Abstract
GATA-3 is a zinc finger transcription factor that is expressed in T cell lineages as well as in the nervous system during development. In this study, we report that forced expression of GATA-3 resulted in an increased number of dopamine β-hydroxylase (DBH)-expressing neurons in primary neural crest stem cell (NCSC) culture, suggesting that the DBH gene may be a downstream target gene of GATA-3. GATA-3 robustly transactivates the promoter function of the noradrenaline (NA)-synthesizing DBH gene, via two specific upstream promoter domains; one at -62 to -32 bp and the other at -891 to -853 bp. Surprisingly, none of these domains contain GATA-3 binding sites but encompass binding motifs for transcription factors Sp1 and AP4, respectively. Protein-protein interaction analyses both in vitro and in vivo and chromatin immunoprecipitation (ChIP) assays showed that GATA-3 effects its transcriptional regulatory function through physical interactions with these transcription factors.
Keywords: GATA-3, Sp1, AP4, siRNA, ChIP assay, dopamine β-hydroxylase, protein-protein interaction
Introduction
Extracellular signaling molecules from tissues surrounding neural plate induce neuronal differentiation and specification during vertebrate development. These signals turn on the expression of downstream transcription factors that regulate target genes required for the determination of general and specific neuronal phenotype. Thus, transcriptional control has been demonstrated as a critical mechanism for specification of individual neurons (e.g. the types of neurotransmitter, neurotransmitter transporter, receptors and channel proteins expressed by a given cell) in vertebrate organisms [1-4].
Numerous studies have identified major signaling molecules (e.g. bone morphogenetic proteins (BMPs) and cAMP) and key fate-determining transcription factors (e.g. Mash1/Cash1, Phox2a, Phox2b, and dHand) that are critical for the NA phenotype specification during sympathetic nervous system (SNS) development [3, 5, 6]. Among the candidate factors, Mash1 and Phox2b (whose expression is induced by BMPs) are considered to be master regulators because both loss- and gain-of-function analyses showed that they are necessary and sufficient for SNS development. Another potential candidate is dHand whose misexpression can induce the NA phenotype in developing chick embryo [7]. The level of tyrosine hydroxylase (TH) and DBH was reduced in conditional dHand knockout mouse suggesting its role in the SNS development [8]. Interestingly, Phox2a was dispensable for SNS development although it was necessary for the development of the central NA cell groups in the locus coeruleus [9]. Thus, the transcriptional code for NA neuron development appears to be somewhat different in the central nervous system (CNS) and peripheral nervous system (PNS).
The zinc finger transcription factor GATA-3, which is a T cell-specific DNA binding factor [10-12], is essential for the development of T cell lineage and is a master regulator for the differentiation of type 2 T helper (Th2) cells (reviewed in [13-15]). GATA-3-/- embryos die between embryonic day 11 and 12 and display massive internal bleeding, marked growth retardation, severe deformities of the brain and spinal cord, and gross fetal liver haematopoiesis aberrations [16]. Surprisingly, NA deficiency was a major cause of its embryonic lethality [17]. Consistent with this, GATA-3 null mutation led to reduced accumulation of TH and DBH mRNAs in the SNS, whereas several other SNS genes were unaffected [17]. Furthermore, the same group recently showed that both TH and DBH expression levels were markedly altered in purified chromaffin cells from GATA-3 mutant mice and were partially restored by sympathoadrenal-specific GATA-3 expression [18]. These in vivo studies suggest that GATA-3 may affect the transcription of both the TH and DBH genes. Indeed, we recently found that GATA-3 is able to activate the transcriptional activity of the TH gene suggesting that it is a direct target of GATA-3 [19]. Interestingly, GATA-3 activated the TH promoter function via interacting with CREB that binds to a proximal TH promoter at -61 to -39 bp.
Here, we addressed whether DBH is another target gene of GATA-3 and, if so, by what mechanism GATA-3 may control DBH gene transcription. We show that forced expression of GATA-3 resulted in increased DBH-expressing neurons among NCSC culture, and GATA-3 robustly transactivates the transcriptional activity of DBH gene via two promoter subdomains. Interestingly, our promoter analysis, protein interaction assays, and ChIP assays show that GATA-3 regulates the DBH gene promoter via distinct protein-protein interactions with the known transcription factors, Sp1 and AP4. Taken together, we propose that GATA-3 may directly regulate transcription of the DBH gene by novel and distinct protein-protein interactions.
Experimental procedure
Overexpression of GATA-3 in NCSC and in situ hybridization (ISH)
Primary culture of trunk region of quail eggs were performed as described [19]. NCSCs of Japanese quail (Conturnix japonica) eggs were infected with 5 MOI (multiplicity of infection) of RCAS-cGATA3 and control viruses in 24 well plates. Neural crest cells were cultured for 5 day, fixed in 4% formaldehyde. DBH expression in fixed NCSC was visualized by ISH. Partial chicken DBH cDNA was amplified with cDNA, which was made from chick embryos, using primer set, 5′-CACCACATAATCATGTATGAGCCA-3′ and 5′-GTGTGGAGCTGGGAGGCGAAGATG-3′. Fixed neural crest cells were hybridized with digoxigenin (DIG)-labeled chick DBH probe and the signal was visualized by treatment with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate. Total cell count was obtained by staining with Hoechst dye. Ten random fields containing stained cells for DBH were selected and counted.
Cell culture and transient transfection assays
Human neuroblastoma SK-N-BE(2)C, HeLa, and 293T cell lines (ATCC) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (Hyclone), 100 μg/ml of streptomycin, and 100 units/ml of penicillin in a CO2 incubator. Transfection was performed by the calcium phosphate method, using effector plasmids expressing GATA-3 along with various DBH- reporter constructs [20]. Each 60-mm dish was transfected with an equimolar amount (0.5 pmol) of each reporter construct, 0.5 μg of pRSV-LUC, 0.1 pmol of the effector plasmid, and pUC19 plasmid to a total of 5 μg DNA. To correct for differences in transfection efficiencies among different DNA precipitates, CAT activity was normalized by that of luciferase. CAT and luciferase activities were assayed as previously described [20].
siRNA transfection and Western blot Analysis
SK-N-BE(2)C cells were plated on polystyrene tissue culture dishes at a density of 1∼2×106 cells/60 mm plate, and transfected with 3 μg of GATA-3 siRNA or control siRNA (Santa Cruz Biotechnology, Santa Cruz, CA) using Lipofectamine Plus reagent (Invitrogen, San Diego, CA) according to the manufacturer’s instructions. After 24h or 72h, the cells were then subjected to the following experiments. For Western blot analysis, SK-N-BE(2)C cells were washed with PBS, harvested, and lysed with RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 50 mM Tris-Cl, pH 7.5). Equal amounts of protein were separated on a 10% SDS polyacrylamide gel and transblotted on polyvinylidene difluoride-nitrocellulose filters. Membranes were incubated with anti-GATA-3 (1:1,000; Santa Cruz Biotechnology), anti-DBH (1:4,000), anti-Sp1 (1:2,000; Santa Cruz Biotechnology), or anti-AP4 (1:2,000, Chemicon, Temecula, CA) and then incubated with horseradish peroxide-conjugated secondary antibody (1:5,000; Santa Cruz Biotechnology). Specific bands were visualized using an Enhanced Chemiluminescence (ECL) detection kit (Amersham Biosciences, Piscataway, NJ). The blots were probed with anti-β-actin antibody (1: 2,000; Sigma-Aldrich, St. Louis, MO) to serve as a control for gel loading.
