Peroxisome biogenesis [1–4], as well as the importance of peroxisomes in human health and disease [5], have been the subject of many reviews. Proteins that control peroxisome biogenesis encompass the processes by which matrix and membrane proteins are assembled into the organelle, as well as those involved in the control of peroxisome size, volume and number [6–9]. The proteins involved in these processes are peroxins that are encoded by PEX genes, which have been studied in many unicellular and multicellular organisms from yeast to man. This review provides an overview of peroxisome matrix and membrane protein biogenesis with an emphasis on recent insights and unanswered questions.
Peroxisomal Matrix Protein Import
Three types of targeting signals direct most proteins to their membrane or matrix locations in peroxisomes. The peroxisome targeting signals (PTSs) used by peroxisomal matrix proteins are called PTS1 and PTS2, while those used by peroxisomal membrane proteins (PMPs) are dubbed mPTSs. Most matrix proteins have only a single PTS1 or PTS2, with rare ones having both (e.g. Pichia pastoris Pex8), in which case they are generally redundant [10]. However, many PMPs have multiple and redundant mPTSs [11]. The PTSs for matrix proteins are recognized by specific cytosolic receptors and/or co-receptors, which escort the cargoes to the peroxisome membrane [3]. Here the matrix proteins and their receptors enter the peroxisome matrix [12–16], where cargoes are released, and the cargo-free receptors are first exported to the peroxisome membrane via a retro-translocation step [13], and then the PTS receptors are recycled back to the cytosol by an ATP-dependent receptor recycling machinery [17]. This receptor recycling step often (e.g. Pex5 and Pex20), but not always (e.g. Pex7), utilizes mono-ubiquitination of the receptors (unusually on a Cys residue near the N-terminus of the protein, rather than on one or more Lys residue/s) [18–21]. During the PTS receptor-recycling step, the monoubiquitin is removed by an unknown deubiquitinating enzyme, so that the recycled receptor is capable of sustaining additional rounds of matrix protein import [3].
The import of proteins into the peroxisome matrix can be divided into the following steps.
1. Cargo recognition and binding
This essentially involves the recognition of the PTS1 or PTS2 by their cognate receptor/co-receptor. The C-terminal tripeptide (SKL, or its conserved variants) that constitutes the PTS1 is recognized by the receptor protein Pex5, which is normally tetrameric [22–24]. Binding of cargo to Leishmania donovani Pex5 has been reported to shift the equilibrium of Pex5 oligomers to the predominantly dimeric state [22]. Pex5 is a two domain protein, with an N-terminal region comprised of sequences required for its recycling [18–21, 25] followed by a series of WxxxF/Y repeats. The WxxxF/Y repeats are required for Pex5 interactions with Pex14, but not all binding sites need to be intact for Pex5 function in vivo [26], and the number of these sites varies in Pex5 proteins from different species. The structure of this N-terminal region of Pex5 is not defined and is reported to be unstructured [27, 28]. This region contains binding sites for Pex13 and Pex14, and in the case of mammalian Pex5L (a longer, alternatively-spliced isoform of Pex5), also for Pex7 [26]. The C-terminal half of Pex5 has 6–7 tetratricopeptide (TPR) repeats whose crystal structure is known and this is the region that interacts with the PTS1 cargo [28–31]. Most cargoes that bind Pex5 do so via this interaction of the TPR repeats on Pex5 with the PTS1, but a few Pex5-dependent cargoes lack canonical PTS1 sequences and may interact with Pex5 by other poorly-defined mechanisms [32]. These might have a PTS that is not yet defined. The dissociation constant of Pex5 for cargo is in the 18–100 nM range [24, 30]. While the C-terminal tripeptide on the cargo is essential for the binding Pex5, there are indeed other contacts between the cargo and Pex5, which may account for the many PTS1 variants that can still function in vivo to achieve peroxisomal matrix targeting [28–31].
In contrast, proteins using the PTS2 sequence, comprised of the internally-located consensus sequence, (R/K)(L/V/I/Q)XX(L/V/I/H/Q)(L/S/G/A/K)X(H/Q)(L/A/F) [33], interact primarily with Pex7 [34–39], apparently as a monomer [36, 40]. Some PTS2 cargoes also interact with a co-receptor, Pex20 [10, 41]. This protein is in the same family as Pex18 and its redundant counterpart, Pex21 in Saccharomyces cerevisiae [10, 41, 42], is found only in fungi, and forms a complex with Pex7 [13, 43, 44]. HpPex20 has been reported to form hexamers and has a weak affinity (Kd= 400 nM) for PTS2 peptides [41]. When bound to one cargo, it has been reported that a dimer of Yarrowia lipolytica Pex20 interacts with a corresponding dimer of thiolase [42]. Mammals and plants do not have Pex20-like proteins, so in this case, all known PTS2 proteins are recognized by Pex7.
A few rare proteins enter the peroxisome with no obvious PTSs. They do so by exploiting a unique feature of peroxisomal matrix protein import, which is that fully folded and oligomeric proteins can traverse the peroxisome membrane, and these proteins hitch a ride in a piggy-back manner by association with some other subunit or protein that does have a PTS1 or a PTS2 [45, 46].
2. Docking of the receptor/cargo complex at the peroxisome membrane
The PTS receptor/cargo complex formed in the cytosol finds its way to the peroxisome membrane where it interacts with a peroxisome-membrane associated docking subcomplex comprised of the conserved proteins Pex13 and Pex14, as well as Pex17, which is not conserved in all organisms [47, 48]. Pex8 is also part of this subcomplex in yeasts [47, 48], but is considered separately below because it is not necessary for the formation or stability of this subcomplex and also it is found only in yeasts [48]. The docking complex components are generally integral membrane proteins, but in a few species Pex14 has been described to be a peripheral protein of the peroxisome membrane [49]. The N-terminal region of Pex14 interacts with Pex5, with 4–6 Pex14 molecules interacting with a single molecule of Pex5, usually through the interactions of conserved and repetitive WxxxF/Y motifs on Pex5 with a conserved motif AX&2FLX7SPX6FLKGKGL/V present in the first 80 amino acids approximately of all Pex14 proteins [23, 50]. The dissociation constant for Pex5-Pex14 interaction is in the low nanomolar range [23, 50], but when Pex5 releases cargo, the affinity of Pex14 for Pex5 is much lower (Kd of 2.75 µM), showing that Pex14 preferentially interacts with cargo-loaded Pex5 [51]. Pex14 tends to form oligomers in vivo or when expressed in, and purified from, E. coli [52–54]. Two domains on Pex14 control its oligomeric state – one favors dimerization while the other drives oligomerization [53]. Interestingly, in the presence of Pex14, the interaction of Pex5 with cargo still occurs, but this binding is about 10-fold weaker than that in the absence of Pex14 [50]. Thus, as the Pex5/cargo complex lands on the peroxisome, the interaction between Pex5 and cargo is weakened, but cargo is presumably not yet released. During this Pex14-Pex5 interaction, Pex14 undergoes conformation changes, especially in its hydrophobic domains as judged by shifts in the environment of Trp residues from non-polar to polar environments [50]. Pex14 has a greater affinity than Pex13 for cargo-loaded Pex5 and when cargo-loaded Pex5 interacts with Pex14 it is in a complex containing Pex13. However, upon cargo release (discussed below) from Pex5, the receptor interacts more tightly with Pex13, and at this stage the interaction between Pex13 and Pex14 is lost transiently [26].
