann H, Coupland SE, Gochman R, Hellmich M, Foerster MH. Alterations in expression of mucin, tenascin-c and syndecan-1 in the conjunctiva following retinal surgery and plaque radiotherapy. Graefes Arch Clin Exp Ophthalmol. 2001;239:488–495. doi: 10.1007/s004170100301. [DOI] [PubMed] [Google Scholar]
- Heit JJ, Apelqvist AA, Gu X, Winslow MM, Neilson JR, Crabtree GR, Kim SK. Calcineurin/NFAT signalling regulates pancreatic beta-cell growth and function. Nature. 2006;443:345–349. doi: 10.1038/nature05097. [DOI] [PubMed] [Google Scholar]
- Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E. NFATc1 balances quiescence and proliferation of skin stem cells. Cell. 2008;132:299–310. doi: 10.1016/j.cell.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Beliakoff J, Li X, Lee J, Li X, Sharma M, Lim B, Sun Z. hZimp7, a novel PIAS-like protein, enhances androgen receptor-mediated transcription and interacts with SWI/SNF-like BAF complexes. Mol Endocrinol. 2005;9:2915–2929. doi: 10.1210/me.2005-0097. [DOI] [PubMed] [Google Scholar]
- Hughes WE, Elgundi Z, Huang P, Frohman MA, Biden TJ. Phospholipase D1 regulates secretagogue-stimulated insulin release in pancreatic beta-cells. J Biol Chem. 2004;279:27534–27541. doi: 10.1074/jbc.M403012200. [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Kimzey AL, Masselon CD, Smith RD, Dunker AK, Ackerman EJ. Aberrant mobility phenomena of the DNA repair protein XPA. Protein Sci. 2001;10:1353–1362. doi: 10.1110/ps.ps.40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Baum J. Basic tear flow. Does it exist? Ophthalmology. 1980;87:920–930. doi: 10.1016/s0161-6420(80)35143-9. [DOI] [PubMed] [Google Scholar]
- Karring H, Thøgersen IB, Klintworth GK, Møller-Pedersen T, Enghild JJ. The human cornea proteome: bioinformatic analyses indicate import of plasma proteins into the cornea. Mol Vis. 2006;12:451–460. [PubMed] [Google Scholar]
- Kitagawa K, Tomosugi N, Tuchida H, Sasaki H. Automated identification and quantification of tear proteins in Sjögren’s syndrome by LC-MS on a hybrid linear ion trap mass spectrometer. 5th International Conference on the Tear Film and Ocular Surface: Basic Science and Clinical Relevance; Taormina, Sicily, Italy. 2007. [Google Scholar]
- Koo BS, Lee DY, Ha HS, Kim JC, Kim CW. Comparative analysis of the tear protein expression in blepharitis patients using two-dimensional electrophoresis. J Proteome Res. 2005;4:719–724. doi: 10.1021/pr0498133. [DOI] [PubMed] [Google Scholar]
- Laurie GW, Olsakovsky LA, Conway BP, McKown RL, Kitagawa K, Nichols JJ. Dry eye and designer ophthalmics. Optom Vis Sci. 2008;85:643–652. doi: 10.1097/OPX.0b013e318181ae73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin MH, Verkman AS. Aquaporin-dependent water permeation at the mouse ocular surface: in vivo microfluorimetric measurements in cornea and conjunctiva. Invest Ophthalmol Vis Sci. 2004;45:4423–4432. doi: 10.1167/iovs.04-0816. [DOI] [PubMed] [Google Scholar]
- Li Y, Hollingworth P, Moore P, Foy C, Archer N, Powell J, Nowotny P, Holmans P, O’Donovan M, Tacey K, Doil L, van Luchene R, Garcia V, Rowland C, Lau K, Cantanese J, Sninsky J, Hardy J, Thal L, Morris JC, Goate A, Lovestone S, Owen M, Williams J, Grupe A. Genetic association of the APP binding protein 2 gene (APBB2) with late onset Alzheimer disease. Hum Mutat. 2005;25:270–277. doi: 10.1002/humu.20138. [DOI] [PubMed] [Google Scholar]