Plasmids construction
The DBH2600CAT, DBH978CAT, DBH262CAT, DBH62CAT, and DBH32CAT reporter constructs contain 2,600, 978, 262, 62, and 32 bp upstream sequences of the human DBH gene, respectively, fused to the bacterial CAT gene [21, 22]. The AvaI/BamHI, EcoNI/BamHI, and BglII/BamHI fragments, which contain upstream sequences of the human DBH gene, of DBH978CAT was cloned at pBLCAT3′ to make DBH925CAT, DBH891CAT, and DBH831CAT, respectively. PCR was performed to amplify 853 bp upstream sequences of the human DBH gene and clone at pBLCAT3′ to make DBH853CAT. Base substitutions at the Sp1 sites and AP4 sites were generated in the context of DBH62CAT and DBH978CAT using the transformer™ Site-directed mutagenesis kit (Clontech, Palo Alto, CA), respectively. Oligonucleotides 5′ - GCT GCC TGG ACC CAC TAT GTT CAG GAC CAG GGC ATA AAT GGC - 3′ and 5′ - GGA GAA GCA GGG CTG CGG CAG AAA ACC CTG AGA GGC TTC AAA TT - 3′ were used to mutate Sp1 and AP4 site, respectively (underlined bases represent mutant sequences). The full length of AP4 was made by RT-PCR from SK-N-BE(2)C mRNA using primers, 5′- AAA AAG GAT CCA TGG AGT ATT TCA TGG TGC CCA CT - 3′ and 5′ - AAA AAG AAT TCT CAG GGA AGC TCC CCG TCC CCC GA - 3′, digested with BamHI and EcoRI, and subcloned into the pcDNA3.1/Zeo, resulting in the AP4-expressing plasmid, pcDNA/AP4.
GST pull-down assays
pGEX-2T (Pharmacia) was used to express GATA-3, SP1, and AP4 which were fused to Glutathione S-transferase (GST). Deletion constructs were made by PCR and cloned in pGEX-2T. For the expression of the GST-fusion proteins, E. coli BL21 (DE3) harboring the expression plasmids were induced by addition of 0.5 mM of isopropyl-β-D-thiogalactopyranoside (IPTG). The proteins were purified according to the manufacturer’s protocol (Pharmacia Biotech). The purified proteins were stored in aliquots containing 10% glycerol at -70°C. For the GST pull-down assay, in vitro translated [35S]-methionine labeled proteins were made using TNT-coupled wheat germ extract system (Promega). Five μg of GST or GST fusion proteins were bound to glutathione Sepharose beads and incubated with the labeled proteins for 2 h at 4°C in binding buffer (50 mM potassium phosphate (pH 7.5), 150 mM KCl, 1 mM MgCl2, 10% glycerol, 1 mM Triton X-100, and protease inhibitor cocktail (Roche)). Beads were washed with binding buffer three times and bound proteins were eluted by boiling in 20 μl of SDS loading buffer (60 mM Tris-HCl (pH 7.0), 2% SDS, 6% glycerol, 0.1M dithiothreitol, 0.01% bromophenol blue). The products were subjected to SDS-polyacrylamide gel electrophoresis, fluorographic reagent, and autoradiography.
In vivo Co-IP
For in vivo Co-IP, 293T cells were harvested 48 h after transfection with GFP-tagged GATA3 and Flag-tagged AP4 constructs driven by a CMV promoter. Cell extracts were treated with either anti-GFP or M2 anti-Flag antibody (1:1,000), and then with Protein A-Sepharose. Bound proteins were analyzed by Western blotting. Detections were performed using anti-GFP (1:5,000), and M2 anti-FLAG (1:5,000) antibodies. For in vivo GST-pull down assay, 293T cells transfected with GST-Sp1 (aa 613 - 716) and Flag-GATA3, which are expressed by CMV promoter, were harvested and treated with glutathione Sepharose beads. The bound proteins were detected with anti-Flag antibody using ECL western blotting system (Amersham).
Electrophoretic mobility shift assay (EMSA)
Sense and antisense oligonucleotides corresponding to the sequences of AP4 binding motif in the DBH promoter, its mutant form, and the AP4 binding site of SV40 [23] were synthesized with the following nucleotide sequences: 5′- CTGCGGCCAGCTGCCCTGA - 3′ and 5′- CTCAGGGCAGCTGGCCGCA - 3′, 5′ - CTGCGGCAGAAAACCCTGA - 3′ and 5′ - CTCAGGGTTTTCTGCCGCA - 3′, 5′ - AAGAACCAGCTGTGGAAT - 3′ and 5′ - CATTCCACAGCTGGTTCT - 3′, respectively (underlined bases represent mutant sequences). Oligonucleotides representing the Sp1 binding site were used as a negative control. EMSA was performed using in vitro translated proteins or nuclear extracts from HeLa or SK-N-BE(2)C as described [24] and supershift assay was performed with AP4 specific antibody (Santa Cruz Biotechnology).