For PTS2 proteins, in yeast it is generally a ternary Pex20/Pex7/PTS2 cargo complex that forms in the cytosol and is delivered to the peroxisome membrane [13, 44]. In S. cerevisiae, the role of Pex20 is substituted by the redundant proteins Pex18 and Pex21, which are related to Pex20 [55]. Mammals and plants lack Pex20, but instead they have an alternatively-spliced longer variant of Pex5 (Pex5L) with which the Pex7-cargo complex interacts, so in this case, a ternary complex is still formed but is comprised of Pex5L/Pex7/PTS2 cargo [40].
Details of the docking interactions of PTS receptors are better studied for Pex5 than for Pex7. Pex14 binds more strongly to cargo-loaded Pex5 (and reducing the affinity of the Pex5-cargo interaction as described above) than to free Pex5 [50], whereas Pex13 interacts more strongly with cargo-free Pex5 [26]. Pex13 has an SH3 domain that interacts with Pex5 in a manner not involving canonical PxxP motifs on Pex5 [56, 57]. Based on these facts, and our own recent finding that Pex14 may be the real peroxisomal translocon (Ma et al., manuscript submitted, see below), we suggest that Pex13-Pex5 interactions may be downstream of cargo release from Pex5.
3. Cargo translocation and release
A major unresolved question in the field is what peroxins are needed directly for the protein translocation step across the peroxisome membrane. Based on studies of mutants blocked in the import of both PTS1 and PTS2 proteins, it was believed that components of the docking and RING subcomplexes, as well as the proteins that bridge these subcomples (Pex3 and Pex8) formed the importomer – a term that was initially assumed to be synonymous with the translocon [47, 58]. However, further studies, especially with Pex8 entry into peroxisomes, have revealed some surprising and important simplifications.
Pex8 is the only intraperoxisomal peroxin at steady-state and is needed for both PTS1 and PTS2 import. It enters peroxisomes via either the PTS1 or the PTS2 pathways, which are redundant [10]. However, its entry into peroxisomes, by either the PTS1 o the PTS2 pathways, is not dependent on the components of the RING subcomplex [10] or of the receptor recycling machinery (Ma et al., manuscript submitted). Additionally, our recent data show that even Pex13 (an evolutionarily-conserved peroxin) and Pex17 (a peroxin not conserved in mammals) are not completely essential for Pex8 import, although they make the process more efficient (Ma et al., manuscript submitted). These results suggest that, at least for Pex8, Pex14 alone may be the minimal translocon in the peroxisome membrane. It is unclear at present whether PTS receptors are also part of the translocon, but it should be noted here that Pex5 has been proposed to insert into lipid bilayers and potentially form pores [59].
Unlike membrane-associated translocons in other subcellular organelles that translocate only unfolded polypeptides, as a consequence of which targeting signal receptors cannot traverse the membrane, folded and oligomeric proteins do go across the peroxisome membrane. In fact, the PTS receptors/co-receptors enter the matrix, following what has been called the extended shuttle model for receptor dynamics [12–16]. Here they are resistant to externally-added proteases, and in mutants that block the next step, receptor export, Pex5 and Pex20 accumulate inside peroxisomes and are inaccessible to the cytosolic machinery that ubiquitinates these receptors during receptor recycling (described below) [3]. It is presumed that receptor/co-receptor entry into peroxisomes takes cargo into the matrix where cargo must be released.
How cargo is released is still an open question. We alluded earlier to the fact that the docking of Pex5-cargo to Pex14 loosens the receptor cargo interaction 10-fold [22]. The oligomeric state of Pex5 after interaction with Pex14 is unknown, but it has been shown that the intraperoxisomal protein, Pex8, interacts with Pex5, forming a 1:1 complex, and that this interaction facilitates cargo release [24]. This is based on studies of Pex5-cargo interaction, in vitro, in the presence and absence of Pex8, which is not quite the same as the situation in vivo. The involvement of Pex8, which has both a PTS1 and PTS2 signal, and interacts with Pex5 and Pex20, in cargo release was attractive because one could imagine a role for Pex8 in competitively using its own PTSs to cause cargo release, but nature does not adhere to the simplest human solutions – deletion of the PTS1 on Pex8 had no affect on the delivery and release of PTS1 cargo into and inside the peroxisome matrix [10]!
It is also known that slightly acidic pH causes the Pex5 tetramer to dissociate into the monomeric state, which interestingly does not bind cargo [24], so it is also plausible that an acidic pH inside peroxisomes might aid cargo release. However, measurements of intraperoxisomal pH have reported peroxisomes to be acidic (pH 5.8–6.0) [60], neutral (pH 6.9–7.1) [61] and basic (pH 8.2 for mammalian and yeast peroxisomes) [62, 63], and PTS1 protein import is unimpaired in mutant fibroblasts in which the intraperoxisomal pH is 6.8 [64], making it difficult to assess the relevance of peroxisomal pH in cargo release within peroxisomes.
4. Export or retro-translocation of cargo-free receptors to the peroxisomal membrane
Studies with the PTS2 co-receptor, Pex20, have shown that in the absence of the components of the RING subcomplex, Pex20 accumulates inside peroxisomes, defining the RING subcomplex proteins as playing some role in the export of Pex20 to the peroxisome membrane [13]. Indeed interactions have been reported between both Pex5 and Pex20 with Pex12 [19, 65]. Whether these RING peroxins constitute a retro-translocon for receptor export or somehow reverse the directionality of the same translocon that lets cargo into peroxisomes is unknown.
5. Recycling of cargo-free receptors from peroxisome membranes to the cytosol
If PTS receptors enter peroxisomes, they might need signals and specific proteins for receptor recycling back to the cytosol [12]. This is indeed true. Recently, a Cys residue, near the N-terminus of Pex5 and Pex20 was identified as being required for receptor recycling in yeast and mammalian systems [18–21]. Deletion of residues 1–17 in the N-terminal region of human PEX5 affects its recycling from peroxisomes to the cytosol [25, 66]. Deletion of the first 19 residues in P. pastoris Pex20 also leads to loss-of function of the protein due to its accumulation in peroxisomes [19]. These represent cis-acting sequences whose presence on the receptors is necessary for their recycling.
The receptor export step would deliver Pex5 and Pex20 to the cytosolic face of the peroxisome membrane in the cargo-free state. It is plausible that this cargo-free state of the PTS receptor that has just completed a round of matrix protein import is distinct from that of cytosolic PTS receptors that have not yet bound cargo. Note that Pex5 is normally tetrameric when cargo is absent, which is likely its state in the cytosol [22–24]. Pex20 is probably hexameric [41]. Cargo-bound Pex5 and Pex20 are dimeric [22, 42]. However, while in the peroxisome, where Pex8 is located, Pex5 forms a 1:1 complex with Pex8 [22–24], but the oligomeric state of Pex20 when it binds Pex8 is unknown at present. This suggests that the cargo-free Pex5 that has just completed a round of import may arrive at the peroxisome membrane in a monomeric state and the same may be true for Pex20. At this stage Pex5, and probably Pex20, are mono-ubiquitinated on a Cys residue by the E2 enzyme Pex4 [20, 21, 67–69], which is held on the peroxisome membrane by association with Pex22 [70]. It is likely that one or more of the RING peroxins (most likely Pex12, which interacts with both proteins) play a role as an E3 ligase for this monoubiquitination reaction [71].
