Abstract
The protein and lipid composition of eukaryotic plasma membranes is highly dynamic and regulated according to need. The sphingolipid-responsive Pkh kinases are candidates for mediating parts of this regulation, as they affect a diverse set of plasma membrane functions, such as cortical actin patch organization, efficient endocytosis, and eisosome assembly. Eisosomes are large protein complexes underlying the plasma membrane and help to sort a group of membrane proteins into distinct domains. In this study, we identify Nce102 in a genome-wide screen for genes involved in eisosome organization and Pkh kinase signaling. Nce102 accumulates in membrane domains at eisosomes where Pkh kinases also localize. The relative abundance of Nce102 in these domains compared with the rest of the plasma membrane is dynamically regulated by sphingolipids. Furthermore, Nce102 inhibits Pkh kinase signaling and is required for plasma membrane organization. Therefore, Nce102 might act as a sensor of sphingolipids that regulates plasma membrane function.
Introduction
The plasma membrane is highly dynamic and crucial for communication of cells with their environment. It transduces numerous extracellular signals and transports molecules in and out of the cell. To accommodate these diverse tasks, it is highly organized, and plasma membrane processes are tightly coordinated, both spatially and temporally (Simons and Toomre, 2000; Anderson and Jacobson, 2002; Simons and Vaz, 2004). How the abundance of most lipids and proteins in the plasma membrane is regulated is largely unknown.
In the yeast Saccharomyces cerevisiae, the plasma membrane is laterally organized into spatial domains that have different protein and lipid composition. One type of domain harbors several integral membrane proteins, such as the arginine transporter Can1 and members of the Sur7 family of proteins and was accordingly termed membrane compartment occupied by Can1 (MCC). MCCs were suggested to contain a distinct lipid composition enriched in ergosterol, as visualized by staining with filipin, a fluorescent marker binding this lipid (Malinska et al., 2003; Grossmann et al., 2007).
Yeast mother cells possess 25–45 MCCs that can be visualized as discrete foci in the plasma membrane. MCCs are mutually exclusive with a second domain, marked by the plasma membrane ATPase Pma1, termed membrane compartment occupied by Pma1 (MCP; Malinska et al., 2003, 2004). The organization of the plasma membrane in distinct spatial domains is at least in part mediated by large protein complexes termed eisosomes (Walther et al., 2006). Eisosomes lie underneath each MCC forming a punctate, distributed pattern in the cell cortex. When the gene encoding the major eisosome component Pil1 is deleted, MCCs and all remaining eisosome proteins investigated so far coalesce into one or a few punctae per cell (Walther et al., 2006; Grossmann et al., 2007). In addition to concentrating at such eisosome remnants, a significant fraction of MCC transmembrane proteins, such as Sur7, localizes uniformly in the plasma membrane. Similarly, the ergosterol marker filipin looses its normal punctate eisosome localization in Δpil1 cells, distributes more evenly over the plasma membrane surface, and enriches at eisosome remnants (Grossmann et al., 2007).
One possible function of MCCs and eisosomes is to regulate protein and lipid abundance by sorting them into distinct, spatially separated pools where they are stabilized or from which they can be endocytosed selectively. Consistent with this notion, disruption of MCCs leads to increased turnover of some proteins normally localized there (Grossmann et al., 2008). The precise molecular function of eisosomes is still unclear, but it was suggested that they regulate sites of endocytosis based on their colocalization with endocytic intermediates visualized by the lipophilic dye FM4-64 and a hexose transporter GFP fusion protein (Walther et al., 2006). A clue of how eisosomes might be regulated is provided by the discovery that Pkh kinases localize at eisosomes and that Pil1 and Lsp1 are Pkh kinase substrates (Zhang et al., 2004; Walther et al., 2007; Luo et al., 2008).
Pkh kinases regulate physiology and plasma membrane functions such as actin patch organization, endocytosis, and eisosome assembly (Inagaki et al., 1999; Sun et al., 2000; Friant et al., 2001; deHart et al., 2002; Liu et al., 2005; Grosshans et al., 2006; Daquinag et al., 2007; Walther et al., 2007; Luo et al., 2008). These responses are mediated by their targets, including Ypk1 and Ypk2 (homologues of the mammalian serum glucocorticoid kinase), Sch9 (homologue of human AKT kinase), Pkc1 (atypical protein kinase C), and myosin-I. In addition, Pkh kinase phosphorylation of Pil1 regulates the assembly state of eisosomes (Walther et al., 2007; Luo et al., 2008).
Pkh kinases are regulated by sphingoid long chain bases such as phytosphingosine (PHS) and dihydrosphingosine, which are precursors in sphingolipid synthesis (Zhang et al., 2004). However, it is not known how Pkh kinases sense and respond to long chain bases. Pkh kinases and several other kinases of the signaling module are regulated by levels of long chain bases in vitro, but whether this is relevant in vivo and whether it is the only way to control Pkh kinase activity is not clear (Zhang et al., 2004; Liu et al., 2005).