Chromatin immuno-precipitation (ChIP) assay
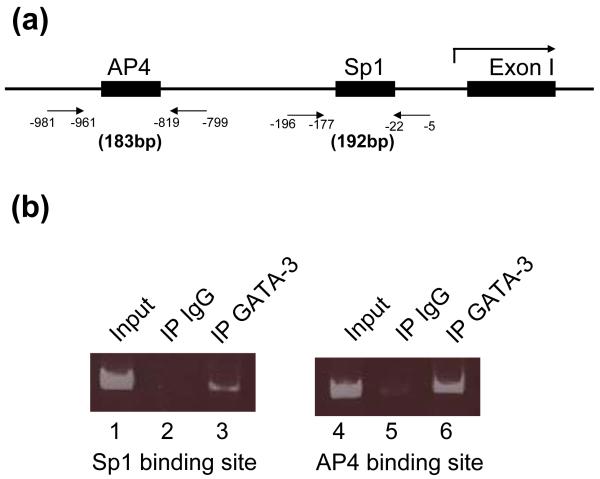
ChIP assay was performed according to manufacturer’s protocol (Upstate, Lake Placid, NY). Briefly, 3 × 106 cells of SK-N-BE(2)C cells were plated in 100 mm plates and cultured for 24 h. Cells were cross-linked with 1% formaldehyde for 10 min and harvested in the presence of protease inhibitor (EDTA-free Complete, Roche). These cells were then lysed and sonicated to generate 200-500 bp DNA fragments. One tenth of the lysates was used for input control. The remaining lysates were divided by half and treated with 1 μg of mouse monoclonal anti-GATA3 antibody (HG3-31, Santa Cruz Biotechnology, Santa Cruz, CA) or normal rabbit IgG as a negative control overnight at 4°C. After incubating Salmon sperm DNA/Protein A agarose slurry to immunoprecipitated complexes, the precipitates were extensively washed, eluted in elution buffer (1% SDS, 0.1M NaHCO3). The cross linked protein-DNA complexes were reversed by incubating NaCl. The DNA was recovered by ethanol precipitation following phenol extraction and resuspended in 40 μl of distilled water. PCR was performed to detect specifically bound DNA using 2X PCR Premix IN buffer (Epicentre, Madison, WI) using 5 μl of the resuspended sample as a template at 94°C 30sec, 55°C 30sec, 72°C 30sec for 40 cycles with primer sets in 25 μl reaction volume. Primers for GATA-3 and Sp1 binding sites are 5′- TTCAACTCCCACTGATGACG -3′ (-196 to -177 bp) and 5′- TCTCTGGTCCCACCTGGC -3′ (-5 to -22 bp). Primers for GATA-3 and AP4 binding sites are 5′- CAGTCTACTTGCGGGAGAGG -3′ (-981 to -961 bp) and 5′- AAACCCACAGCTTCCTCTTG -3′(-799 to -819 bp) (the numbers in the parenthesis indicate nucleotide position in the DBH promoter).
Results
Forced expression of GATA-3 increases the number of DBH-expressing cells in primary NCSC culture
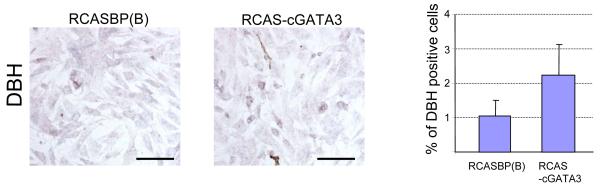
GATA-3 is expressed in sympathetic ganglia (SG) of mouse embryos [25], and its null mutation leads to reduced accumulation of TH and DBH mRNA in the SNS [17]. We recently showed that TH may be a direct target gene of GATA-3 [19]. To address whether GATA-3 can directly affect fate induction of NCSC lineages, we performed a gain-of-function analysis of GATA-3 by infecting quail primary NCSC with a chick GATA3-expressing avian virus construct, RCAS-cGATA3. NCSCs were infected with 5 MOI of RCAS-cGATA3 or RCASBP(B) viruses and cultured for 5 days. Forced expression of GATA-3 led to an approximately 2-fold increase of DBH-expressing cells, as examined by ISH (Fig. 1). The total number of NCSCs was not detectably changed under these conditions (data not shown & [19]).
Fig. 1.
GATA-3 overexpression increases the number of DBH expressing cells in neural crest cell culture. Primary neural crest cell cultures were infected with RCASBP(B) alone or RCAS-cGATA3 viruses. ISH were performed to detect DBH expression. DBH positive cells are represented as a percentage. Data are presented as mean ±SEM from four independent experiments. Bar: 300 μm.
GATA-3 directly activates the transcriptional activity of the DBH gene
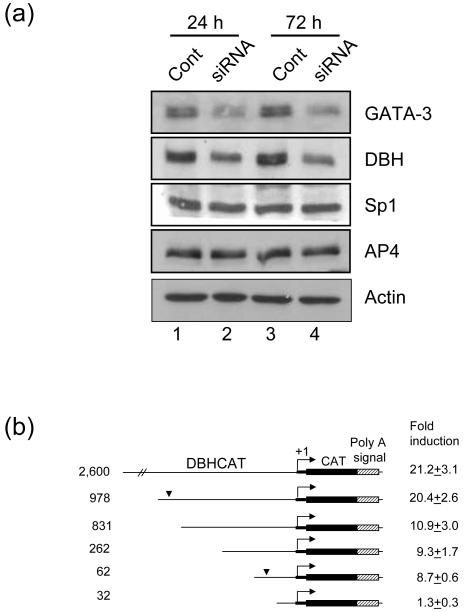
Next we suppressed expression of GATA-3 by siRNA in SK-N-BE(2)C, where both GATA-3 and DBH are expressed, to address whether DBH is downstream of GATA-3. By GATA-3 specific siRNA treatment, expression of GATA-3 and DBH was reduced, while that of general transcriptipn factors, Sp1 and AP4, was not changed (Fig. 2a lane 2, 4). This siRNA experiment suggests that expression of DBH is regulated by GATA-3.
Fig. 2.
GATA-3 regulates expression of DBH. (a) The expression of DBH is reduced by GATA-3 siRNA. SK-N-BE(2)C cells were transfected with GATA-3 siRNA (lane 2, 4) or control siRNA (lane 1, 3) using Lipofectamine. After 24 (lane 1, 2) or 72 h (lane 3, 4) of transfection, cells were harvested and Western blot analyses were performed. The expression of GATA-3, DBH, Sp1, AP4 was detected with their specific antibodies as indicated. β-actin was detected as a loading control. (b) GATA-3 transactivation of serially deleted DBH promoter. HeLa cells were cotransfected with DBH-CAT reporter plasmids and pcDNA/GATA3 or empty vector at a molar ratio of 0.2. Fold induction by effector plasmid cotransfection is presented as mean ± SEM value from six to nine independent samples. The numbers on the left of the diagram represent the size of the DBH promoter. The bent arrow represents the DBH transcription start site. The bold thick line denotes the 5′ untranslated sequences and the thin line denotes the 5′ upstream sequences of the DBH gene. The arrowhead represents the GATA-3 response region of the DBH promoter.