The monoubiquitinated Pex5 and Pex20 are then recognized, by unknown mechanisms involving the AAA-ATPases, Pex1 and Pex6 [17], held on the peroxisome membrane in association with Pex15 in yeast and Pex26 in mammals. These ATPases use ATP hydrolysis to pull the PTS receptors into the cytosol [17]. The last steps of receptor recycling must involve deubiquitination and oligomerization of the PTS receptors, but the deubiquitinating enzyme (DUB) is unknown at present. By analogy with the ER-associated degradation (ERAD), of misfolded proteins, a DUB in the OTU family may be involved.
In the absence of one or more of the components (Pex1, Pex4, Pex6, Pex22 an Pex15/26) of the receptor-recycling machinery, one or more lysines near the N-terminus of Pex5 and Pex20 are polyubiquitaned by the RADAR machinery [13, 67–69]. This polyubiquitination uses a different E2 (ubc4 or ubc5, in yeast) and E3 ligase activity provided by one of the RING peroxins [71]. The net result of this polyubiquination is that the proteasome degrades this cargo-free receptor that is blocking the peroxisome surface.
General comments on behavior and dynamics of Pex5 and Pex20
Despite the fact that these receptors/co-receptors involved in the PTS1 and PTS2 pathways have very little sequence similarity, there are remarkable similarities in their behavior and dynamics during the matrix protein import cycle. Both proteins are oligomeric and can bind cargo (directly or indirectly for Pex20), which causes their higher-order oligomeric state to become dimeric. Both interact with Pex14 first, followed probably by downstream interactions with Pex8, Pex13 and Pex12. Both peroxins enter and exit peroxisomes, using similar machineries [3]. Following a round of import, they both face a choice of either monoubiquitination and recycling back to the cytosol in a manner dependent on the peroxisomal receptor recycling machinery, or are subject to RADAR and proteolytic turnover by similar mechanisms. The amino acid residues that are monoubiquitinated (on a Cys residues near their N termini, but not yet proven definitively for Pex20) or polyubiquitinated (on one or more Lys) are in conserved domains. This similarity in behavior may have made it possible during evolution to dispense with the PEX20 gene in plants and mammals, and to facilitate PTS2 protein import by having Pex7 interact instead with an extra exon in Pex5 that has the Pex20 domain which allows it to interact with cargo-loaded Pex7 [40, 72, 73].
Peroxisomal Membrane Protein Import
The involvement of the endoplasmic reticulum (ER) in PMP biogenesis
The targeting of peroxisomal membrane proteins and the origin of peorxisomes are two tightly associated questions. The prevailing view within the peroxisome field in the past two decades was that, like mitochondria and chloroplasts, peroxisomes proliferate by growth and division of pre-existing organelles [74, 75]. According to this growth and division model, all peroxisomal membrane, as well as matrix, proteins are synthesized on free ribosomes and post-translationally targeted directly from the cytoplasm to peroxisomes. However, the growth and division model could not explain one puzzling question: how could mutants like pex3, pex16, and pex19 that completely lack peroxisomal membrane structures regain peroxisomes when the corresponding wild-type gene is reintroduced into these cells [76–79]? This question has been addressed, at least partially, by the recent de novo biogenesis model, which proposes that new peroxisomes are derived from the ER. Several groups have demonstrated that when the PEX3 gene is reintroduced into pex3 cells, Pex3 first inserts into the ER and then escapes from the ER via small vesicles, which later mature into peroxisomes [80–82]. Based on the de novo biogenesis model, many, if not all, peroxisomal membrane proteins (PMPs) are indirectly sorted to peroxisomes via the ER [4]. A growing list of PMPs from various organisms that have been demonstrated to be sorted to peroxisomes via the ER is shown in Table 1.
Table 1.
Peroxisomal membrane proteins currently known to be targeted to peroxisomes via the ER
| Name | Species | PMP features | Initial ER localization | Characteristics | References |
|---|---|---|---|---|---|
| Pex2 | Yl | Multi-spanning membrane protein | gER | RING-finger protein required for matrix protein import, member of the RING subcomplex | [103] |
| Pex3 | Sc | Amino-terminally anchored protein | gER | Co-operates with Pex19 in PMP biogenesis and/or assembly | [80, 82] |
| Hp | [104] | ||||
| Pex10 | At | Multi-spanning membrane protein | pER | RING-finger protein required for matrix protein import, member of the RING subcomplex | [102] |
| Pex13 | Mm | Multi-spanning membrane protein | pER | Member of the docking subcomplex | [105] |
| Pex15 | Sc | Tail-anchored protein | n.d | Membrane anchor for Pex6 | [85] |
| Pex16 | Yl | Multi-spanning membrane protein | gER | Integral PMP required for membrane biogenesis | [103] |
| At | [106, 107] | ||||
| Cs* | [83] | ||||
| Pex30 | Pp | Multi-spanning membrane protein | n.d | Dysferlin domain-containing protein required for peroxisome growth and division | [8] |
| Pex31 | Pp | Multi-spanning membrane protein | n.d | Dysferlin domain-containing protein required for peroxisome growth and division | [8] |
| PMP34 | Cs* | Multi-spanning membrane protein | pER | Peroxisomal ATP transporter | [83] |
| PMP70 | Mm | Multi-spanning membrane protein | pER | ATP-binding transporter | [105] |
| APX | At | Tail-anchored protein | pER | scavenger of reactive oxygen species | [108] |
| Nt | pER | [102] |
gER: general ER; pER: perioxisomal domain of ER; n.d: targeted to ER but specific region not defined. Cs*: COS-7 cells derived from the kidney cells of African green monkey. At: Arabidopsis thaliana; Hp: Hansenula polymorpha; Mm: Mus musculus; Pp: Pichia pastoris; Sc: Saccharomyces cerevisiae; Yl: Yarrowia lipolytica; Nt: Nicotiana tabacum.
Role of ER-derived vesicles in peroxisome growth and division
The current accepted view is that peroxisomes can arise de novo from the ER-derived vesicles, as well as from the fission of pre-existing peroxisomes [4, 83, 84]. However, it is still under debate whether de novo formation operates continuously or only switches on under unusual conditions in mutant cells lacking peoxisomes since different results were obtained in studies of lower and higher eukaryotic organisms.
As shown in S. cereivisiae using pulse-chase experiments and a mating assay, peroxisomes proliferate by division and do not form de novo in wildtype cells. In such cells, ER-derived vesicles provide pre-existing peroxisomes with peroxiosmal membrane proteins and lipids by fusion, which enables the subsequent growth and division of pre-existing peroxisomes [84]. It was shown that several peroxisomal membrane proteins such as Pex2, Pex15 and Pex16 underwent posttranslational glycosylation while passing through the ER [85, 86]. Proper folding of some proteins relies on glycosylation [87, 88]. It is still not known whether the glycosylation occurring in the ER results in proper folding or stabilization of Pex2, Pex15 and Pex16. If the peroxisomal importomer and receptor recycling machinery are only able to assemble on ER-derived vesicles, the fusion of ER-derived vesicles with the pre-existing peroxisomes would then provide the driving force for peroxisomal growth and division.
It has been proposed that the ER is one of the major resources of peroxisomal membrane lipids [89, 90]. However, the ER-derived vesicles are unlikely play a major role in supplying young peroxisomes with phospholipids. A recent report suggests that lack of Sec proteins required for vesicular trafficking from the ER does not affect lipid transfer between these two organelles [91]. Instead, it was shown that lipids are directly transferred from the ER to peroxisomes by a non-vesicular pathway, possibly through physical contact.
