Abstract
HIV type 1 (HIV-1) drug resistance mutations were selected during antiretroviral therapy successfully suppressing plasma HIV-1 RNA to <50 copies/ml. New resistant mutant subpopulations were identified by clonal sequencing analyses of viruses cultured from blood cells. Drug susceptibility tests showed that biological clones of virus with the mutations acquired during successful therapy had increased resistance. Each of the five subjects with new resistant mutants had evidence of some residual virus replication during highly active antiretroviral therapy (HAART), based on transient episodes of plasma HIV-1 RNA > 50 copies/ml and virus env gene sequence changes. Each had received a suboptimal regimen before starting HAART. Antiretroviral-resistant HIV-1 can be selected from residual virus replication during HAART in the absence of sustained rebound of plasma HIV-1 RNA.
Resistance mutations selected by a new antiretroviral drug regimen have not been identified in HIV type 1 (HIV-1) isolated from peripheral blood mononuclear cells while plasma HIV-1 RNA levels remained suppressed to <50 copies/ml (1–4). However, residual virus replication may occur during such successful highly active antiretroviral therapy (HAART) (4–9), as is evident when HIV-1 RNA transiently rises to >50 copies/ml. This prompted speculation that HIV-1 may replicate during successful HAART in body sites lacking adequate exposure to antiretrovirals for selection of drug-resistant mutants (3). The present study evaluated this concept by attempting to detect minor subpopulations of new, HAART-selected variants in replication-competent viruses isolated from blood cells while plasma HIV-1 RNA was <50 copies/ml (1, 2, 10). Viruses in which resistance mutations had not previously been found by less intensive analyses (1, 2) were studied.
Methods
Study Population and Virus Isolates.
Specimens were from 10 patients who did not have sustained rebound of plasma HIV-1 RNA over 96 to 123 weeks of HAART (1, 2, 10). Two had treatment initiated during acute HIV-1 infection (patients 23 and 24; ref. 10) and eight during established infection. The study was designed to detect minorities of resistant virus in the same specimens in which resistance had not been identified earlier (1, 2). Treatment histories and baseline resistance mutations are indicated in Table 1. Patients A, B, C, K, L, and M had pooled virus isolated (1). Patients 9, 11, 23, and 24 had HIV-1 biologically cloned by limiting dilution of purified blood resting CD4+ T lymphocytes (2).
Table 1.
Summary of resistance mutations selected during successful therapy
| Pt. | Previous drugs* | Current drugs* | Baseline mutations† | Clone no. | New mutations† (Clone no.) |
|---|---|---|---|---|---|
| Analyzed by molecular cloning | |||||
| M | ZDV | ZDV + 3TC + IDV | RT 41L, 44D, 67N, 70R, 215Y | 42 | RT 118I (3) |
| C | ZDV | ZDV + 3TC + IDV | RT 67N, 70R, 219Q | 98 | PR 54V; RT 184V (2) |
| K | ddI, ZDV | ZDV + 3TC + IDV | PR {71V}; RT 41L, 74V, 215Y | 39 | PR 46I, 77I; RT 118I, 210W (2) |
| L | ZDV | ZDV + 3TC + IDV | RT 67N, 219Q | 26 | PR 46V, I;RT 69D, 70R (6) |
| A | ZDV | ZDV + 3TC + IDV | RT 70R | 17 | None |
| B | ZDV | ZDV + 3TC + IDV | RT 70R | 36 | None |
| Analyzed by biological cloning | |||||
| 9 | RTV, RTV + ZDV | DV + 3TC + RTV + SQ | Gag (p7p1)V; PR 54V, 77I, 82A; RT 70R (on-HAART) | 65 (6 times) | PR {10I, 71V}, 90M (6) |
| 11 | None | d4T + RTV + SQV | – | 4 | None |
| 23 | None | ZDV + 3TC + IDV | – | 16 | None |
| 24 | None | ZDV + 3TC + NFV | – | 12 | None |
ZDV, zidovudine; 3TC, lamivudine; IDV, indinavir; RTV, ritonavir; SQV, saquinavir; d4T, stavudine; and NFV, nelfinavir.
Baseline resistance mutations were identified in ref. 1. New on-therapy resistance mutations selected by drugs given only during the current successful regimen are in bold: lamivudine selected (italics and bold) and protease inhibitor selected (bold, not italicized). Brackets signify natural polymorphisms, which may also be drug-selected. Each position was not interrogated in every clone.
PCR Amplification and Molecular Cloning.
A proofreading polymerase (XL rTth DNA PCR, PE Biosystems, Foster City, CA) was used for all amplifications. For molecular cloning, CD8 cell-depleted culture supernatant fluids from ref. 1 were centrifuged at 21,000 × g for 90 min. Extracted virion RNA (QiAmp Viral RNA kit, Qiagen, Chatsworth, CA) was reverse transcribed (Superscript II, GIBCO/BRL) with a specific primer before nested PCR. Final amplicons spanning NL4–3 nucleotides 1,988–4,255 (primers Apa1988–3R4226) (11) were cloned (CloneAmp, GIBCO/BRL). For biological clones from ref. 2, HIV-1 DNA extracted from clone-infected peripheral blood mononuclear cell (Puregene, Gentra Systems) was amplified [gag-pol: primers 5CAI1964B-3CAI4155LIG (11) spanning NL4–3 nucleotides 1,964–4,156 and including gag p7/p1 and p1/p6 cleavage sites to beyond reverse transcriptase (RT) codon 350; env C2-V3-C3: Primers spanning NL4–3 nucleotides 6,835–6,855 and 7,368–7,347]. Amplicons were purified (QIAquick, Qiagen).