To address whether DBH is another immediate downstream target of GATA-3, we performed cotransfection assays using the DBH2.6CAT reporter gene construct. The reporter gene assays showed that GATA-3 robustly transactivates the DBH promoter activity in HeLa cells (>20-fold) (Fig. 2b). To locate the promoter domain(s) responsible for transactivation function of GATA-3, we performed cotransfection assays using various deletional reporter constructs of the DBH promoter in HeLa cells. As shown in Fig. 2b, this deletion analysis identified two upstream subdomains of the DBH gene that might be important for transactivation by GATA-3; one at -62 to -32 bp and the other at -978 to -831 bp. Interestingly, we could not identify any sequences having homology with the known GATA-3 binding consensus motif in the proximal and distal subdomains (see below).
The Sp1-binding site is required for the activation of the DBH proximal promoter by GATA-3
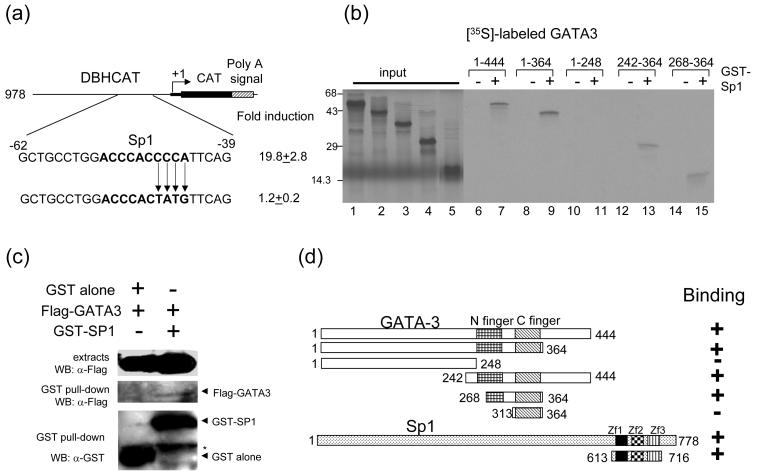
The only identifiable cis-regulatory element residing within the proximal subdomain between -62 and -32 bp of the DBH promoter is the Sp1-binding site (Fig. 3a). We have previously shown that this motif is in fact interacting with the transcription factor Sp1 and is critical for the basal promoter function of the DBH gene [22]. These observations suggest the possibility that GATA-3 regulates the DBH promoter function through this Sp1-binding site. To address this, we performed cotransfection assays using the mutant reporter construct DBH978(Sp1m)CAT, which contains base substitution mutations within the Sp1 site. As shown in Fig. 3a, the transactivation function of GATA-3 was almost completely abolished in this mutant construct. Thus, we conclude that the Sp1 site of the proximal DBH promoter is essential for the transactivation function of GATA-3.
Fig. 3.
GATA-3 physically interacts with Sp1. (a) Mutation of Sp1 binding site in DBH978CAT diminishes reporter gene activation by GATA-3. The bold characters represent the Sp1 binding site. (b) Localization of the interaction domains of GATA-3 and Sp1. In vitro translated [35S]-methionine labeled GATA-3 proteins were incubated with GST (lanes 6, 8, 10, 12, and 14) or with full-length GST-Sp1 (lanes 7, 9, 11, 13, and 15) bound to glutathione Sepharose beads. [35S]-methionine labeled input proteins are shown (lanes 1 (aa 1-144), 2 (aa 1-364), 3 (aa 1-248), 4 (aa 242-364) and 5 (aa 268-364)). The numbers at the top of the figure represent the amino acid residues of GATA3. (c) Interaction between GATA-3 and Sp1 in vivo. 293T cells were cotransfected with the Sp1 (aa 613 - 716), and empty vector or GATA-3 as indicated on the top. The cell lysates were precipitated with glutathione-Sepharose beads. Monoclonal anti-FLAG was used to detect the FLAG-tagged GATA-3 proteins. The blot was stripped and re-probed with the anti-GST to detect the precipitated Sp1proteins (bottom). Top, crude cell extracts; middle, immunoprecipitates; bottom, GST-Sp1. Asterisk indicates non-specific bands containing the heavy chain of IgG. (d) Summary of the interaction results of GATA3 with Sp1. Three zinc finger motifs (Zf1-3) of Sp1 are indicated.
Since the proximal domain responsible for transactivation by GATA-3 contains the Sp-1 binding site but not GATA-3 binding site, we speculated that this transactivation might occur via the protein-protein interaction between GATA-3 and Sp1 factors. To test this possibility we incubated in vitro translated [35S]-labeled GATA-3 proteins with GST-Sp1 fusion proteins bound to glutathione Sepharose beads. The full-length GATA-3 protein interacts with GST-Sp1 fusion proteins, while it does not interact with GST protein alone (Fig. 3b lanes 6, 7). To determine the minimal interaction domain of GATA-3, deleted fragments of GATA-3 were [35S]-labeled and incubated with GST-Sp1 or GST protein. This analysis showed that the minimal domain required for the interaction with Sp1 was amino acid residue 268-364 (Fig. 3b & d). This minimal region contains the C-terminal finger and the incomplete N-terminal finger of GATA-3. Interestingly, the GATA-3 deletion protein containing only the C finger region (aa 313 - 364) did not interact with Sp1 (Fig. 3d). Similar results were observed using GST-Sp1 deletion construct, containing three zinc finger motifs (aa 613 - 716) (data not shown).
Interaction between GATA-3 and Sp1 was confirmed by in vivo GST pull down assay. 293T cells transfected with Flag tagged GATA-3 and GST tagged Sp1 (aa 613 - 716) were lysed and incubated with glutathione Sepharose beads. Weak but clear in vivo interaction was detected between GATA-3 and Sp1 (Fig. 3c). Taken together, these results support the possibility that these protein-protein interactions may occur in vivo.
GATA-3 transactivates the distal human DBH promoter through a putative AP4-binding sequence motif
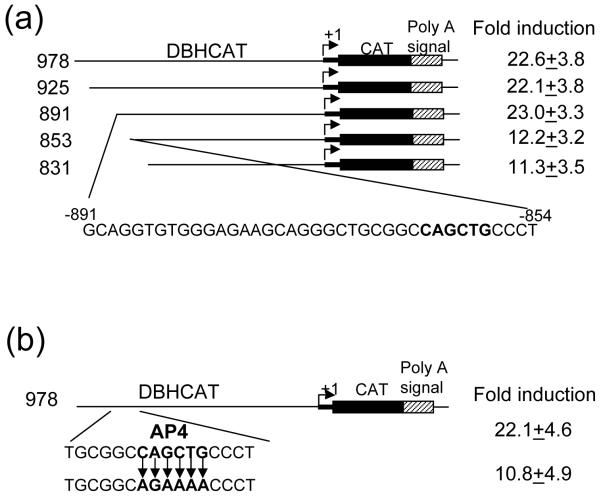
Through the use of additional deletion constructs we identified important nucleotide sequences located between -891 and -853 bp in the distal upstream domain of the DBH promoter (Fig. 4a). However, our DNase I footprinting and EMSA analyses showed that these sequences do not interact with GATA-3 (data not shown). These observations led us to hypothesize that GATA-3 may transactivate this distal upstream DBH promoter without direct DNA binding, similar to the proximal promoter. The GATA-3 response sequence of the distal DBH promoter was subjected to analysis to identify interacting transcription factor(s). The Transcription Factor Binding Site Search program (http://www.cbrc.jp/research/db/TFSEARCH.html) predicted the presence of an E-box motif (CANNTG) with a potential AP4 binding site (Fig. 4a). This sequence motif is an as-yet-unidentified cis-regulatory element in the DBH gene promoter. To test the functionality of this putative motif, we mutated the AP4 binding site by site-directed mutagenesis. The fold-induction of DBH978(AP4m)CAT by GATA-3 was similarly diminished by half compared to that of wild type (Fig. 4b).