The ER-derived vesicles mature into peroxisomes only in S. cereivisiae cells lacking peroxisomes. For example, in peroxisomal inheritance defective Δinp2 cells, peroxisomes formed from ER-derived vesicles in daughter cells are capable of importing peroxisomal cargoes [84].
However, the situation is different in mammalian cells. Based on live cell imaging approaches, it was shown that peroxisomes form de novo independent of pre-existing ones [83]. Therefore, the ER-derived vesicles mature into peroxisomes and contribute to the peroxisome proliferation even under normal physiological conditions. It remains to be investigated why ER-derived vesicles do not mature into peroxisomes in the presence of pre-existing peroxisomes in S. cerevisiae cells or whether mammalian cells have special mechanisms to orchestrate de novo formation and division of peroxisomes.
Anterograde movement of peroxisomal membrane peroxins
The ER-to-peroxisome pathway is a complicated process, which is not fully understood [92]. Based on the available data from evolutionarily diverse organisms, we divide the ER-to-peroxisome pathway into four distinct steps: (i) Targeting of PMPs to the ER; (ii) segregation of PMPs from secretory and ER-resident membrane proteins; (iii) selective incorporation of the PMPs from the ER into ER-derived vesicles; (iv) fusion of these ER-derived pre-peroxisomal vesicles with the pre-existing peroxisomes (in yeast) or subsequent maturation of these pre-peroxisomal vesicles into mature organelles (in mammalian cells).
Exactly how PMPs are targeted to the ER is unknown. It should be noted that the peroxisome membrane has two classes of PMPs – the tail-anchored variety, such as ScPex15, as well as regular membrane proteins with single- or multiple-membrane spanning domains (e.g. Pex2).
Pex3, Pex16 and Pex19 are proposed to be involved in the early stages of the ER-to-peroxisome pathway and are among the earliest PMPs that initially target to the ER [76–79]. In mammalian cells, Pex16 is inserted co-translationally into the general ER (evenly distributed throughout the entire ER) and serves as the initial scaffold for recruiting at least Pex3 and PMP34 from the cytoplasm [83]. Afterwards, Pex16 with the recruited PMPs moves into the pre-peroxisomal temeplate and segregates from the secretory and ER-resident membrane proteins. Y. lipolytica Pex16, which is known to be involved in peroxisome proliferation, is initially targeted to the general ER as well [86, 93]. However, whether it functions exactly like its mammalian homolog is still not clear.
A slightly different process exists in other lower eukaryotic cells that do not have a Pex16 homolog. In S. cerevisiae, Pex3 is initially targeted to the general ER and then segregates to the pre-preoxisomal template followed by recruitment of Pex19, which is required for the exit of PMPs from the ER [80, 82]. Pex19 is thought to act as a PMP receptor that co-operates with Pex3 for the import of PMP to peroxisomes [94], but the validity of this view really depends on whether or not most PMPs can be targeted to the ER in a Pex19-independnt step. If so, there could be a distinct role for Pex19 in facilitating the formation of the ER-derived vesicles containing the pre-inserted PMPs. A resolution of this question will come when we know how and whether all PMPs are targeted to the ER and to what extent this ER membrane insertion step requires Pex19.
The mechanism underlying the segregation of PMPs from secretory and ER-resident membrane proteins is among the least understood in the ER-to-peroxisome targeting pathway. However, some clues are emerging: (i), the ER-derived vesicles in Y. lipolytica have enriched ergosterol-and ceramide-rich domains, which may be used as a tool to segregate PMPs from secretory and ER-resident membrane proteins [90, 95]; (ii), Pex19, through its interaction with PMPs, may functions as a chaperone to assemble PMP complexes and facilitate the movement of PMPs to an ER specialized subdomain, similar to a mechanism that has been proposed for the assembly of the importomer complexes in the peroxisome membrane in P. postoris [96].
SRP54, Sec238, Pex1 and Pex6 in Y. lipolytica were found to be required for the exit of PMPs from the ER. Lack of any of the above proteins in Y. lipolytica resulted in accumulation of Pex2 and Pex16 in the ER [86], indicating that these mutants may be blocked in the formation of ER-derived vesicles, extraction of PMPs from the ER or maturation of the ER-derived vesicles. Pex1 and Pex6 belong to AAA ATPase family and have been found to be predominantly associated with small vesicles that are distinct from mature peroxisomes in P. pastoris [97]. Therefore, Pex1 and Pex6 were proposed to be required for the fusion of small vesicles, which mature into large peroxisomes at the end. Later, it was demonstrated that in Y. lipolytica the fusion of small pre-peroxisomal vesicles, P1 and P2, was depended on Pex1 and Pex6 [98]. However, subsequent fusion processes did not rely on Pex1 and Pex6, indicating new factors exist and need to be discovered. Two other peroxins, PpPex30p and PpPex31p, which belong to the dysferlin domain-containing protein family, may also contribute to the fusion of ER-derived vesicles in a similar manner to that of their homologues [8, 99, 100].
Possibility of retrograde movement of proteins from peroxisomes to the ER
Based on the vesicle-mediated trafficking events in the secretory pathway, proteins required for anterograde trafficking might need to be retrieved by retrograde trafficking. So far, the peroxisome-to-ER sorting pathway has only been observed in TBSV (Tomato bushy stunt virus)-infected BY2 cells [101]. When p33, one out of five of TBSV encoded proteins, was expressed alone, it was targeted first to peroxisomes from the cytosol and then to a specialized subdomain of the ER together with at least two PMPs, PMP22 and ascorbate peroxidase (APX). Similar to the Golgi-to-ER targeting pathway, the peroxisome-to-ER targeting of p33 depended on ADP-ribosylation factor 1, indicating peroxisome-derived vesicles belong to coat protein complex I (COPI) coated vesicles. If the peroxisome-to-ER pathway does exist, akin to the Golgi-to-ER retrograde movement, it might also function in the retrieval of resident ER membrane proteins that might be mis-sorted to pre-peroxisomal vesicles [90, 101, 102]. Although it still not known whether the peroxisome-to-ER retrograde transport exists under normal physiological conditions in plant and/or in other organisms, this possibility has been alluded to in P. pastoris [8].
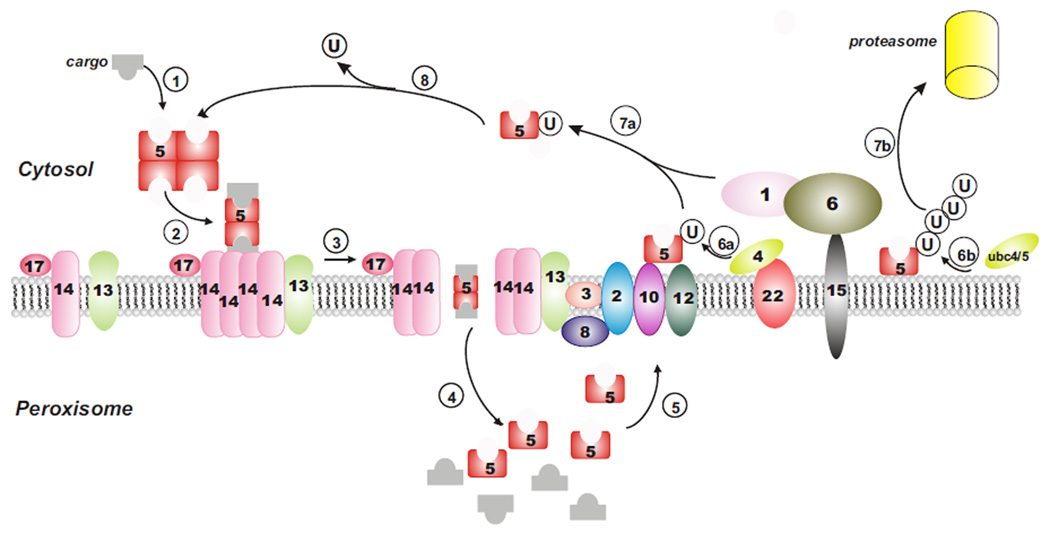
Figure 1. The import of peroxisomal matrix proteins.