HIV-1 Genotyping.
Each biological clone gag-pol amplicon from patients 9, 11, 23, and 24 was cycle sequenced, as were env amplicons from patients 9, 23, and 24 (BigDye, ABI 377, PE Biosystems; Sequencher 3.1, Gene Codes, Ann Arbor, MI).
Gag-pol plasmid clones (≥15) from patients A, B, C, K, L, and M were genotyped; ≥3 were sequenced. Others were genotyped by a more rapid method, either line probe assay [LiPa HIV-1 RT, Innogenetics, Atlanta, GA (ref. 12) for RT codons 41, 69, 70, 74, 75, 184, and 215] or HIV site-specific sequencing (HIV-SSS) (13). HIV-SSS interrogated specific nucleotides (in brackets) in protease codons 10 [1st nucleotide], 30 [1], 46 [1, 3], 63 [2], 82 [1, 2], and 90 [1]; reverse transcriptase (RT) codons 41 [1], 70 [2], 74 [1], 103 [3], 151 [1], 181 [2], 184 [1], 190 [2], and 215 [2]; and the known gag p7/p1 cleavage site changes (14–16). Each mutation identified by LiPa or HIV-SSS was confirmed by cycle sequencing of that clone.
Molecular clones were genotyped as follows: patient A (3 sequenced, 14 by HIV-SSS); patient B (3 sequenced, 33 by HIV-SSS); patient C (13 and 2 sequenced in protease and RT, respectively, 83 by LiPA for RT); patient K (5 sequenced, 34 by HIV-SSS); patient L (3 protease clones by sequencing and 5 by HIV-SSS for protease, 3 RT clones by sequencing and 5 by HIV-SSS for RT, 10 clones by LiPA for RT); patient M (3 sequenced, 39 by HIV-SSS). Molecular clones were initially genotyped without knowledge of the patient from which they were derived. Patient C had previously been noted to have a minority-resistant HIV subpopulation at RT codon 184 in peripheral blood mononuclear cell RNA at an earlier time point in a separate previous study (28). Therefore, after the specimen matching patient C's dominant virus genotype was identified, genotyping was undertaken on more molecular clones than studied from other specimens. Sequences included protease and RT codons to RT codon 230.
Biological clones were sequenced as follows: patient 9 (64 each for gag-pol and env; one additional pol sequence from ref. 2); patient 11 (4 each for gag-pol only); patient 23 (16 for gag-pol and 14 for env); and patient 24 (12 each for gag-pol and env). Sequences of gag-pol included gag p7/p1 and p1/p6 cleavage sites through, in most cases, RT codon 230.
Genetic Analyses.
Phylogenetic analyses used two-parameter Kimura algorithm (dnadist) or maximum likelihood (dnaml) in phylip (17, 18). Neighbor-joining trees (19) were plotted with treeview version 1.5.3 (20). Bootstrap analyses were performed on neighbor-joining trees by using up to 1,000 resamplings (17). Mann-Whitney U tests evaluated significance of differences in nucleotide distances.
Plasma RNA Measurements.
Plasma HIV-1 RNA levels were determined by an assay with a limit of detection of either 200 copies/ml or 50 copies/ml (Amplicor, Roche Diagnostics).
Drug Susceptibility Assay.
IC50 values were measured in a single-round recombinant virus assay (21). Each biological clone from patient 9 with at least a primary resistance mutation in protease codon 82 (PR V82A or F) or a secondary resistance mutation in codon 54 (PR I54V) was tested. IC50 values >2.5-fold above that of wild-type virus indicate reduced susceptibility (21).‡‡
Results
Resistant Mutants Developed During Successful Therapy.
Viruses cultured from blood cells of 10 subjects receiving successful HAART underwent analyses by using either molecular or biological clones (Table 1). Each subject started at least two drugs which had not been used previously: lamivudine and a protease inhibitor (PI). No subject experienced sustained viral load rebound in plasma over 46 to 100 weeks of follow-up after the on-therapy specimen was obtained. Five subjects' viruses developed lamivudine or PI resistance mutations during successful HAART (Table 1). Each had previously experienced failure of an antiretroviral treatment. Two subjects' viruses had resistance mutations selected by a current PI; one had mutations selected by lamivudine, and two had mutations selected by both a current PI and lamivudine.
There was no evidence of sequence cross-contamination from another source to account for the identified resistance mutations. In rooted trees of protease sequences of these subjects' isolates and laboratory strains (NL4–3, HxB2, JRFL, and 8–96), all viruses from each subject formed individual clusters with significant bootstrap values. Clones with new resistance mutations were related only to other clones from the same subject in trees of protease, RT, and env C2-V3-C3 sequences.