Fig. 4.
GATA-3 transactivates the DBH promoter through AP4 binding site. (a) GATA-3 transactivation analysis of serially deleted human DBH promoter. (b) The AP4 binding site (CAGCTG) was mutated. Cotransfections were performed in HeLa cells and the same symbols are used as in Fig. 2.
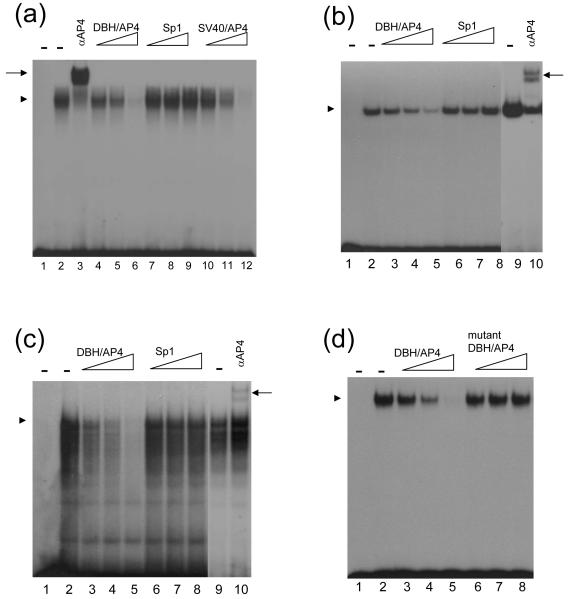
To further investigate the functionality of the putative AP4 site in the distal DBH promoter, we addressed whether it interacts with the transcription factor AP4. In vitro translated AP4 protein formed a complex with the oligonucleotide (DBH/AP4) containing the putative AP4 binding site present in DBH promoter (Fig. 5a, lane 2). This complex was completely abolished by incubation with a 1,000 molar excess of cold oligonucleotide (Fig. 5a, lane 6) or other AP4 binding motif, such as the one found on SV40 enhancer [23] (Fig. 5a, lane 12). However, the AP4 protein-DBH/AP4 DNA complex was not affected by addition of nonspecific Sp1 sequences (Fig. 5a, lanes 7-9). These results demonstrate the specificity of the complex. These specific complexes were also formed by incubation with nuclear extracts from HeLa or SK-N-BE(2)C cells (Fig, 5b & c). Formation of the specific complex was not inhibited when the mutant DBH/AP4 oligonucleotide was used as competitor (Fig. 5d). Furthermore, the identity of AP4 as the interacting protein with the E-box motif was confirmed by antibody supershift assays of the in vitro translated protein (Fig. 5a, lane 3), HeLa extracts (Fig. 5b, lane 10), and SK-N-BE(2)C extracts (Fig. 5c, lane 10). These binding assays show that the ubiquitously expressed AP4 protein physically interacts with the upstream DBH promoter subdomain at -863 to -858 bp and regulates DBH promoter function.
Fig. 5.
AP4 binds to the E-box motif of the DBH upstream promoter. Oligonucleotide DBH/AP4 containing the E-box (CANNTG) motif of the DBH promoter was radiolabeled and used as a probe. (a) Two μl of the in vitro translated AP4 proteins were incubated with the radiolabeled probe (lane 2) and 0.2 μg of AP4 antibody was added to the reaction (lane 3). For competition, 50-fold (lanes 4, 7, 10), 200-fold (lanes 5, 8, 11), or 1,000-fold (lanes 6, 9, 12) molar excess of unlabeled oligonucleotides DBH/AP4 (lanes 4-6), Sp1 (lanes 7-9), and SV40/AP4 (10-12) were added to the reaction mixture before the addition of radiolabeled probe. The same amount of in vitro translated protein using an empty vector was incubated with the probe and did not generate any complex (lane 1). (b, c) The 32P-labeled DBH/AP4 probe was incubated with 3 μg of HeLa (b) or 1 μg of SK-N-BE(2)C (c) nuclear extracts (lane 2, 9). For competition, 50-fold (lanes 3, 6), 200-fold (lanes 4, 7), or 1,000-fold (lanes 5, 8) molar excess of unlabeled oligonucleotides DBH/AP4 (lanes 3-5) and Sp1 (lanes 6-8) were added to the reaction mixture before the addition of radiolabeled probe. AP4 specific antibody was incubated with the probe and nuclear extracts (lane 10). In lane 1, the probe was incubated without nuclear extracts. (d) The 32P-labeled DBH/AP4 probe was incubated with 2 μl of in vitro translated AP4 proteins (lane 2). For competition, 50-fold (lanes 3, 6), 200-fold (lanes 4, 7), or 1,000-fold (lanes 5, 8) molar excess of unlabeled oligonucleotide wild type DBH/AP4 (lanes 3-5) and mutant type DBH/AP4 (lanes 6-8) were used. The same amounts of in vitro translated proteins using an empty vector were incubated with the probe and did not generate any complex (lane 1). Specific protein-DNA complex and supershifted complex were indicated by an arrow head and an arrow, respectively.