In this study, we visually screened for genes that affect eisosome organization either directly or through altering Pkh kinase activity. We identified the transmembrane protein Nce102 as part of the sphingolipid–Pkh signaling network. Our functional experiments suggest that Nce102 might act as a sphingolipid sensor that modulates Pkh kinase activity to regulate plasma membrane organization and function.
Results
Nce102 is required for normal eisosome organization
To identify genes required for eisosome assembly and organization, we screened by fluorescence microscopy a systematic gene deletion collection into which we introduced GFP-labeled Pil1 fusion protein (Pil1-GFP; Fig. S1 a; Tong et al., 2001). To determine the effect of individual mutations on eisosomes, we grew the library in 96-well plates to mid-log phase and imaged cells with an automated microscope. Visual inspection of the images led to identification of 88 genes that affect eisosomes (Fig. 1 a).
Figure 1.
A functional genomic screen reveals genes required for eisosome organization. (a) Genes with a Pil1 organization phenotype shown in functional groups. (b) Nce102 is required for normal eisosome organization. Images of Pil1-GFP expressed in wild-type (wt; left) or Δnce102 cells (right) are shown. (c) Number of eisosomes per mother cell (left), cytoplasmic Pil1-GFP fluorescence (middle), and Pil1-GFP fluorescence per eisosome (right). Error bars indicate SD. Bar, 5 µm.
To obtain quantitative data for the identified mutants, we collected confocal images and quantitated the number of eisosomes per cell, the cytosolic fluorescence signal representing unassembled Pil1-GFP, the integrated fluorescence on the cell surface, the size of individual eisosomes, and the percentage of cell surface covered with eisosomes (Fig. S1 a). The relative values for these parameters were used to cluster the genes according to the similarity of their phenotypes (Fig. S1 b).
For example, deletion of genes encoding subunits of the NatC complex involved in N-terminal protein acetylation (MAK3, MAK10, and MAK31) showed a similar phenotype characterized by fewer eisosomes, increased cytoplasmic Pil1-GFP, and little change in the size of remaining eisosomes.
In addition, several hits (YMR031C, YMR086W, and MSC3) are good candidates for genes encoding previously not recognized eisosome components because they localize in a punctuate pattern at the cell periphery similar to eisosomes, and at least Ymr031c and Ymr086c were copurified with Pil1 (Huh et al., 2003; Grossmann et al., 2008; Deng et al., 2009).
In this study, we focused on Nce102 because it is the only hit in our screen previously found as a transmembrane plasma membrane protein (Cleves et al., 1996). To confirm the role of Nce102 in eisosome formation, we investigated the localization of Pil1-GFP in Δnce102 compared with wild-type cells. Consistent with the systematic genome-wide screen, confocal images showed a clear reduction of eisosome number and a wider spacing of remaining eisosomes (Fig. 1 b). Quantitation revealed that deletion of NCE102 results in a twofold decrease of eisosome number per mother cell and concomitant increase of cytosolic signal (Fig. 1 c).
Nce102 is required for membrane organization
Eisosomes organize MCCs, and mutation of PIL1 results in mislocalization of all tested MCC components (Walther et al., 2006; Grossmann et al., 2007, 2008). Because NCE102 deletion has a strong effect on eisosomes, we asked whether NCE102 is also required for plasma membrane organization. Indeed, compared with wild-type cells in which the MCC marker Sur7-GFP was organized in distinct domains, Δnce102 cells showed only few clusters and more uniform localization of Sur7-GFP throughout the plasma membrane (Fig. 2 a). Furthermore, we observed a consistent but less pronounced phenotype on the MCP marker Pma1, which localized more uniformly in Δnce102 cells compared with its normal localization in structured plasma membrane domains (Fig. 2 b).
Figure 2.
NCE102 is required for normal plasma membrane organization. (a and b) Sur7-GFP (a) and Pma1-GFP (b) were expressed in wild-type (wt) or Δnce102 cells, and representative confocal top (bottom) and mid sections (top) are shown. (c) Wild-type and Δnce102 cells expressing Pil1-GFP (green) were pulse labeled with FM4-64 (red) for 1 min, and representative images are shown. Boxes indicate the area magnified in the bottom panels. (d) Numbers of FM4-64 intermediates per cell (n > 50) from images as in c are shown as a histogram. Bars, 5 µm.
Next, we asked whether Nce102 influences the formation of endocytic intermediates formed as foci at eisosomes by using the lipophilic tracer FM4-64 (Vida and Emr, 1995; Walther et al., 2006). FM4-64 foci formed after short chase periods of ∼1 min and colocalized with eisosomes in both wild-type and Δnce102 cells (Fig. 2 c). However, when we quantitated the number of foci, we found their number greatly reduced in Δnce102 cells (median = 1 foci/cell; Fig. 2 d, right) compared with wild-type cells (median = 4 foci/cell; Fig. 2 d, left). Together, this shows that normal plasma membrane organization requires Nce102.