- Lonyai A, Kodama S, Burger D, Faustman DL. Fetal Hox11 expression patterns predict defective target organs: a novel link between developmental biology and autoimmunity. Immunol Cell Biol. 2008;86:301–309. doi: 10.1038/icb.2008.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Beck SL, Raab RW, McKown RL, Coffman GL, Utani A, Chirico WJ, Rapraeger AC, Laurie GW. Heparanase deglycanation of syndecan-1 is required for binding of the epithelial-restricted prosecretory mitogen lacritin. J Cell Biol. 2006;74:1097–1106. doi: 10.1083/jcb.200511134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P, Wang N, McKown RL, Raab RW, Laurie GW. Focus on Molecules: Lacritin. Exp Eye Res. 2008;86:457–458. doi: 10.1016/j.exer.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majczak G, Lilla S, Garay-Malpartida M, Markovic J, Medrano FJ, de Nucci G, Belisario JE. Prediction and biochemical characterization of intrinsic disorder in the structure of proteolysis-inducing factor/dermcidin. Genet Mol Res. 2007;6:1000–1011. [PubMed] [Google Scholar]
- Mannan AU, Krawen P, Sauter SM, Boehm J, Chronowska A, Paulus W, Neesen J, Engel W. ZFYVE27 (SPG33), a novel spastin-binding protein, is mutated in hereditary spastic paraplegia. Am J Hum Genet. 2006;79:351–357. doi: 10.1086/504927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoussakis MN, Boiu S, Korkolopoulou P, Kapsogeorgou EK, Kavantzas N, Ziakas P, Patsouris E, Moutsopoulos HM. Rates of infiltration by macrophages and dendritic cells and expression of interleukin-18 and interleukin-12 in the chronic inflammatory lesions of Sjögren’s syndrome: correlation with certain features of immune hyperactivity and factors associated with high risk of lymphoma development. Arthritis Rheum. 2007;56:3977–3988. doi: 10.1002/art.23073. [DOI] [PubMed] [Google Scholar]
- Merner ND, Hodgkinson KA, Haywood AF, Connors S, French VM, Drenckhahn JD, Kupprion C, Ramadanova K, Thierfelder L, McKenna W, Gallagher B, Morris-Larkin L, Bassett AS, Parfrey PS, Young TL. Arrhythmogenic right ventricular cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic disorder caused by a missense mutation in the TMEM43 gene. Am J Hum Genet. 2008;82:809–821. doi: 10.1016/j.ajhg.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens F, Fletcher CD, Antonescu CR, Coindre JM, Colecchia M, Domanski HA, Downs-Kelly E, Fisher C, Goldblum JR, Guillou L, Reid R, Rosai J, Sciot R, Mandahl N, Panagopoulos I. Clinicopathologic and molecular genetic characterization of low-grade fibromyxoid sarcoma, and cloning of a novel FUS/CREB3L1 fusion gene. Lab Invest. 2005;85:408–415. doi: 10.1038/labinvest.3700230. [DOI] [PubMed] [Google Scholar]
- Murakami T, Fujihara T, Nakamura M, Nakata K. P2Y(2) receptor elicits PAS-positive glycoprotein secretion from rabbit conjunctival goblet cells in vivo. J Ocul Pharmacol Ther. 2003;9:345–352. doi: 10.1089/108076803322279390. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Walkup RD, Tochigi A, Shearer TR, Azuma M. Establishment of an appropriate animal model for lacritin studies: cloning and characterization of lacritin in monkey eyes. Exp Eye Res. 2007;85:651–658. doi: 10.1016/j.exer.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Nasser NJ. Heparanase involvement in physiology and disease. Cell Mol Life Sci. 2008;65:1706–1715. doi: 10.1007/s00018-008-7584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozyildirim AM, Wistow GJ, Gao J, Wang J, Dickinson DP, Frierson HF, Jr, Laurie GW. The lacrimal gland transcriptome is an unusually rich source of rare and poorly characterized gene transcripts. Invest Ophthalmol Vis Sci. 2005;46:1572–1580. doi: 10.1167/iovs.04-1380. [DOI] [PubMed] [Google Scholar]
- Patel VN, Knox SM, Likar KM, Lathrop CA, Hossain R, Eftekhari S, Whitelock JM, Elkin M, Vlodavsky I, Hoffman MP. Heparanase cleavage of perlecan heparan sulfate modulates FGF10 activity during ex vivo submandibular gland branching morphogenesis. Development. 2007;134:4177–4186. doi: 10.1242/dev.011171. [DOI] [PubMed] [Google Scholar]