GATA-3 physically interacts with AP4
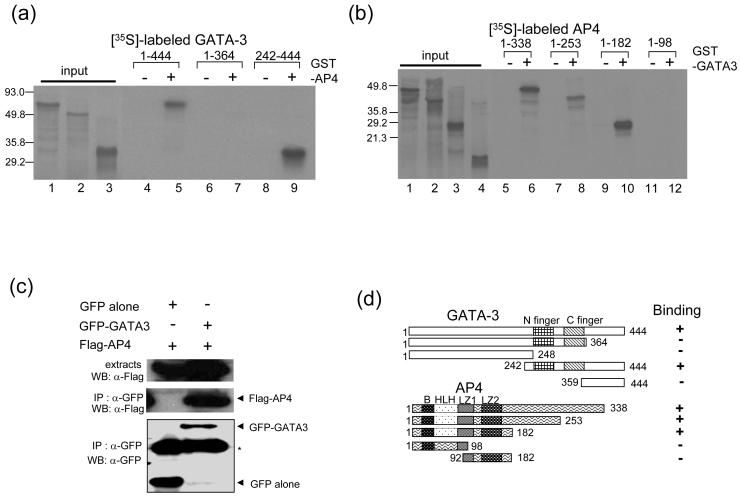
Next, we addressed the direct interaction between AP4 and GATA-3. We used in vitro translated [35S]-labeled full length GATA-3 and incubated it with purified GST or GST-AP4 bound to glutathione Sepharose beads. GST-AP4 retained [35S]-labeled GATA-3 on the glutathione Sepharose beads, while GST did not (Fig. 6a, lanes 4, 5). The GATA-3 deletion construct containing both zinc finger motifs and the C terminal region (aa 242-444) interacts with GST-AP4 (Fig. 6a, lanes 8, 9), while a smaller deletion construct (aa 359-444) failed to interact with GST-AP4 (data not shown; Fig. 6d). The full-length and deletion construct of AP4 (aa 1-253 and 1-182) interact with GST-GATA-3, while the deletion constructs (aa 1-98 and aa 92-182) failed to interact with GATA-3 (Fig. 6b). These results show that the N terminal portion of AP4 encompassing the basic region, helix-loop-helix, and leucine zipper is essential for interaction with GATA-3 (Fig. 6d). To test if GATA-3 interacts with this transcription factor in vivo, we used co-immunoprecipitation (Co-IP) assays. We co-transfected 293T cells with GFP tagged GATA-3 and Flag tagged AP4 to 293T cells. Expression of tagged transcription factors was confirmed by Western blot analysis. Cell lysates were precipitated with αGFP antibody and Western blot analysis performed with αFlag antibody to detect the presence of tagged proteins. In vivo interaction between GATA-3 and AP4 (Fig. 6c) was robustly detected.
Fig. 6.
GATA-3 physically interacts with AP4. (a, b) Determination of the interaction domains of GATA-3 and AP4. (a) In vitro translated [35S]-methionine labeled GATA-3 proteins were incubated with GST (lanes 4, 6, and 8) or full length GST-AP4 (lanes 5, 7, and 9), bound to glutathione Sepharose beads. [35S]-methionine labeled input proteins are shown (lanes 1 (aa 1-144), 2 (aa 1-364), and 3 (aa 242-444)). (b) In vitro translated [35S]-methionine labeled AP4 proteins were incubated with GST (lanes 5, 7, 9, and 11) or full-length GST-GATA3 (lanes 6, 8, 10, and 12) bound to glutathione Sepharose beads. [35S]-methionine labeled input proteins are shown (lanes 1 (aa 1-338), 2 (aa 1-253), 3 (aa 1-182), and 4 (aa 1-98)). (c) Interaction between GATA-3 and AP4 in vivo. 293T cells were cotransfected with the empty vector, GATA-3, and the AP4 as indicated on the top. The cell lysates were precipitated with an anti-GFP. Monoclonal anti-FLAG was used to detect the FLAG-tagged AP4 proteins. The blot was stripped and re-probed with the anti-GFP to detect the precipitated GATA-3 protein (bottom). Top, crude cell extracts; middle, immunoprecipitates; bottom, GFP-GATA3. Asterisk indicates non-specific bands containing the heavy chain of IgG. (d) Schematic representation of interactions between GATA-3 and AP4. The basic (B) region, helix-loop-helix (HLH) domain, and two leucine zipper domains (LZ1, 2) are indicated.
Next, we performed ChIP assays to determine whether the protein-protein interaction occurs on the DBH promoter in vivo. GATA-3 expressing SK-N-BE(2)C cells were cross-linked by 1% formaldehyde, and 200-500 bp DNAs were generated by sonication. Following immuno-precipitation, specifically bound DNA fragments were purified and PCR was performed using specific primer sets (Fig. 7). ChIP assay using GATA-3 specific antibody detected expected products which containing Sp1 (Fig. 7b, lane 3) and AP4 (Fig 7b, lane 6) binding sites in the DBH promoter. While ChIP assay using normal rabbit IgG showed very weak bands (Fig 7b, lanes 2, 5). These ChIP assays results further support protein-protein interaction between GATA-3 and Sp1 (or AP4).
Fig. 7.
ChIP analysis indicates in vivo interaction of GATA-3 with SP1 and AP4 on the DBH promoter. (a) Schematic drawing of primers to detect Sp1 and AP4 binding sites by PCR. The first exon of human DBH gene is shown (Exon I). The numbers indicate nucleotide position in the DBH promoter. (b) The protein-DNA complexes were immunoprecipitated using antibodies against GATA-3 (lane 3, 6). As a negative control, rabbit IgG was used (lane 2, 5). Lane 1 and 4 show input DNAs. PCR was performed with primer sets shown (a) as described in experimental procedure. One twelfth (lanes 1, 4) or one third (lanes 2, 3, 5, 6) of the reaction PCR products were run in 7% polyacrylamide gel.
Discussion
Cell-type specific gene expression is achieved by interactions between general transcription factor(s) and cell-type specific transcription factor(s). Our results indicate that the transcription factor GATA-3 activates the expression of NA specific gene, DBH, by interacting with general transcription factors, Sp1 and AP4. Transcription factors belonging to the GATA family contain two C4-type zinc finger domains and bind to the DNA motif WGATAR [26]. Among the six members of the GATA family, GATA-2/-3 are the only proteins expressed in neuronal cells [27]. GATA3 is best known as a master regulator of type 2 T helper (Th2) cell differentiation [28, 29].
We recently reported that GATA-3 directly transactivates the transcriptional activity of the TH gene, suggesting that it is a direct target of GATA-3 [19]. Our gain-of-function and knock down experiment suggested that DBH is regulated by GATA-3 (Figs. 1 & 2). Here, we hypothesized that the DBH gene is another immediate downstream target of GATA-3. Alternatively, GATA-3 can indirectly regulate DBH gene expression by regulating other transcription factors, e.g., Phox2a and 2b, which are known to transactivate DBH gene expression. We favor the first possibility because GATA-3 was able to transactivate the DBH promoter function (Figs. 3 & 4). Furthermore, our DBH promoter deletion and site-directed mutational analyses mapped the subdomain(s) that may mediate its responsiveness to GATA-3. Two domains of the DBH promoter at -62 to -32 bp and at -891 to -853 bp were critical for transactivation in response to GATA-3.