(1) Cargoes are bound by a soluble receptor/s (Pex5 for PTS1 cargoes, Pex7 and PTS2 co-receptors for PTS2 cargoes, not depicted). (2) The receptor-cargo complex docks at the peroxisome membrane with the docking subcomplex. (3) The translocon is assembled and the receptor-cargo complex translocates into the peroxisome matrix. (4) The receptor-cargo complex is disassembled in the peroxisome matrix, causing cargo release. (5) Receptors are exported to the peroxisome membrane. (6a) Receptors are mono-ubiquitinated by Pex4 (for recycling) or (6b) poly-ubiquitinated by ubc4/5 (for degradation by the RADAR pathway). (7a) Receptors are recycled to the cytosol by the action of the AAA ATPases, Pex1 and Pex6, or (7b) degraded via the RADAR pathway involving the proteasome. (8) Receptors are deubiquitinated and utilized for the next round of import.
Acknowledgements
This work was supported by an NIH MERIT award (DK41737) to SS.
References
- 1.Brown LA, Baker A. Shuttles and cycles: transport of proteins into the peroxisome matrix. Mol. Membr. Biol. 2008;25:363–375. doi: 10.1080/09687680802130583. [DOI] [PubMed] [Google Scholar]
- 2.Eckert JH, Erdmann R. Peroxisome biogenesis. Rev. Physiol. Biochem. Pharmacol. 2003;147:75–121. doi: 10.1007/s10254-003-0007-z. [DOI] [PubMed] [Google Scholar]
- 3.Leon S, Goodman JM, Subramani S. Uniqueness of the mechanism of protein import into the peroxisome matrix: transport of folded, co-factor-bound and oligomeric proteins by shuttling receptors. Biochim. Biophys. Acta. 2006;1763:1552–1564. doi: 10.1016/j.bbamcr.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 4.Tabak HF, van der Zand A, Braakman I. Peroxisomes: minted by the ER. Curr. Opin. Cell Biol. 2008;20:393–400. doi: 10.1016/j.ceb.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW. Peroxisome biogenesis disorders. Biochim. Biophys. Acta. 2006;1763:1733–1748. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Vizeacoumar FJ, Torres-Guzman JC, Bouard D, Aitchison JD, Rachubinski RA. Pex30p, Pex31p, and Pex32p form a family of peroxisomal integral membrane proteins regulating peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:665–677. doi: 10.1091/mbc.E03-09-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vizeacoumar FJ, Torres-Guzman JC, Tam YY, Aitchison JD, Rachubinski RA. YHR150w and YDR479c encode peroxisomal integral membrane proteins involved in the regulation of peroxisome number, size, and distribution in Saccharomyces cerevisiae. J. Cell Biol. 2003;161:321–332. doi: 10.1083/jcb.200210130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan M, Rachubinski DA, Joshi S, Rachubinski RA, Subramani S. Dysferlin domain-containing proteins, Pex30p and Pex31p, localized to two compartments, control the number and size of oleate-induced peroxisomes in Pichia pastoris. Mol. Biol. Cell. 2008;19:885–898. doi: 10.1091/mbc.E07-10-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan M, Rayapuram N, Subramani S. The control of peroxisome number and size during division and proliferation. Curr. Opin. Cell Biol. 2005;17:376–383. doi: 10.1016/j.ceb.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L, Leon S, Subramani S. Two independent pathways traffic the intraperoxisomal peroxin PpPex8p into peroxisomes: mechanism and evolutionary implications. Mol. Biol. Cell. 2006;17:690–699. doi: 10.1091/mbc.E05-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones JM, Morrell JC, Gould SJ. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J. Cell Biol. 2004;164:57–67. doi: 10.1083/jcb.200304111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dammai V, Subramani S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell. 2001;105:187–196. doi: 10.1016/s0092-8674(01)00310-5. [DOI] [PubMed] [Google Scholar]
- 13.Leon S, Zhang L, McDonald WH, Yates J, 3rd, Cregg JM, Subramani S. Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J. Cell Biol. 2006;172:67–78. doi: 10.1083/jcb.200508096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nair DM, Purdue PE, Lazarow PB. Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J. Cell Biol. 2004;167:599–604. doi: 10.1083/jcb.200407119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salomons FA, Kiel JA, Faber KN, Veenhuis M, van der Klei IJ. Overproduction of Pex5p stimulates import of alcohol oxidase and dihydroxyacetone synthase in a Hansenula polymorpha Pex14 null mutant. J. Biol. Chem. 2000;275:12603–12611. doi: 10.1074/jbc.275.17.12603. [DOI] [PubMed] [Google Scholar]
- 16.Miyata N, Fujiki Y. Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol. Cell. Biol. 2005;25:10822–10832. doi: 10.1128/MCB.25.24.10822-10832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Platta HW, Grunau S, Rosenkranz K, Girzalsky W, Erdmann R. Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 2005;7:817–822. doi: 10.1038/ncb1281. [DOI] [PubMed] [Google Scholar]
- 18.Grou CP, Carvalho AF, Pinto MP, Wiese S, Piechura H, Meyer HE, Warscheid B, Sa-Miranda C, Azevedo JE. Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J. Biol. Chem. 2008;283:14190–14197. doi: 10.1074/jbc.M800402200. [DOI] [PubMed] [Google Scholar]
- 19.Leon S, Subramani S. A conserved cysteine residue of Pichia pastoris Pex20p is essential for its recycling from the peroxisome to the cytosol. J. Biol. Chem. 2007;282:7424–7430. doi: 10.1074/jbc.M611627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Platta HW, El Magraoui F, Schlee D, Grunau S, Girzalsky W, Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 2007;177:197–204. doi: 10.1083/jcb.200611012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams C, van den Berg M, Sprenger RR, Distel B. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 2007;282:22534–22543. doi: 10.1074/jbc.M702038200. [DOI] [PubMed] [Google Scholar]
- 22.Madrid KP, De Crescenzo G, Wang S, Jardim A. Modulation of the Leishmania donovani peroxin 5 quaternary structure by peroxisomal targeting signal 1 ligands. Mol. Cell. Biol. 2004;24:7331–7344. doi: 10.1128/MCB.24.17.7331-7344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schliebs W, Saidowsky J, Agianian B, Dodt G, Herberg FW, Kunau WH. Recombinant human peroxisomal targeting signal receptor PEX5. Structural basis for interaction of PEX5 with PEX14. J. Biol. Chem. 1999;274:5666–5673. doi: 10.1074/jbc.274.9.5666. [DOI] [PubMed] [Google Scholar]
- 24.Wang D, Visser NV, Veenhuis M, van der Klei IJ. Physical interactions of the peroxisomal targeting signal 1 receptor Pex5p, studied by fluorescence correlation spectroscopy. J. Biol. Chem. 2003;278:43340–43345. doi: 10.1074/jbc.M307789200. [DOI] [PubMed] [Google Scholar]