Nce102 localizes to the plasma membrane at MCC
To answer how Nce102 functions, we first investigated its subcellular localization. In agreement with a recent study that identified Nce102 as an MCC component, we found Nce102-GFP localizing in the plasma membrane and accumulating in foci reminiscent of MCCs (Fig. 3 a; Grossmann et al., 2008). This notion was further confirmed by colocalization of Nce102 with plasma membrane markers but not, for example, with cortical ER markers (unpublished data).
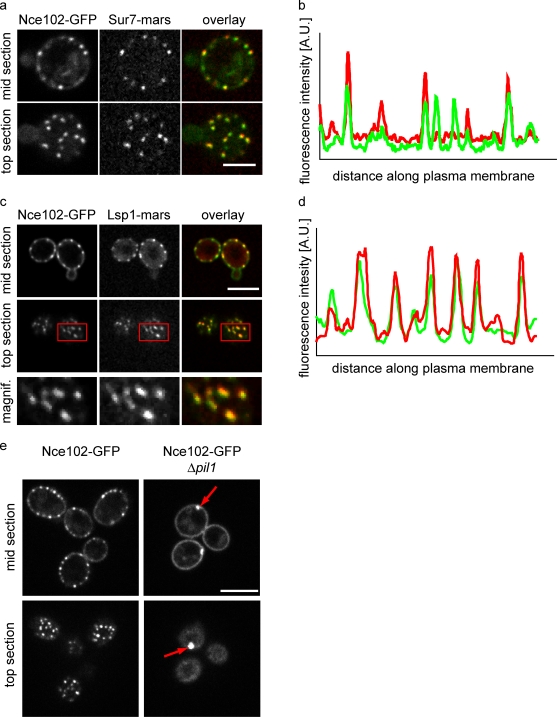
Figure 3.
Nce102 localizes to both MCC and non-MCC domains in the plasma membrane. (a) Images from cells expressing Nce102-GFP (left; green in overlay) and Sur7-mars (middle; red in the overlay) are shown. (b) Intensity profiles of Nce102-GFP and Sur7-mars along the plasma membrane. (c) Images from cells expressing Nce102-GFP (left; green in overlay) and Lsp1-mars (middle; red in the overlay) are shown. Boxes indicate the area magnified in the bottom panels. (d) Intensity profiles of Nce102-GFP and Lsp1-mars along the plasma membrane. (e) Pil1 is required for normal Nce102 distribution. Wild-type (left) and Δpil1 (right) cells expressing Nce102-GFP are shown. Arrows highlight eisosome remnants. Bars, 5 µm.
To test whether Nce102 foci are MCCs, we investigated their localization in respect to Sur7 tagged with the red fluorophore RFP-mars (Fischer et al., 2004), an MCC marker. Nce102 foci completely overlapped with Sur7-marked MCCs (Fig. 3, a and b) with significant levels of Nce102-GFP also diffusely localized in the remainder of the plasma membrane. As MCCs are membrane domains located over eisosomes, we additionally investigated Nce102-GFP localization in respect to eisosomes marked by Lsp1-mars and found that Nce102-GFP foci and eisosomes colocalize perfectly (Fig. 3, c and d). Because Nce102 is a multipass transmembrane domain protein, it is most likely located in the MCC and partially in the remainder of the membrane.
One hallmark of proteins localizing to eisosomes or MCCs is that they collapse to one or a few eisosome remnants in Δpil1 cells (Walther et al., 2006; Grossmann et al., 2007). Indeed, Nce102-GFP localization in Δpil1 cells also closely resembles that of MCC proteins such as Sur7. Most of Nce102-GFP localizes to one or a few eisosome remnants in Δpil1 cells, whereas the remaining portion shows a uniformly distributed signal throughout the plasma membrane (Fig. 3 e).
Nce102 acts negatively on Pil1 phosphorylation
Because eisosome organization is sensitive to Pil1 phosphorylation, we expected to find genes in our screen that directly affect the architecture, assembly, or stability of eisosomes and genes involved in signaling that modifies Pil1 phosphorylation. In fact, the Δnce102 phenotype on Pil1-GFP localization closely resembles the phenotype of pil1(4D) mutant cells, bearing a phosphomimicking mutant of Pil1-GFP in which four residues that are phosphorylated in the wild-type protein are mutated to aspartate (S45D, S59D, S230D, and T233D; Fig. 1 b and Fig. S4 c; Walther et al., 2007).
Therefore, we tested genetically whether Nce102 acts on eisosomes by altering Pil1 phosphorylation that could then indirectly modulate eisosome assembly. If Δnce102 effect is mediated by phosphorylation, we expect that a pil1(4A)-GFP mutant in which four residues that are required for the effect of Pkh kinases on eisosome assembly are changed to alanine (S45A, S59A, S230A, and T233A; Walther et al., 2007) blocks the effect of NCE102 deletion. However, if Nce102's effect on eisosomes is independent of Pil1 phosphorylation state, we expect to see similar effects of Δnce102 on wild-type Pil1 and pil1(4A). Indeed, pil1(4A)-GFP localization was indistinguishable between Δnce102 and wild-type cells, showing slightly more eisosome pil1(4A) assembly at the plasma membrane compared with wild-type Pil1 (Fig. 4 b). Therefore, pil1(4A) is epistatic to Δnce102. Because Pil1 needs to get phosphorylated on residues mutated in pil1(4A) to develop the Δnce102 eisosome phenotype, we conclude that Nce102 acts upstream of Pil1 phosphorylation.
