Abstract
Human acute promyelocytic leukemias (APLs) are associated with chromosomal translocations that replace the NH2 terminus of wild-type retinoic acid receptor (RAR) α with portions of the promyelocytic leukemia protein (PML) or promyelocytic leukemia zinc-finger protein (PLZF). The wild-type RARα readily forms heterodimers with the retinoid X receptors (RXRs), and these RAR/RXR heterodimers appear to be the principal mediators of retinoid signaling in normal cells. In contrast, PML-RARα and PLZF-RARα display an enhanced ability to form homodimers, and this enhanced homodimer formation is believed to contribute to the neoplastic properties of these chimeric oncoproteins. We report here that the DNA recognition specificity of the RXRα/RARα heterodimer, which is presumed to be the dominant receptor species in normal cells, differs from that of the PML-RARα and PLZF-RARα homodimers, which are thought to prevail in the oncogenic cell. We suggest that differences in target gene recognition by the normal and oncogenic RARα proteins may contribute to the leukemogenic phenotype.
Introduction
Nuclear hormone receptors are eukaryotic transcription factors that regulate vertebrate cell differentiation, morphogenesis, and development (1– 6). The family of nuclear hormone receptors includes the steroid receptors, T3Rs,3 RXRs, and RARs (2). Nuclear hormone receptors regulate transcription by binding to specific DNA sequences denoted as hormone response elements and modulating the expression of adjacent target genes. T3Rs and RARs bind DNA as protein dimers, either as homodimers or as heterodimers with RXRs (1 – 6). As a consequence, a prototypic hormone response element consists of two conserved “half-sites,” with each half-site representing the DNA sequence contacted by one receptor monomer, and DNA recognition by nuclear hormone receptors depends on the sequence, orientation, and spacing of these two half-sites (1–11). Intriguingly, once bound to a response element, many nuclear hormone receptors can either repress or activate target gene expression, depending on the nature of the DNA binding site, the hormone status, and the cell type (1– 6). These bimodal transcriptional properties are mediated, in part, by the ability of the nuclear hormone receptors to physically recruit auxiliary proteins, denoted corepressors and coactivators, to the target promoter. These auxiliary factors in turn interact with the general transcriptional machinery and with the chromatin template to enhance or suppress gene transcription (12–16).
Mutant nuclear hormone receptors are involved in several forms of neoplastic diseases. For example, aberrant forms of RARα are found in over 95% of patients with APL (17–25). These aberrant proteins are the result of chromosomal translocations wherein a portion of the NH2-terminal region of RARα is replaced with novel NH2-terminal sequences (Refs. 19–25; Fig. 1B). Although the location of the breakpoint in the RARα sequence is highly conserved in these leukemias, the nature of the novel NH2 terminus can differ. The clinically most common form of APL is associated with a t(15;17) chromosomal translocation, resulting in expression of a PML-RARα chimeric receptor (17–21). Less frequently observed are t(11;17), t(5;17), or t(11;17) chromosomal translocations, which result in PLZF-RARα, NPM-RARα, or NuMA-RARα chimeric receptors, respectively (18 – 21). Intriguingly, the PML, PLZF, NPM, and NuMA open reading frames do not share significant sequence homology with one another and have distinct functions in the normal organism (18 – 21). The PML-RARα, PLZF-RARα, NPM-RARα, and NuMA-RARα chimeras themselves appear to play a central role in the etiology of APL, although other factors may also contribute. When introduced into transgenic mice, for example, PML-RARα and PLZF-RARα constructs induce myeloproliferative disorders that can advance to neoplasias similar in phenotype to those observed in human patients (26 – 29).
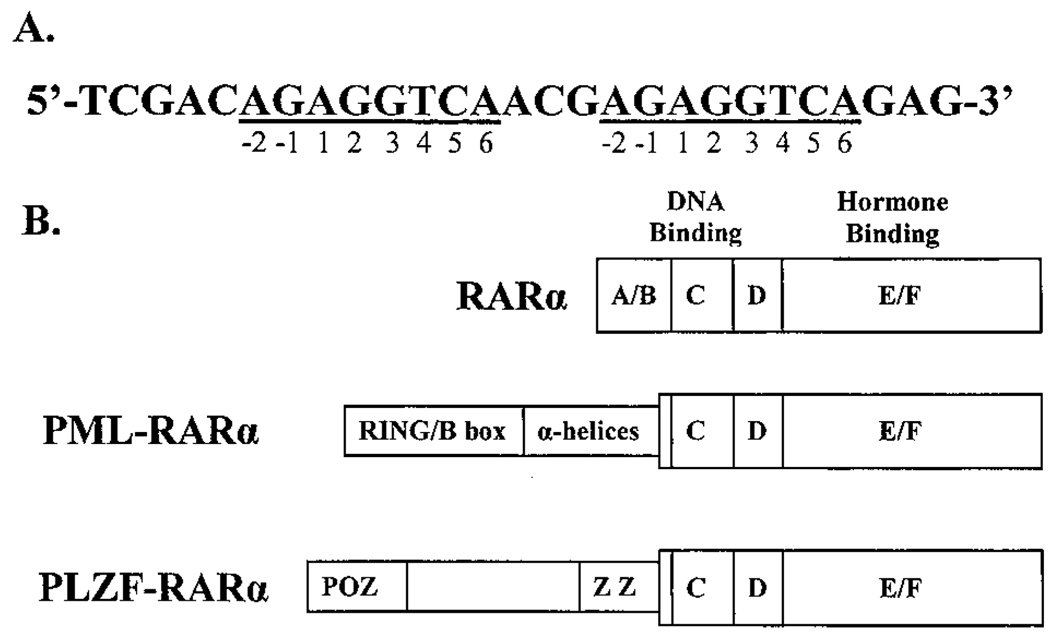
Fig. 1.
Consensus DNA recognition sequence for RARα and schematic representation of the human RARα, PML-RARα, and PLZF-RARα proteins. A, consensus DNA recognition sequence for RARα, as adapted from Kurokawa et al. (8). The sequence consists of two 8-base half-sites (underlined). The bases in each half-site are numbered from −2 to 6. B, schematic representations of human RARα, PML-RARα, and PLZF-RARα. The proteins are depicted from the NH2 to the COOH terminus. Structural domains in RARα are labeled A–F as described previously (1 – 6), and regions involved in DNA binding or hormone binding are indicated. Putative structural motifs in PML or PLZF, retained in the fusion proteins, are a cysteine-rich RING/B-box motif (RING/B box), an α-helical coiled-coil domain (α-helices), a POZ domain (POZ), and a series of zinc-finger motifs (Z; Refs. 12–16).
The PML-RARα and PLZF-RARα chimeras can function as dominant negative inhibitors of normal RARα function, apparently due to defects in the ability of these chimeric receptors to release corepressor under physiological hormone concentrations (28, 30–33). These defects in corepressor release are closely associated with the leukemogenic phenotype, suggesting that repression of normal RAR function plays an important role in oncogenesis. However, several observations suggested to us that the DNA recognition properties of PML-RARα and PLZF-RARα might be different from those of wild-type RARα and that, in addition to defects in corepressor release, the chimeric oncoproteins might also display a distinct target gene specificity from that of the normal RARα progenitor. First, we and others have noted that the NH2 terminus of the nuclear hormone receptors, which is lost in the leukemogenic RARα variants, plays an important role in defining the DNA recognition properties of the normal nuclear hormone receptors (34 – 39). Second, it has been noted that the chimeric RARα oncoproteins exhibit an enhanced ability to bind to DNA as homodimers, rather than heterodimers with RXR, and that this enhanced homodimerization appears to correlate with oncogenesis (40 – 43). This enhanced homodimerization by RAR chimeras has been implicated in the aberrant corepressor interaction properties of these oncogenic receptor derivatives (43), but any alteration in the dimerization properties of the chimeric RAR oncoproteins is also likely to influence their DNA recognition properties.
In the present study, we show that the in vitro DNA binding specificities of PML-RARα and PLZF-RARα were indeed modestly altered from that of RARα when these receptors were tested as homodimers. More significantly, perhaps, we found that the heterodimeric interaction of RARα with RXRα conferred an enhanced binding to a broader range of DNA sequences relative to that seen for the corresponding homodimers. The wild-type RARα is believed to function in cells almost exclusively as a heterodimer with RXR (44–47) and would therefore be expected to display this broadened range of DNA recognition characteristic of the RXRα/RARα heterodimer. In contrast, PML-RARα and PLZF-RARα have been proposed to function in leukemogenesis as homodimers or perhaps as higher order homo-oligomers (40 – 43, 48), indicating that PML-RARα and PLZF-RARα in cells would exhibit the more restrictive DNA recognition specificity that we observe for homodimers in vitro. Results from our in vivo transactivation studies are consistent with this proposal: transcriptional regulation by RARα is enhanced by cointroduction of RXRα; whereas transcriptional regulation by PML-RARα is impaired by cointroduction of RXRα. Our results therefore suggest that not all genes regulated by RXRα/RARα in normal cells may be recognized or subject to repression by the chimeric receptor homodimers found in APL. PML-RARα and PLZF-RARα homodimers may therefore participate in oncogenesis by aberrantly regulating only a subset of the total gene repertoire normally controlled by RXRα/RARα heterodimers.
Results
The DNA Binding Specificities of PML-RARα and PLZF-RARαHomodimers Are Similar but not Identical to Those of RARα Homodimers
Given the role of the NH2 terminus in DNA recognition by the normal nuclear hormone receptors, it was plausible that the novel NH2 termini present on the aberrant RARαs involved in APL might confer an altered DNA half-site recognition on these chimeric receptors. In prior experiments on T3Rs, RARs, and retinoid orphan receptors, alterations in the structure of the receptor NH2 terminus were manifested as changes in the ability of the receptor to recognize DNA elements bearing nonconsensus half-sites (34 – 39). We therefore tested the ability of RARα, PML-RARα, and PLZF-RARα to bind to an assortment of different oligonucleotide probes, each bearing a distinct half-site sequence. Many nuclear hormone receptors recognize extended half-sites that are 8 or more bases in length (reviewed in Ref. 1). A P-box recognition helix in the receptor’s zinc-finger domain contacts the major groove of the DNA over bp 1–6 in the half-site, whereas a more COOH-terminal A-box, which is also present in many nuclear receptors, can make additional contacts in the minor groove with bases located at positions −2 and −1 of the half-site [Fig. 1A; we use this numbering system to permit comparison with earlier work based on the premise of a hexanucleotide half-site (1)]. We therefore began our experiments with an optimized consensus half-site sequence for RARα derived by a PCR selection procedure and consisting of an AGAGGTCA sequence (8), and we systematically altered each position to each of the alternative three bases. Each of the 25 possible permutated half-sites was synthesized as a direct repeat element with a 5-bp spacer, an arrangement found both in the experimentally defined consensus sequence and in many naturally occurring response elements for RARα (Fig. 1A). Corresponding bp changes were made concurrently in both half-sites in the direct repeat element.