- 25.Costa-Rodrigues J, Carvalho AF, Gouveia AM, Fransen M, Sa-Miranda C, Azevedo JE. The N terminus of the peroxisomal cycling receptor, Pex5p, is required for redirecting the peroxisome-associated peroxin back to the cytosol. J. Biol. Chem. 2004;279:46573–46579. doi: 10.1074/jbc.M406399200. [DOI] [PubMed] [Google Scholar]
- 26.Otera H, Setoguchi K, Hamasaki M, Kumashiro T, Shimizu N, Fujiki Y. Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol. Cell. Biol. 2002;22:1639–1655. doi: 10.1128/MCB.22.6.1639-1655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho AF, Costa-Rodrigues J, Correia I, Costa Pessoa J, Faria TQ, Martins CL, Fransen M, Sa-Miranda C, Azevedo JE. The N-terminal half of the peroxisomal cycling receptor Pex5p is a natively unfolded domain. J. Mol. Biol. 2006;356:864–875. doi: 10.1016/j.jmb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Stanley WA, Wilmanns M. Dynamic architecture of the peroxisomal import receptor Pex5p. Biochim. Biophys. Acta. 2006;1763:1592–1598. doi: 10.1016/j.bbamcr.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 29.Gatto GJ, Jr, Geisbrecht BV, Gould SJ, Berg JM. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 2000;7:1091–1095. doi: 10.1038/81930. [DOI] [PubMed] [Google Scholar]
- 30.Stanley WA, Filipp FV, Kursula P, Schuller N, Erdmann R, Schliebs W, Sattler M, Wilmanns M. Recognition of a functional peroxisome type 1 target by the dynamic import receptor Pex5p. Mol. Cell. 2006;24:653–663. doi: 10.1016/j.molcel.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanley WA, Pursiainen NV, Garman EF, Juffer AH, Wilmanns M, Kursula P. A previously unobserved conformation for the human Pex5p receptor suggests roles for intrinsic flexibility and rigid domain motions in ligand binding. BMC Struct. Biol. 2007;7:24. doi: 10.1186/1472-6807-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Klei IJ, Veenhuis M. PTS1-independent sorting of peroxisomal matrix proteins by Pex5p. Biochim. Biophys. Acta. 2006;1763:1794–1800. doi: 10.1016/j.bbamcr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 33.Petriv OI, Tang L, Titorenko VI, Rachubinski RA. A new definition for the consensus sequence of the peroxisome targeting signal type 2. J. Mol. Biol. 2004;341:119–134. doi: 10.1016/j.jmb.2004.05.064. [DOI] [PubMed] [Google Scholar]
- 34.Braverman N, Steel G, Obie C, Moser A, Moser H, Gould SJ, Valle D. Human PEX7 encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nat. Genet. 1997;15:369–376. doi: 10.1038/ng0497-369. [DOI] [PubMed] [Google Scholar]
- 35.Elgersma Y, Elgersma-Hooisma M, Wenzel T, McCaffery JM, Farquhar MG, Subramani S. A mobile PTS2 receptor for peroxisomal protein import in Pichia pastoris. J. Cell Biol. 1998;140:807–820. doi: 10.1083/jcb.140.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghys K, Fransen M, Mannaerts GP, Van Veldhoven PP. Functional studies on human Pex7p : Subcellular localisation and interaction with proteins containing a peroxisome targeting signal type 2 and other peroxins. Biochem. J. 2002;365:41–50. doi: 10.1042/BJ20011432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marzioch M, Erdmann R, Veenhuis M, Kunau WH. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 1994;13:4908–4918. doi: 10.1002/j.1460-2075.1994.tb06818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pilar AV, Madrid KP, Jardim A. Interaction of Leishmania PTS2 receptor peroxin 7 with the glycosomal protein import machinery. Mol. Biochem. Parasitol. 2008;158:72–81. doi: 10.1016/j.molbiopara.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Zhang JW, Lazarow PB. Peb1p (Pas7p) is an intraperoxisomal receptor for the NH2-terminal, type 2, peroxisomal targeting sequence of thiolase: Peb1p itself is targeted to peroxisomes by an NH2-terminal peptide. J. Cell Biol. 1996;132:325–334. doi: 10.1083/jcb.132.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukai S, Fujiki Y. Molecular mechanisms of import of peroxisome-targeting signal type 2 (PTS2) proteins by PTS2 receptor Pex7p and PTS1 receptor Pex5pL. J. Biol. Chem. 2006;281:37311–37320. doi: 10.1074/jbc.M607178200. [DOI] [PubMed] [Google Scholar]
- 41.Otzen M, Wang D, Lunenborg MG, van der Klei IJ. Hansenula polymorpha Pex20p is an oligomer that binds the peroxisomal targeting signal 2 (PTS2) J. Cell Sci. 2005;118:3409–3418. doi: 10.1242/jcs.02463. [DOI] [PubMed] [Google Scholar]
- 42.Titorenko VI, Smith JJ, Szilard RK, Rachubinski RA. Pex20p of the yeast Yarrowia lipolytica is required for the oligomerization of thiolase in the cytosol and for its targeting to the peroxisome. J. Cell Biol. 1998;142:403–420. doi: 10.1083/jcb.142.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Einwächter H, Sowinski S, Kunau WH, Schliebs W. Yarrowia lipolytica Pex20p, Saccharomyces cerevisiae Pex18p/Pex21p and mammalian Pex5pL fulfil a common function in the early steps of the peroxisomal PTS2 import pathway. EMBO Rep. 2001;2:1035–1039. doi: 10.1093/embo-reports/kve228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sichting M, Schell-Steven A, Prokisch H, Erdmann R, Rottensteiner H. Pex7p and Pex20p of Neurospora crassa function together in PTS2-dependent protein import into peroxisomes. Mol. Biol. Cell. 2003;14:810–821. doi: 10.1091/mbc.E02-08-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thoms S, Debelyy MO, Nau K, Meyer HE, Erdmann R. Lpx1p is a peroxisomal lipase required for normal peroxisome morphology. FEBS J. 2008;275:504–514. doi: 10.1111/j.1742-4658.2007.06217.x. [DOI] [PubMed] [Google Scholar]
- 46.Yang X, Purdue PE, Lazarow PB. Eci1p uses a PTS1 to enter peroxisomes: either its own or that of a partner, Dci1p. Eur. J. Cell Biol. 2001;80:126–138. doi: 10.1078/0171-9335-00144. [DOI] [PubMed] [Google Scholar]
- 47.Agne B, Meindl NM, Niederhoff K, Einwachter H, Rehling P, Sickmann A, Meyer HE, Girzalsky W, Kunau WH. Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol. Cell. 2003;11:635–646. doi: 10.1016/s1097-2765(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 48.Hazra PP, Suriapranata I, Snyder WB, Subramani S. Peroxisome remnants in pex3Δ cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic. 2002;3:560–574. doi: 10.1034/j.1600-0854.2002.30806.x. [DOI] [PubMed] [Google Scholar]
