Abstract
Background information
Protein degradation via the UPS (ubiquitin–proteasome system) plays critical roles in muscle metabolism and signalling pathways. The present study investigates temporal requirements of the UPS in muscle using conditional expression of mutant proteasome β subunits to cause targeted inhibition of proteasome function.
Results and conclusions
The Drosophila GeneSwitch system was used, with analyses of the well-characterized larval somatic body wall muscles. This method acutely disrupts proteasome function and causes rapid accumulation of polyubiquitinated proteins, specifically within the muscle. Within 12 h of transgenic proteasome inhibition, there was a gross disorganization of muscle architecture and prominent muscle atrophy, progressing to the arrest of all co-ordinated movement by 24 h. Progressive muscle architecture changes include rapid loss of sarcomere organization, loss of nuclei spacing/patterning, vacuole formation and the accumulation of nuclear and cytoplasmic aggregates at the ultrastructural level. At the neuromuscular junction, the highly specialized muscle membrane folds of the subsynaptic reticulum were rapidly lost. Within 24 h after transgenic proteasome inhibition, muscles contained numerous autophagosomes and displayed highly elevated expression of the endoplasmic reticulum chaperone GRP78 (glucose-regulated protein of 78 kDa), indicating that the loss of muscle maintenance correlates with induction of the unfolded protein response. Taken together, these results demonstrate that the UPS is acutely required for maintenance of muscle and neuromuscular junction architecture, and provides a Drosophila genetic model to mechanistically evaluate this requirement.
Keywords: autophagy, Drosophila, muscle, proteasome, ubiquitin
Introduction
The UPS (ubiquitin–proteasome system) mediates protein turnover through a sequential cascade of the E1, E2 and E3 ubiquitin ligases, which catalyse the attachment of the 76-amino-acid protein ubiquitin to lysine residues on a protein for targeting to the degradative proteasome (Hicke, 2001; Glickman and Ciechanover, 2002). The UPS is important for muscle cell function at several levels (Argiles and Lopez-Soriano, 1996; Jackman and Kandarian, 2004). In high catabolic states, such as cancer, renal failure and starvation, UPS up-regulation mediates increased degradation of muscle proteins, resulting in muscle atrophy (Lecker et al., 2006). Under other conditions, impairment in specific UPS pathways can also lead to muscle breakdown and degeneration. For example, studies have shown that mutations of the E3 ligase, parkin, or its upstream effector PINK1 (PTEN-induced kinase 1), lead to adult indirect flight muscle degeneration in Drosophila models of Parkinson’s disease (Greene et al., 2003; Clark et al., 2006; Park et al., 2006). Similarly, UPS impairment is implicated in the pathophysiology of several degenerative myopathies, including myofibrillary myopathies and IBM (inclusion body myositis) (Ferrer et al., 2004; Fratta et al., 2005). Thus bidirectional alteration of UPS activity has been correlated with the maintenance of the musculature.
We hypothesized that, during muscle growth and contractile function, a high level of proteasome function would be critical for the maintenance of muscle architecture. Due to its genetic pliability and well-characterized musculature, Drosophila provides an excellent in vivo genetic model system to study the targeted effects of muscle proteasome inhibition. The larval body wall musculature is composed of a reiterated set of 30 defined muscles in each abdominal hemisegment (Bate, 1990; Baylies et al., 1998; Beckett and Baylies, 2006). The development of these muscles is highly stereotyped, with each muscle cell achieving a specific orientation, a standard number of nuclei and innervation at NMJs (neuromuscular junctions) of exacting size and placement (Bate, 1990; Abmayr et al., 1995; Beckett and Baylies, 2006). Transgenic expression of mutant proteasome β subunits has been used to specifically inhibit the Drosophila proteasome (Schweisguth, 1999; Belote and Fortier, 2002; Speese et al., 2003; Neuburger et al., 2006). These mutant proteasome subunits can be spatially targeted only to muscles using the GAL4-UAS system (where UAS stands for up-stream activating sequence) (Brand and Perrimon, 1993), and temporally targeted using the conditional GS (GeneSwitch) system (Osterwalder et al., 2001). This provides a methodology to investigate the temporal requirement of proteasome function specifically within the developing musculature.
We report here that targeted transgenic proteasome inhibition in the muscle during an acute period in third instar larval development leads to rapid muscle wasting. This muscle atrophy is accompanied by loss of the striated sarcomeric architecture, erosion of the NMJ SSR (subsynaptic reticulum), formation of nuclear rod-like and cytoplasmic fibrillar aggregates, and initiation of an autophagocytic process that is probably triggered by the UPR (unfolded protein response). These results demonstrate that proteasome function is acutely required for the maintenance of muscle cytoarchitecture.
Results
Targeted expression of mutant proteasome subunits blocks protein degradation
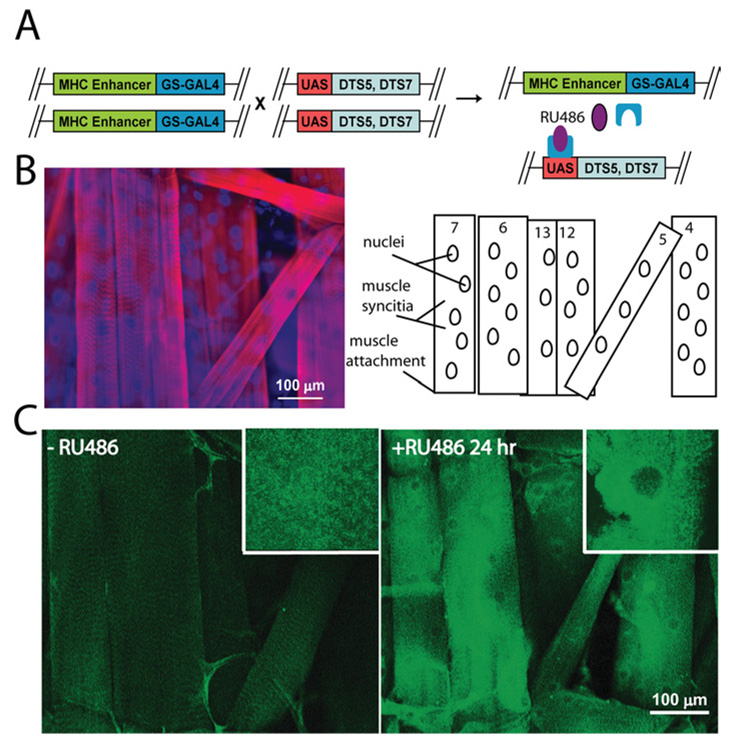
The DTS (dominant temperature-sensitive) mutations DTS5 and DTS7 result from single amino acid substitutions in the β6 and β2 subunits of the 20S proteasome respectively (Saville and Belote, 1993; Smyth and Belote, 1999). The mutant subunits act in a dominant-negative manner to interfere with proteasome function; in many previous studies, these mutant subunits have been shown to inhibit UPS-mediated protein degradation of known proteasome substrates (Schweisguth, 1999; Speese et al., 2003; Neuburger et al., 2006). Transgenic co-expression of both proteasome subunit mutants in the Drosophila eye using the GAL4-UAS system has a synergistic effect in blocking proteasome function (Belote and Fortier, 2002). Both mutant subunits were therefore co-expressed in the musculature in the present study, and this double mutant strain will hence be referred to as DTS. To target DTS-mediated proteasome inhibition spatially in muscle and temporally under experimental control, the GS inducible expression system was used under control of the muscle-specific MHC (myosin heavy chain) promoter (Figure 1A) (Osterwalder et al., 2001).