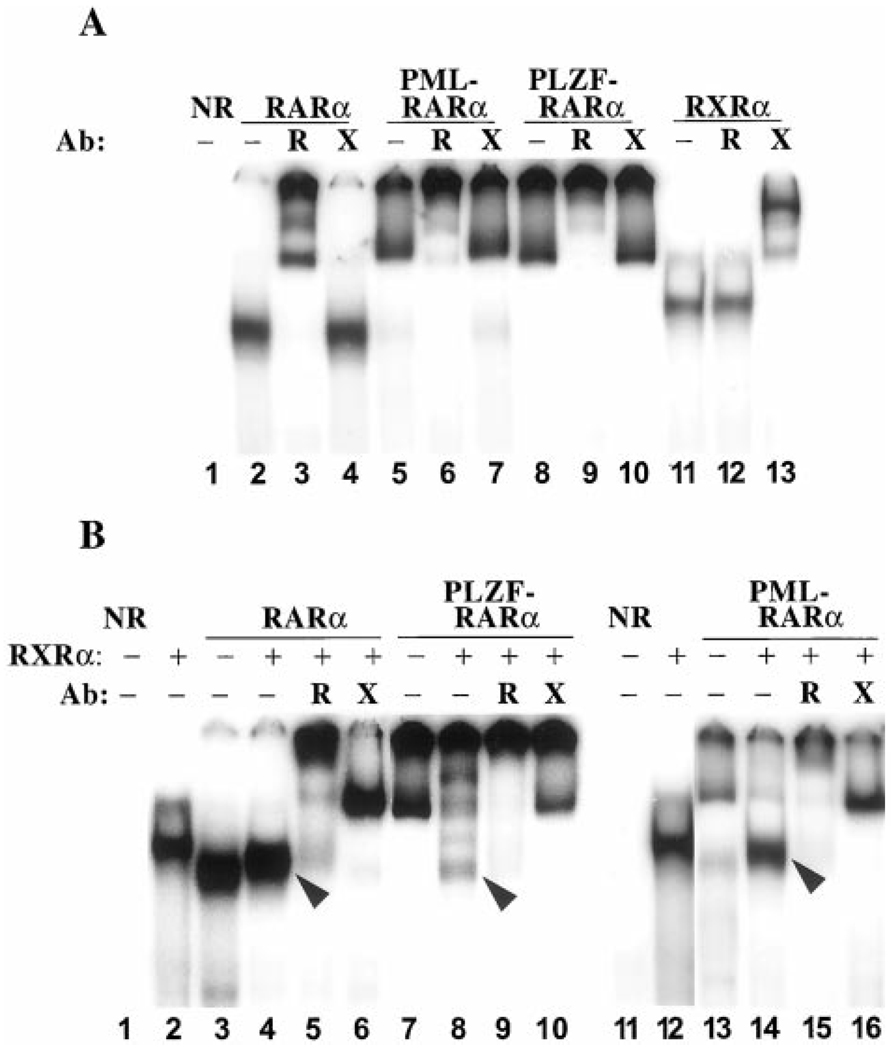
In our first series of experiments, we tested the ability of human RARα to bind to our panel of consensus and variant half-site DNA elements by using a gel electrophoretic mobility shift assay. Using RARα and the consensus AGAGGTCA repeat element as a probe, a single specific protein-DNA complex was detected by this methodology (Fig. 2A, Lane 2). This complex was specifically supershifted using anti-RARα antibodies and not by anti-RXRα antibodies (Fig. 2A, Lanes 3 and 4), whereas an equivalent complex was not detected with otherwise comparable protein preparations lacking RARα (Fig. 2A, Lane 1). We therefore identify this protein- DNA complex as representing a RARα/RARα homodimer bound to the DNA. This characterization is also consistent with the apparent mobility of the complex in the gel, the alteration in mobility of the complex on formation of a RXRα/RARα heterodimer (Fig. 2B), and the inability of RARα to bind detectably to DNA elements bearing only a single half-site (i.e., as a protein monomer).4
Fig. 2.
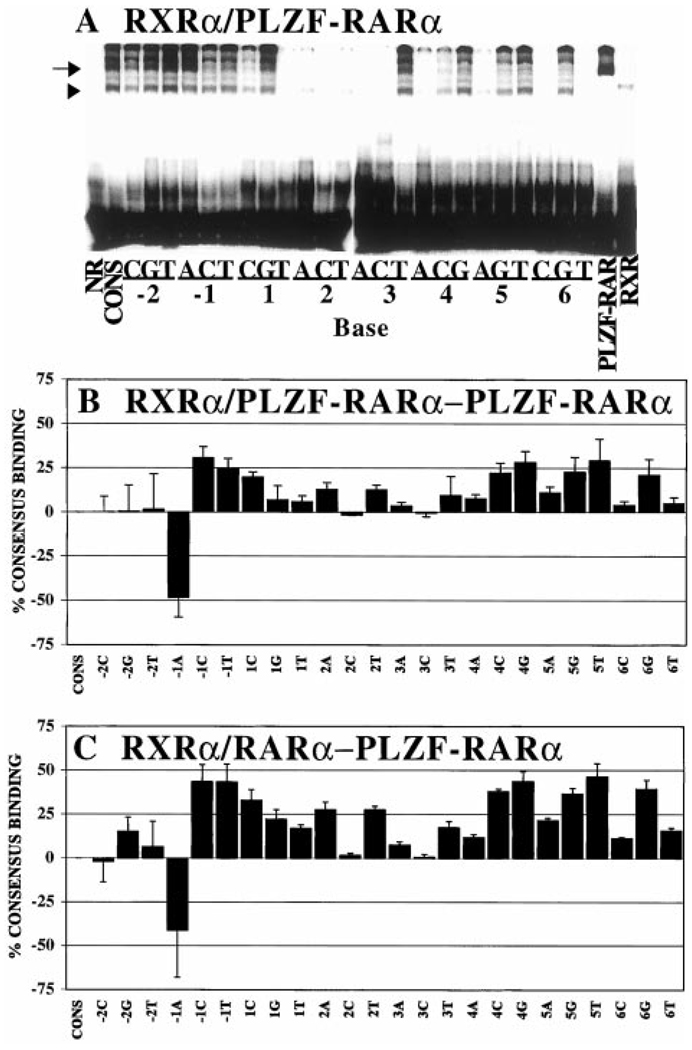
DNA binding by RARα, PML-RARα, PLZF-RARα, or RXRα as characterized by electrophoretic mobility shift assay. RARα, PML-RARα, PLZF-RARα, and/or RXRα, isolated from baculovirus-infected Sf9 cells, were incubated with the radiolabeled consensus oligonucleotide presented in Fig. 1A, and the resulting protein-DNA complexes were resolved by native PAGE and visualized by PhosphorImager analysis. Lane NR (no receptor) represents an otherwise identical electrophoretic mobility shift experiment using extracts of Sf9 cells infected by a nonrecombinant baculovirus. The various RARα derivatives were tested either alone or together with the RXRα heterodimer partner, as indicated above the panels. Please note that 2–4-fold lower concentrations of receptor were used in the heterodimer assays compared with the homodimer assays (see “Materials and Methods”). The identities of these protein-DNA complexes were further probed by supershift experiment using RXRα (X) - or RARα (R) -specific antibodies, also as indicated above each panel (Ab). Representative electrophoretograms are presented; comparable results were obtained in triplicate experiments. A, DNA binding by receptor homodimers. B, DNA binding by heterodimers. Arrowheads point to presumed heterodimers of RXRα/RARα, RXRα/PML-RARα, and RXRα/PLZF-RARα.
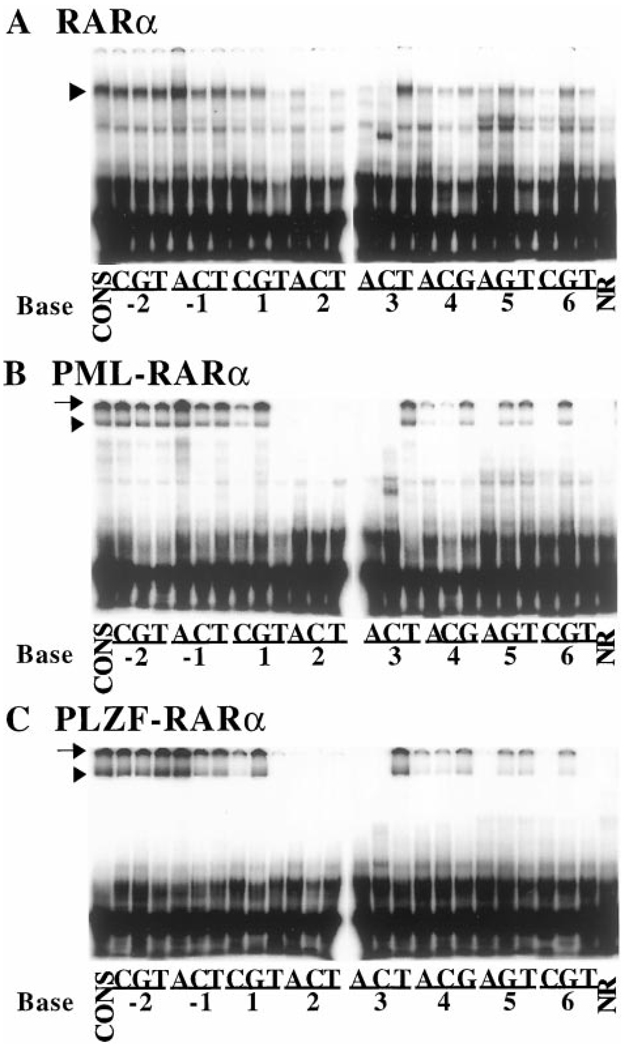
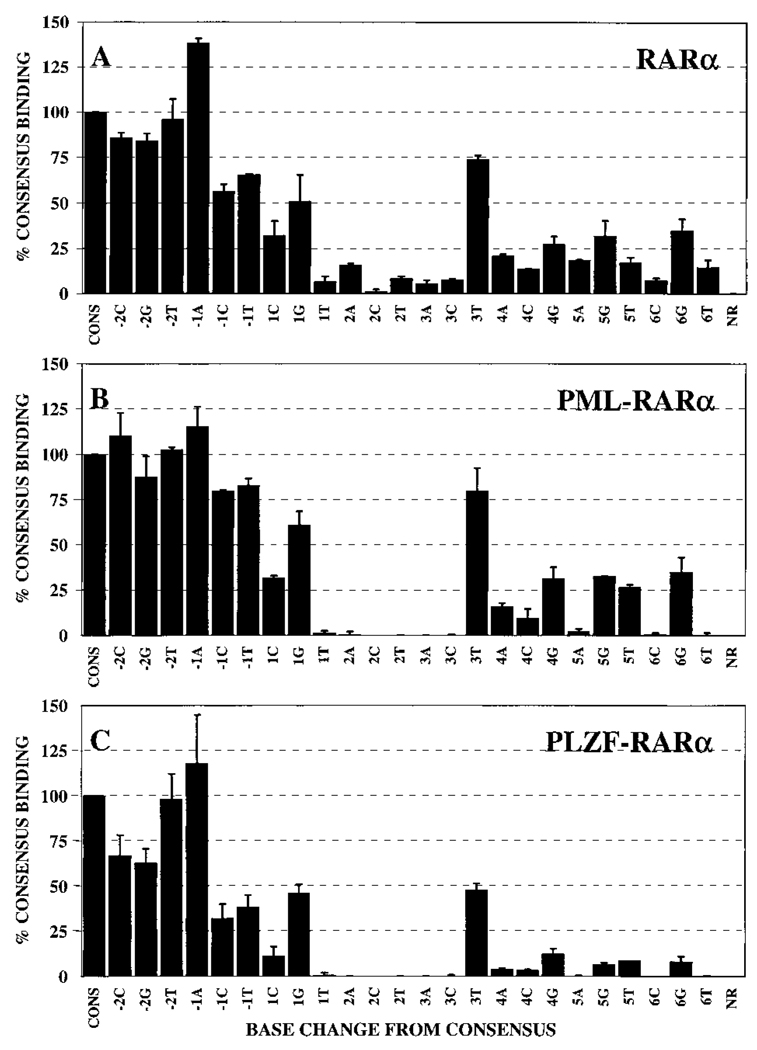
Notably, a surprising number of our single base substitutions of the AGAGGTCA consensus half-site had little or no effect on the ability of RARα to bind to the corresponding response element (Fig. 3A). This ability of RARα to accept nonconsensus sequences was particularly evident for substitutions at the −2 and −1 positions of the half-site, all of which were bound with at least 50% of the ability of the consensus half-site element (Fig. 3A; quantified in Fig. 4A). Indeed, under our conditions, a −1A half-site substitution element was bound somewhat better than was the consensus element itself. In contrast, base substitutions at the +2 position were uniformly destabilizing, with any departure from consensus at this location significantly reducing RARα binding (Fig. 3A and Fig. 4A). Substitutions at the remaining +1, +3, +4, +5, and +6 positions displayed an intermediate phenotype: certain substitutions at these positions exerted relatively minimal effects on DNA binding by RARα (e.g., the +3T element); whereas other substitutions at the same sites exhibited strongly destabilizing effects on RARα binding (e.g., the +3A or +3C elements). The specific activities of the different probes were comparable to one another, and free probe was in excess in all reactions.