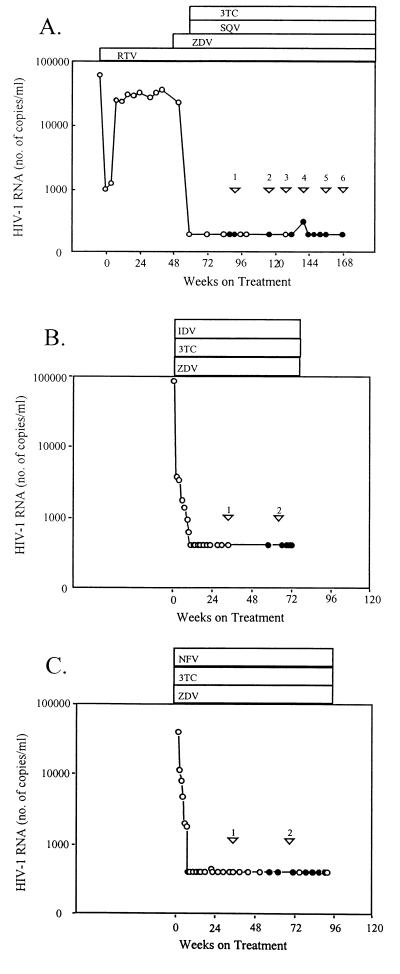
Longitudinal clonal analyses of infectious biological clones obtained from patient 9 documented that resistance mutations were selected during the successful therapy. Patient 9 had previously experienced virological failure of nonsuppressive ritonavir monotherapy (Fig. 1A). Plasma HIV-1 RNA levels became undetectable after starting HAART by adding zidovudine, saquinavir, and lamivudine to ritonavir (400 mg twice daily). Although a preHAART specimen was not available, comparisons were made over six time points on-therapy (Fig. 1A and Table 2). No lamivudine resistance mutations developed. PI resistance mutations are depicted in Table 2 (in italics). New PI resistance mutations in patient 9's viruses (bold) appeared in an otherwise wild-type background in two biological clones. Clones 3A and 6I had only the primary ritonavir resistance mutation V82F (Table 2). A larger number of patient 9 biological clones accumulated new mutations in a preexisting background of other PI-selected mutations (Table 2). Eight of 16 clones at the first time point, 9 months after first achieving suppression to <50 copies/ml, had protease mutations: the primary ritonavir resistance mutation PR V82A, the secondary ritonavir resistance mutation PR I54V, and two resistance-associated natural polymorphisms, PR L63P and V77I. The suggestion that this quadruple mutation pattern (underlined in Table 2) was selected by earlier ritonavir monotherapy (2) was supported by higher levels of resistance to ritonavir than to other PIs (Table 2). New protease mutations after the first time point were characteristic of saquinavir selection pressure, as expected when ritonavir is used primarily to enhance saquinavir blood levels. The primary saquinavir resistance mutations PR L90 M, as well as secondary saquinavir resistance mutations L10I and A71V, and the gag cleavage site mutation gag A(p7/p1)V, accumulated as additions to the quadruple mutation background (clones 1K, 1L, 2A, 4B, 5L, 5O, 6B, 6E; Table 2). The mutation in the gag p7/p1 cleavage site seen at the first, fourth, and fifth on-HAART time points may be selected by saquinavir (23), indinavir (14), or certain investigational PIs (15, 16). This, and other mutations, have been speculated to improve replicative fitness in compensation for effects of certain protease mutations (24).
Figure 1.
Plasma HIV-1 RNA levels and antiretroviral regimens for patients 9 (A), 23 (B), and 24 (C). ( ) When viruses were analyzed. RNA assays with a lower detection limit of 50 (●) or 400 (○) copies/ml.
Table 2.
Longitudinal analysis of resistance in HIV biologic clones (Patient 9)
| RT
|
gag
|
Protease
|
IC50,
μM, [fold X WT]
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 70 | P7/P1 | 10 | 14 | 16 | 24 | 54 | 63 | 71 | 77 | 82 | 90 | Ritonavir | Saquinavir | |
| NL4-3 | Lys | Ala | Leu | Lys | Gly | Leu | Ile | Leu | Ala | Val | Val | Leu | Mean WT* = 0.0144 | WT* = 0.0021 |
| 1I | Arg | – | – | – | – | – | – | – | – | – | – | – | ||
| 1J | Arg | – | – | – | – | – | – | – | – | – | – | – | ||
| 1M | Arg | – | – | – | – | – | – | – | – | – | – | – | ||
| 1A | – | – | – | Arg | – | – | – | – | – | – | – | – | ||
| 1B | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 1E | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 1N | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 1O | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 1P | – | ND | – | ND | Ala | ND | Val | Pro | – | Ile | Ala | – | ND | ND |
| 1C | – | – | – | – | Ala | – | Val | Pro | – | Ile | Ala | – | 0.2248 [15.6] | 0.0035 [1.7] |
| 1D | – | – | – | – | – | – | Val | Pro | – | Ile | Ala | – | 0.2366 [16.4] | 0.0026 [1.2] |
| 1F | – | – | – | – | Ala | – | Val | Pro | – | Ile | Ala | – | 0.0430 [3.0] | 0.0036 [1.7] |
| 1G | – | – | – | – | Ala | Ile | Val | Pro | – | Ile | Ala | – | 0.4005 [27.8] | 0.0050 [2.4] |
| 1H | – | – | – | – | –/Ala | –/Ile | –/Val | –/Pro | – | –/Ile | –/Ala | – | 0.0380 [2.6] | 0.0027 [1.3] |