GATA family transcription factors bind to GATA-containing sequence motifs of the target genes [26]. Indeed, all known target genes of GATA-3 appear to be regulated via direct DNA-binding to GATA-containing sequence motifs [13-15]. Strikingly, our previous results showed that GATA-3 regulates promoter activity of TH by interaction with CREB without direct binding to the promoter [19]. Furthermore, two DBH promoter subdomains responsive to GATA-3, one at -62 to -32 and the other at -891 to -853, also do not contain any GATA-3 recognition motif. The proximal subdomain contains an Sp1-binding site that appears to be important for the responsiveness of the proximal DBH promoter to GATA-3. Indeed, our GST-pull down assays demonstrated that GATA-3 could interact with Sp1 (Fig. 3). For this interaction, a short fragment of GATA-3 (aa 268-364) encompassing the C-terminal finger and a portion of the N-terminal finger was sufficient. This result is in agreement with a previous study [30] showing that the GATA-1 fragment including the C-terminal finger and part of the N-terminal finger (aa 208-304) was able to interact with Sp1. In addition, we found that the three finger motifs (aa 613 to 716) of Sp1 were sufficient for full interaction with GATA-3 (Fig. 3d). Taken together, our results support the model that Sp1 may contribute to DBH transcription activation by GATA-3 via protein-protein interaction.
Another GATA-3 responsive subdomain was identified at -891 to -853 bp in the distal human DBH promoter. Interestingly, this region also did not have any functional GATA-3-binding site(s), as examined by EMSA, DNase I footprinting, site-directed mutation, and transfection analyses (Fig. 4 and data not shown). Instead, we have identified a functional AP4 binding site in this region. AP4, a basic helix-loop-helix (bHLH) transcription factor, is ubiquitously expressed and contains multiple dimerization domains with the binding motif CAGCTG [23]. Mutation of this AP4 site diminished the responsiveness to GATA-3 (Fig. 4), suggesting the latter transactivates the DBH promoter via additional physical interaction with AP4. Indeed, our GST-pull down, in vivo Co-IP and ChIP assays (Figs. 6 & 7) supported that GATA-3 physically interacts with AP4 in the DBH promoter. The C-terminal half (aa 242-444) of GATA-3 and the N-terminal residue (aa 1-182) of AP4 were sufficient for physical interaction with each other.
Critical signaling molecules and fate-determining transcription factors are beginning to be identified in sympathetic neuron development. Gene knockout studies showed that Phox2a is genetically downstream of Mash1 and Phox2b [31-34]. Consistent with this, promoter function analyses showed that Phox2b directly activates the transcriptional activity of Phox2a [24, 35]. However, it is not known whether Mash1 controls Phox2a and other targets genes (e.g., panneuronal genes) directly or indirectly. While Phox2a/2b seem to directly activate DBH gene expression [22, 36-38], the mechanism of TH regulation by Phox2a/2b is not clearly resolved [39, 40]. Our present study demonstrates that GATA-3 can robustly transactivate the transcriptional activity of the DBH gene, strongly suggesting that DBH is an immediate downstream target of GATA-3. Transactivation of DBH promoter does not appear to require GATA-3 binding to the promoter. Instead, our results suggest that GATA-3 regulates DBH gene transcription by physically interacting with general transcription factors, Sp1 and AP4. Taken together with our previous report showing that GATA-3 regulates the TH gene via interacting with CREB [19], these results support the possibility that GATA-3 may regulate the NA phenotype specification without direct DNA binding, which is quite different from its mechanism of gene activation during T cell differentiation. This novel mechanism of GATA-3 gene activation warrants further investigation.
Acknowledgements
The authors thank J. Engel for chick GATA-3 clone, R. Ratan for the Sp1 expressing plasmids, C. Tabin for RCASBP vectors and pSlax13, and O. Andrisani for NCSC culture. This work was supported by NIH grants (MH48866 and DC006501).
References
- 1.Anderson DJ. Lineages and transcription factors in the specification of vertebrate primary sensory neurons. Curr Opin Neurobiol. 1999;9:517–524. doi: 10.1016/S0959-4388(99)00015-X. [DOI] [PubMed] [Google Scholar]
- 2.Edlund T, Jessell TM. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- 3.Goridis C, Rohrer H. Specification of catecholaminergic and serotonergic neurons. Nat Rev Neurosci. 2002;3:531–541. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- 4.Marquardt T, Pfaff SL. Cracking the transcriptional code for cell specification in the neural tube. Cell. 2001;106:651–654. doi: 10.1016/s0092-8674(01)00499-8. [DOI] [PubMed] [Google Scholar]
- 5.Brunet JF, Pattyn A. Phox2 genes - from patterning to connectivity. Curr Opin Genet Dev. 2002;12:435–440. doi: 10.1016/s0959-437x(02)00322-2. [DOI] [PubMed] [Google Scholar]
- 6.Goridis C, Brunet JF. Transcriptional control of neurotransmitter phenotype. Curr Opin Neurobiol. 1999;9:47–53. doi: 10.1016/s0959-4388(99)80006-3. [DOI] [PubMed] [Google Scholar]
- 7.Howard MJ, Stanke M, Schneider C, Wu X, Rohrer H. The transcription factor dHAND is a downstream effector of BMPs in sympathetic neuron specification. Development. 2000;127:4073–4081. doi: 10.1242/dev.127.18.4073. [DOI] [PubMed] [Google Scholar]
- 8.Morikawa Y, D’Autreaux F, Gershon MD, Cserjesi P. Hand2 determines the noradrenergic phenotype in the mouse sympathetic nervous system. Dev Biol. 2007;307:114–126. doi: 10.1016/j.ydbio.2007.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morin X, Cremer H, Hirsch MR, Kapur RP, Goridis C, Brunet JF. Defects in sensory and autonomic ganglia and absence of locus coeruleus in mice deficient for the homeobox gene Phox2a. Neuron. 1997;18:411–423. doi: 10.1016/s0896-6273(00)81242-8. [DOI] [PubMed] [Google Scholar]