Fig. 3.
The DNA binding specificities of RARα, PML-RARα, and PLZF-RARα homodimers, as determined by gel electrophoretic mobility shift assay. A panel of radiolabeled oligonucleotide probes representing a series of single base substitutions relative to the consensus sequence were individually incubated with RARα (A), PML-RARα (B), or PLZF-RARα (C), and the resulting protein-DNA complexes were resolved by native PAGE and visualized by a PhosphorImager methodology. The nature of the base substitution in each oligonucleotide is indicated under the panel; in each case, the number refers to the position of the substituted base in the consensus half-site as described in the Fig. 1 legend, and the letter refers to the identity of the base substitution at that particular position (see “Materials and Methods” for further details of this nomenclature). Binding of each RARα derivative to the consensus sequence (CONS) was tested in Lane 1 of each panel. NR refers to negative controls using the consensus DNA element and nuclear extracts lacking retinoid receptors. Representative electrophoretograms are presented; comparable results were obtained in repeat experiments, which are quantified and presented in Fig. 4.
Fig. 4.
Half-site recognition properties of homodimers of RARα, PML-RARα, or PLZF-RARα: quantification. Gel electrophoretic mobility shift assays using our panel of consensus and nonconsensus half-site elements were performed as described in the Fig. 3 legend. The protein-DNA complexes representing homodimers of RARα (A), PML-RARα (B), or PLZF-RARα (C) were quantified for each half-site derivative by Phosphor- Imager methodology. Binding of RARα, PML-RARα, or PLZF-RARα to each probe is presented relative to the binding of the same protein to the consensus sequence (AGAGGTCA), which was set as 100%. The average obtained from two or more independent experiments is shown, as is the SD.
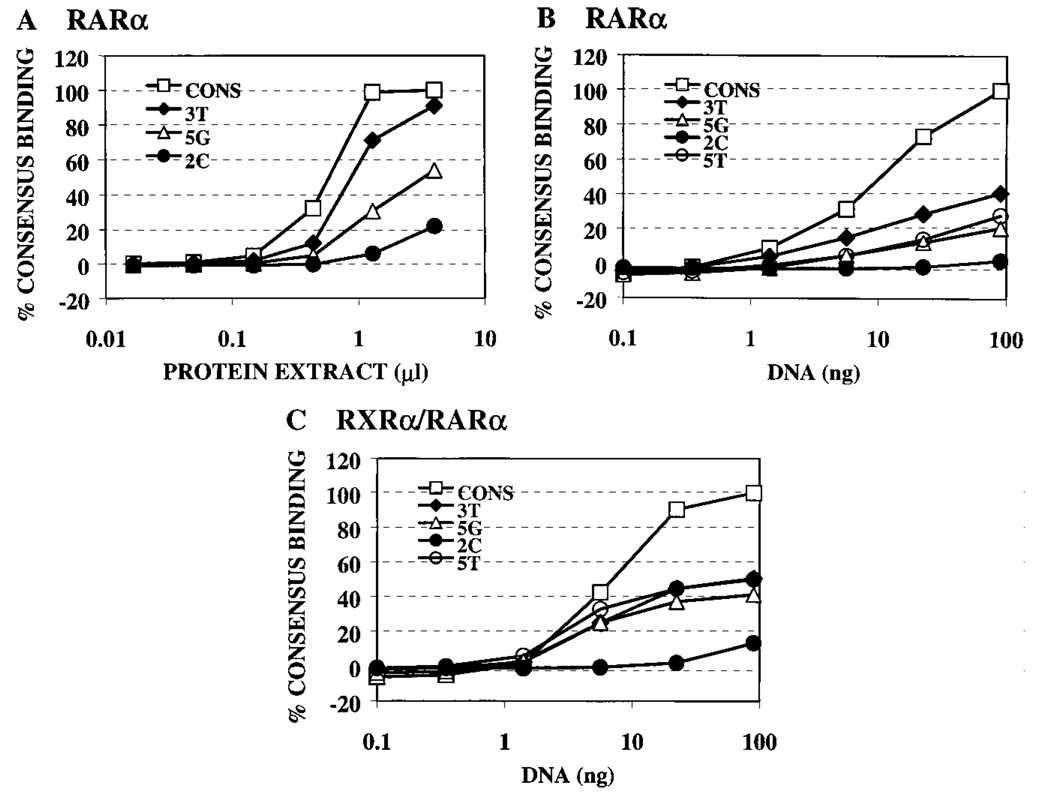
These results were obtained from single point assays. To better validate our conclusions, we performed titrations varying the amount of receptor or of DNA probe used in the mobility shift assay and testing representative DNA elements that displayed strong, medium, or weak binding by RARα in our single point assay. In all cases, these titrations paralleled the results obtained by single point assays: RARα bound very weakly to the 2C element, bound with intermediate strength to the 5G and 5T elements, bound stronger still to the 3T element, and bound best to the consensus element (Fig. 5, A and B). We conclude that RARα, in addition to binding with high efficiency to the consensus AGAGGTCA element, is able to also recognize a select series of half-site sequences that differ from the consensus by single-bp substitutions.
Fig. 5.
RARα binding to strong, intermediate, or weak DNA sequences confirmed by titration assays. The ability of RARα or RXRα/RARα to bind to representative weak (2C), intermediate (5G and 5T), or strong (3T) binding DNA sequences was assayed in titration assays. Fixed amounts of radiolabeled consensus, 3T, 5G, or 2C probes were incubated with increasing amounts of RARα protein extract (A) or, conversely, with a constant amount of RARα protein (B), or RXRα/ RARα proteins (C) were incubated with increasing amounts of radiolabeled consensus, 3T, 5G, 5T, or 2C DNA probes. The protein-DNA complexes that formed were subsequently resolved by electrophoresis and quantified. Binding to the different DNA sequences is graphed as a percentage of the maximal binding to the consensus DNA sequence after subtracting background obtained with nonrecombinant protein extract. The results for each panel represent the data obtained from a single experiment, with the different probes assayed in parallel.
We next performed a similar analysis of DNA binding by the PML-RARα and PLZF-RARα proteins. Using the same electrophoretic mobility shift assay conditions as described for RARα, both PML-RARα and PLZF-RARα generated specific protein-DNA complexes on the consensus AGAGGTCA direct repeat element that were not observed using control nuclear extracts lacking the corresponding receptors (Fig. 2A, Lanes 5, 8, and 1, respectively). In contrast to RARα, however, PML-RARα and PLZF-RARα each generated two distinct protein-DNA complexes (Fig. 2A, Lanes 5 and 8), one of which entered the gel and migrated at the position expected for the corresponding receptor homodimer (Fig. 3, B and C, arrowheads), and the other of which remained near the very top of the electrophoretogram (Fig. 3, B and C, arrows). The presence of a PML-RARα or PLZF-RARα complex migrating at or near the top of the electrophoretic resolving space has been reported by other researchers and is due to the formation of high order oligomeric receptor complexes greater than dimer in size (e.g., Refs. 41 and 49–51). We therefore report our results for the more readily resolved homodimeric complexes, the identity of which was confirmed by supershift with RARα-directed antisera (Fig. 2A, Lanes 6 and 9). Nonetheless, when quantified, the amount of high molecular weight complex closely paralleled the amount of the homodimeric species for the different response elements (data not shown); therefore, interpretation of our results would not differ were the high molecular weight complexes included in our analysis.
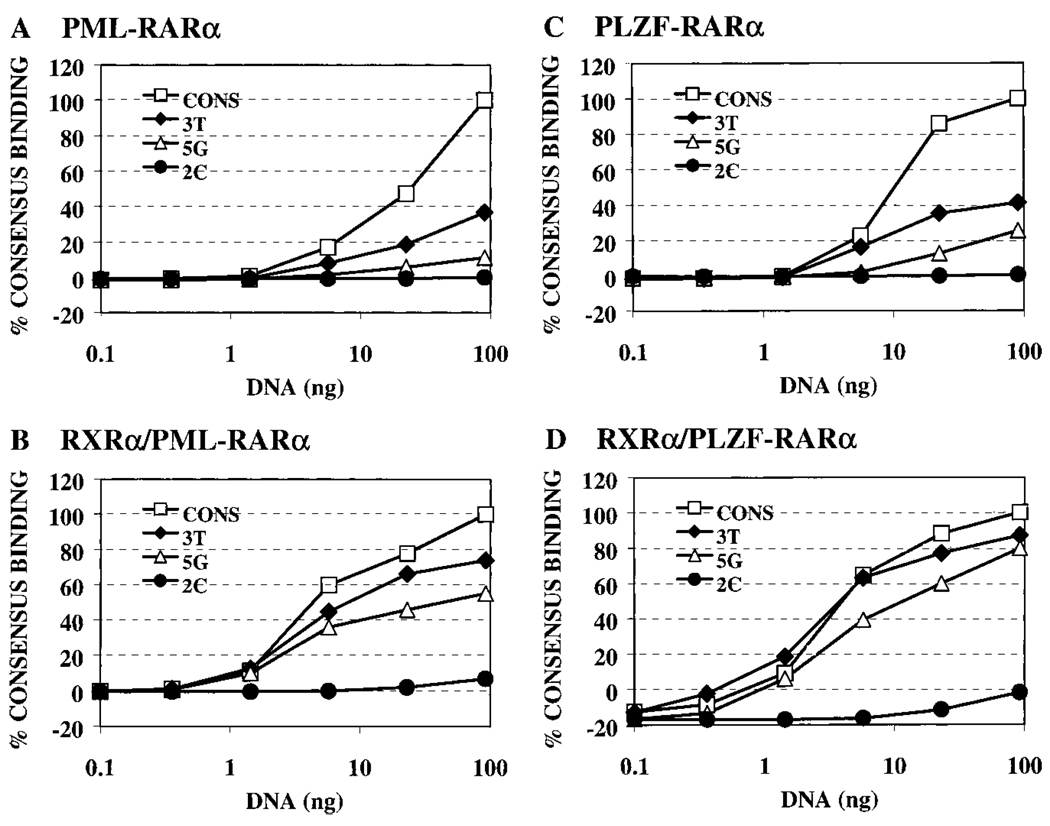
Overall, the ability of the PML-RARα and PLZF-RARα proteins to recognize the different half-site elements generally paralleled the pattern observed with the wild-type RARα (Fig. 3 and Fig. 4, compare A, B, and C). Thus, substitutions at the −2 and −1 positions had relatively modest effects on the binding of either PML-RARα or PLZF-RARα to the DNA, whereas any base change from the consensus sequence at the −2 position severely destabilized the ability of both PML-RARα and PLZF-RARα to bind to the corresponding DNA probes (Fig. 3, B and C; quantified in Fig. 4, B and C). Also in common with RARα, the ability of PML-RARα and of PLZF-RARα to bind to half-sites bearing substitutions at the +1, +3, +4, +5, and +6 positions depended on the precise nature of the substituted base. The pattern observed in these single point analyses was recapitulated in titration experiments with the representative consensus, 2C, 3T, and 5G DNA elements (Fig. 6, A and C).
Fig. 6.
PML-RARα and PLZF-RARα binding to strong, intermediate, or weak DNA sequences confirmed by titration assays. The ability of PML-RARα, PLZF-RARα, RXRα/PML-RARα, or RXRα/PLZF-RARα to bind to representative weak (2C), intermediate (5G), or strong (3T) binding DNA sequences was assayed in titration assays. Fixed amounts of PML-RARα (A), RXRα/ PML-RARα (B), PLZF-RARα (C), or RXRα/ PLZF-RARα (D) were incubated with increasing amounts of radiolabeled consensus, 3T, 5G, or 2C DNA probes. The protein- DNA complexes that formed were subsequently resolved by electrophoresis and quantified. Binding to the different DNA sequences is graphed as a percentage of the maximal binding to the consensus DNA sequence after subtracting background obtained with nonrecombinant protein extract. The results for each panel represent the data obtained from a single experiment, with the different probes assayed in parallel.