| 1K | – | Val | – | – | Ala | Ile | Val | Pro | – | Ile | Ala | – | 0.4144 [28.8] | 0.0038 [1.8] |
| 1L | – | Val | – | – | –/Ala | –/Ile | –/Val | –/Pro | – | –/Ile | –/Ala | – | 0.0152 [1.1] | 0.0016 [0.8] |
| Mutants mean: 0.1961 [13.6] | 0.0033 [1.6] | |||||||||||||
| 2B | Arg | – | – | – | – | – | – | – | – | – | – | – | ||
| 2C | – | – | – | Arg | – | – | – | – | – | – | – | – | ||
| 2E | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 2F | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 2H | – | – | – | Arg | – | – | – | – | – | – | – | – | ||
| 2I | Arg | – | – | – | – | – | – | – | – | – | – | – | ||
| 2K | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 2G | – | – | – | – | Ala | – | Val | – | – | Ile | Ala | – | 0.0820 [5.7] | 0.0022 [1.1] |
| 2J | – | – | – | Arg | – | – | Val | Pro | – | – | – | – | 0.0131 [0.9] | 0.0016 [0.8] |
| 2D | – | – | – | – | Ala | – | Val | Pro | – | Ile | Ala | – | 0.2115 [14.7] | 0.0034 [1.6] |
| 2A | – | – | – | – | Ala | – | Val | Pro | – | Ile | Ala | Met | 0.1019 [7.1] | 0.0024 [1.1] |
| Mutants mean: 0.1021 [7.1] | 0.0024 [1.1] | |||||||||||||
| 3B | Arg | – | – | – | – | – | – | – | – | – | – | – | ||
| 3C | Arg | – | – | – | – | – | – | – | – | – | – | – | ||
| 3D | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 3E | – | – | – | Arg | – | – | – | – | – | – | – | – | ||
| 3F | Arg | – | – | – | – | – | – | – | – | – | – | – | ||
| 3G | Arg | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 3H | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 3A | – | – | – | – | Glu | – | – | – | – | – | Phe | – | 0.0154 [1.1] | 0.0013 [0.6] |
| 4A | Arg | – | – | – | – | – | – | – | – | – | – | – | ||
| 4B | – | – | Ile | Arg | – | – | Val | Pro | Val | Ile | Ala | Met | 0.438 [30.4] | 0.0058 [2.8] |
| 5B | – | – | – | Arg | – | – | – | – | – | – | – | – | ||
| 5C | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 5D | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 5E | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 5F | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 5G | – | – | – | Arg | – | – | – | – | – | – | – | – | ||
| 5K | Arg | – | – | – | – | – | – | – | – | – | – | – | ||
| 5M | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 5H | – | – | – | Arg | – | – | – | – | – | Ile | – | – | ||
| 5A | – | – | – | – | Glu | – | – | – | – | Ile | – | – | ||
| 5I | – | – | – | – | Glu | – | Val | Pro | – | Ile | Ala | – | 0.2164 [15.0] | 0.0034 [1.6] |
| 5J | – | – | – | – | Glu | – | Val | Pro | – | Ile | Ala | – | 0.2352 [16.3] | 0.0036 [1.7] |
| 5N | – | – | – | – | Glu | – | Val | Pro | – | Ile | Ala | – | 0.1330 [9.2] | 0.0022 [1.1] |
| 5L | – | Val | – | – | Glu | Ile | Val | Pro | – | Ile | Ala | – | 0.6882 [47.8] | 0.0052 [2.5] |
| 5O | – | Val | Ile | – | Glu | Ile | Val | Pro | – | Ile | Ala | – | 0.7864 [5.5] | 0.0072 [3.4] |
| Mutants mean: 0.4118 [28.6] | 0.0043 [2.1] | |||||||||||||
| 6D | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 6F | Arg | – | – | – | – | – | – | – | – | – | – | – | ||
| 6G | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 6H | – | – | – | Arg | – | – | – | – | – | – | – | – | ||
| 6J | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 6L | – | – | – | – | – | – | – | – | – | – | – | – | ||
| 6M | – | – | – | – | Glu | – | – | – | – | – | – | – | ||
| 6N | – | – | – | Arg | – | – | – | – | – | – | – | – | ||
| 6K | – | – | – | Arg | – | – | – | – | – | Ile | – | – | ||
| 6C | – | – | – | – | Glu | – | – | – | – | Ile | – | – | ||
| 6I | – | – | – | – | Glu | – | – | – | – | – | Phe | – | 0.0731 [5.1] | 0.0009 [0.4] |
| 6B | – | Val | – | – | Ala | Ile | Val | Pro | – | Ile | Ala | – | 0.4384 [30.4] | 0.0037 [1.8] |
| 6E | – | – | Ile | Arg | – | – | Val | Pro | Val | Ile | Ala | Met | 0.3398 [23.6] | 0.0056 [2.7] |
| Mutants mean: 0.2838 [19.7] | 0.0034 [1.6] | |||||||||||||
IC50s of wild-type virus clones: 1M, 0.0184 μM ritonavir (RTV), 0.0024 μM saquinavir (SQV); 3B, 0.012 μM RTV, 0.0017 μM SQV; 5D, 0.0127 μM RTV, 0.0021 μM SQV; mean, 0.0144 μM RTV, 0.0021 μM SQV.