Figure 4.
Nonphosphorylatable Pil1 is resistant to Δnce102 . (a and b) Representative top and mid sections of wild-type (wt; left) or Δnce102 cells (right) expressing Pil1-GFP (a) or nonphosphorylatable pil1(4A) fused to GFP (b) are shown. Bars, 5 µm.
When combined with pil1(4D)-GFP, Δnce102 cells showed a similar, albeit slightly more severe phenotype than NCE102 pil1(4D)-GFP cells (Fig. S4 c), indicating that phosphorylation at additional sites can enhance eisosome disassembly. Previous work identified at least seven additional phosphorylation sites in Pil1, which may mediate this effect (Zhang et al., 2004; Walther et al., 2007; Luo et al., 2008). Together, pil1(4A) epistasis on Δnce102 and enhancement of the pil1(4D) phenotype by Δnce102 suggest that Nce102 negatively regulates Pil1 phosphorylation.
Nce102 inhibits Pil1 phosphorylation
To substantiate these findings, we determined whether the phosphorylation state of Pil1 is altered in Δnce102 cells. To this end, we purified Pil1 fused to a tandem affinity purification (TAP) tag from wild-type and metabolically heavy lysine-labeled Δnce102 cells (stable isotope labeling by amino acids in cell culture [SILAC]; Fig. S2 a; Ong et al., 2002) and analyzed it by two strategies. First, we mixed proteins of Pil1-TAP eluates 1:1 and separated them by SDS-PAGE, resulting in a clearly visible doublet of Pil1, where the upper band represents phosphorylated Pil1 (Walther et al., 2007). Separate analysis of the two bands by liquid chromatography (LC) mass spectrometry (MS)/MS revealed that contaminant proteins present in both bands have a 1:1 ratio of protein from wild-type and Δnce102 sample, as determined by comparing mean peptide peak intensities. In contrast, Pil1 from Δnce102 was 30% enriched in the upper phospho-Pil1 band (1.28 ratio, Pil1 heavy vs. light) and correspondingly decreased in the lower band (0.59 ratio, Pil1 heavy vs. light; Fig. 5 a; and Fig. S2, c and d).
Figure 5.
Pil1 phosphorylation is increased in Δnce102 cells. (a) Pil1-TAP was purified from SILAC-labeled wild-type and Δnce102 cells, mixed in equal amounts, resolved by SDS-PAGE, and two resulting Pil1 bands were separately analyzed by MS. The contaminant Nsr1 was present in a 1:1 ratio from both strains in each band. Pil1 was1.3-fold enriched in the upper band (red) and decreased to a ratio 0.6 in the lower band (black). (b) Pil1-TAP purified from SILAC-labeled wild-type and Δnce102 cells was analyzed by MS. Unphosphorylated Pil1 peptide amino acids 222–248 is shown in black, and the T233 phosphopeptide is shown in red. (c) Pil1-TAP purified from SILAC-labeled untreated cells or treated with myriocin for 1 h was analyzed as in b. Representative data of three experiments are shown. Ratio H/L denotes the mean ratio of the abundance of heavy- versus light-labeled peptides.
In a second approach, we directly mixed, digested, and analyzed Pil1 pull-down eluates by LC-MS/MS (Fig. S2 a). From this approach, we identified many unphosphorylated peptides and, for example, the phosphorylated peptide harboring T233 of Pil1 from both wild-type and Δnce102 cells. Importantly, the phosphopeptide was more than threefold more abundant coming from the heavy lysine-labeled Δnce102 samples compared with the wild-type control (Fig. 5 b; and Fig. S2, e and f). The total amount of Pil1 was equal in both experiments because unphosphorylated peptides were present in a 1:1 ratio (Fig. S2 f). Together, these data demonstrate that Pil1 was present in a roughly equimolar ratio in both pull-down eluates but that phosphorylation at T233 was more than threefold increased in Δnce102 cells. Therefore, the results of the two biochemical approaches converge, demonstrating that Pil1 phosphorylation is increased in Δnce102 cells.
Because Pil1 phosphorylation is inhibited by sphingoid bases (Zhang et al., 2004; Walther et al., 2007), we tested whether NCE102 deletion results in a similar effect on Pil1 phosphorylation as sphingolipid depletion. To this end, we measured the relative abundance of the Pil1 T233 phosphopeptide in cells where synthesis of sphingolipids was blocked for 1 h by myriocin, a drug targeting the serine palmitoyl transferase that catalyzes the rate-limiting step of sphingolipid synthesis, compared with wild-type cells. In this experiment, we detected a twofold increase of Pil1 phosphorylation at T233 after myriocin treatment compared with wild-type cells (Fig. 5 c and Fig. S2 g). This shows that the block of sphingolipid synthesis by myriocin and NCE102 deletion has a similar effect on Pil1 phosphorylation.