We conclude that homodimers of PML-RARα and PLZF-RARα have DNA recognition properties similar to those of RARα. However, close inspection of the PhosphorImager scan (Fig. 3) and quantification of the data (Fig. 4) revealed several consistent differences in the DNA binding properties of PML-RARα and PLZF-RARα relative to those of the wild-type RARα. For example, wild-type RARα exhibited an enhanced ability to bind to elements bearing substitutions at the +2 and +3 positions, as well as to the +SA and +6T elements, compared with PML-RARα and PLZF-RARα (Fig. 4). Interestingly, differences between the abilities of PML-RARα and PLZF-RARα to recognize individual DNA sequences were also discernible and were manifested primarily as a reduced ability of PLZF-RARα to bind to several of the base substitutions tested.
RXRα/RARα Heterodimers Recognize a Distinct Panel of Half-Site Sequences Compared to RXRα or RARα Homodimers
RARs can form heterodimers with other members of the nuclear hormone receptor family. RXRs are particularly important heterodimer partners for RARs, and RXR/RAR heterodimers exhibit both an enhanced overall DNA binding affinity and enhanced transcriptional regulatory properties when compared with homodimers of the same receptors (1 – 6, 44–47). PML-RARα and PLZF-RARα have been shown to retain at least some of this ability to heterodimerize with RXRs (40–42), leading us to examine the effect of this heterodimerization on DNA recognition by these different receptor variants.
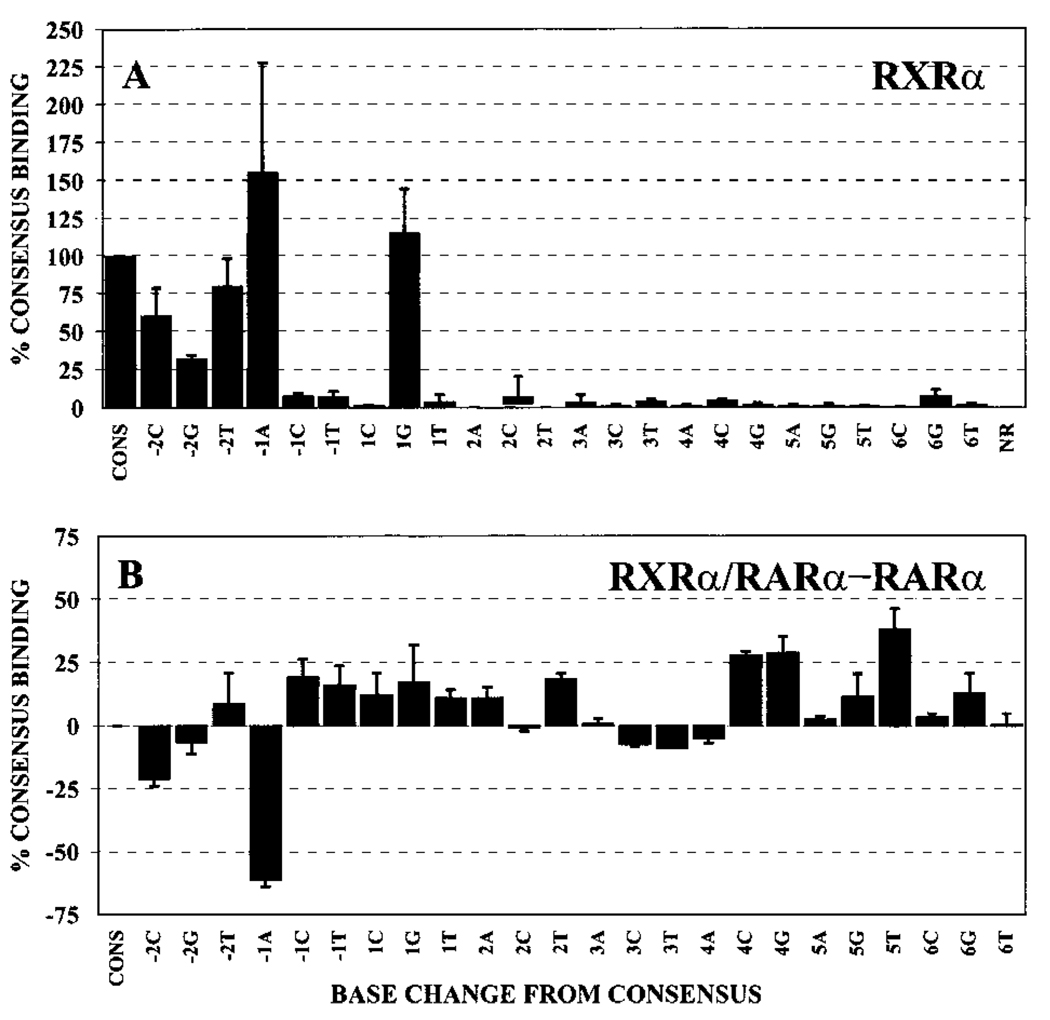
We first tested the ability of RXRs to bind to our different DNA probes in the absence of RARs. Although RXR homodimers bind with highest avidity to DNA elements composed of a DR-1 spacing, we were able to detect a modest binding of RXRα to our DR-5 consensus element at high receptor concentrations (Fig. 2A, compare Lane 11 with the nonrecombinant extract, Lane 1). We interpret this complex as being a RXRα/RXRα homodimer, and it was shifted in mobility by anti-RXRα antibodies but not by the control anti-RARα antibodies (Fig. 2A, Lanes 12 and 13). Significantly, the DNA recognition properties of RXRα homodimers were very different from the pattern seen for RARα, PML-RARα, and PLZF-RARα homodimers. For example, whereas RARα and its oncoprotein derivatives could accommodate a variety of base substitutions at positions +3, +4, +5, and +6 of the DNA half-site, these same substitutions virtually eliminated binding by RXRα (compare Fig. 4A and Fig. 7A). Thus, different nuclear hormone receptors can display very different DNA recognition specificities under the conditions used here.
Fig. 7.
The half-site recognition properties of RXRα homodimers and a comparison of the DNA recognition properties of RXRα/RARα heterodimers relative to RARα homodimers. A, gel electrophoretic mobility shift assays, using our panel of consensus and nonconsensus half-site elements, were performed using RXRα. The protein-DNA complexes representing homodimers of RXRα were quantified for each half-site derivative by PhosphorImager methodology. Binding of RXRα to each probe is presented relative to the binding of the same protein to the consensus sequence (AGAGGTCA), which was set as 100%. The average obtained from two or more independent experiments is shown, as is the SD. B, the ability of RARα to bind to the different half-site DNA elements as a homodimer and as a heterodimer with RXRα (data in Fig. 4 and Fig. 8) was graphed as a difference plot (the binding of a given oligonucleotide probe by the heterodimer minus the binding of the same oligonucleotide probe by the analogous homodimer). Positive numbers indicate that the RXRα/RARα heterodimer binds the oligonucleotide better than does the RARα homodimer; negative numbers indicate that the homodimer binds better. The average obtained from multiple experiments, and the SD, are shown. Binding of homodimers and heterodimers to the different half-site sequences was defined relative to binding of the same receptor species to the consensus sequence; by this analysis, the difference between heterodimer and homodimer binding to the consensus sequences is defined as zero.
Mixing of RXRα with RARα resulted in the appearance of a strong electrophoretic shift complex migrating at a position distinct from that of either the RARα homodimer or the RXRα homodimer (Fig. 2B, compare Lane 4 with Lanes 2 and 3). This novel complex migrated at a position consistent with that of a receptor heterodimer and contained both RARα and RXRα, as demonstrated by its ability to be supershifted by the appropriate antisera (Fig. 2B, Lanes 5 and 6). This RXRα/RARα heterodimer exhibited two features distinct from those of the corresponding homodimers: (a) consistent with prior work, the overall affinity of the RXRα/RARα heterodimer for the consensus AGAGGTCA repeat element was greater than that of the corresponding homodimers (Fig. 2B, the RARα and RXRα protein extracts were used at a 4-fold lower concentration in the heterodimer lane than in the homodimer lanes but generated comparable levels of complex); and (b) distinct from this increase in absolute affinity for the consensus element, the RXRα/RARα heterodimer also displayed reproducible changes in its relative ability to bind to the different half-site variants compared with either the RARα/RARα or RXRα/RXRα homodimers (compare Fig. 4A, Fig. 7A, and Fig. 8A; the same results are expressed as a difference plot in Fig. 7B). It is important to note that binding to the consensus RARE was defined as 100% for each experiment; therefore, differences in the ability of homodimers and heterodimers to bind to the individual DNA variants represent differences in relative avidity for the different DNA sequences and are distinct from the overall increased affinity displayed by the heterodimer for the consensus DNA sequence. For example, although the RXRα/RARα heterodimer bound to the consensus DNA sequence better than did the RARα/RARα homodimer, the heterodimer actually bound comparatively less well to the −1A element than did the homodimer (compare Fig. 7A, 7B, and Fig. 8A).
Fig. 8.
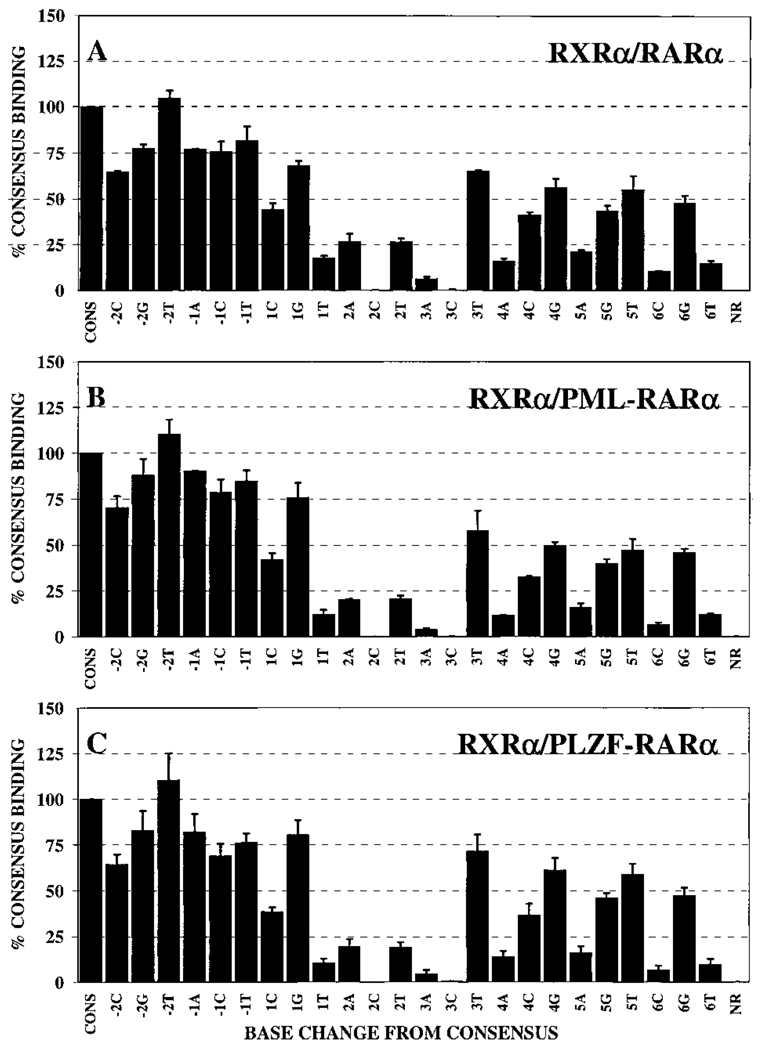
Half-site recognition properties of heterodimers of RXRα/RARα, RXRα/PML-RARα, or RXRα/PLZF-RARα. Gel electrophoretic mobility shift assays, using our panel of consensus and nonconsensus half-site elements, were performed as described in the Fig. 3 legend but in the presence of the heterodimer partner RXRα. The protein-DNA complexes representing heterodimers of RXRα/RARα (A), RXRα/PML-RARα (B), or RXRα/PLZF-RARα (C) were quantified for each half-site derivative by PhosphorImager methodology. Binding of each heterodimer to the different nonconsensus probes was normalized relative to the binding of the same heterodimer to the consensus sequence (AGAGGTCA), which was set as 100%. The average obtained from two or more independent experiments is shown, as is the SD.