Figure 1. Induction of DTS mutant proteasome subunits in muscle causes rapid accumulation of polyubiquitinated proteins.
(A) The GS GAL4-UAS transgenic method used to conditionally induce expression of the two mutant proteasome β subunits (DTS5 and DTS7) with the MHC GAL4 driver within the musculature specifically. In the MHC-GS-GAL4/UAS DTS5, DTS7 line, GS-GAL4 (blue) binds to the UAS promoter only in the presence of RU486 (purple). (B) A third instar larvae abdominal muscle hemisegment immunostained with phalloidin to label actin (red) and DAPI to label nuclei (blue). The right-hand panel shows the architecture of the specific subset of muscles of the ventral internal abdominal hemisegment. (C) Representative confocal muscle images of polyubiquitin immunostaining from uninduced (−RU486) and induced (+RU486) DTS-expressing larvae. Third instar larvae were labelled with an antibody (FK-1) specifically against polyubiquitinated proteins. Following RU486 induction of proteasome mutant expression, polyubiquitinated proteins progressively accumulate in muscle cytoplasm, particularly in perinuclear regions (insets).
The Drosophila larval musculature is formed during embryonic development by myoblast fusion, leading to formation of 30 unique multinucleated muscles per abdominal hemisegment (Bate, 1990; Beckett and Baylies, 2006). A subset of internal ventral longitudinal muscles, including muscles 4, 5, 6, 7, 12 and 13, have been particularly well characterized (Figure 1B). Each of these muscles, individually identifiable by position and size, contain a muscle-specific number of nuclei that are evenly distributed along the length of the syncitial cell (Figure 1B). Proteasome inhibition was temporally targeted during the early third instar stage of larval development by transferring age-matched larvae to RU486-containing food (+RU486) or RU486-free food (−RU486) for 24 h at 29°C. To assess the specific effect of DTS expression on UPS function in muscle, both sets of larvae were stained with an antibody against polyubiquitinated proteins (FK-1 antibody; Figure 1C). A marked accumulation of polyubiquitinated proteins was observed in the 24 h +RU486-treated compared with 24 h −RU486 treated controls (Figure 1C). As expected, polyubiquitinated proteins accumulated only in the targeted-muscles, but not in other tissues. Polyubiquitinated proteins accumulated throughout the muscle cell, but the accumulation was most striking in perinuclear regions where a confluent staining pattern was observed (Figure 1C, inset). These results demonstrate the ability to interfere with proteasome function in vivo, only in muscle cells and in a temporally controlled manner.
Targeted proteasome mutant expression causes rapid muscle atrophy
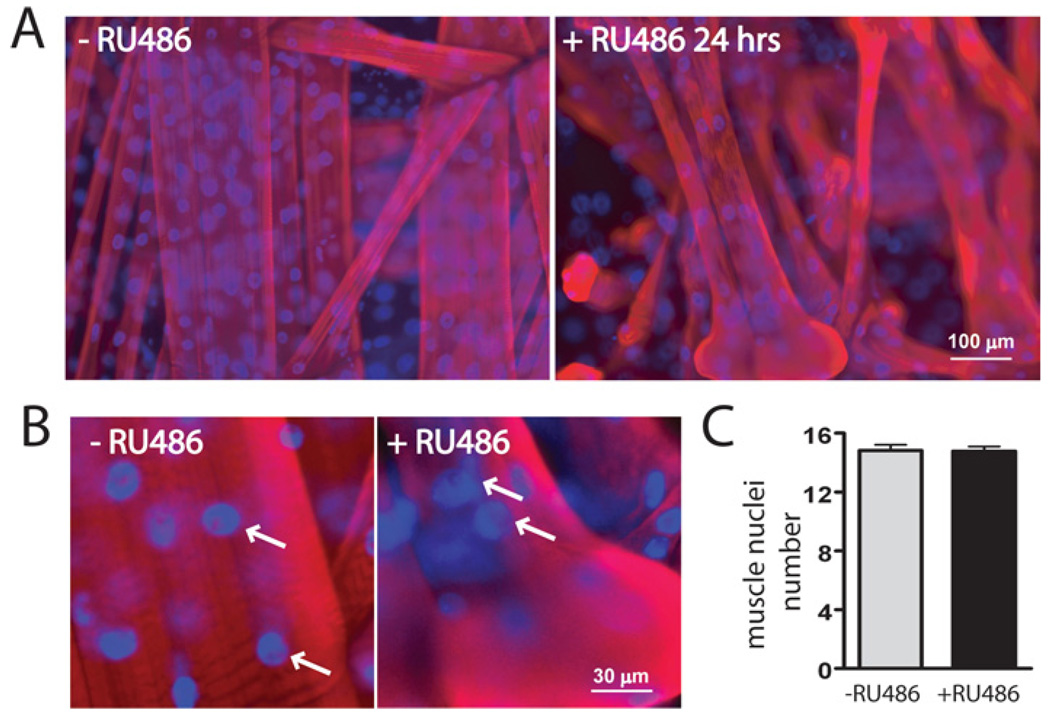
When genetic proteasome inhibition was induced for 24 h (+RU486), third instar larvae became progressively movement impaired, eventually stopped moving and failed to pupate. The −RU486 control larvae moved robustly and developed normally. Likewise, feeding of RU486 to heterozygous DTS/+ or GS-MHC-GAL4/+ animals had no detectable effect on movement, morphology or larval development, similar to previous reports (Osterwalder et al., 2001). To directly assay the impact on the musculature, phalloidin (actin) and DAPI (4′,6-diamidino-2-phenylindole) (nuclear) staining was used to examine muscle architecture (Figure 2). Phalloidin labelling revealed striking muscle atrophy after 24 h induction of the DTS proteasome mutants (Figure 2A). Although muscle cell integrity and attachments were maintained, all muscles showed a dramatic reduction in cellular volume, a loss of the regular rectangular cell architecture and often displayed bulging at the tendon insertion sites. DAPI staining also revealed a dramatic shift in nuclei location within the muscle (Figure 2A). In controls (−RU486), nuclei were periodically distributed along the length of the myotube. In contrast, inhibition of the proteasome (+RU486) results in a collapse of this regular patterning, with nuclei clustered at the end of muscles near the tendon insertion sites (Figure 2B). Interestingly, this nuclear clustering phenotype recapitulates a pattern seen early in embryonic muscle development (Bate, 1990). Despite the prominent muscle atrophy and loss of nuclear spacing/localization, there was no change in the number of nuclei per muscle (−RU486, 14.8 ± 0.38, n=18; +RU486, 14.8 ± 0.31, n=24; Figure 2C). Thus conditional inhibition of the proteasome disrupts muscle architecture within 24 h, but does not cause cellular degeneration.
Figure 2. DTS mutant proteasome expression causes muscle atrophy and nuclear clustering at tendon attachment sites.