Bold indicates IC50s at least 2.5-fold above mean wild-type IC50. Clones tested at the first time point had IC50s 1.2–6.1-fold above control for indinavir and 1.1–4.4-fold above control for nelfinavir.
Mutations Developing During Successful Therapy Confer Increasing Resistance.
Drug-susceptibility assays confirmed that increased resistance to saquinavir and ritonavir was conferred by the new mutations selected during patient 9's therapy (Table 2). A 2.5-fold or greater increase in saquinavir IC50 was noted only in four clones with additional saquinavir resistance mutations in the background of the preexisting quadruple mutations (clones 4B, 5L, 5O, and 6E, Table 2; IC50s ≥ 2.5-fold above the mean wild-type IC50 are in bold). Clones with wild-type protease sequence had similar levels of susceptibility to all PIs, within a 2.0-fold range of each other and a wild-type control laboratory strain (Table 2 legend). V82F by itself also did not increase IC50, as in earlier reports (25). The specificity of the decreased susceptibility to that expected from the identified genotypes was confirmed by IC50s of the 20 tested clones to other drugs in addition to the PIs (zidovudine, stavudine, didanosine, lamivudine, zalcitabine, abacavir, adefovir, delavirdine, nevirapine, and efavirenz; not shown).
Evidence of Residual Replication: Plasma Viral Load.
To evaluate whether the identified resistance mutations were related to magnitude of residual replication during successful HAART, subjects were categorized into three levels of apparent residual virus replication based on both initial decline in plasma HIV-1 RNA levels and later transient increases as in ref. 7. The most apparent residual replication was observed in subjects M and C who had slow initial declines in viral load and more than one transient episode of low level, detectable plasma HIV-1 RNA (>50 copies/ml) during therapy. Subjects K, L, and 9 (Fig. 1A) were in an intermediate category; each had at least one transient episode of plasma HIV-1 RNA >50 copies/ml despite steep initial declines in viral load. The five other subjects (A, B, 11, 23, and 24) had no detected episodes of plasma HIV-1 RNA >50 copies/ml and initial steep declines in plasma viral load. Viral load responses of subjects 23 and 24 (10, 26) are representative of this group, which promptly achieved consistent antiretroviral suppression (Fig. 1 B and C).
The specific lamivudine- and/or PI-resistant mutations identified in viruses from the subjects with any detected episodes of plasma HIV-1 RNA >50 copies/ml were consistent with greater resistance in viruses from subjects with more apparent residual replication (patients M and C) than in viruses from subjects with less replication (patients K, L, and 9). The subjects with the greatest estimated residual replication, M and C, had the only dominant resistance mutation [RT V118I, which causes low-level lamivudine resistance (27)] and the only mutation conferring high-level resistance (RT M184V), respectively. [The 1 of 98 molecular clones from patient C's on-therapy virus with RT M184V, which confers high level (>100-fold) lamivudine resistance, is consistent with earlier detection of a RT codon 184 mutant subpopulation in HIV RNA in peripheral blood mononuclear cells at an earlier time point on-therapy (28)].
Minority subpopulations with resistance mutations causing only lower-level resistance were identified in viruses from the subjects in the intermediate category (K, L, and 9) who had less apparent residual replication. Patient K's on-therapy isolate had a primary resistance mutation selected by indinavir, PR M46I (29), linked to RT V118I (27) and to a zidovudine resistance mutation (RT L210W) not identified in baseline plasma, as well as to all of the mutations present in baseline plasma (Table 1). Patient L had primary indinavir resistance mutations appear on-therapy: PR M46V in one and PR M46I in a second protease-containing molecular clone. Viruses from patient 9 had minority subpopulations containing primary or secondary saquinavir resistance mutations (L10I, A71V, and L90 M). Each of these mutations in protease [PR L10I, M46V, or I, A71V, L90M (ref. 25) and Table 2], and reverse transcriptase (RT V118I) (27) confer only low-level resistance in vitro. Viruses from the subjects with no detected episodes of plasma HIV-1 RNA >50 copies/ml did not have new resistant mutant subpopulations identified.
Evidence of Residual Replication: env Genetic Analyses.
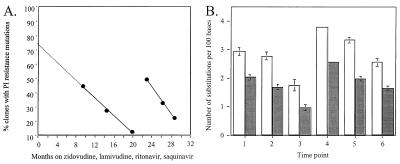
An increase in protease mutations and resistance in patient 9's infectious, full-length biological clones was temporally associated with both transiently detectable plasma HIV-1 RNA levels and increased env genetic diversity (Fig. 2 A and B). Plasma HIV-1 RNA was 306 copies/ml at the fourth virus sampling time point after being <50 at all earlier and subsequent measures, including one week later (Fig. 1A). The proportion of clones with any resistance-associated protease substitution increased only between the third and fifth time points (Fig. 2A). Env genetic distances also significantly increased only from the third to fifth time point (P < 0.001, Fig. 2B). The number of PI-selected mutations linked together in single genes also increased only at, and after, the fourth time point. At the first two times, only a single mutation was added to the baseline quadruple mutation background (clones 1K, 1L, and 2A; Table 2). In contrast, two or three additional mutations were linked together at, and after, the fourth time point (clones 4B, 5O, 6B, and 6E; Table 2). The mean saquinavir and ritonavir IC50s of the subpopulation with protease mutants also increased only at, and after, the fourth time point (Table 2).