With the exception of the −1A element, the RXRα/RARα heterodimer typically displayed an enhanced ability to bind to nonconsensus half-site sequences relative to the RARα/RARα homodimer; this broadened recognition specificity was most clearly observed for the +4C, +4G, and +5T probes, with more subtle effects on recognition at other positions also detected (Fig. 7A, 7B, and Fig. 8A). These differences in sequence specificity between homo- and heterodimers were reproducible from experiment to experiment. We also confirmed the results of these single point assays by performing titration experiments using several representative DNA elements (Fig. 5, B and C; note the enhanced relative binding of the +5T and +5G DNA elements by the heterodimer compared with that of the RARα/RARα homodimer). It is noteworthy that the DNA binding pattern of the RXRα/RARα heterodimer was not simply a summation of the individual specificities of the RXRα and RARα homodimers; for example, the enhanced recognition of the +4C, +4G, and +5T elements by the RXRα/RARα heterodimer was not predictable from the specificities of either of the corresponding homodimers (compare Figs. 4A, Fig. 7A, and Fig. 8A).
RXRα/PML-RARα and RXRα/PLZF-RARα Heterodimers Display a Broader DNA Recognition Specificity than Do the Corresponding Homodimers but Form Inefficiently Compared with RXRα/RARα Heterodimers
We next extended our analyses of heterodimer formation to the chimeric RARα derivatives associated with APL. Addition of RXRα to PML-RARα resulted in the formation of a novel complex that was not observed with the parental receptors when tested individually (Fig. 2B, Lane 14, arrowhead) and was supershifted with antisera to either RXRα or PML-RARα (Fig. 2B, Lanes 15 and 16). In keeping with the assignments of others (40), we interpret this complex as representing a RXRα/PML-RARα heterodimer. Notably, PML-RARα formed heterodimers less efficiently than did RARα, and a significant amount of residual PML-RARα homodimer complex was observed even when PML-RARα was mixed with large amounts of RXR (Fig. 2B; data not shown). In the case of PLZF-RARα, addition of RXRα resulted in the formation of three different presumptive RXRα/PLZF-RARα heteromeric species (Fig. 2B, Lane 8) that could be quantitatively supershifted with antibodies to either RXRα or RARα determinants (Fig. 2B, Lanes 9 and 10). Based on the relative migration, the antibody supershift, and the analysis of analogous complexes by other researchers (41, 42), the fastest migrating heteromeric complex (Fig. 2B, Lane 8, arrowhead) was identified as a RXRα/PLZF-RARα heterodimer. This heterodimerization of PLZF-RARα with RXRα on the consensus element was also inefficient, resulting in a mixture of homo- and heterodimers under the conditions used here (Fig. 2B and Fig. 9B). This enhanced ability of PML-RARα and PLZF-RARα to form homodimers, as compared with RARα, has been reported by other researchers (40–42).
Fig. 9.
Comparison of DNA half-site recognition by retinoid receptor heterodimers versus homodimers: difference plots. A, EMSA analysis of DNA binding by PLZF-RARα homodimers and heterodimers. Preparations of RXRα and PLZF-RARα were mixed and tested for the ability to bind to the different response elements, as indicated. A PhosphorImager image of the resulting electrophoretogram is presented. Under these conditions, both PLZF-RARα homodimers (arrow) and RXRα/PLZF-RARα heterodimers (arrowhead) are observed. B, difference plot comparing RXRα/PLZF-RARα heterodimers with PLZF-RARα homodimers. The data in A was also quantified and graphed as a difference plot. Positive numbers indicate that the RXRα/PLZF-RARα heterodimer binds the oligonucleotide better than does the PLZF-RARα homodimer; negative numbers indicate that the homodimer binds better. The average obtained from multiple experiments and the SD are shown. C, difference plot comparing RXRα/ RARα heterodimers with PLZF-RARα homodimers. The ability of RXRα/RARα heterodimers to bind to the different DNA elements (Fig. 8A) was compared with that of PLZF-RARα homodimers (Fig. 4C). Positive numbers indicate that the RXRα/RARα heterodimer binds the oligonucleotide better than does the PLZF-RARα homodimer; negative numbers indicate that the homodimer binds better. The average obtained from multiple experiments and the SD are shown.
The RXRα/PML-RARα and RXRα/PLZF-RARα heterodimers that did form under these conditions exhibited the broadened ability to bind to nonconsensus half-sites that was observed for RXRα/RARα heterodimers (compare Fig. 8, B and C, with Fig. 4, B and C). This enhancement in the recognition of nonconsensus half-sites, such as the +3T and +5G elements, was also observed in titration experiments (Fig. 6). We conclude that the DNA binding specificities of RXRα/RARα, RXRα/PML-RARα, and RXRα/PLZF-RARα heterodimers are very similar to one another but are distinct from and typically more accepting of nonconsensus half-site sequences than the DNA recognition specificities of the corresponding receptor homodimers.
The difference in the half-site recognition specificity of receptor homodimers and heterodimers was particularly evident for PLZF-RARα, which formed a mixed population of homo- and heterodimers on the consensus element even in the presence of high levels of RXR (Fig. 2B, compare Lane 8 with Lane 7 and note the absence of a supershift of the homodimer when using RXR-directed antibodies; Lane 10). This feature served as an internal control, permitting an accurate comparison of the formation of PLZF-RARα homodimers versus RXRα/PLZF-RARα heterodimers on each DNA sequence variant under identical conditions in the same binding reaction (Fig. 9A; compare the strong homodimer formation on the −1A element, for example, to the strong heterodimer formation on the 5T element). This type of experiment was also quantified and is presented as a difference plot (relative binding of the heterodimer minus the relative binding of homodimers) for the different response elements tested (Fig. 9B). We conclude that heterodimerization with RXRα PML-RARα, and PLZF-RARα from that of the corresponding receptor homodimers. Given that both PML-RARα and PLZF-RARα may function in oncogenesis as homodimers (40, 41, 43), our results also imply that these aberrant receptor homodimers have a DNA recognition specificity distinct from that of the RXRα/RARα heterodimers, which are the prevailing species in the normal cell. A comparison of the DNA binding specificities of RXR/RAR heterodimers with that of PLZF-RAR homodimers is presented as a difference plot to illustrate this phenomenon (Fig. 9C).
Transactivation in Vivo by RARα and PML-RARα Largely Parallels but Is not Identical to Their Ability to Bind DNA Elements in Vitro
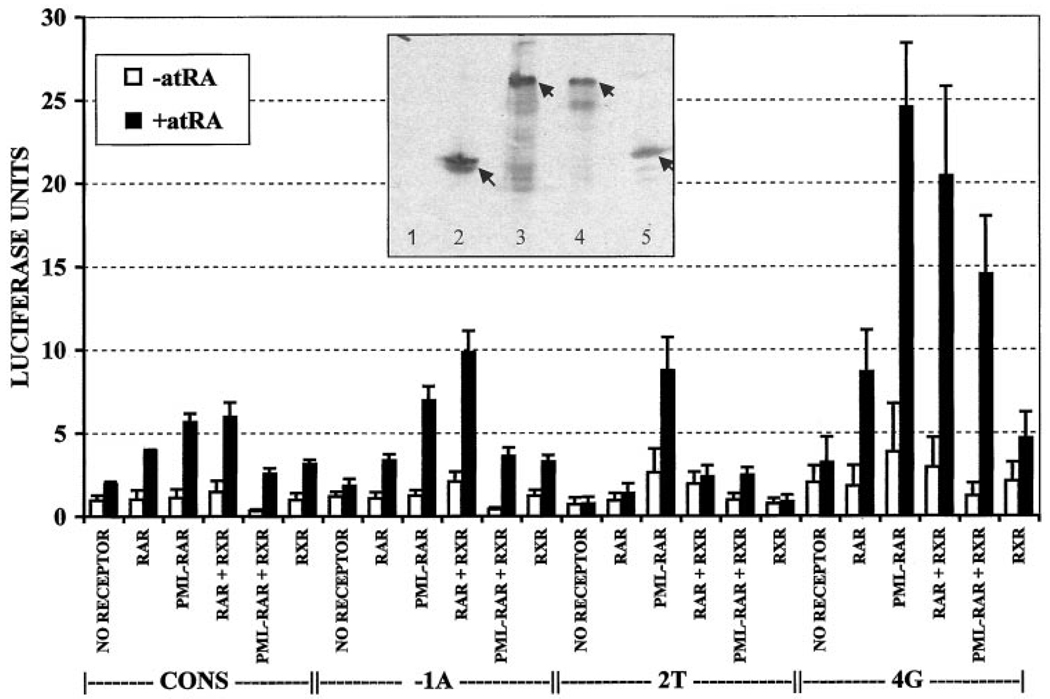
We next created a series of reporter constructs containing representative DNA response elements (i.e., those that displayed strong, medium, or weak receptor binding in vitro) and tested the ability of RARα, PML-RARα, and PLZF-RARα to modulate transcription in transfected cells. At physiological hormone conditions (10−9 to 10−8 m all-trans retinoic acid), PML-RARα and PLZF-RARα are unable to release from corepressor and function as dominant negative inhibitors, interfering with gene activation by the wild-type RARα(Refs. 40 and 50–53). However, in the presence of higher concentrations of all-trans retinoic acid (10−7 to 10−6 m), PML-RARα releases from corepressor and activates reporter gene expression, whereas PLZF-RARα remains a dominant negative even at high hormone concentrations (30, 31, 52, 53). A characterization of the DNA recognition properties of PML-RARα or PLZF-RARα as dominant negative inhibitors (i.e., assaying the ability to interfere with RARα at nanomolar hormone concentrations) would require interpreting a complex interaction between the DNA recognition properties of the positive-acting RARα and those of the interfering, chimeric receptor. To allow us to characterize the autonomous DNA recognition properties of PML-RARα, we therefore used 10−7 m hormone and measured the ability of PML-RARα to activate transcription from the different response element reporters. PLZF-RARα, as anticipated, failed to activate transcription of any of the reporter constructs tested even at micromolar hormone concentrations and was therefore excluded from the analysis (data not shown).
CV-1 cells have low but detectable levels of endogenous RAR activity that can be observed as a hormone-dependent activation of the consensus reporter element in the absence of exogenous receptor (Fig. 10, NO RECEPTOR). As anticipated, introduction of exogenous RARα into these cells further enhanced the ability of retinoid hormone to activate the consensus reporter (Fig. 10, RAR). Consistent with the receptor binding properties we observed in our in vitro DNA binding studies (Fig. 4A), the −1A DNA element functioned as a RARα response element at levels comparable with that of the consensus element, whereas the 2T element, which was poorly bound by RARα in vitro, was poorly activated by RARα in our transfection experiments (Fig. 10). Also consistent with our in vitro DNA binding studies, cointroduction of a RXRα expression vector in our transfection experiments enhanced the ability of RARα to activate the reporter construct, and this was evident on all four DNA elements tested (Fig. 10, RAR + RXR). These results suggest that RXRα/RARα heterodimers are indeed stronger transcriptional activators than the corresponding homodimers and that heterodimer formation with RXRα can broaden the half-site recognition properties of RARα. Despite this general correlation between DNA binding in vitro and transactivation in vivo, some modest differences were noted. For example, the 4G element, which was less efficient than the consensus element at binding RARα in vitro, was equal or better than the consensus element in mediating transcriptional activation in the transfected cells. Other researchers have previously noted similar discrepancies between DNA binding and transcriptional activation by nuclear hormone receptors when tested on different response elements (e.g., Refs. 54–57).
Fig. 10.
Transactivation in vivo from representative DNA elements. CV-1 cells were transiently transfected with RARα, PML-RARα, PLZF-RARα, and/or RXRα or no receptor as well as with luciferase reporters containing the consensus, −1A, 2T, or 4G DNA elements and incubated with or without all-trans retinoic acid. Luciferase activity, relative to a β-galactosidase reporter used as an internal standard, was calculated for each experiment. Presented in the graph are the averages from three independent experiments, as well as the SD. The DNA element in the luciferase reporter (CONS, −1A, 2T, or 4G) is indicated below the graph. □, without all-trans retinoic acid; ■, with all-trans retinoic acid. Inset, extracts of the transfected cells were also analyzed by immunoblotting to confirm that the different receptors were expressed at comparable levels (65). A mixture of RAR-directed (sc55) and RXR-directed (4RX-1D1Z) antibodies was used to visualize the blot. The cells were transfected as follows: Lane 1, empty pSG5 vector; Lane 2, pSG5-RARα; Lane 3, pSG-5 PML-RARα; Lane 4, pSG5 PLZF-RARα; and Lane 5, pSG5 RXRα.