(A) Representative images of ventral abdominal muscle hemisegments stained with phalloidin (red) and DAPI (blue) from uninduced (−RU486) and induced (+RU486) third instar larvae. Phalloidin staining reveals prominent muscle atrophy after 24 h of mutant proteasome expression. DAPI staining reveals the loss of the regular spacing of nuclei along the muscle length. (B) Higher magnification images of muscle 6 show that nuclei (arrows) cluster at insertion sites after the 24-h induction of DTS proteasome mutants. (C) Quantification of nuclei number in muscle 6 revealed no significant differences in the number of muscle nuclei without or with RU486 induction of proteasome mutant expression after 24 h.
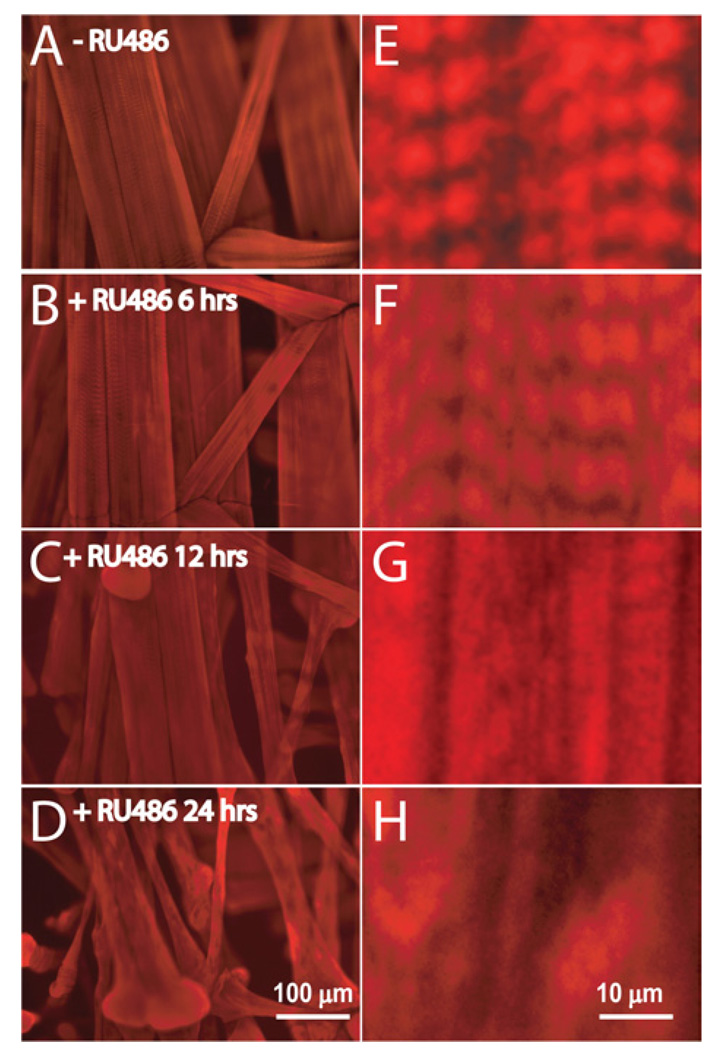
To examine the time course of the effects of proteasome inhibition, muscle structures were examined at timed intervals following the RU486-feeding induction of DTS proteasome mutant expression (Figure 3). Control muscles (−RU486) maintained a regular rectangular architecture (Figure 3A) and a clearly delineated striated sarcomeric arrangement (Figure 3E). At 6 h following proteasome inhibition (+RU486), the muscles appeared indistinguishable from controls with no evident morphological changes (Figures 3B and 3F). However, by 12 h, muscles clearly began to atrophy, developing thin ribbon-like bodies and bulges at the tendon insertion sites (Figure 3C). This coincided with a loss of the muscle striation pattern. The sarcomeric boundaries were no longer clearly defined, although there was still intense phalloidin actin staining along the length of the muscle (Figure 3G). At 24 h after induction of DTS proteasome mutant expression, there was a complete loss of the normal striated sarcomeric banding pattern. The residual expression was characterized by an aberrant clumping pattern, suggesting the presence of intracellular aggregates (Figure 3H). A similar loss of the striated banding pattern was seen when muscles were immunostained with antibody against MHC (Supplementary Figure 1 at http://www. biolcell.org/boc/099/boc0990615add.htm). This loss of sarcomeric banding indicates a major disruption of the muscle architecture.
Figure 3. Progressive muscle atrophy and loss of the sarcomeric striation pattern with increasing duration of transgenic proteasome inhibition.
(A–D) Representative images of phalloidin-labelled actin of the ventral abdominal muscle hemisegments for control (−RU486) and 6, 12 and 24 h RU486-induced larvae (+RU486). (E–H) Higher magnification images show the progressive loss of striated muscle banding beginning at 12 h after induction of proteasome block.
Proteasome inhibition causes protein aggregate formation and autophagocytosis
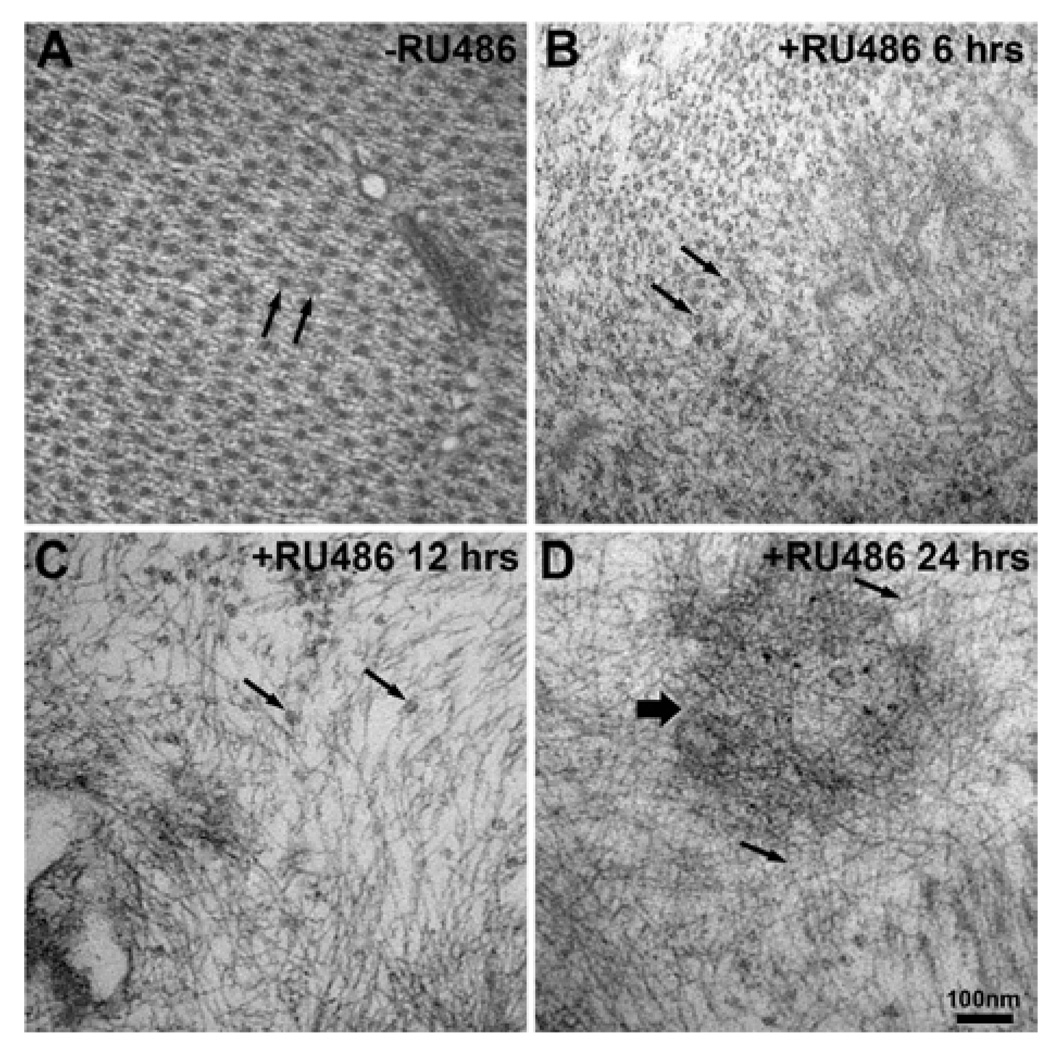
To assess muscle architecture in detail, a comprehensive ultrastructural analysis was performed using transmission EM (electron microscopy) of control (−RU486) and proteasome inhibited (+RU486) temporally staged larval muscle. In controls, muscle actin and myosin form long filaments that alternate, anchored at the Z-line, forming the contractile framework of a sarcomere. In cross-section, this architecture gives a repeating pattern of alternating thin diameter actin and thick diameter myosin filaments (Figure 4A). This regular sarcomeric pattern was mostly maintained after 6 h induction of DTS proteasome mutant expression, consistent with the light microscopy analyses (Figure 3), but there were some discontinuous patches in which the regular pattern was locally disrupted (Figure 4B). By 12 h, the regular contractile framework of a sarcomere was widely disrupted. Although individual sarcomeres were still evident, they were no longer packed in the regular crystalline array characteristic of control muscle (Figure 4C). After 24 h of proteasome inhibition, little sarcomere structure was detectable. Instead, protein filaments were often observed clustered in large tangled aggregates, which are never observed in control muscle (Figure 4D).