- 10.Ho IC, Vorhees P, Marin N, Oakley BK, Tsai SF, Orkin SH, Leiden JM. Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J. 1991;10:1187–1192. doi: 10.1002/j.1460-2075.1991.tb08059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joulin V, Bories D, Eleouet JF, Labastie MC, Chretien S, Mattei MG, Romeo PH. A T-cell specific TCR delta DNA binding protein is a member of the human GATA family. EMBO J. 1991;10:809–1816. doi: 10.1002/j.1460-2075.1991.tb07706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko LJ, Yamamoto M, Leonard MW, George KM, Ting P, Engel JD. Murine and human T-lymphocyte GATA-3 factors mediate transcription through a cis-regulatory element within the human T-cell receptor delta gene enhancer. Mol Cell Biol. 1991;11:778–2784. doi: 10.1128/mcb.11.5.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong C, Flavell RA. Control of T helper cell differentiation--in search of master genes. Sci STKE. 2000;2000:PE1. doi: 10.1126/stke.2000.49.pe1. [DOI] [PubMed] [Google Scholar]
- 14.Ho IC, Glimcher LH. Transcription: tantalizing times for T cells. Cell. 2002;109(Suppl):S109–120. doi: 10.1016/s0092-8674(02)00705-5. [DOI] [PubMed] [Google Scholar]
- 15.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 16.Pandolfi PP, Roth ME, Karis A, Leonard MW, Dzierzak E, Grosveld FG, Engel JD, Lindenbaum MH. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet. 1995;11:40–44. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- 17.Lim KC, Lakshmanan G, Crawford SE, Gu Y, Grosveld F, Engel JD. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat Genet. 2000;25:09–212. doi: 10.1038/76080. [DOI] [PubMed] [Google Scholar]
- 18.Moriguchi T, Takako N, Hamada M, Maeda A, Fujioka Y, Kuroha T, Huber RE, Hasegawa SL, Rao A, Yamamoto M, Takahashi S, Lim KC, Engel JD. Gata3 participates in a complex transcriptional feedback network to regulate sympathoadrenal differentiation. Development. 2006;133:3871–3881. doi: 10.1242/dev.02553. [DOI] [PubMed] [Google Scholar]
- 19.Hong SJ, Huh Y, Chae H, Hong S, Lardaro T, Kim KS. GATA-3 regulates the transcriptional activity of tyrosine hydroxylase by interacting with CREB. J Neurochem. 2006;98:773–781. doi: 10.1111/j.1471-4159.2006.03924.x. [DOI] [PubMed] [Google Scholar]
- 20.Hong SJ, Chae H, Kim KS. Promoterless luciferase reporter gene is transactivated by basic helix-loop-helix transcription factors. Biotechniques. 2002;33:1236–1238. doi: 10.2144/02336bm11. [DOI] [PubMed] [Google Scholar]
- 21.Ishiguro H, Kim KT, Joh TH, Kim KS. Neuron-specific expression of the human dopamine beta-hydroxylase gene requires both the cAMP-response element and a silencer region. J Biol Chem. 1993;268:17987–17994. [PubMed] [Google Scholar]
- 22.Kim HS, Seo H, Yang C, Brunet JF, Kim KS. Noradrenergic-specific transcription of the dopamine beta-hydroxylase gene requires synergy of multiple cis-acting elements including at least two Phox2a-binding sites. J Neurosci. 1998;18:8247–8260. doi: 10.1523/JNEUROSCI.18-20-08247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mermod N, Williams TJ, Tjian R. Enhancer binding factors AP-4 and AP-1 act in concert to activate SV40 late transcription in vitro. Nature. 1988;332:557–561. doi: 10.1038/332557a0. [DOI] [PubMed] [Google Scholar]
- 24.Hong SJ, Kim CH, Kim KS. Structural and functional characterization of the 5′ upstream promoter of the human Phox2a gene: possible direct transactivation by transcription factor Phox2b. J Neurochem. 2001;79:1225–1236. doi: 10.1046/j.1471-4159.2001.00672.x. [DOI] [PubMed] [Google Scholar]
- 25.George KM, Leonard MW, Roth ME, Lieuw KH, Kioussis D, Grosveld F, Engel JD. Embryonic expression and cloning of the murine GATA-3 gene. Development. 1994;120:2673–2686. doi: 10.1242/dev.120.9.2673. [DOI] [PubMed] [Google Scholar]
- 26.Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates) Curr Opin Genet Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- 27.Simon MC. Gotta have GATA. Nat Genet. 1995;11:9–11. doi: 10.1038/ng0995-9. [DOI] [PubMed] [Google Scholar]
- 28.Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 29.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 30.Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch MR, Tiveron MC, Guillemot F, Brunet JF, Goridis C. Control of noradrenergic differentiation and Phox2a expression by MASH1 in the central and peripheral nervous system. Development. 1998;125:599–608. doi: 10.1242/dev.125.4.599. [DOI] [PubMed] [Google Scholar]
- 33.Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 1997;124:4065–4075. doi: 10.1242/dev.124.20.4065. [DOI] [PubMed] [Google Scholar]
- 34.Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:66–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- 35.Flora A, Lucchetti H, Benfante R, Goridis C, Clementi F, Fornasari D. Sp proteins and Phox2b regulate the expression of the human Phox2a gene. J. Neurosci. 2001;21:7037–7045. doi: 10.1523/JNEUROSCI.21-18-07037.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seo H, Hong SJ, Guo S, Kim HS, Kim CH, Hwang DY, Isacson O, Rosenthal A, Kim KS. A direct role of the homeodomain proteins Phox2a/2b in noradrenaline neurotransmitter identity determination. J Neurochem. 2002;80:905–916. doi: 10.1046/j.0022-3042.2002.00782.x. [DOI] [PubMed] [Google Scholar]
- 37.Swanson DJ, Adachi M, Lewis EJ. The homeodomain protein Arix promotes protein kinase A-dependent activation of the dopamine beta-hydroxylase promoter through multiple elements and interaction with the coactivator cAMP-response element-binding protein-binding protein. J Biol Chem. 2000;275:2911–2923. doi: 10.1074/jbc.275.4.2911. [DOI] [PubMed] [Google Scholar]
- 38.Yang C, Kim HS, Seo H, Kim CH, Brunet JF, Kim KS. Paired-like homeodomain proteins, Phox2a and Phox2b, are responsible for noradrenergic cell-specific transcription of the dopamine beta-hydroxylase gene. J Neurochem. 1998;71:1813–1826. doi: 10.1046/j.1471-4159.1998.71051813.x. [DOI] [PubMed] [Google Scholar]
- 39.Yang C, Kim HS, Seo H, Kim KS. Identification and characterization of potential cis-regulatory elements governing transcriptional activation of the rat tyrosine hydroxylase gene. J Neurochem. 1998;71:1358–1368. doi: 10.1046/j.1471-4159.1998.71041358.x. [DOI] [PubMed] [Google Scholar]
- 40.Zellmer E, Zhang Z, Greco D, Rhodes J, Cassel S, Lewis EJ. A homeodomain protein selectively expressed in noradrenergic tissue regulates transcription of neurotransmitter biosynthetic genes. J Neurosci. 1995;15:8109–8120. doi: 10.1523/JNEUROSCI.15-12-08109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]