Reflecting its DNA binding properties in vitro, PML-RARα mediated strong transcriptional activation from both the consensus and the −1A elements (Fig. 10, PML-RAR). In fact, activation of the reporter construct when PML-RARα was introduced alone was consistently higher than that observed for RARα; this was particularly notable for the 2T and 4G elements, which mediated reporter gene activation at levels significantly greater than those anticipated from our in vitro DNA binding studies (compare Fig. 4B with Fig. 10). Immunoblot analysis confirmed that the levels of expression of RARα and PML-RARα in the transfected cells were very similar (Fig. 10, inset). These results are in agreement with prior work indicating that at high hormone concentrations PML-RARα can function as a stronger transcriptional activator than RARα (51). In notable contrast with the results observed for RARα, cointroduction of RXRα resulted in a decrease, not an increase, in the ability of PML-RARα to activate transcription, and this too extended to all four DNA elements tested (Fig. 10). These results are consistent with our own studies and with prior suggestions that PML-RARα may function as a homodimer and not as a heterodimer in cells (40, 41, 43).
Discussion
Wild-Type RARα Can Recognize a Spectrum of Distinct Half-site Sequences
One conclusion from the experiments reported here is that the wild-type RARα homodimer is able to recognize a wide range of response elements representing single-base substitutions of the AGAGGTCA consensus sequence. In fact, only the G at the +2 position of the consensus half-site proved to be absolutely required for RARα binding, with any other base substitution at the +2 position incompatible with efficient RARα recognition. In contrast, other positions in the half-site were able to accept at least one base substitution with relatively minimal effect on RARα binding. The effect of substitutions at the +1, +3, +4, +5, and +6 positions was dependent on the precise base introduced, with certain substitutions at these sites compatible with, and other substitutions highly disruptive of, DNA recognition by RARα. Conversely, base substitutions at the −2 and −1 position were quite permissive, with virtually every substitution at these positions compatible with RARα binding.
The ability to bind to a spectrum of half-site sequences might appear inconsistent with structural studies indicating that very specific amino acid-bp contacts can occur between nuclear hormone receptors and their consensus DNA response elements. However, binding to nonconsensus sequences appears, at least in part, to be accommodated by relatively modest rearrangements of receptor conformation (58–61). This ability of RARα to recognize a spectrum of nonconsensus artificial half-sites in our experiments is also consistent with the diversity of half-site sequences found in naturally occurring retinoic acid response elements (47). Our artificially permuted half-site sequences emphasize, however, that not all single base variations from the consensus sequence are compatible with response element function, and they help define the set of half-site sequences that are acceptable RARα binding sites. As such, our results complement prior work defining optimal response element sequences and may assist in the identification of authentic response elements in the promoters of retinoid-regulated genes.
The DNA Recognition Specificities of PML-RARα and PLZF-RARα Homodimers Are Similar but not Identical to That of RARα Homodimers
The nature of the NH2 terminus of the T3Rs and retinoid orphan receptor α receptors plays an important role in the ability of these receptors to recognize nonconsensus DNA half-sites (34 –39). In fact, as observed here, the replacement of the normal RARα NH2 terminus with the novel PML or PLZF sequences does result in reproducible, modest alterations in the DNA recognition properties of these chimeric receptors, relative to those of RARα. These changes were manifested principally as alterations in the recognition of base substitutions at the +2, +3, +4, +5, and +6 positions of the half-site, and notably, the PML-RARα and PLZF-RARα recognition patterns differed somewhat from one another, as well as differing from that of the wild-type RARα. The precise molecular mechanisms by which the receptor NH2 terminus participates in DNA recognition by nuclear hormone receptors have not yet been fully determined. Available evidence suggests that the receptor NH2 terminus may act by influencing the three-dimensional structure of the receptor, thereby affecting the conformation of and precise DNA contacts made by the regions of the receptor that interact directly with the response element (34, 37–39). The modest differences in DNA recognition by PLZF-RARα relative to RARα and PML-RARα may therefore reflect subtle effects on receptor conformation that are conferred by the different NH2 termini of these different receptor derivatives.
Heterodimerization with RXRα Expands the Half-Site Repertoire
Many nuclear hormone receptors efficiently heterodimerize with RXRs, and the resulting receptor heterodimers exhibit enhanced DNA binding and transcriptional regulatory properties compared with the parental homodimers. RXR/RAR heterodimers have been proposed to be the physiologically dominant receptor species in the biology of retinoid signaling in many cell types (44–47). We report here that heterodimer formation with RXRα not only enhances the absolute affinity of RARα for DNA but also alters the relative ability of RARα to bind to a variety of nonconsensus half-site elements. These changes in relative specificity for the different half-site sequences are distinct from the overall increase in affinity of the RXR/RAR or RXR/ T3R heterodimers for DNA. For example, it is striking that recognition of certain half-site substitutions is relatively unaffected by heterodimerization with RXR, whereas recognition of other response elements is significantly enhanced, and that the DNA binding specificity of the heterodimer is not a simple additive combination of the specificities of the parental homodimers. These results suggest that heterodimerization with RXR may actually alter the bp contacts made by the RAR or T3R partner. This hypothesis is supported by demonstrations that RXR/T3R heterodimers generate DNA footprint patterns that differ from those generated by T3R/T3R homodimers (62).
PML-RARα and PLZF-RARα share the ability of RARα to bind DNA both as homodimers and as heterodimers with RXRα (40–42). Also in common with RARα, the formation of RXRα heterodimers by PML-RARα and PLZF-RARα expands the half-site repertoire of DNA binding compared with the corresponding homodimers. Interestingly, however, PLZF-RARα and PML-RARα retain the ability to form homodimers even at high RXRα concentrations. The enhanced homodimerization capability of these oncoproteins has been mapped to protein-protein interaction domains located within the PML- and PLZF-derived sequences (41, 49). The capacity for enhanced protein dimerization is a hallmark of the chimeric RARαs found in APL and has been implicated in mediating the oncogenic phenotype (17–21, 43). Thus, in contrast to RARα, which is believed to function in cells exclusively as a RXR/RARα heterodimer (44–47), PML-RARα and PLZF-RARα have the potential to exert at least some of their functions in the neoplastic cell as receptor homodimers or as still higher order oligomers (43, 48). Our own experiments indicate that this enhanced ability of PML-RARα and PLZF-RARα to form homodimers would further shift the DNA recognition specificities of these chimeric oncoproteins away from those of the RXR/RAR heterodimers that are prevalent in normal cells; the implications of this observation for leukemogenesis are discussed below.
Transactivation from DNA Elements Generally Correlates with but Is Not Identical to Binding by Receptors in Vitro
We found that the ability of RARα and PML-RARα to activate transcription from a given DNA response element in transfection experiments generally correlated with the ability of these receptors to bind to the response element in vitro. Typically, transcriptional activation was weak with DNA elements that were bound poorly by receptor in vitro, whereas transcriptional activation was strong from DNA elements that were bound with high avidity by receptor in vitro. However, there are some exceptions to this generic rule. Both RARα and PML-RARα displayed better activation on the 4G element than would have been expected from our in vitro binding studies, and, in addition, PML-RARα exhibited an unusually strong ability to activate transcription from the 2T element, which was bound very poorly by any of the RAR derivatives in vitro. Similar discrepancies between in vitro binding and in vivo transcriptional activation have been reported in previous studies, and response element sequences that bind nuclear hormone receptors poorly in vitro may nonetheless serve as efficient response elements in vivo (54 – 57). This “uncoupling” between DNA avidity and transcriptional activation may reflect differences in the ability of nuclear hormone receptors to activate transcription once bound to different response elements. Alternatively, posttranslational modifications of the receptor in vivo may confer novel sequence binding properties not seen with unmodified receptor in vitro, or response elements may bind additional transcription factors in vivo that assist in the binding or function of the nuclear hormone receptor.
Implications for the Role of PML-RARα and PLZF-RARα in APL
It has been proposed that the chimeric RARα proteins involved in APL function by perturbing normal retinoid signaling, leading to a block in differentiation and in an accumulation of leukemic cells at a promyelocytic stage. This hypothesis is consistent with observations that retinoids can regulate differentiation of normal myeloid cells and that supraphysiological levels of retinoic acid can induce differentiation of leukemic cells bearing the PML-RARα translocation, driving the corresponding leukemias into clinical remission (21). Also consistent with these ideas, one consequence of the translocations that produce the PML-RARα and PLZF-RARα proteins is an impairment in the ability of these chimeric receptors to release corepressor on addition of retinoic acid (28, 30–33). Thus, PML-RARα and PLZF-RARα can function as dominant negative inhibitors of normal retinoid signaling, and this dominant negative activity correlates with a number of important aspects of the leukemogenic phenotype.
The results described here, however, raise the possibility that the panel of target genes that are subject to this aberrant regulation by PML-RARα and by PLZF-RARα may be only a subset of the full panel of genes that represent the targets of regulation by RARα in normal cells. Three hypotheses can be advanced in this regard, depending on the nature of the RARs actually operative in the leukemic cell.
RARα, PML-RARα, and PLZF-RARα may all exert their functions in the cell as homodimeric species. If this is true, then given their overlapping but not identical DNA recognition specificities as homodimers, PML-RARα and PLZF-RARα would be expected to function as dominant negatives on many of the response elements that are regulated by normal RARα. This hypothesis, however, appears unlikely: RARα does not appear to homodimerize in solution, and RARα homodimers are relatively inefficient at binding to DNA in vitro. Instead, analysis both in vitro and by genetic dissection in vivo suggests that RAR functions principally as a RXRα/RARα heterodimer (44–47).
Alternatively, RARα, PML-RARα, and PLZF-RARα may all carry out their roles as heterodimers with RXRα. Arguing against this hypothesis, however, are the observations that both PML-RARα and PLZF-RARα bind DNA efficiently as homodimers, that PML-RARα also forms stable homodimers in solution, that the addition of RXR can actually impair transcriptional regulation by PML-RARα, and that the ability of PML-RARα to form homodimers appears to be both necessary and sufficient for its ability to block myeloid differentiation (40–43).
A third hypothesis, which we favor, is that PML-RARα and PLZF-RARα homodimers play an important role in the leukemic cell and act by interfering with the actions of RXRα/RARα heterodimers. Significantly, the DNA recognition properties of PML-RARα and PLZF-RARα homodimers are similar to one another but different from that of RXRα/RARα heterodimers. Therefore, our data would suggest that PML-RARα and PLZF-RARα homodimers, given their more narrow DNA binding specificity, may be able to target only a subset of the response elements that are recognized by RXRα/RARα heterodimers. This prediction appears to account for observations that the dominant negative effects of PML-RARα and PLZF-RARα are promoter specific (49 –51). Importantly, NuMA, NPM, PML, and PLZF, although otherwise structurally unrelated, all have dimerization domains that are retained in the fusion protein (18, 19), which may suggest that homodimerization is an important common factor in the function of all of the RARα-fusion oncoproteins. Of course, reality may represent an amalgamation of these reductionalist views, and a mixture of receptor homodimers, heterodimers, and heterotetramers may participate in conferring the leukemic cell phenotype.