Figure 4. Progressive disruption of muscle architecture with increasing duration of proteasome inhibition.
Electron micrographs of transverse sections through Drosophila third instar larval muscles. (A) Representative image from a control (−RU486) larvae showing the alternating crystalline array of actin (small diameter) and myosin (large diameter; thin arrows) filaments. (B) After RU486-induced (6 h) proteasome mutant expression, the sarcomeric structure is mostly maintained, but some focal regions of disorganized filaments appear. (C) After 12 h, most of the crystalline sarcomeric structure has been replaced by loosely organized myosin and actin filaments. (D) After 24 h, there is diffuse sarcomeric disorganization throughout the muscle, with aggregations of electron dense filamentous tangles (thick arrow).
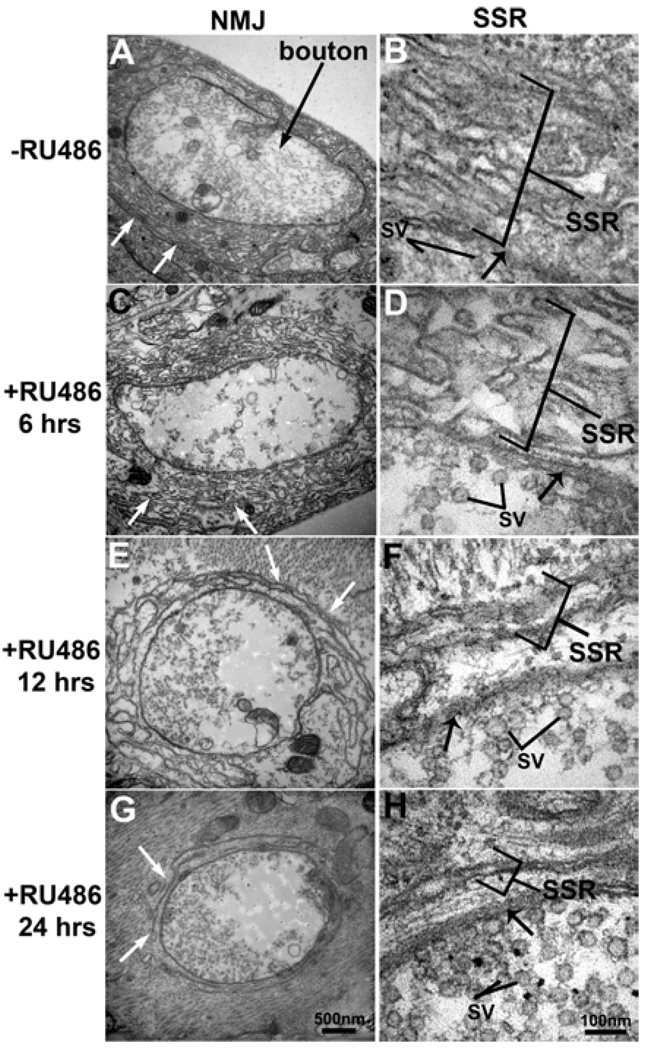
Similarly dramatic ultrastructural changes occur at the interface of the muscle and neuron at the NMJ. In control animals, type I presynaptic boutons are deeply embedded within the muscle, surrounded by an elaborate maze of muscle membrane folds called the SSR (Figure 5) (Lahey et al., 1994; Guan et al., 1996). The SSR is a postsynaptically differentiated specialization which probably serves to increase the muscle’s synaptic receptive and signalling domain to keep pace with the rapidly growing muscle volume during larval development. Inhibition of the muscle proteasome causes a rapid disassembly of the SSR. In controls and RU486-treated animals up to 6 h, an elaborate and extensive SSR was maintained around all type INMJ domains (Figures 5A–5D). In contrast, proteasome inhibition for 12 h resulted in a loss of both the complexity of membrane folds and the SSR area (Figures 5E and 5F). After 24 h of proteasome mutant subunit induction, there was a near complete loss of SSR at most NMJ boutons, although presynaptic bouton architecture, including active zone T-bars and synaptic vesicle pools, was not detectably altered (Figures 5G and 5H). Thus proteasome function in the muscle is required to maintain the post-synaptic membrane morphological specialization of the SSR at the NMJ.
Figure 5. Progressive reduction in NMJ SSR with increasing duration of transgenic proteasome inhibition.
Electron micrographs of NMJ boutons in control larvae (−RU486, A, B) and in proteasome-mutant-expressing larvae (+RU486, C–H). (A) In control NMJ, the convoluted membrane layers of the SSR (white arrows) encompass the synaptic bouton. (B) A higher magnification image shows the multiple membrane layers of the postsynaptic SSR and the adjacent presynaptic terminal membrane (black arrow). For reference, synaptic vesicles (SV) are labelled in the presynaptic bouton. (C, D) After RU486-induced proteasome mutant expression (6 h), there is no change in the arrangement of the SSR around the synaptic bouton. (E, F) After 12 h, the SSR area is reduced and fewer membrane convolutions are observed, however, the presynaptic structure remains unchanged. (G, H) After 24 h, there is dramatic loss of the SSR, with an increase in thin 2 nm fibres between the membranes (black arrows).