- 49.Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel JA, Veenhuis M, Kunau WH. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- 50.Madrid KP, Jardim A. Peroxin 5-peroxin 14 association in the protozoan Leishmania donovani involves a novel protein-protein interaction motif. Biochem. J. 2005;391:105–114. doi: 10.1042/BJ20050328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Urquhart AJ, Kennedy D, Gould SJ, Crane DI. Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J. Biol. Chem. 2000;275:4127–4136. doi: 10.1074/jbc.275.6.4127. [DOI] [PubMed] [Google Scholar]
- 52.Azevedo JE, Schliebs W. Pex14p, more than just a docking protein. Biochim. Biophys. Acta. 2006;1763:1574–1584. doi: 10.1016/j.bbamcr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Cyr N, Madrid KP, Strasser R, Aurousseau M, Finn R, Ausio J, Jardim A. Leishmania donovani Peroxin 14 undergoes a marked conformational change following association with Peroxin 5. J. Biol. Chem. 2008;283:31488–31499. doi: 10.1074/jbc.M803529200. [DOI] [PubMed] [Google Scholar]
- 54.Oliveira ME, Reguenga C, Gouveia AM, Guimaraes CP, Schliebs W, Kunau WH, Silva MT, Sa-Miranda C, Azevedo JE. Mammalian Pex14p: membrane topology and characterisation of the Pex14p-Pex14p interaction. Biochim. Biophys. Acta. 2002;1567:13–22. doi: 10.1016/s0005-2736(02)00635-1. [DOI] [PubMed] [Google Scholar]
- 55.Stein K, Schell-Steven A, Erdmann R, Rottensteiner H. Interactions of Pex7p and Pex18p/Pex21p with the peroxisomal docking machinery: implications for the first steps in PTS2 protein import. Mol. Cell. Biol. 2002;22:6056–6069. doi: 10.1128/MCB.22.17.6056-6069.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bottger G, Barnett P, Klein AT, Kragt A, Tabak HF, Distel B. Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner. Mol. Biol. Cell. 2000;11:3963–3976. doi: 10.1091/mbc.11.11.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams C, Distel B. Pex13p: docking or cargo handling protein? Biochim. Biophys. Acta. 2006;1763:1585–1591. doi: 10.1016/j.bbamcr.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 58.Rayapuram N, Subramani S. The importomer--a peroxisomal membrane complex involved in protein translocation into the peroxisome matrix. Biochim. Biophys. Acta. 2006;1763:1613–1619. doi: 10.1016/j.bbamcr.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 59.Kerssen D, Hambruch E, Klaas W, Platta HW, de Kruijff B, Erdmann R, Kunau WH, Schliebs W. Membrane association of the cycling peroxisome import receptor Pex5p. J. Biol. Chem. 2006;281:27003–27015. doi: 10.1074/jbc.M509257200. [DOI] [PubMed] [Google Scholar]
- 60.Nicolay K, Veenhuis M, Douma AC, Harder W. A 31P NMR study of the internal pH of yeast peroxisomes. Arch. Microbiol. 1987;147:37–41. doi: 10.1007/BF00492902. [DOI] [PubMed] [Google Scholar]
- 61.Jankowski A, Kim JH, Collins RF, Daneman R, Walton P, Grinstein S. In situ measurements of the pH of mammalian peroxisomes using the fluorescent protein pHluorin. J. Biol. Chem. 2001;276:48748–48753. doi: 10.1074/jbc.M109003200. [DOI] [PubMed] [Google Scholar]
- 62.Dansen TB, Wirtz KW, Wanders RJ, Pap EH. Peroxisomes in human fibroblasts have a basic pH. Nat. Cell Biol. 2000;2:51–53. doi: 10.1038/71375. [DOI] [PubMed] [Google Scholar]
- 63.van Roermund CW, de Jong M, van Marle IJLJ, Dansen TB, Wanders RJ, Waterham HR. The peroxisomal lumen in Saccharomyces cerevisiae is alkaline. J. Cell Sci. 2004;117:4231–4237. doi: 10.1242/jcs.01305. [DOI] [PubMed] [Google Scholar]
- 64.Dansen TB, Pap EHW, Wanders RJ, Wirtz KW. Targeted fluorescent probes in peroxisome function. Histochem. J. 2001;33:65–69. doi: 10.1023/a:1017927728892. [DOI] [PubMed] [Google Scholar]
- 65.Harper CC, Berg JM, Gould SJ. PEX5 binds the PTS1 independently of Hsp70 and the peroxin PEX12. J. Biol. Chem. 2003;278:7897–7901. doi: 10.1074/jbc.M206651200. [DOI] [PubMed] [Google Scholar]
- 66.Oliveira ME, Gouveia AM, Pinto RA, Sa-Miranda C, Azevedo JE. The Energetics of Pex5p-mediated peroxisomal protein import. J. Biol. Chem. 2003;278:39483–39488. doi: 10.1074/jbc.M305089200. [DOI] [PubMed] [Google Scholar]
- 67.Kiel JA, Emmrich K, Meyer HE, Kunau WH. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J. Biol. Chem. 2005;280:1921–1930. doi: 10.1074/jbc.M403632200. [DOI] [PubMed] [Google Scholar]
- 68.Kragt A, Voorn-Brouwer T, van den Berg M, Distel B. The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J. Biol. Chem. 2005;280:7867–7874. doi: 10.1074/jbc.M413553200. [DOI] [PubMed] [Google Scholar]
- 69.Platta HW, Girzalsky W, Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p. Biochem. J. 2004;384:37–45. doi: 10.1042/BJ20040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koller A, Snyder WB, Faber KN, Wenzel TJ, Rangell L, Keller GA, Subramani S. Pex22p of Pichia pastoris, essential for peroxisomal matrix protein import, anchors the ubiquitin-conjugating enzyme, Pex4p, on the peroxisomal membrane. J. Cell Biol. 1999;146:99–112. doi: 10.1083/jcb.146.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams C, van den Berg M, Geers E, Distel B. Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem. Biophys. Res.Commun. 2008;374:620–624. doi: 10.1016/j.bbrc.2008.07.054. [DOI] [PubMed] [Google Scholar]
- 72.Braverman N, Dodt G, Gould SJ, Valle D. An isoform of Pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum. Mol. Genet. 1998;7:1195–1205. doi: 10.1093/hmg/7.8.1195. [DOI] [PubMed] [Google Scholar]
- 73.Matsumura T, Otera H, Fujiki Y. Disruption of the interaction of the longer isoform of Pex5p, Pex5pL, with Pex7p abolishes peroxisome targeting signal type 2 protein import in mammals. Study with a novel Pex5-impaired Chinese hamster ovary cell mutant. J. Biol. Chem. 2000;275:21715–21721. doi: 10.1074/jbc.M000721200. [DOI] [PubMed] [Google Scholar]
- 74.Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 75.Purdue PE, Lazarow PB. Peroxisome biogenesis. Annu. Rev. Cell Dev Biol. 2001;17:701–752. doi: 10.1146/annurev.cellbio.17.1.701. [DOI] [PubMed] [Google Scholar]
- 76.Hohfeld J, Veenhuis M, Kunau WH. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J. Cell Biol. 1991;114:1167–1178. doi: 10.1083/jcb.114.6.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsuzono Y, Kinoshita N, Tamura S, Shimozawa N, Hamasaki M, Ghaedi K, Wanders RJ, Suzuki Y, Kondo N, Fujiki Y. Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc. Natl. Acad. Sci. U. S. A. 1999;96:2116–2121. doi: 10.1073/pnas.96.5.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sacksteder KA, Jones JM, South ST, Li X, Liu Y, Gould SJ. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J. Cell Biol. 2000;148:931–944. doi: 10.1083/jcb.148.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.South ST, Gould SJ. Peroxisome synthesis in the absence of preexisting peroxisomes. J. Cell Biol. 1999;144:255–266. doi: 10.1083/jcb.144.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 81.Kragt A, Voorn-Brouwer T, van den Berg M, Distel B. Endoplasmic reticulum-directed Pex3p routes to peroxisomes and restores peroxisome formation in a Saccharomyces cerevisiae pex3Δ strain. J. Biol. Chem. 2005;280:34350–34357. doi: 10.1074/jbc.M505432200. [DOI] [PubMed] [Google Scholar]