Nce102 acts on Pil1 via Pkh kinases
Several studies show that Pil1 phosphorylation is mediated by Pkh kinases (Zhang et al., 2004; Walther et al., 2007; Luo et al., 2008). Therefore, we asked whether the effect of Nce102 on Pil1 phosphorylation occurs through the Pkh kinase pathway. First, we determined the epistatic relationship between Pkh kinase mutants and Δnce102 using a yeast strain that harbors a deletion of PKH2 and a temperature-sensitive allele of PKH1 (pkhts; Friant et al., 2001). Already at the permissive temperature, these mutations lead to a strong over-assembly effect on eisosomes. Specifically, pkhts strains display large elongated threads of Pil1 at the plasma membrane with more of the membrane covered with Pil1 (Fig. 6; Walther et al., 2007). This phenotype is also very severe at the restrictive temperature, with most of the membrane covered with Pil1 (Fig. S4 e).
Figure 6.
Mutation of Pkh kinases is epistatic to Δnce102. Optical top and mid sections of wild-type (wt; top left), Δnce102 (top right), pkhts (bottom left), or pkh1ts Δnce102 (bottom right) cells expressing Pil1-GFP are shown. Bars, 5 µm.
Combining pkhts and the Δnce102 mutations, we found that double-mutant cells show a Pil1-GFP phenotype indistinguishable from the pkhts phenotype at both 24 (Fig. 6) and 37°C (Fig. S4 e). This indicates that Pkh kinases are required to obtain the Δnce102 phenotype and shows that Nce102 acts upstream of Pkh kinases, inhibiting them in a linear pathway.
Nce102 acts downstream of long chain bases in sphingoid base signaling
Our data suggest that Nce102 acts on Pil1 phosphorylation by inhibiting Pkh kinases. Because Pkh kinases are regulated by sphingoid bases and both, a block of sphingolipid synthesis and Δnce102, lead to increased Pil1 phosphorylation, Nce102 itself may be controlled by sphingolipids, acting upstream of the Pkh kinases. Alternatively, Nce102 could be required for efficient synthesis of sphingolipids that are then sensed by the kinases by some other route. One more possibility is that Nce102 acts independently of sphingolipids to regulate Pkh kinases.
To distinguish between these possibilities, we tested whether Nce102 is required for the synthesis of sphingoid bases. If this was the case, addition of exogenous sphingoid base would rescue the Δnce102 phenotype. Exogenously added sphingoid bases can enter the cell and rescue a sphingoid base synthesis defect because addition of 5 µM PHS suppressed the Pil1-GFP phenotype of the lcb1-100 mutant (which impairs sphingolipid synthesis) both at the permissive and restrictive temperatures (Fig. 7 a). In contrast, the eisosome defect of Δnce102 cells was not rescued by addition of PHS under conditions that rescued the lcb1-100 mutant phenotype (Fig. 7 a) or by a 10-fold higher concentration of PHS (unpublished data). Quantitation of eisosome number and cytosolic Pil1 levels confirmed the visual evaluation (Fig. 7 b).
Figure 7.
Nce102 and sphingolipids act in the same pathway. (a) lcb1-100 but not Δnce102 is suppressed by sphingoid bases. Wild-type (wt), lcb1-100, or Δnce102 cells expressing Pil1-GFP were analyzed with or without addition of 5 µM PHS, and representative images are shown. (b) Eisosome number per mother cell (left) and cytosolic Pil1-GFP fluorescence (right) are shown. (c) Myriocin has no additive effect on Δnce102 cells. Wild-type and Δnce102 cells were treated for 1 h with 5 µM myriocin, and representative images are shown. (d) Quantitation of eisosome number per mother cell (left) and cytoplasmic Pil1-GFP fluorescence (right) of cells treated as in c. Error bars indicate SD. Bars, 5 µm.
These data show that Nce102 is not required for sphingoid base synthesis but rather acts downstream of it. Because PHS has no additive effect on Δnce102 cells, sphingolipids and Nce102 act in the same pathway (Fig. 7 b).
To further test this hypothesis, we investigated the interaction between myriocin treatment and Δnce102. If both act independent from each other, we expect an additive effect between them. However, this is not the case. As shown in images and quantitation of eisosomes, myriocin has no further effect on eisosomes in Δnce102 cells, confirming that sphingolipid signaling and Nce102 act in the same pathway on eisosomes (Fig. 7, c and d).
We previously showed that Pil1 hyperphosphorylation caused by decreased sphingolipid synthesis causes eisosome disassembly (Walther et al., 2007). If our hypothesis is true and Nce102 is an inhibitor of Pil1 phosphorylation acting downstream of sphingolipids, we predict that overexpression of Nce102 will rescue eisosome disassembly when sphingolipid synthesis is decreased. To test this, we expressed Nce102-mars from the inducible Gal promoter in cells that have Pil1-GFP–marked eisosomes. When these cells are grown on raffinose, Nce102-mars is not expressed, and eisosomes appear normal, as these cells also express endogenous Nce102 (Fig. 8). When these cells grow on galactose, Nce102-mars is overexpressed (Fig. 8). Strikingly, when we blocked sphingolipid synthesis in these cells by treating them with myriocin, the normal effect of disassembling eisosomes apparent in control cells was completely blocked (Fig. 8). This shows that increasing Nce102 levels blocks the effect of inhibiting sphingolipid synthesis on eisosomes.