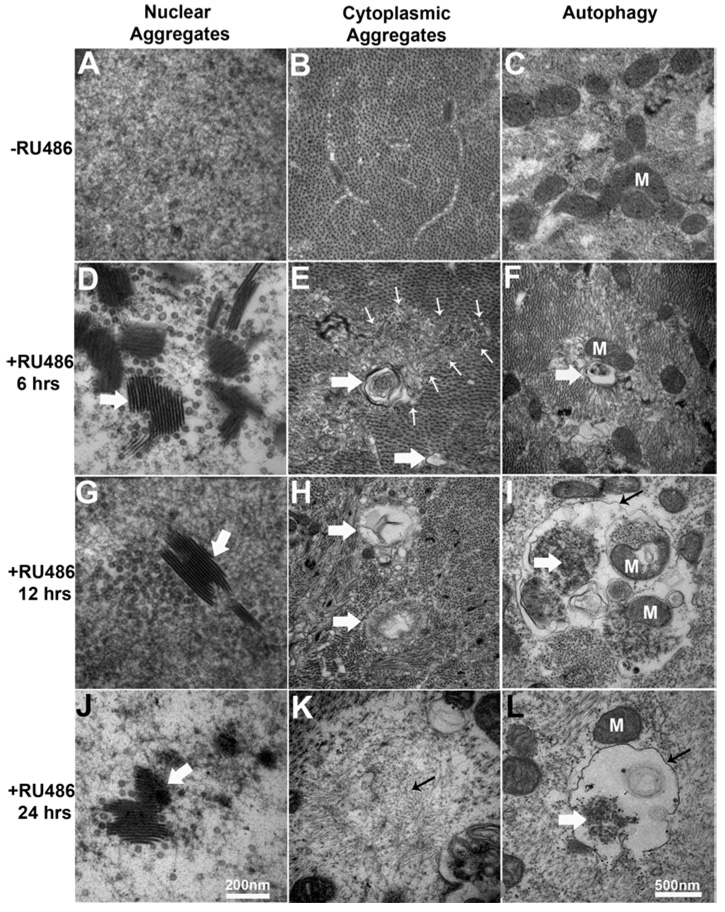
As shown above, proteasome inhibition does not cause loss of nuclei at a light microscope level (DAPI staining; Figure 2). Similarly, ultrastructural analyses indicate that overall nuclear structure and the nuclear envelope remain intact, as do other prominent organelles (e.g.mitochondria) following 24 h of RU486 induction (Figure 6). More detailed ultrastructural examinations, however, revealed the progressive appearance of nuclear aggregates and rod-like inclusions following proteasome inhibition. These aberrant structures first appear as early as 6 h after induction of proteasome subunit mutants, but become increasingly prominent at the 12 and 24 h time points (Figures 6A, 6D, 6G and 6J). In the cytoplasm, there was progressive accumulation of fibrillar aggregates that coincided with loss of sarcomeric architecture (Figure 4, Figure 6B, 6E, 6H and 6K). There was also accumulation of membrane-rimmed vacuoles that had the ultrastructural characteristics of autophagosomes (Yu et al., 2004; Fernandez et al., 2005). These autophagosomes were rare, but apparent after 6 h of RU486 induction, and became increasingly abundant at 12 and 24 h (Figures 6C, 6F, 6I and 6L). At both later time points, phagosomes were observed in the process of engulfing the tangled cytoplasmic aggregates, as well as mitochondria. After 24 h of transgenic proteasome inhibition, many of these autophagocytic vesicles showed characteristics of secondary lysosomes, displaying ultrastructural evidence of lysis of their contents (Figure 6L) (Selcen et al., 2004; Yu et al., 2004; Fernandez et al., 2005). These observations provide a mechanistic basis for the gross muscle atrophy and loss of architectural definition caused by loss of proteasome function.
Figure 6. Aggregate accumulation and progression of autophagy with increasing duration of transgenic proteasome inhibition.
(A) Electron micrograph showing intra-nuclear electron-dense chromatin in a control (−RU486) muscle nucleus. (D, G, J) Layered, rod-shaped inclusions (white arrows) found in muscle nuclei at 6, 12 and 24 h after RU486 induction of proteasome mutant expression respectively. The diameters of the rod-shaped structures measured 8–12 nm, and these were often associated with electron-lucent spheres in the area of 32–36 nm diameter. (B) A transverse EM image of muscle cytoplasm from control shows the regular crystalline array of alternating actin and myosin filaments. (E) After RU486-induced proteasome mutant expression (6 h), small disruptions of this regular myofilament appear (small white arrows). These areas sometimes contain membrane-rimmed vacuoles (large white arrows). (H) After 12 h, there are many more vacuoles (large white arrows), often filled with fibrillar electron-dense material. (K) After 24 h, large disorganized fibrillar aggregates (small back arrow) form in regions of vacuolization. (C) An EM image of muscle cytoplasm from a control larva, with multiple mitochondria (M). (F) After induction (6 h), small membrane-rimmed vacuoles form (white arrow). (I) After induction (12 h), clear autophagic vacuoles form (small black arrow), in this example containing partially digested mitochondria (M) and myofilament arrays (large white arrow). (L) After induction (24 h), autophagosomes and secondary autolysosomes (small black arrow) are frequent. The material of the autolyso some is amorphous (large white arrow), with only remnants of digested material remaining.
Inhibition of the proteasome causes a UPR
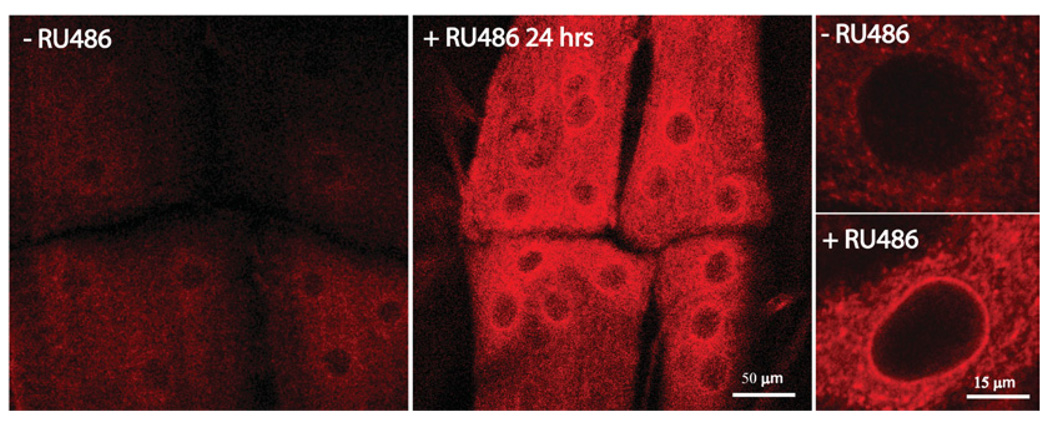
Cytoplasmic aggregate formation followed by autophagocytosis is frequently observed when the cellular ability to clear unfolded or misfolded proteins through ERAD (endoplasmic reticulum-associated degradation) is overwhelmed (Johnston et al., 1998; Friedlander et al., 2000; Kostova and Wolf, 2003; Iwata et al., 2005; Yorimitsu et al., 2006). To test for evidence of a UPR, the musculature of control and experimental animals was probed for the endoplasmic reticulum chaperone protein, GRP78 (glucose-regulated protein of 78 kDa), a well-characterized marker of the UPR (Vattemi et al., 2004; Schroder and Kaufman, 2005). In controls (−RU486), GRP78 was expressed diffusely at low levels in the larval muscles (Figure 7). In contrast, transgenic proteasome inhibition in the muscles resulted in a marked up-regulation of GRP78 expression after 24 h (Figure 7). GRP78 accumulated in a punctate pattern, with the punctae coalescing in perinuclear regions. These regions of coalescence were most prominent surrounding clusters of nuclei that formed at muscle attachment sites after induction of proteasome inhibition (Figure 7). Interestingly, this GRP78 subcellular pattern was nearly identical to that observed for polyubiquitinated proteins that accumulate after RU486 induction of proteasome inhibition (Figure 1C). Thus an overwhelming UPR provides a likely mechanism for the rapid atrophy of the musculature following proteasome inhibition.