Figure 2.
(A) Percentage of biological clones from patient 9 with at least one protease inhibitor resistance mutation. Solid lines indicate best fit (r2 > 0.99). Extrapolation (dashed line) estimates proportion of virus population with ritonavir-selected mutations at HAART baseline. (B) Genetic variability of patient 9's biological clones in env C2-V3-C3, by using Jukes-Cantor (open bars) and Kimura measures (solid bars) (17). Each time point shown is based on at least eight clones. Second and third time points were each significantly less than the first (P < 0.05 and P < 0.001, respectively). The third time point was significantly less than the second (P < 0.001). The fifth time point had significantly greater env diversity than the third (P < 0.001), but was not significantly different from the first. The sixth time point was significantly less diverse than the fifth (P < 0.01). Statistical analyses were the same with each measure of genetic distance. Bars indicate standard error.
Increased env sequence diversity was detected only in on-therapy viruses with new resistant mutant subpopulations, corroborating the relation between extent of residual replication and development of resistance mutations. Divergence of env C2-V3 sequences from that of the baseline plasma, consistent with ongoing residual replication, was noted in on-therapy viruses from patients M, C, and K, but not in those from patients A, B, and L in an earlier report (7). However, the on-therapy virus from patient L did show some sequence change in env, consistent with recovery from cells infected before emergence of the viruses in preHAART therapy plasma. The on-therapy isolate had nucleoside resistance mutations not present in baseline plasma RNA (RT T69D or K70R; Table 1), and less genetic distance to the most recent common ancestor than did baseline plasma RNA (Fig. 2 and Table 3 in ref. 7), as well as new PR mutations.
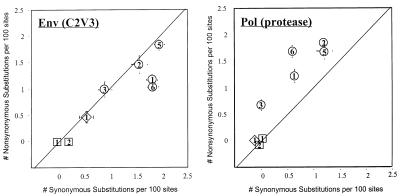
Selection Pressure on Protease, Not env.
Analyses of ratios of nonsynonymous to synonymous substitutions in biological clones of virus containing both pol and env genes from patient 9 indicated selection pressure on the protease gene during residual replication. The ratio of nonsynonymous to synonymous substitutions in the protease genes was greater than one at each time point (Fig. 3 Right). No selection pressure was evident in env genes of those same clones by this criterion at any time point (Fig. 3 Left). There was also a nonrandom distribution of env diversity among patient 9 clones. Biological clones with protease resistance mutations (PR V82A or F, or multiple resistance-associated substitutions) from patient 9 had more diverse env C2-V3-C3 sequences than other clones (not shown). No selection pressure was evident in protease, or env, genes of patient 23 or 24 virus clones (Fig. 3), which also lacked env diversity and resistance mutations.
Figure 3.
Non-synonymous to synonymous substitution ratios for C2-V3 region env and protease sequences from virus biological clones from patients 9 (○), 23 (□), and 24 (◊). Mean genetic distances and standard error are plotted. The number in each symbol represents the time point.
Discussion
Replication-competent, drug-resistant virus can be selected by a successful HAART regimen in the absence of sustained rebound of plasma HIV-1 RNA above 50 copies/ml. This was documented during combination antiretroviral treatment currently recommended as optimal and in subjects for whom a previous suboptimal regimen had failed. Thus, residual virus replication during HAART (4–9) can be exposed to enough drug pressure to select resistant mutants.
This study was designed to improve detection of minor subpopulations of resistant virus in the previously analyzed specimens (1, 2) and not to determine prevalence of resistance development during successful therapy. Several factors may have improved identification of resistance over earlier results (1, 2). Clonal analyses improved detection of minority subpopulations. One newly characterized resistance mutation (27) was not known to confer lamivudine resistance when it was first found to dominate the on-therapy virus sequence (1). Additional time points were analyzed from some subjects (2).
The data indicate that the identified mutations were selected by drug therapy in vivo, and were not artifactual. None of the new lamivudine or PI resistance mutations (PR M46V or I, PR I54V, PR L90M, RT M184V, and RT V118I) are found in untreated patients (30). Analyses excluded laboratory contamination. There was excellent concordance of IC50s and genotypes determined from separate amplifications of biologically cloned viruses (Table 2), which is not consistent with in vitro misincorporation causing the observed mutations. Mutants were identified only in patients, and at time points, where viral loads and env sequence analyses indicated residual replication. This suggests that stochastic differences in sampling seem less likely than in vivo biological differences over time. Phylogenetic analyses reported earlier also did not support biases from cell sampling or virus culture (7).