Materials and Methods
Source of Proteins
Human RARα, human PML-RARα, human PLZF-RARα, and human RXRα proteins were obtained as nuclear extracts from Sf9 insect cells that were infected with the appropriate recombinant baculoviruses. Briefly, full-length clones of the human RARα, PML-RARα, PLZF-RARα, and RXRα genes were independently cloned into the baculovirus transfer vector pVL1393 as NotI/NotI, Blunt/EcoRI, EcoRI/EcoRI, or EcoRI/EcoRI fragments, respectively (63).4 Recombinant plasmids or a nonrecombinant pVL1393 plasmid was individually transfected into Sf9 cells by lipofection together with a suitable linearized baculovirus genome; stocks of virus originating by recombination with the transfer vector in vivo were identified and isolated by plaque purification by use of the BaculoGold system (PharMingen). The plaque-purified, recombinant viruses were subsequently amplified in Sf9 cells, and nuclear extracts were prepared from the infected cells (36). Expression of the correct proteins was confirmed by SDS-PAGE analysis. The functionality of the baculovirus-encoded RARα, PML-RARα, PLZF-RARα, and RXRα proteins was confirmed by electrophoretic mobility shift assays, and their identity was confirmed by using appropriate antibodies to supershift the corresponding protein- DNA complexes (see “Gel Electrophoretic Mobility Shift Assays” below).
Oligonucleotide Probes
The oligonucleotide probes used in the electrophoretic mobility shift assays were chemically synthesized (Operon Technologies, Inc.). Each oligonucleotide probe was made as two complementary strands with 4-base overhangs on each end to permit radiolabeling and/or subsequent molecular cloning. The consensus sequence (8) consisted of the following two oligonucleotides (the half-site sequences are underlined): 5′-TCGACAGAGGTCAACGAGAGGTCAGAG-3′ and 5′-TCGACTCTGACCTCTCGTTGACCTCTG-3′.
Systematic variations of these consensus oligonucleotides were designed such that one base at a time, concurrently in both half-sites, was altered to all other possible bases. As an example, base changes at position +2 (Fig. 1A) were synthesized as the following oligonucleotides (only the upper strand oligonucleotide is depicted): +2A, 5′-TCGACAGAAGTCAACGAGAAGTCAGAG-3′; +2C, 5′-TCGACAGACGTCAACGAGACGTCAGAG-3′; and +2T, 5′-TCGACAGATGTCAACGAGATGTCAGAG-3′ (half-sites are underlined, substitutions from consensus are in bold).
The two strands were annealed, and the probes were radiolabeled by a fill-in reaction with the Klenow fragment of Escherichia coli DNA polymerase in the presence of [α-32P]dGTP.
Gel Electrophoretic Mobility Shift Assays
Standard gel electrophoretic mobility shift assays were performed as described previously (34). Nuclear protein extracts containing RARα, PML-RARα, or PLZF-RARα, with or without RXRα, were incubated in 13 µl of binding buffer [3% glycerol, 11 mm Tris-HCl (pH 7.5), 15 mg/ml BSA, 77 mm KCl, 2 mm MgCl2, 154 µg/ml poly(deoxyinosine-deoxycytosine) containing 40,000–60,000 cpm (approximately 10–20 ng) of α-32P-labeled oligonucleotide) for 25 min at room temperature. Receptors were titrated so as to bind approximately 25% of the consensus probe at maximum; this represented approximately 12.5 ng of RARα, 6–8 ng of PML-RARα, 1–2 ng of PLZF-RARα, and 2.5–5 ng of RXRα when the receptors were assayed as homodimers and approximately 3 ng of RARα, 6–12.5 ng of PML-RARα (with the exception of the experiment in Fig. 2, which used 2 ng), 0.4–0.8 ng of PLZF-RARα, and 0.6 – 1.2 ng of RXRα when the receptors were used as heterodimers. The protein-DNA complexes that formed were subsequently resolved by electrophoresis on a 5% nondenaturing polyacrylamide gel at 200 V for 75 min in 0.25 X Tris-borate EDTA buffer. The electrophoretogram was then dried, visualized, and quantified by PhosphorImager analysis (Molecular Dynamics STORM system). Supershifts were performed in a similar manner, with the addition of either 0.5 µl/lane of RXRα-specific antibody 4RX-1D12 (1.0 µg/µl IgG; provided by P. Chambon, Pasteur Institute, Strassbourg, France) or 1.5 µl/lane of RARα-specific antibody sc551 (1.5 µg/µl IgG; Santa Cruz Biotechnology, Inc.). No detectable cross-reaction of the RXRα-specific antibody with RARα, PML-RARα, or PLZF-RARα or of the RARα-specific antibody with RXRα was observed (Fig. 2B).
Titration Experiments
Titration experiments were performed in two different ways: either a constant amount of radiolabeled DNA probe was titrated with differing amounts of protein nuclear extract (0.02, 0.05, 0.1, 0.4, 1.3, or 4 µl; approximately 50 ng/µl protein); or a constant amount of protein was titrated with differing amounts of a radiolabeled DNA probe (0, 0.35, 1.4, 5.6, 23, or 90 ng). The protein-DNA complexes that formed were subsequently resolved by gel electrophoretic mobility shift assays, as described above.
Transient Transfections
Representative DNA elements were cloned into a thymidine kinase promoter/reporter vector for use in transient transfection assay. Oligonucleotides consisted of two retinoic acid response elements, each comprised of the consensus, −1A, 2T, or 4G DR-5 sequence. Oligonucleotides were made as two complementary strands bearing 4-base overhangs compatible with Sa/I and XhoI sites. For example, the plus oligonucleotide for the consensus sequence was composed of the following sequence (half-sites are underlined): 5′-TCGACAGAGGTCAACGAGAGGTCAGAGCTCAGAGGTCAACGAGAGGTCAGAG-3′.
These oligonucleotides were then individually cloned into the XhoI and Sa/I sites of a thymidine kinase/luciferase reporter construct (64), and appropriate recombinant vectors were screened by restriction mapping and confirmed by DNA sequence analysis.
Transient transfections of CV-1 cells were performed in 12-well tissue culture plates with 9 × 104 cells/well. Transfections were performed using a liposome/LipofectAMINE plus technology (Life Technologies, Inc.); 20 ng of a pSG5-plasmid (either an empty vector or a vector encoding RARα, PML-RARα, PLZF-RARα, or RXRα) were transfected per well, together with 100 ng of the luciferase reporter plasmid, 100 ng of a pCH110-promoter-lacZ reporter plasmid (used as an internal control), and 280 ng of pUC18 (to standardize the total amount of transfected DNA to 500 ng/well). Three h after transfection, all-trans retinoic acid was added (or not added) to 400 nm, and the cells were subsequently incubated for an additional 24 h. Thereafter, the cells were harvested, and the luciferase and β-galactosidase activities were determined as described previously (30). Transfected cell extracts were also analyzed by immunoblotting to determine the levels of expression of the different nuclear receptor derivatives.
Acknowledgments
We thank A. Dejean, S. Kogan, and R. Evans for the generous gifts of molecular clones of the receptors used in these studies; M. d. M. Vivanco Ruiz for the ptk-luc vector from which we constructed our transfection reporter constructs; and P. Chambon for providing the RXRα-specific antibodies. We thank S. Lee-Bond for initiating the construction of the baculovirus vector clones of human RARα, PML-RARα, and PLZF-RARα used in these experiments. We are also grateful to Behnom Farboud for assistance in transient transfection assays.
Footnotes
Supported by USPHS/NIH Grants CA53394 and DK54064.
The abbreviations used are: T3R, thyroid hormone receptor; RAR, retinoic acid receptor; RXR, retinoid X receptor; ROR, retinoid orphan receptor; DR-1, direct repeat-1; PML, promyelocytic leukemia protein; PLZF, promyelocytic leukemia zinc-finger protein; NuMA, nuclear mitotic apparatus protein; APL, acute promyelocytic leukemia; NPM, nucleophosmin.
Unpublished results.
References
- 1.Glass CK. Differential recognition of target genes by nuclear receptor monomers, dimers and heterodimers. Endocr. Rev. 1994;15:391–407. doi: 10.1210/edrv-15-3-391. [DOI] [PubMed] [Google Scholar]
- 2.Williams GR, Franklyn JA. Physiology of the steroid-thyroid hormone nuclear receptor superfamily. Bailliére’s Clin. Endocrinol. Metab. 1994;8:241–266. doi: 10.1016/s0950-351x(05)80251-4. [DOI] [PubMed] [Google Scholar]
- 3.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 5.Kastner P, Mark M, Chambon P. Nonsteroid nuclear receptors: what are genetic studies telling us about their role in real life? Cell. 1995;83:859–869. doi: 10.1016/0092-8674(95)90202-3. [DOI] [PubMed] [Google Scholar]
- 6.Tsai M-J, O’Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 7.Katz RW, Koenig RJ. Nonbiased identification of DNA sequences that bind thyroid hormone receptor α1 with high affinity. J. Biol. Chem. 1993;268:19392–19397. [PubMed] [Google Scholar]
- 8.Kurokawa R, Yu VC, Naar A, Kyakumoto S, Han Z, Silverman S, Rosenfeld MG, Glass CK. Differential orientations of the DNA-binding domain and carboxy-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Genes Dev. 1993;7:1423–1435. doi: 10.1101/gad.7.7b.1423. [DOI] [PubMed] [Google Scholar]
- 9.Naar AM, Boutin J-M, Lipkin SM, Yu VC, Holloway JM, Glass CK, Rosenfeld MG. The orientation and spacing of core DNA-binding motifs dictate selective transcriptional responses to three nuclear receptors. Cell. 1991;65:1267–1279. doi: 10.1016/0092-8674(91)90021-p. [DOI] [PubMed] [Google Scholar]
- 10.Forman BM, Casanova J, Raaka BM, Ghysdael J, Samuels HH. Half-site spacing and orientation determines whether thyroid hormone and retinoic acid receptors and related factors bind to DNA response elements as monomers, homodimers, or heterodimers. Mol. Endocrinol. 1992;6:429–442. doi: 10.1210/mend.6.3.1316541. [DOI] [PubMed] [Google Scholar]
- 11.Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polak M. New data on nuclear receptor cofactors suggest a control of transcriptional repression by hormone-dependent chromatin remodelling. Eur. J. Endocrinol. 1997;137:455–456. doi: 10.1530/eje.0.1370455. [DOI] [PubMed] [Google Scholar]
- 13.Shibata H, Spencer TE, Onate SA, Jenster G, Tsai SY, Tsai M-J, O’Malley BW. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent Prog. Horm. Res. 1997;52:141–165. [PubMed] [Google Scholar]
- 14.Torchia J, Glass C, Rosenfeld MG. Co-activators and corepressors in the integration of transcriptional responses. Curr. Opin. Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 15.Chen JD, Li H. Coactivation and corepression in transcriptional regulation by steroid/nuclear hormone receptors. Crit. Rev. Eukaryotic Gene Expression. 1998;8:169–190. doi: 10.1615/critreveukargeneexpr.v8.i2.40. [DOI] [PubMed] [Google Scholar]
- 16.Horwitz KB, Jackson TA, Bain DL, Richer JK, Takimoto GS, Tung L. Nuclear receptor coactivators and corepressors. Mol. Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 17.Lavau C, Jansen J, Dejean A. The t(15;17) translocation in acute promyelocytic leukemia. Pathol. Biol. 1995;43:188–196. [PubMed] [Google Scholar]
- 18.Chen Z, Wang Z-Y, Chen S-J. Acute promyelocytic leukemia: cellular and molecular basis of differentiation and apoptosis. Pharmacol. Ther. 1997;76:141–149. doi: 10.1016/s0163-7258(97)00090-9. [DOI] [PubMed] [Google Scholar]
- 19.Kalantry S, Delva L, Gaboli M, Gandini D, Giorgio M, Hawe N, He L-Z, Peruzzi D, Rivi R, Tribioli C, Wang Z-G, Zhang H, Pandolfi PP. Gene rearrangements in the molecular pathogenesis of acute promyelocytic leukemia. J. Cell. Physiol. 1997;173:288–296. doi: 10.1002/(SICI)1097-4652(199711)173:2<288::AID-JCP38>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 20.Quignon F, Chen Z, de Thé H. Retinoic acid and arsenic: towards oncogene-targeted treatments of acute promyelocytic leukaemia. Biochim. Biopohys. Acta. 1997;1333:M53–M61. doi: 10.1016/s0304-419x(97)00025-5. [DOI] [PubMed] [Google Scholar]