Figure 7. UPR marker GRP78 rapidly accumulates in muscles following transgenic proteasome inhibition.
Representative images of ventral longitudinal muscles 6/7, including muscle attachment sites for abdominal hemisegments A2 and A3 stained with an antibody against GRP78 in control (−RU486) and proteasome inhibition-induced (+RU486 24 h) third instar larvae. In the control, GRP78 is seen in a diffuse cytoplasmic punctate pattern. Following induction of proteasome inhibition (+RU486), nuclei cluster near muscle attachment sites, surrounded by intense perinuclear GRP78 staining. Images of individual nuclei (right-hand panels) show the perinuclear coalescence of GRP78 staining.
Discussion
The primary finding of the present study is that conditionally induced proteasome inhibition in muscles rapidly causes muscle wasting, formation of rod-like nuclear and fibrillar cytoplasmic aggregates and autophagocytosis. The present study was conducted using mutant proteasome subunits that dominantly block proteasome function, and the effects assessed in the well-characterized Drosophila larval musculature system. The transgenic approach employed here has the benefits of in vivo analyses, specific targeting of proteasome inhibition only within the muscle cells and tight temporal control of the period of proteasome inhibition via induction with the transcriptional activator RU486. Within 6–12 h of RU486 induction of proteasome mutants, there were early indications of muscle atrophy and loss of muscle contractile machinery organization, and ultrastructural evidence of loss of muscle sarcomeric architecture, vacuolization and cytoplasmic aggregate formation. These changes became progressively more severe until, by 24 h, animals were unable to move, showed both gross and ultrastructural muscle disorganization, and had perinuclear accumulation of both polyubiquitinated proteins and a well-characterized indicator of the UPR machinery, GRP78.
In addition, transgenic proteasome inhibition led to a rapid loss of the NMJ SSR. Although the specific function of the SSR is not known, it has been hypothesized to serve to amplify voltage-triggered ionic flux and to be an important organizing centre for postsynaptic densities. The SSR is known to be maintained through the function of DLG (discs large), a muscle membrane-associated PDZ-domain-containing scaffolding protein which shows prominent enrichment in the postsynaptic SSR (Lahey et al., 1994; Guan et al., 1996). Although loss of synaptic DLG after proteasome inhibition could explain SSR loss, preliminary experiments suggest that DLG and synaptic function are relatively maintained in the face of dramatic muscle atrophy (K.F.Haas and K. Broadie, unpublished data). It has been shown that the SSR development correlates with the size of the muscle (Lahey et al., 1994). Thus a similar mechanism could be acting to reduce the SSR during the rapid muscle atrophy caused by proteasome inhibition. In addition to the reorganization of muscle nuclei, this loss of SSR suggests a regression to architectures characteristic of earlier stages in muscle development.
In mammalian studies in vitro (Lang-Rollin et al., 2004; Goldbaum et al., 2006), proteasome inhibition has been shown to trigger apoptotic degeneration in muscle cells. Similarly, in parkin mutant-expressing flies, defects in the function of the UPS lead to gross muscle degeneration (Greene et al., 2003). In contrast, in the present study, there was no ultrastructural evidence of an apoptotic degenerative process observed in the study by (Greene et al. 2003), as the nuclear envelope, chromatin pattern and mitochondrial architecture were maintained after proteasome inhibition. Proteasome inhibition for up to 24 h caused severe muscle atrophy, but no detectable loss of muscle cells. Individual muscle cells, likewise, showed no loss of nuclei and ultrastructural analyses indicated preservation of nuclear architecture. Similarly, there was no ultrastructural evidence of compromised mitochondrial architecture. Instead, proteasome inhibition resulted in progressive loss of sarcomere organization with the concurrent appearance of tangled cytoplasmic protein aggregates, as well as the progressive appearance of autophagocytic vacuoles engaged in engulfing these aggregates in addition to cytoplasmic organelles, especially mitochondria. Most tellingly, proteasome inhibition caused the progressive accumulation of polyubiquitinated proteins in punctate perinuclear aggregates, as well as the accumulation of the endoplasmic reticulum chaperone protein GRP78 in the same apparent structures. Taken together, these results strongly suggest that acute proteasome inhibition in the musculature triggers a rapid UPR that may provide a mechanistic cause for the rapid dissolution of muscle architecture and cellular atrophy.
One primary UPS function is to mediate ERAD, a critical pathway for protein quality control that clears unfolded and misfolded proteins (Kostova and Wolf, 2003; Meusser et al., 2005). Studies in muscle culture systems suggest that the UPS ERAD pathway may account for >60% of protein degradation, with the remainder of protein degradation processed through other UPS and lysosomal pathways (Purintrapiban et al., 2003). When the ERAD pathway is overwhelmed, aggregates of unfolded proteins trigger an autophagocytic response, leading to aggresome formation, phagocytosis of protein aggregates and lysosomal degradation (Friedlander et al., 2000; Iwata et al., 2005; Yorimitsu et al., 2006). Thus the results of the present study are consistent with defects caused by disruption of the UPS-dependent ERAD pathway.
The progressive autophagic muscle degradation after genetic proteasome inhibition in this Drosophila model is similar to the sequence of changes identified in degenerative myopathies. A hereditary form of IBM is caused by mutations in p97/VCP (valosin-containing protein), a chaperone protein involved in the translocation and trafficking of unfolded and misfolded proteins to the proteasome (Weihl et al., 2006). In muscle biopsies from patients with IBM and myofibrillar myopathies, there is an accumulation and co-localization of multiple proteins involved in the UPR (Ferrer et al., 2004; Vattemi et al., 2004). EM analysis of muscle biopsies from patients with myofibrillar myopathies show a very similar progression to the one described in the present study, with loss of normal myofibrillar architecture, accumulation of cytoplasmic filamentous material, aggregation of membranous organelles in vacuoles and autophagic degradation of organelles and fibrillar aggregates (Selcen et al., 2004). Moreover, the rod-like nuclear aggregates described in the present study are similar to nuclear and cytoplasmic tubulofilamentous aggregates that have been described previously for IBM(Fidzianska and Glinka, 2006).