Figure 8.
Overexpression of Nce102 suppresses eisosomes disassembly after sphingolipid synthesis block. Nce102-mars (right) controlled by the Gal promoter was either not expressed in raffinose-containing medium or induced in galactose-containing medium in Pil1-GFP (left) cells, which were incubated for 1 h with 5 µM myriocin as indicated. Bar, 5 µm.
Together, our genetic experiments thus place Nce102 in a linear pathway inhibiting Pkh kinases and suggest a relationship: sphingoid bases → Nce102–| Pkh1/2 → Pil1 phosphorylation → eisosome disassembly. In this scenario, Nce102 would function as part of a sensor relay for sphingoid base or sphingolipid levels.
Nce102 localization responds to changes in sphingolipid levels
Because Nce102 negatively regulates Pkh kinases that localize to eisosomes, we next tested whether Nce102 distribution between MCCs at eisosomes and the rest of the plasma membrane is affected by sphingolipid levels. To this end, we determined Nce102-GFP localization after blocking sphingolipid synthesis. Strikingly, after a 1 h incubation of cells with myriocin, the punctate pattern of Nce102-GFP localization in MCCs disappeared, and the protein distributed diffusely across the plasma membrane (Fig. 9 a). Consistently, in intensity plots of surface images, myriocin-treated cells show a rather flat distribution of the Nce102-GFP signal, whereas control samples show many Nce102-GFP peaks corresponding to MCCs at eisosomes (Fig. 9 b). Relocalization of Nce102-GFP could be reversed by addition of exogenous PHS for a short time (15 min). In this case, the MCC pattern of Nce102-GFP localization reappeared, showing an even more pronounced pattern of Nce102-GFP foci than normal (Fig. 9 b).
Figure 9.
Nce102-GFP localization depends on sphingolipid levels. (a) Nce102-GFP was imaged under normal growth conditions (control), after addition of 5 µM myriocin for 1 h (myriocin), after sequential treatment with 5 µM myriocin for 1 h and 50 µM PHS for 15 min (myriocin→PHS), or after addition of 50 µM PHS for 15 min (PHS). Boxes indicate the area magnified in the bottom panels. (b) Fluorescence intensities of the area are shown plotted against xy image coordinates. (c) Nce102 redistribution is not dependent on new protein synthesis. Nce102-GFP cells were treated with myriocin or myriocin and PHS successively as in a after 10-min preincubation and continued presence of cycloheximide (CHX). (d) Nce102 partitions into detergent-resistant membranes dependent on sphingoid bases. Untreated Nce102-TAP–expressing cells and cells treated as in a were lysed in buffer containing 1% Triton X-100 and analyzed by gradient centrifugation and Western blotting against TAP (left). The same blots were probed with Pma1 antibodies (right). (e) pil1(4A)-GFP is resistant to disassembly after myriocin treatment. pil1(4A)-GFP–expressing cells were imaged after 1-h 5 µM myriocin incubation. (f) Redistribution of Nce102-GFP after myriocin treatment is independent of eisosome disassembly. Cells expressing pil1(4A) and Nce102-GFP were imaged before (left) and after 1 h treatment with 5 µM myriocin (right). (g and h) Sur7-mars does not behave like Nce102 after myriocin treatment. Wild-type Pil1 (g) or pil1(4A) (h) cells expressing Sur7-mars were treated with myriocin and imaged. wt, wild type. Bars, 5 µm.
Nce102 relocalization could be caused by redistribution of existing protein in the plasma membrane or new protein synthesis under conditions in which myriocin blocks the efficient incorporation of Nce102 in MCCs. To distinguish these possibilities, we added myriocin after a preincubation with the translation-blocking drug cycloheximide. During persistent translation block, Nce102 localized normally to MCCs and the remainder of the membrane (Fig. 9 c). Moreover, it still relocalized from MCCs after myriocin treatment, indicating that this phenomenon is not dependent on protein synthesis (Fig. 9 c). Similarly, when PHS was added to cells in which translation was blocked, Nce102 relocalization occurred normally (Fig. 9 c).
To test whether this effect is specific to the availability of sphingoid bases or a more general response to sphingolipid levels, we added aureobasidin to cells for 1 h. This drug inhibits complex sphingolipid synthesis from ceramide downstream of the formation of sphingoid bases (Nagiec et al., 1997). We observed the same Nce102-GFP redistribution in the plasma membrane after aureobasidin or myriocin addition (Fig. 9 a and Fig. S4). Consistent with the downstream block of sphingolipid synthesis by aureobasidin, addition of exogenous sphingoid bases did not result in reformation of the punctate Nce102 pattern (Fig. S4).