- Pioszak AA, Xu HE. Molecular recognition of parathyroid hormone by its G protein-coupled receptor. Proc Natl Acad Sci U S A. 2008;105:5034–5039. doi: 10.1073/pnas.0801027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D, Weremowicz S, Chin K, Seth P, Keshaviah A, Lahti-Domenici J, Bae YK, Monitto CL, Merlos-Suarez A, Chan J, Hulette CM, Richardson A, Morton CC, Marks J, Duyao M, Hruban R, Gabrielson E, Gelman R, Polyak K. A neural survival factor is a candidate oncogene in breast cancer. Proc Natl Acad Sci U S A. 2003;100:10931–10936. doi: 10.1073/pnas.1932980100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proikas-Cezanne T, Ruckerbauer S, Stierhof YD, Berg C, Nordheim A. Human WIPI-1 puncta-formation: a novel assay to assess mammalian autophagy. FEBS Lett. 2007;581:3396–3404. doi: 10.1016/j.febslet.2007.06.040. [DOI] [PubMed] [Google Scholar]
- Ramachandran P, Boontheung P, Xie Y, Sondej M, Wong DT, Loo JA. Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J Proteome Res. 2006;5:1493–1503. doi: 10.1021/pr050492k. [DOI] [PubMed] [Google Scholar]
- Rolando M, Zierhut M. The ocular surface and tear film and their dysfunction in dry eye disease. Surv Ophthalmol. 2001;45:S203–210. doi: 10.1016/s0039-6257(00)00203-4. [DOI] [PubMed] [Google Scholar]
- Runge S, Thøgersen H, Madsen K, Lau J, Rudolph R. Crystal Structure of the Ligand-bound Glucagon-like Peptide-1 Receptor Extracellular Domain. J Biol Chem. 2008;283:11340–11347. doi: 10.1074/jbc.M708740200. [DOI] [PubMed] [Google Scholar]
- Sanghi S, Kumar R, Lumsden A, Dickinson D, Klepeis V, Trinkaus-Randall V, Frierson HF, Jr, Laurie GW. cDNA and genomic cloning of lacritin, a novel secretion enhancing factor from the human lacrimal gland. J Mol Biol. 2001;310:127–139. doi: 10.1006/jmbi.2001.4748. [DOI] [PubMed] [Google Scholar]
- Schaumberg DA, Buring JE, Sullivan DA, Dana MR. Hormone replacement therapy and dry eye syndrome. JAMA. 2001;286:2114–2119. doi: 10.1001/jama.286.17.2114. [DOI] [PubMed] [Google Scholar]
- Shafat I, Vlodavsky I, Ilan N. Characterization of mechanisms involved in secretion of active heparanase. J Biol Chem. 2006;281:23804–23811. doi: 10.1074/jbc.M602762200. [DOI] [PubMed] [Google Scholar]
- Shirane M, Nakayama KI. Protrudin induces neurite formation by directional membrane trafficking. Science. 2006;314:818–821. doi: 10.1126/science.1134027. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Guerlain SA, Pearson WR. Visualization of near-optimal sequence alignments. Bioinformatics. 2004;20:953–958. doi: 10.1093/bioinformatics/bth013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern ME, Gao J, Siemasko KF, Beuerman RW, Pflugfelder SC. The role of the lacrimal functional unit in the pathophysiology of dry eye. Exp Eye Res. 2004;78:409–416. doi: 10.1016/j.exer.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Stevanin G, Santorelli FM, Azzedine H, Coutinho P, Chomilier J, Denora PS, Martin E, Ouvrard-Hernandez AM, Tessa A, Bouslam N, Lossos A, Charles P, Loureiro JL, Elleuch N, Confavreux C, Cruz VT, Ruberg M, Leguern E, Grid D, Tazir M, Fontaine B, Filla A, Bertini E, Durr A, Brice A. Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus scallosum. Nat Genet. 2007;39:366–372. doi: 10.1038/ng1980. [DOI] [PubMed] [Google Scholar]
- Sullivan DA. Tearful relationships? Sex, hormones, the lacrimal gland, and aqueous-deficient dry eye. Ocul Surf. 2004;2:92–123. doi: 10.1016/s1542-0124(12)70147-7. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi M, Ricciuto J, Tisdale A, Gipson IK, Mantelli F, Argüeso P. Antiadhesive character of mucin O-glycans at the apical surface of corneal epithelial cells. Invest Ophthalmol Vis Sci. 2008;49:197–203. doi: 10.1167/iovs.07-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki-Utsunomiya K, Hadano S, Otomo A, Kunita R, Mizumura H, Osuga H, Ikeda JE. ALS2CL, a novel ALS2-interactor, modulates ALS2-mediated endosome dynamics. Biochem Biophys Res Commun. 2007;354:491–497. doi: 10.1016/j.bbrc.2006.12.229. [DOI] [PubMed] [Google Scholar]