- 82.Tam YY, Fagarasanu A, Fagarasanu M, Rachubinski RA. Pex3p initiates the formation of a preperoxisomal compartment from a subdomain of the endoplasmic reticulum in Saccharomyces cerevisiae. J Biol Chem. 2005;280:34933–34939. doi: 10.1074/jbc.M506208200. [DOI] [PubMed] [Google Scholar]
- 83.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J. Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Motley AM, Hettema EH. Yeast peroxisomes multiply by growth and division. J. Cell Biol. 2007;178:399–410. doi: 10.1083/jcb.200702167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Elgersma Y, Kwast L, van den Berg M, Snyder WB, Distel B, Subramani S, Tabak HF. Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S. cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J. 1997;16:7326–7341. doi: 10.1093/emboj/16.24.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Titorenko VI, Rachubinski RA. The endoplasmic reticulum plays an essential role in peroxisome biogenesis. Trends Biochem. Sci. 1998;23:231–233. doi: 10.1016/s0968-0004(98)01226-2. [DOI] [PubMed] [Google Scholar]
- 87.Hardesty B, Kramer G. Folding of a nascent peptide on the ribosome. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:41–66. doi: 10.1016/s0079-6603(00)66026-9. [DOI] [PubMed] [Google Scholar]
- 88.Hardesty B, Tsalkova T, Kramer G. Co-translational folding. Curr. Opin. Struct. Biol. 1999;9:111–114. doi: 10.1016/s0959-440x(99)80014-1. [DOI] [PubMed] [Google Scholar]
- 89.Lazarow PB. Peroxisome biogenesis: advances and conundrums. Curr. Opin. Cell Biol. 2003;15:489–497. doi: 10.1016/s0955-0674(03)00082-6. [DOI] [PubMed] [Google Scholar]
- 90.Titorenko VI, Mullen RT. Peroxisome biogenesis: the peroxisomal endomembrane system and the role of the ER. J. Cell Biol. 2006;174:11–17. doi: 10.1083/jcb.200604036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Raychaudhuri S, Prinz WA. Nonvesicular phospholipid transfer between peroxisomes and the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15785–15790. doi: 10.1073/pnas.0808321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Titorenko VI, Rachubinski RA. Spatiotemporal dynamics of the ER-derived peroxisomal endomembrane system. Int. Rev. Cell Mol. Biol. 2008;Chapter 5:191–244. doi: 10.1016/S1937-6448(08)01605-5. [DOI] [PubMed] [Google Scholar]
- 93.Eitzen GA, Szilard RK, Rachubinski RA. Enlarged peroxisomes are present in oleic acid-grown Yarrowia lipolytica overexpressing the PEX16 gene encoding an intraperoxisomal peripheral membrane peroxin. J. Cell Biol. 1997;137:1265–1278. doi: 10.1083/jcb.137.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Platta HW, Erdmann R. The peroxisomal protein import machinery. FEBS Lett. 2007;581:2811–2819. doi: 10.1016/j.febslet.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 95.Boukh-Viner T, Guo T, Alexandrian A, Cerracchio A, Gregg C, Haile S, Kyskan R, Milijevic S, Oren D, Solomon J, Wong V, Nicaud JM, Rachubinski RA, English AM, Titorenko VI. Dynamic ergosterol-and ceramide-rich domains in the peroxisomal membrane serve as an organizing platform for peroxisome fusion. J. Cell Biol. 2005;168:761–773. doi: 10.1083/jcb.200409045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Snyder WB, Koller A, Choy AJ, Subramani S. The peroxin Pex19p interacts with multiple, integral membrane proteins at the peroxisomal membrane. J Cell Biol. 2000;149:1171–1178. doi: 10.1083/jcb.149.6.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Faber KN, Heyman JA, Subramani S. Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol. Cell. Biol. 1998;18:936–943. doi: 10.1128/mcb.18.2.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Titorenko VI, Rachubinski RA. Peroxisomal membrane fusion requires two AAA family ATPases, Pex1p and Pex6p. J. Cell Biol. 2000;150:881–886. doi: 10.1083/jcb.150.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Doherty KR, McNally EM. Repairing the tears: dysferlin in muscle membrane repair. Trends Mol. Med. 2003;9:327–330. doi: 10.1016/s1471-4914(03)00136-9. [DOI] [PubMed] [Google Scholar]
- 100.Washington NL, Ward S. FER-1 regulates Ca2+-mediated membrane fusion during C. elegans spermatogenesis. J. Cell Sci. 2006;119:2552–2562. doi: 10.1242/jcs.02980. [DOI] [PubMed] [Google Scholar]
- 101.McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell. 2005;17:3513–3531. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mullen RT, Trelease RN. The ER-peroxisome connection in plants: development of the "ER semi-autonomous peroxisome maturation and replication" model for plant peroxisome biogenesis. Biochim. Biophys. Acta. 2006;1763:1655–1668. doi: 10.1016/j.bbamcr.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 103.Titorenko VI, Rachubinski RA. Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol. Cell. Biol. 1998;18:2789–2803. doi: 10.1128/mcb.18.5.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haan GJ, Baerends RJ, Krikken AM, Otzen M, Veenhuis M, van der Klei IJ. Reassembly of peroxisomes in Hansenula polymorpha pex3 cells on reintroduction of Pex3p involves the nuclear envelope. FEMS Yeast Res. 2006;6:186–194. doi: 10.1111/j.1567-1364.2006.00037.x. [DOI] [PubMed] [Google Scholar]
- 105.Geuze HJ, Murk JL, Stroobants AK, Griffith JM, Kleijmeer MJ, Koster AJ, Verkleij AJ, Distel B, Tabak HF. Involvement of the endoplasmic reticulum in peroxisome formation. Mol. Biol. Cell. 2003;14:2900–2907. doi: 10.1091/mbc.E02-11-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Karnik SK, Trelease RN. Arabidopsis peroxin 16 coexists at steady state in peroxisomes and endoplasmic reticulum. Plant Physiol. 2005;138:1967–1981. doi: 10.1104/pp.105.061291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Karnik SK, Trelease RN. Arabidopsis peroxin 16 trafficks through the ER and an intermediate compartment to pre-existing peroxisomes via overlapping molecular targeting signals. J. Exp. Bot. 2007;58:1677–1693. doi: 10.1093/jxb/erm018. [DOI] [PubMed] [Google Scholar]
- 108.Lisenbee CS, Karnik SK, Trelease RN. Overexpression and mislocalization of a tail-anchored GFP redefines the identity of peroxisomal ER. Traffic. 2003;4:491–501. doi: 10.1034/j.1600-0854.2003.00107.x. [DOI] [PubMed] [Google Scholar]