Together, these data suggest an equilibrium of two pools of Nce102-GFP localizing to MCCs at eisosomes or to spaces in between that is shifted by levels of sphingolipids. Consistent with this notion, addition of PHS to untreated cells shifted more Nce102-GFP into punctate structures, rendering them more pronounced (Fig. 9, a and b).
In the yeast plasma membrane, many proteins are sensitive to extraction by detergents to a different level, and Nce102 was previously described operationally as an abundant component of detergent-resistant membranes (Bagnat et al., 2000). To test whether Nce102 distribution between MCCs and the remainder of the plasma membrane observed by microscopy is reflected in its partitioning between different membrane environments, we analyzed its solubility in a buffer containing 1% Triton X-100 at 4°C. Proteins partitioning into sphingolipid/ergosterol-rich domains show lower solubility in this buffer compared with proteins embedded in mostly phospholipid bilayers. This difference can be tested in a velocity sedimentation gradient. Nce102 from untreated cells was present in most fractions of the gradient, indicating that it is localized both to detergent-resistant and other membranes (Fig. 9 d). When treated with myriocin, Nce102 partitioning changed toward the bottom of the gradient, indicating higher solubility. After short treatment of these cells with PHS, Nce102 redistributed to more detergent-insoluble membrane fractions at the top of the gradient. Pma1, a marker for MCP membranes, did not significantly alter its migration in the gradient after either myriocin or PHS treatment.
Together, these results show that relative levels of Nce102 in different plasma membrane domains are regulated by sphingolipids and that this redistribution is highly sensitive to changes in membrane composition because it can be observed after small perturbations that do not change Pma1 behavior. These data are consistent with the hypothesis that Nce102 sphingolipid-mediated redistribution is the mechanism by which Nce102 regulates Pil1 phosphorylation. In this model, Nce102 is recruited to MCCs at eisosomes under conditions of sufficient sphingolipids to repress Pkh kinase activity toward Pil1 and released from there when sphingolipid levels drop, for example, after inhibition of their synthesis by myriocin or aureobasidin. To test this, we determined the localization of Nce102-mars relative to Pkh kinases. Pkh2-GFP localizes in few very dim spots that colocalize with eisosomes and Nce102 foci (Fig. S5, a and b). Importantly, when we blocked sphingolipid synthesis with myriocin, Nce102-mars relocalized from these foci, leaving Pkh2-GFP behind (Fig. 5, c and d).
If Nce102 acts as a plasma membrane sphingolipid sensor, then its partitioning should be upstream and independent of the assembly state of eisosomes. Alternatively, Nce102 redistribution could be a consequence of eisosome disassembly after inhibition of sphingolipid synthesis. To test this directly, we used pil1(4A) mutant cells in which Pil1 cannot be sufficiently phosphorylated. Eisosomes are therefore resistant to activation of Pkh kinases (Walther et al., 2007). As expected, when cells that express pil1(4A)-GFP as their sole copy of Pil1 are incubated with myriocin, eisosomes remain stable because Pil1(4A) does not get sufficiently phosphorylated to disassemble (Fig. 9 e). To determine whether Nce102-GFP redistribution is caused by disassembly of eisosomes, we investigated the localization of Nce102-GFP in pil1(4A) cells. We found it to distribute normally in both MCCs and the remainder of the plasma membrane (Fig. 9 f). When we added myriocin to these cells, eisosomes remained assembled as a result of the pil1(4A) mutation, yet Nce102-GFP relocalized to become diffusely distributed throughout the plasma membrane (Fig. 9 f). To test whether this is a general property of MCC proteins, we performed similar experiments with cells expressing Sur7-mars. When we treated these cells with myriocin, the intensity of Sur7 signal in MCC decreased as Pil1 disassembled (Fig. 9 g; not depicted), but this could be completely blocked by expressing Pil1(4A) (Fig. 9 h). Thus, Nce102-GFP has a distinct behavior from other MCC components and redistributes in the plasma membrane dependent on sphingolipids but independently of the eisosome assembly state.
Discussion
We report a comprehensive screen for genes involved in Pil1-GFP localization. Recently, a similar screen using an MCC reporter identified 27 genes (Grossmann et al., 2008). Compared with that screen, we found roughly three times more genes affecting Pil1 localization (n = 88). It remains to be seen whether this difference is caused by different thresholding during the screen or biological differences between MCCs versus eisosomes. Because Pil1 and MCC components colocalize and PIL1 is required for normal MCC formation, it is surprising that there is little overlap between the two screens: only NCE102, SUR4, MNN10, and few biological processes were found in both. Examples include retrograde transport from the Golgi apparatus to the ER (VPS52 and VPS54 in the MCC screen; VPS51 and VPS53 in the Pil1 screen). Therefore, these genes and processes likely have a global effect on plasma membrane organization affecting MCCs and eisosomes. Genes that were identified only in one screen likely perform more specific functions; future experiments will, for example, show whether YMR086W, YMR031C, or genes encoding the NatC complex are important for eisosomes but not MCCs.