In vitro models of sporadic IBM, combining over expression of amyloid-β or amyloid-β precursor protein with proteasome inhibition, display formation of fibrillar aggregates and autophagic muscle cell degeneration (Fratta et al., 2005; Wojcik et al., 2006). The inhibition of proteasome function at a time of a high level of protein synthesis during Drosophila larval development is probably triggering a similar response. This Drosophila genetic system offers the advantage of a readily malleable in vivo preparation to further study the-mechanisms of muscle autophagy and the UPR, processes critical to understanding the pathophysiology of degenerative myopathies.
Materials and methods
Drosophila stocks
All stocks were maintained at 25°C on standard corn-meal medium supplemented with dried yeast. For the inducible UAS transgene expression to interfere with proteasome function, the GS system was utilized (Osterwalder et al., 2001). The homozygous UAS-DTS5 2B(2), DTS7 1B(3) line [generously provided by Dr John Belote, Department of Biology, Syracuse University, Syracuse NY, U.S.A. (Belote and Fortier, 2002)] was crossed with the homozygous muscle-specific MHC GS-GAL4 line [generously provided by Dr Haig Keshishian, Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT, U.S.A. (Osterwalder et al., 2001)] to generate heterozygote GS-MHC-GAL4/UAS-DTS 5,7 flies containing a single copy of the UAS transgene and a single copy of the GAL4 driver. The GS system uses a RU486-dependent-GAL4-progesterone-receptor fusion protein. For induction of the UAS transgene, staged third instar larvae were transferred into corn-meal medium containing 15 µg/ml RU486 (Sigma–Aldrich) in 4% ethanol or 4% ethanol alone (−RU486 control) for defined periods at 29°C for up to 24 h.
Immunohistology
Preparations were fixed and immunohistologically stained, as described previously (Broadie and Bate, 1993; Broadie et al., 1995; Rohrbough et al., 2000). Third instar larvae were dissected along the dorsal midline and secured flat with histoacryl glue in standard saline, fixed for 30–45 min with 4% paraformaldehyde, then washed in 0.1% Triton X-100 in PBS containing 0.5% BSA. All antibody dilutions were in 0.1% Triton X-100 in PBS. Primary antibodies were applied overnight at 4°C (FK1, 1:1000, monoclonal, Biomol; GRP78, 1:50, mouse monoclonal, Santa Cruz Biotechnology; MHC, 1:500, rabbit polyclonal, Professor Daniel P. Kiehart, Department of Biology, Duke University, Durham, NC, U.S.A.). Preparations were incubated with a fluorescently tagged secondary antibody (1:300). Phalloidin and DAPI staining was performed for 5 min at room temperature (21°C). Images were collected with a Zeiss LSM 510 META confocal microscope and presented using Adobe Photoshop and Illustrator. In all cases, control and experimental matched pairs were fixed side by side on the same coverslip, and processed, imaged and quantified using identical methods.
EM
Age-matched third instar larvae were collected for the control (−RU486) condition, and after 6, 12, and 24 h of RU486 induction of proteasome mutant expression. Preparation of Drosophila tissue for ultrastructural analysis was performed as previously described (Pan et al., 2004; Rohrbough et al., 2004). Briefly, third instar larvae where dissected in standard saline and immediately fixed in ice-cold (2%) glutaraldehyde in 0.05 M PBS. Anterior and posterior extremities were excised and the larvae were transferred into fresh (2%) glutaraldehyde for 2 h. The specimens were then washed two times in phosphate-buffered sucrose, transferred into 1% OsO4 in distilled water for 2 h, and washed three times in distilled water. Preparations were then stained en bloc in 1% aqueous uranyl acetate for 1 h and then washed three times in distilled water. Water was replaced within the tissue gradually by using a graded series of ethanol (30–100%), followed by propylene oxide for 30 min as a transitional solvent to interface with the embedding media. Propylene oxide was gradually replaced with propylene oxide/Araldite (1:1, then 1:3) until specimens were completely infiltrated with 100% Araldite embedding media, standard infiltration methods. For selective sectioning, specimens were further dissected to isolate the ventral longitudinal muscle group containing muscles 7, 6, 13 and 12 (Bate, 1990). Ultrathin sections of approx. 50–60 nm were obtained using a Leica Ultracut UCT 54 ultramicrotome, and the sections were transferred into formvar-coated slot grids and examined using a Phillips CM10 TEM, operating at 80 V. Digital images were taken with 2-megapixelAMT CCD(charge-coupled-device) side-mounted camera.
Statistical Analysis
Statistical analyses were performed using a two-tailed Student’s t test. The significance level was set at P < 0.05. Results are expressed as the means ± S.E.M. for all groups.
Supplementary Material
Acknowledgements
We are particularly grateful to Dr John Belote (Department of Biology, Syracuse University, Syracuse NY, U.S.A.) for providing the UAS-DTS line and to Dr Haig Keshishian (Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT, U.S.A.) for providing the GSMHC-GAL4 line. We thank the Iowa Developmental Studies Hybridoma Bank for providing the DLG antibodies used in this study. This work was supported by NIH (National Institutes of Health) grants (NS048882 to K.F.H. and NS41740 to K.B.).