- Takegahara N, Takamatsu H, Toyofuku T, Tsujimura T, Okuno T, Yukawa K, Mizui M, Yamamoto M, Prasad DV, Suzuki K, Ishii M, Terai K, Moriya M, Nakatsuji Y, Sakoda S, Sato S, Akira S, Takeda K, Inui M, Takai T, Ikawa M, Okabe M, Kumanogoh A, Kikutani H. Plexin-A1 and its interaction with DAP12 in immune responses and bone homeostasis. Nat Cell Biol. 2006;8:615–622. doi: 10.1038/ncb1416. [DOI] [PubMed] [Google Scholar]
- Tauber J, Davitt WF, Bokosky JE, Nichols KK, Yerxa BR, Schaberg AE, LaVange LM, Mills-Wilson MC, Kellerman DJ. Double-masked, placebo-controlled safety and efficacy trial of diquafosol tetrasodium (INS365) ophthalmic solution for the treatment of dry eye. Cornea. 2004;23:784–792. doi: 10.1097/01.ico.0000133993.14768.a9. [DOI] [PubMed] [Google Scholar]
- Tomosugi N, Kitagawa K, Takahashi N, Sugai S, Ishikawa I. Diagnostic potential of tear proteomic patterns in Sjögren’s syndrome. J Proteome Res. 2005;4:820–825. doi: 10.1021/pr0497576. [DOI] [PubMed] [Google Scholar]
- Tsai PS, Evans JE, Green KM, Sullivan RM, Schaumberg DA, Richards SM, Dana MR, Sullivan DA. Proteomic analysis of human meibomian gland secretions. Br J Ophthalmol. 2006;90:372–377. doi: 10.1136/bjo.2005.080846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubels JL, Wertz JT, Ingersoll KE, Jackson RS, 2nd, Aupperlee MD. Down-regulation of androgen receptor expression and inhibition of lacrimal gland cell proliferation by retinoic acid. Exp Eye Res. 2002;75:561–571. doi: 10.1006/exer.2002.2054. [DOI] [PubMed] [Google Scholar]
- Van Goethem G, Martin JJ, Van Broeckhoven C. Progressive external ophthalmoplegia and multiple mitochondrial DNA deletions. Acta Neurol Belg. 2002;102:39–42. [PubMed] [Google Scholar]
- van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, Wapenaar MC, Barnardo MC, Bethel G, Holmes GK, Feighery C, Jewell D, Kelleher D, Kumar P, Travis S, Walters JR, Sanders DS, Howdle P, Swift J, Playford RJ, McLaren WM, Mearin ML, Mulder CJ, McManus R, McGinnis R, Cardon LR, Deloukas P, Wijmenga C. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang N, Xie J, Walton SC, McKown RL, Raab RW, Ma P, Beck SL, Coffman GL, Hussaini IM, Laurie GW. Restricted epithelial proliferation by lacritin via PKCalpha-dependent NFAT and mTOR pathways. J Cell Biol. 2006;174:689–700. doi: 10.1083/jcb.200605140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Zeng H, Zhang J, Okamoto CT, Warren DW, Wood RL, Bachmann M, Mircheff AK. Stimulation with carbachol alters endomembrane distribution and plasma membrane expression of intracellular proteins in lacrimal acinar cells. Exp Eye Res. 1999;69:651–661. doi: 10.1006/exer.1999.0742. [DOI] [PubMed] [Google Scholar]
- Zcharia E, Philp D, Edovitsky E, Aingorn H, Metzger S, Kleinman HK, Vlodavsky I, Elkin M. Heparanase regulates murine hair growth. Am J Pathol. 2005;166:999–1008. doi: 10.1016/S0002-9440(10)62321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A, Alizadeh BZ, Bevova M, van Leeuwen MA, Coenen MJ, Franke B, Franke L, Posthumus MD, van Heel DA, van der Steege G, Radstake TR, Barrera P, Roep BO, Koeleman BP, Wijmenga C. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 2007;81:1284–1288. doi: 10.1086/522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Beuerman RW, Foo Y, Liu S, Ang LP, Tan DT. Characterisation of human tear proteins using high-resolution mass spectrometry. Ann Acad Med Singapore. 2006;35:400–407. [PubMed] [Google Scholar]