Each of the viruses with new resistance mutations was from a subject with some apparent residual replication, as well as failure of a prior treatment. The extent of resistance was related to the magnitude of apparent residual replication. Lack of detection of resistance in some subjects was in keeping with such a relationship, but does not exclude the possibility of resistance in such patients. Indeed, in the present study, more clones were analyzed from the subjects receiving their second, rather than their first, antiretroviral regimen (Table 1). However, another report did not identify resistance in latent reservoir virus from patients receiving their initial antiretroviral therapy, even when episodes of plasma HIV-1 RNA >50 copies/ml were detected (4). This suggests the hypothesis that preexisting genetic diversity in drug-selected genes above some threshold level, as well as episodes of detectable residual replication, each may be necessary for emergence of resistance in the absence of sustained rebound of plasma HIV-1 RNA. There also is an appreciable failure rate of initial HAART regimens and the development of resistance has not yet been studied during successful rescue regimens after initial HAART failure. Further study will be needed to test whether virus may be more genetically diverse, and resistance may be more likely to develop, during a successful HAART regimen after either suboptimal therapy or an initial HAART regimen failure, than during HAART which initiates antiretroviral therapy.
Mutational patterns may also differ based on extent of previous treatment. Although only solitary RT codon 184 mutations are seen in rebounding plasma HIV-1 RNA from most patients early during failure of an initial triple combination regimen including a PI, multiple mutations in RT and/or protease can be seen in some of those patients (31–33). Preliminary data suggest that multiple resistance mutations are detected more commonly in rebounding plasma HIV RNA from patients who began HAART after extensive previous suboptimal treatment (unpublished). This is consistent with the result in the present study that mutations other than RT M184V were detected during successful therapy in pretreated patients.
The infrequent detection here of mutations conferring high-level resistance (e.g., RT M184V) while viremia remained suppressed suggests the hypothesis that sustained plasma viral load rebound may be associated only with higher levels of resistance. The lack of rebound in any of the five subjects with new resistance mutations is consistent with previous modeling, suggesting that resistant mutants arising in a cellular or tissue compartment with decreased drug levels may be suppressed by higher systemic drug levels (34). If both wild-type and PI-resistant clones were replicating under PI selection pressure in suboptimal drug levels, however, a slight replicative advantage for a resistant mutant would be expected to lead to an increasing proportion of a resistant mutant over many replication cycles in drug (35). Therefore, the lack of rise to dominance of the PI-resistant mutant subpopulation in patient 9's latent virus reservoir (Table 2) suggests that competition was limited either because replication was only intermittent, or the variants were not present in the same local compartment (36).
The lack of selective pressure on env could be caused by an intrinsic defect in HIV-1 specific immunity, inadequate HIV-1 antigenic stimulation, or other mechanisms. Because acutely infected patients 23 and 24 each had HIV-specific immune responses (ref. 26 and unpublished data), an intrinsic immune defect can be excluded in those cases. Protease, but not env, selection pressure in patient 9 (Fig. 3 A and B) suggests that residual replication during HAART is sufficient to allow selection of resistant mutants by drugs, but not adequate to select immune escape mutants. This hypothesis will require testing.
It is important to emphasize that the mutations identified here have not yet been associated with sustained viral load rebound over prolonged follow-up. Therefore, relevance of this biological phenomenon for patient management is not yet clear. However, this genetic evidence that residual virus replication is under drug selection pressure in the absence of sustained rebound of plasma HIV-1 RNA levels strengthens the rationale for pilot clinical studies of strategies to limit accumulation of resistance mutations in viruses cultured from blood cells during successful therapy. Such strategies may include more potent drug regimens, better drug exposure in all cells, use of more sensitive assays of plasma RNA, or “proactively switching” regimens before sustained plasma HIV-1 RNA rebound occurs (22).
Acknowledgments
We thank Lijia Shi for expert technical assistance. Diane Havlir's care of the San Diego patients is appreciated. This work was supported by National Institutes of Health Grants AI 29193, AI 43222, AI 07387, AI 01696, AI 27670, AI 38858, AI 36214 (Center for AIDS Research), AI 29164, the Research Center for AIDS and HIV Infection of the San Diego Veterans Affairs Medical Center, the Swiss National Science Foundation (84AD-046176). J.M.P. was supported by a postdoctoral fellowship from the Spanish Ministry of Education and a contract from the Fundacio per la Recerca Biomedica Germans Trias I Pujol.
Abbreviations
- HAART
highly active antiretroviral therapy
- HIV-1
HIV type 1
- RT
reverse transcriptase
- HIV-SSS
HIV site-specific sequencing
- PI
protease inhibitor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF292712–AF292932).
Hellmann, N., Johnson, P. & Petropoulos, C., 39th Interscience Conference on Antimicrobial Agents and Chemotherapy, Sept. 26–29, 1999, San Francisco, CA, abstr. 418.