Abbreviations used
- DAPI
4′,6-diamidino-2-phenylindole
- DLG
discs large
- DTS
dominant temperature-sensitive
- EM
electron microscopy
- ERAD
endoplasmic reticulum-associated degradation
- GRP78
glucose-regulated protein of 78 kDa
- GS
GeneSwitch
- IBM
inclusion body myositis
- MHC
myosin heavy chain
- NMJ
neuromuscular junction
- SSR
subsynaptic reticulum
- UAS
upstream activating sequence
- UPR
unfolded protein response
- UPS
ubiquitin–proteasome system
References
- Abmayr SM, Erickson MS, Bour BA. Embryonic development of the larval body wall musculature of Drosophila melanogaster. Trends. Genet. 1995;11:153–159. doi: 10.1016/s0168-9525(00)89030-7. [DOI] [PubMed] [Google Scholar]
- Argiles JM, Lopez-Soriano FJ. The ubiquitin-dependent proteolytic pathway in skeletal muscle: its role in pathological states. Trends Pharmacol. Sci. 1996;17:223–226. doi: 10.1016/0165-6147(96)10021-3. [DOI] [PubMed] [Google Scholar]
- Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bate M, Gomez MR. Myogenesis: a view from Drosophila. Cell. 1998;93:921–927. doi: 10.1016/s0092-8674(00)81198-8. [DOI] [PubMed] [Google Scholar]
- Beckett K, Baylies MK. The development of the Drosophila larval body wall muscles. Int. Rev. Neurobiol. 2006;75:55–70. doi: 10.1016/S0074-7742(06)75003-6. [DOI] [PubMed] [Google Scholar]
- Belote JM, Fortier E. Targeted expression of dominant negative proteasome mutants in Drosophila melanogaster. Genesis. 2002;34:80–82. doi: 10.1002/gene.10131. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Broadie K, Bate M. Innervation directs receptor synthesis and localization in Drosophila embryo synaptogenesis. Nature. 1993;361:350–353. doi: 10.1038/361350a0. [DOI] [PubMed] [Google Scholar]
- Broadie K, Prokop A, Bellen HJ, Okane CJ, Schulze KL, Sweeney ST. Syntaxin and synaptobrevin function downstream of vesicle docking in Drosophila. Neuron. 1995;15:663–673. doi: 10.1016/0896-6273(95)90154-x. [DOI] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seo JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Figarella-Branger D, Meyronet D, Cassote E, Tong S, Pellissier JF. Electron microscopy in neuromuscular disorders. Ultrastruct. Pathol. 2005;29:437–450. doi: 10.1080/01913120500323175. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Martin B, Castano JG, Moreno D, Lucas JJ, Olive M. Proteasomal expression, induction of immunoproteasome subunits, and local MHC class I presentation in myofibrillar myopathy and inclusion body myositis. J. Neuropathol. Exp. Neurol. 2004;63:484–498. doi: 10.1093/jnen/63.5.484. [DOI] [PubMed] [Google Scholar]
- Fidzianska A, Glinka Z. Rimmed vacuoles with β-amyloid and tau protein deposits in the muscle of children with hereditary myopathy. Acta Neuropathol. 2006;112:185–193. doi: 10.1007/s00401-006-0079-3. [DOI] [PubMed] [Google Scholar]
- Fratta P, Engel WK, McFerrin J, Davies KJA, Lin SW, Askanas V. Proteasome inhibition and aggresome formation in sporadic inclusion-body myositis and in amyloid-β precursor protein-overexpressing cultured human muscle fibers. Am. J. Pathol. 2005;167:517–526. doi: 10.1016/s0002-9440(10)62994-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Goldbaum O, Vollmer G, Richter-Landsberg C. Proteasome inhibition by MG-132 induces apoptotic cell death and mitochondrial dysfunction in cultured rat brain oligodendrocytes but not in astrocytes. Glia. 2006;53:891–901. doi: 10.1002/glia.20348. [DOI] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan B, Hartmann B, Kho YH, Gorczyca M, Budnik V. The Drosophila tumor suppressor gene, dlg, is involved in structural plasticity at a glutamatergic synapse. Curr. Biol. 1996;6:695–706. doi: 10.1016/s0960-9822(09)00451-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston J, Kopito R. HDAC6 and microtubules are required for autophagic degradation of aggregated huntington. J. Biol. Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am. J. Physiol. Cell Physiol. 2004;287:C834–C843. doi: 10.1152/ajpcell.00579.2003. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostova Z, Wolf DH. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahey T, Gorczyca M, Jia XX, Budnik V. The Drosophila tumor-suppressor gene DLG is required for normal synaptic bouton structure. Neuron. 1994;13:823–835. doi: 10.1016/0896-6273(94)90249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang-Rollin I, Vekrellis K, Wang Q, Rideout HJ, Stefanis L. Application of proteasomal inhibitors to mouse sympathetic neurons activates the intrinsic apoptotic pathway. J. Neurochem. 2004;90:1511–1520. doi: 10.1111/j.1471-4159.2004.02684.x. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Neuburger PJ, Saville KJ, Zeng J, Smyth KA, Belote JM. A genetic suppressor of two dominant temperature-sensitive lethal proteasome mutants of Drosophila melanogaster is itself a mutated proteasome subunit gene. Genetics. 2006;173:1377–1387. doi: 10.1534/genetics.106.057976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. U.S.A. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Zhang YQ, Woodruff E, Broadie K. The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Curr. Biol. 2004;14:1863–1870. doi: 10.1016/j.cub.2004.09.085. [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Purintrapiban J, Wang MC, Forsberg NE. Degradation of sarcomeric and cytoskeletal proteins in cultured skeletal muscle cells. Comp. Biochem. Physiol. B, Biochem. Mol. Biol. 2003;136:393–401. doi: 10.1016/s1096-4959(03)00201-x. [DOI] [PubMed] [Google Scholar]
- Rohrbough J, Grotewiel MS, Davis RL, Broadie K. Integrin-mediated regulation of synaptic morphology, transmission, and plasticity. J. Neurosci. 2000;20:6868–6878. doi: 10.1523/JNEUROSCI.20-18-06868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrbough J, Rushton E, Palanker L, Woodruff E, Matthies HJG, Acharya U, Acharya JK, Broadie K. Ceramidase regulates synaptic vesicle exocytosis and trafficking. J. Neurosci. 2004;24:7789–7803. doi: 10.1523/JNEUROSCI.1146-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville KJ, Belote JM. Identification of an essential gene, l(3)73Ai, with a dominant temperature-sensitive lethal allele, encoding a Drosophila proteasome subunit. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8842–8846. doi: 10.1073/pnas.90.19.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Schweisguth F. Dominant-negative mutation in the β2 and β6 proteasome subunit genes affect alternative cell fate decisions in the Drosophila sense organ lineage. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11382–11386. doi: 10.1073/pnas.96.20.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcen D, Ohno K, Engel AG. Myofibrillar myopathy: clinical, morphological and genetic studies in 63 patients. Brain. 2004;127:439–451. doi: 10.1093/brain/awh052. [DOI] [PubMed] [Google Scholar]
- Smyth KA, Belote JM. The dominant temperature-sensitive lethal DTS7 of Drosophila melanogaster encodes an altered 20S proteasome-type subunit. Genetics. 1999;151:211–220. doi: 10.1093/genetics/151.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese SD, Trotta N, Rodesch CK, Aravamudan B, Broadie K. The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Curr. Biol. 2003;13:899–910. doi: 10.1016/s0960-9822(03)00338-5. [DOI] [PubMed] [Google Scholar]
- Vattemi G, Engel WK, McFerrin J, Askanas V. Endoplasmic reticulum stress and unfolded protein response in inclusion body myositis muscle. Am. J. Pathol. 2004;164:1–7. doi: 10.1016/S0002-9440(10)63089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihl CC, Dalal S, Pestronk A, Hanson PI. Inclusion body myopathy-associated mutations in p97/VCP impair endoplasmic reticulum-associated degradation. Hum. Mol. Genet. 2006;15:189–199. doi: 10.1093/hmg/ddi426. [DOI] [PubMed] [Google Scholar]
- Wojcik S, Engel WK, McFerrin J, Paciello O, Askanas V. In cultured human muscle fibers (CHMFs) amyloid-β precursor protein (Aβ-PP) and proteasome inhibition increase αB-crystallin (αBC). Relevance to sporadic inclusion-body myositis (s-IBM) Neuromuscul. Disord. 2006;16:657. doi: 10.1016/j.nmd.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WH, Kumar A, Peterhoff C, Shapiro Kulnane L, Uchiyama Y, Lamb BT, Cuervo AM, Nixon RA. Autophagic vacuoles are enriched in amyloid precursor protein-secretase activities: implications for β-amyloid peptide over-production and localization in Alzheimer’s disease. Int. J. Biochem. Cell Biol. 2004;36:2531–2540. doi: 10.1016/j.biocel.2004.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.