- 21.Grimwade D, Solomon E. Characterization of the PML/RARα rearrangement associated with t(15;17) acute promyelocytic leukaemia. Curr. Top. Microbiol. Immunol. 1997;220:81–112. doi: 10.1007/978-3-642-60479-9_6. [DOI] [PubMed] [Google Scholar]
- 22.de The H, Chomienne C, Lanotte M, Degos L, Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor α gene to a novel transcribed locus. Nature (Lond.) 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 23.Alcalay M, Zangrilli D, Pandolfi PP, Longo L, Mencarelli A, Giacomucci A, Rocchi M, Biondi A, Rambaldi A, Coco FL, Diverio D, Donti E, Grignani F, Pelicci PG. Translocation breakpoint of acute promyelocytic leukemia lies within the retinoic acid receptor α locus. Proc. Natl. Acad. Sci. USA. 1991;88:1977–1981. doi: 10.1073/pnas.88.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borrow J, Goddard AD, Sheer D, Solomon E. Molecular analysis of acute promyelocytic leukemia breakpoint cluster region on chromosome 17. Science (Washington DC) 1990;249:1577–1580. doi: 10.1126/science.2218500. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, Brand NJ, Chen A, Chen S-J, Tong J-H, Wang Z-Y, Waxman S, Zelent A. Fusion between a novel Krúppel-like zinc finger gene and the retinoic acid receptor-α locus due to a variant t(11;17) translocation asociated with acute promyelocytic leukaemia. EMBO J. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He L-Z, Tribioli C, Rivi R, Peruzzi D, Pelicci PG, Soares V, Cattoretti G, Pandolfi PP. Acute leukemia with promyelocytic features in PML/RARα transgenic mice. Proc. Natl. Acad. Sci. USA. 1997;94:5302–5307. doi: 10.1073/pnas.94.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grisolano JL, Wesselschmidt RL, Pelicci PG, Ley TJ. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RARα under control of cathepsin G regulatory sequences. Blood. 1997;89:376–387. [PubMed] [Google Scholar]
- 28.He L-Z, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi PP. Distinct interactions of PML-RARα and PLZF-RARα with co-repressors determine differential responses to RA in APL. Nat. Genet. 1998;18:126–135. doi: 10.1038/ng0298-126. [DOI] [PubMed] [Google Scholar]
- 29.Brown D, Kogan S, Lagasse E, Weissman I, Alcalay M, Pelicci PG, Atwater S, Bishop JM. A PMLRARα transgene initiates murine acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA. 1997;94:2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong S-H, David G, Wong C-W, Dejean A, Privalsky ML. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor α (RARα) and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong C-W, Privalsky ML. Components of the SMRT corepressor complex exhibit distinctive interactions with the POZ domain oncoproteins PLZF, PLZF-RARα and BCL-6. J. Biol. Chem. 1998;273:27695–27702. doi: 10.1074/jbc.273.42.27695. [DOI] [PubMed] [Google Scholar]
- 32.Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr., Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature (Lond.) 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- 33.Grignani F, Matteis SD, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, Seiser C, Grignani F, Lazar MA, Minucci S, Pelicci PG. Fusion proteins of the retinoic acid receptor-α recruit histone deacetylase in promyelocytic leukaemia. Nature (Lond.) 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- 34.Wong C-W, Privalsky ML. Role of the N terminus in DNA recognition by the v-erb A protein, an oncogenic derivative of a thyroid hormone receptor. Mol. Endocrinol. 1995;9:551–562. doi: 10.1210/mend.9.5.7565803. [DOI] [PubMed] [Google Scholar]
- 35.Smit-McBride Z, Privalsky ML. DNA sequence specificity of the v-Erb A oncoprotein/thyroid hormone receptor: role of the P-box and its interaction with more N-terminal determinants of DNA recognition. Mol. Endocrinol. 1994;8:819–828. doi: 10.1210/mend.8.7.7984144. [DOI] [PubMed] [Google Scholar]
- 36.Chen H, Smit-McBride Z, Lewis S, Sharif M, Privalsky ML. Nuclear hormone receptors involved in neoplasia: Erb A exhibits a novel DNA sequence specificity determined by amino acids outside of the zinc-finger domain. Mol. Cell. Biol. 1993;13:2366–2376. doi: 10.1128/mcb.13.4.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giguére V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of RORα, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 38.McBroom LDB, Flock G, Giguère V. The nonconserved hinge region and distinct amino-terminal domains of the RORα orphan nuclear receptor isoforms are required for proper DNA bending and RORα-DNA interactions. Mol. Cell. Biol. 1995;15:796–808. doi: 10.1128/mcb.15.2.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Judelson C, Privalsky ML. DNA recognition by normal and oncogenic thyroid hormone receptors. J. Biol. Chem. 1996;271:10800–10805. doi: 10.1074/jbc.271.18.10800. [DOI] [PubMed] [Google Scholar]
- 40.Perez A, Kastner P, Sethi S, Lutz Y, Reibel C, Chambon P. PMLRAR homodimers: distinct DNA binding properties and heteromeric interactions with RXR. EMBO J. 1993;12:3171–3182. doi: 10.1002/j.1460-2075.1993.tb05986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong S, Zhu J, Reid A, Strutt P, Guidez F, Zhong H-J, Wang Z-Y, Licht J, Waxman S, Chomienne C, Chen Z, Zelent A, Chen S-J. Amino-terminal protein-protein interaction motif (POZ-domain) is responsible for activities of the promyelocytic leukemia zinc finger-retinoic acid receptor-α fusion protein. Proc. Natl. Acad. Sci. USA. 1996;93:3624–3629. doi: 10.1073/pnas.93.8.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Licht JD, Shaknovich R, English MA, Melnick A, Li J-Y, Reddy JC, Dong S, Chen S-J, Zelent A, Waxman S. Reduced and altered DNA-binding and transcriptional properties of the PLZF-retinoic acid receptor-α chimera generated in t(11;17)-associated acute promyelocytic leukemia. Oncogene. 1996;12:323–336. [PubMed] [Google Scholar]
- 43.Lin RJ, Evans RM. Acquisition of oncogenic potential by RAR chimeras in acute promyelocytic leukemia through formation of homodimers. Mol. Cell. 2000;5:821–830. doi: 10.1016/s1097-2765(00)80322-6. [DOI] [PubMed] [Google Scholar]
- 44.Kastner P, Mark M, Ghyselinck N, Krezel W, Dupe V, Grondona JM, Chambon P. Genetic evidence that the retinoic signal is transduced by heterodimeric RXR/RAR functional units during mouse development. Development (Camb.) 1997;124:313–326. doi: 10.1242/dev.124.2.313. [DOI] [PubMed] [Google Scholar]
- 45.Leid M, Kastner P, Lyons R, Nakshatri H, Saunders M, Zacharewski T, Chen J-Y, Staub A, Garnier J-M, Mader S, Chambon P. Purification, cloning, and RXR identity of the HeLa cell factor with which RAR or TR heterodimerizes to bind target sequences efficiently. Cell. 1992;68:377–395. doi: 10.1016/0092-8674(92)90478-u. [DOI] [PubMed] [Google Scholar]
- 46.Kastner P, Grondona JM, Mark M, Gansmuller A, LeMeur M, Decimo D, Vonesch J-L, Dolle P, Chambon P. Genetic analysis of RXRα developmental function: convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell. 1994;78:987–1003. doi: 10.1016/0092-8674(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 47.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- 48.Minucci S, Maccarana M, Cioce M, De Luca P, Gelmetti V, Segalla S, Di Croce L, Giavara S, Matteucci C, Gobbi A, Bianchini A, Colombo E, Schiavoni I, Badaracco G, Hu X, Lazar MA, Landsberger N, Nervi C, Pelicci PG. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol. Cell. 2000;5:811–820. doi: 10.1016/s1097-2765(00)80321-4. [DOI] [PubMed] [Google Scholar]
- 49.Kastner P, Perez A, Lutz Y, Rochette-Egly C, Gaub M-P, Durand B, Lanotte M, Berger R, Chambon P. Structure, localization and transcriptional properties of two classes of retinoic acid receptor α fusion proteins in acute promyelocytic leukemia (APL): structural similarities with a new family of oncoproteins. EMBO J. 1992;11:629–642. doi: 10.1002/j.1460-2075.1992.tb05095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RARα fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 51.Kakizuka A, Miller WH, Jr., Umesono K, Warrell RP, Jr., Frankel SR, Murty VVVS, Dmitrovsky E, Evans RM. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 52.Grignani F, Ferrucci PF, Testa U, Talamo G, Fagioli M, Alcalay M, Mencarelli A, Grignani F, Peschle C, Nicoletti I, Pelicci PG. The acute promyelocytic leukemia-specific PML-RARα fusion protein inhibits differentiation and promotes survival of myeloid precursor cells. Cell. 1993;74:423–431. doi: 10.1016/0092-8674(93)80044-f. [DOI] [PubMed] [Google Scholar]
- 53.Ruthardt M, Testa U, Nervi C, Ferrucci PF, Grignani F, Puccetti E, Grignani F, Peschle C, Pelicci PG. Opposite effects of the acute promyelocytic leukemia PML-retinoic acid receptor α (RARα) and PLZF-RARαfusion proteins on retinoic acid signalling. Mol. Cell. Biol. 1997;17:4859–4869. doi: 10.1128/mcb.17.8.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lieberman BA, Bona BJ, Edwards DP, Nordeen SK. The constitution of a progesterone response element. Mol. Endocrinol. 1993;7:515–527. doi: 10.1210/mend.7.4.8388996. [DOI] [PubMed] [Google Scholar]
- 55.Claessens F, Celis L, De Vos P, Peeters B, Heyns W, Verhoeven G, Rombauts W. Intronic androgen response elements of prostatic binding protein genes. Biochem. Biophys. Res. Commun. 1993;191:688–694. doi: 10.1006/bbrc.1993.1272. [DOI] [PubMed] [Google Scholar]
- 56.Tan J-A, Marschke KB, Ho K-C, Perry ST, Wilson EM, French FS. Response elements of the androgen-regulated C3 gene. J. Biol. Chem. 1992;267:4456–4466. [PubMed] [Google Scholar]
- 57.Nelson CC, Hendy SC, Shukin RJ, Cheng H, Bruchovsky N, Koop BF, Rennie PS. Determinants of DNA sequence specificity of the androgen, progesterone, and glucocorticoid receptors: evidence for differential steroid receptor response elements. Mol. Endocrinol. 1999;13:2090–2107. doi: 10.1210/mend.13.12.0396. [DOI] [PubMed] [Google Scholar]
- 58.Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature (Lond.) 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 59.Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature (Lond.) 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 60.Gewirth DT, Sigler PB. The basis for half-site specificity explored through a non-cognate steroid receptor-DNA complex. Nat. Struct. Biol. 1995;2:386–394. doi: 10.1038/nsb0595-386. [DOI] [PubMed] [Google Scholar]
- 61.Schwabe JWR, Chapman L, Rhodes D. The oestrogen receptor recognizes an imperfectly palindromic response element through an alternative side-chain conformation. Structure. 1995;3:201–213. doi: 10.1016/s0969-2126(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 62.Ikeda M, Rhee M, Chin WW. Thyroid hormone receptor monomer, homodimer, and heterodimer (with retinoid-X receptor) contact different nucleotide sequences in thyroid hormone response elements. Endocrinology. 1994;135:1628–1638. doi: 10.1210/endo.135.4.7925126. [DOI] [PubMed] [Google Scholar]
- 63.Chen H, Privalsky ML. Cooperative formation of high-order oligomers by retinoid X receptors: an unexpected mode of DNA recognition. Proc. Natl. Acad. Sci. USA. 1995;92:422–426. doi: 10.1073/pnas.92.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vivanco Ruiz MdM, Bugge TH, Hirschmann P, Stunnenberg HG. Functional characterization of a natural retinoic acid responsive element. EMBO J. 1991;10:3829–3838. doi: 10.1002/j.1460-2075.1991.tb04952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hong S-H, Privalsky ML. The SMRT corepressor is regulated by a MEK-1 kinase pathway: inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol. Cell. Biol. 1991;20:6612–6625. doi: 10.1128/mcb.20.17.6612-6625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]