References
- 1.Wong J K, Hezareh M, Günthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 2.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, et al. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Ramratnam B, Tenner-Racz K, He Y, Vesanen M, Lewin S, Talal A, Racz P, Perelson A S, Korber B T, et al. N Engl J Med. 1999;340:1605–1613. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]
- 4.Ramratnam B, Mittler J E, Zhang L, Boden D, Hurley A, Fang F, Macken C A, Perelson A S, Markowitz M, Ho D D. Nat Med. 2000;6:82–85. doi: 10.1038/71577. [DOI] [PubMed] [Google Scholar]
- 5.Dornadula G, Zhang H, VanUitert B, Stern J, Livornese L J, Ingerman M, Witek J, Kedanis R J, Natkin J, DeSomone J, et al. J Am Med Assoc. 1999;282:627–632. doi: 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 6.Furtado M R, Callaway D S, Phair J P, Kunstman K J, Stanton J L, Macken C A, Perelson A S, Wolinsky S M. N Engl J Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 7.Günthard H F, Frost S D W, Leigh-Brown A J, Ignacio C C, Kee K, Perelson A S, Spina C A, Havlir D V, Hezareh M, Looney D J, et al. J Virol. 1999;73:9404–9412. doi: 10.1128/jvi.73.11.9404-9412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharkey M E, Teo I, Greenough T, Sharova N, Luzuriaga K, Sullivan J L, Bucy R P, Kostrikis L G, Haase A, Veryard C, et al. Nat Med. 2000;6:76–81. doi: 10.1038/71569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yerly S, Rutschmann O T, Opravil M, Marchal F, Hirschel B, Perrin L. J Infect Dis. 1999;180:850–853. doi: 10.1086/314932. [DOI] [PubMed] [Google Scholar]
- 10.Finzi D, Blankson J, Siliciano J D, Margolick J B, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Picado J, Sutton L, De Pasquale M P, Savara A V, D'Aquila R T. J Clin Microbiol. 1999;37:2943–2951. doi: 10.1128/jcm.37.9.2943-2951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuyver L, Wyseur A, Rombout A, Louwagie J, Scarcez T, Verhofstede C, Rimland D, Schinazi R F, Rossau R. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Picado J, Sutton L, Hoes Helfant A, Savara A, D'Aquila R T. Antiviral Ther. 1997;2, Suppl. 1:30. [Google Scholar]
- 14.Zhang Y-M, Imamichi H, Imamichi T, Lane H C, Falloon J, Vasudevachari M B, Salzman N P. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyon L, Croteau G, Thibeault D, Poulin F, Pilote L, Lamarre D. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrillo A, Stewart K D, Sham H L, Norbeck D W, Kohlbrenner W E, Leonard J M, Kempf D J, Molla A. J Virol. 1998;72:7532–7541. doi: 10.1128/jvi.72.9.7532-7541.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 18.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 19.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 20.Page R D. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 21.Petropoulos C J, Parkin N T, Limolo K L, Lie Y S, Wrin T, Huang W, Tian H, Smith D, Winslow G A, Capon D J, et al. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Amato R M, D'Aquila R T, Wein L M. Antiviral Ther. 1998;3:147–158. [PubMed] [Google Scholar]
- 23.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nijhuis M, Schuurman R, de Jong D, Erickson J, Gustchina E, Albert J, Schipper P, Gulnik S, Boucher C A. AIDS. 1999;13:2349–2359. doi: 10.1097/00002030-199912030-00006. [DOI] [PubMed] [Google Scholar]
- 25.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelsi L J, Graham D J, Laird D, Quintero J C, Rhodes A, et al. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg E S, Billingsley J M, Caliendo A M, Boswell S L, Sax P E, Kalams S A, Walker B D. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 27.Hertogs K, Bloor S, DeVroey V, Van den Eynde C, Dehertogh P, Van Cauwenberge A, Sturmer M, Alcorn T, Wegner S, Van Houtte M, et al. Antimicrob Agents Chemother. 2000;44:568–573. doi: 10.1128/aac.44.3.568-573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Günthard H F, Wong J K, Ignacio C C, Guatelli J C, Riggs N L, Havlir D, Richman D D. J Virol. 1998;72:2422–2428. doi: 10.1128/jvi.72.3.2422-2428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirsch M S, Conway B, D'Aquila R T, Johnson V A, Brun-Vezinet F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, et al. J Am Med Assoc. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 30.Shafer R W, Stevenson D, Chan B. Nucleic Acids Res. 1999;27:348–352. doi: 10.1093/nar/27.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Descamps D, Flandre P, Calvez V, Peytavin G, Meiffredy V, Collin G, Delaugerre C, Robert-Delmas S, Bazin B, Aboulker J P, et al. J Am Med Assoc. 2000;283:205–211. doi: 10.1001/jama.283.2.205. [DOI] [PubMed] [Google Scholar]
- 32.Havlir D V, Hellmann N S, Petropoulos C J, Whitcomb J M, Collier A C, Hirsch M S, Tebas P, Sommadossi J-P, Richman D D. J Am Med Assoc. 2000;283:229–234. doi: 10.1001/jama.283.2.229. [DOI] [PubMed] [Google Scholar]
- 33.De Pasquale M P, Murphy R, Kuritzkes D, Martinez-Picado J, Sommadossi J-P, Gulick R, Smeaton L, DeGruttola V, Caliendo A, Sutton L, et al. Antiviral Ther. 1998;3,Suppl. 1:50. [Google Scholar]
- 34.Kepler T B, Perelson A S. Proc Natl Acad Sci USA. 1998;95:11514–11519. doi: 10.1073/pnas.95.20.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coffin J M. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 36.Grossman Z, Polis M, Feinberg M B, Grossman Z, Levi I, Jankelevich S, Yarchoan R, Boon J, DeWolf F, Lange J M A, et al. Nat Med. 1999;5:1099–1104. doi: 10.1038/13410. [DOI] [PubMed] [Google Scholar]