Besides genes globally affecting the plasma membrane, we expected to find genes impacting eisosome organization directly as well as genes that alter sphingolipid signaling, resulting in altered Pil1 phosphorylation and assembly. In the later class, we identified Nce102. Besides it, also SUR4, ISC1, and SAC1 are likely to act on eisosomes through sphingolipid signaling. Deletion of SUR4, required for fatty acid elongation during sphingolipid synthesis (Han et al., 2002; Paul et al., 2006), or deletion of ISC1, the homologue of mammalian neutral sphingomyelinase (Sawai et al., 2000), results in accumulation of complex sphingolipids. In our screen, both Δisc1 and Δsur4 cells showed hyperassembled Pil1-GFP, which is the opposite effect of depleting complex sphingolipids by inhibiting their synthesis (Table S2). In addition, it was recently reported that Δsac1 cells have elevated sphingoid bases and reduced levels of complex sphingolipids (Brice et al., 2009), and we found them to have fewer eisosomes and increased cytoplasmic Pil1-GFP signal (Table S2). We expect that other genes indentified will also turn out to play a role in sphingolipid biology.
In this study, we focused on the function of Nce102, a multispanning plasma membrane protein required for normal eisosomes, MCC formation, and plasma membrane organization. Initially, Nce102 was identified by Cleves et al. (1996) as required for nonclassical export of mammalian galectin from yeast. The mechanism of Nce102 function in this pathway was not investigated. Because the assay used requires solubilization of cell-associated galectin using high pH, it is possible that the basis of the effect could be altered protein extractability from Δnce102 cells because of their altered plasma membrane.
Our experiments revealed that Nce102 has fascinating properties, suggesting that it might act as part of a sphingolipid sensor. Most remarkable, its localization is dynamic and responsive to changes in sphingolipid levels: Nce102 partitions between two domains in the plasma membrane, MCCs overlying eisosomes and the rest of the plasma membrane. We show that the distribution between these two domains is controlled by sphingolipid availability. Nce102 localization to MCCs brings it into close contact with the underlying eisosomes and Pkh kinases (Fig. S5; Walther et al., 2007). Nce102 negatively regulates the activity of these kinases, and, in the presence of sphingolipids, blocks their downstream functions. Conversely, if sphingolipid synthesis is blocked, Nce102 redistributes away from Pkh kinases (Fig. S5), alleviating their inhibition. Therefore, we propose a model that Nce102 acts as part of a sphingolipid-sensing mechanism and that its distribution in the plasma membrane regulates Pkh kinases (Fig. 10). In the simplest hypothesis, Nce102 could simply accomplish repression of the kinases by regulated juxtaposition to them, which is a common scheme in kinase signaling. Based on filipin staining, MCCs were suggested as sites of increased ergosterol concentration in the plasma membrane (Grossmann et al., 2007), and because sterols preferentially interact with sphingolipids, it is likely that sphingolipids are also concentrated there, forming detergent-resistant, liquid-ordered membrane domains or lipid rafts (Simons and Ikonen, 1997; Malinska et al., 2003) where Nce102 was found previously (Bagnat et al., 2000). Thus, it is possible that Nce102 also reacts to ergosterol levels in the plasma membrane. However, we did not observe an effect of nonessential erg mutants or block of sterol synthesis on eisosomes or Nce102 localization (unpublished data). Consistent with our model, Nce102 localizes to eisosome remnants that also show increased filipin staining, likely reflecting increased concentration of ergosterol and possibly sphingolipids (Grossmann et al., 2007). Alternatively, filipin might preferentially report on free sterols not in complex with sphingolipids, and staining of MCCs could actually indicate a lower concentration of sphingolipids in this compartment. This view is supported by a recent observation that filipin staining increases if sphingolipid synthesis is blocked (Jin et al., 2008). A further alternative is that Nce102 could directly bind sphingolipids, changing its affinities to other proteins that help localize it to MCCs and/or switching its activity as a Pkh kinase inhibitor on and off. In either model, Nce102 leaves the MCC when sphingolipid levels there are low, releasing its inhibition of Pkh kinases that now phosphorylate Pil1 and other targets. This is consistent with our observation that Nce102 detergent solubility changes after inhibition of sphingoid base synthesis, indicating that it partitions between different membrane environments. Most likely, this change corresponds to the relocalization of Nce102 from MCCs to the remainder of the membrane observed by microscopy. The interpretation of this result, however, remains vague, as most proteins in the yeast plasma membrane differ only in the degree of their resistance to Triton X-100 detergent extraction. For example, the MCP marker Pma1 was also previously used as a marker for detergent-resistant lipid rafts (Bagnat et al., 2000; Lee et al., 2002; Malinska et al., 2003). The difference in Triton X-100 solubility between Nce102 and Pma1 after myriocin treatment might therefore indicate that these proteins differently partition into such lipid rafts or that the MCP is actually more complex and contains subdomains not easily resolved by light microscopy. Consistent with the later notion, TORC2 (Tor kinase complex 2) is localized at a distinct plasma membrane domain separate from MCC and MCP and is also partially detergent resistant (Aronova et al., 2007; Berchtold and Walther, 2009).
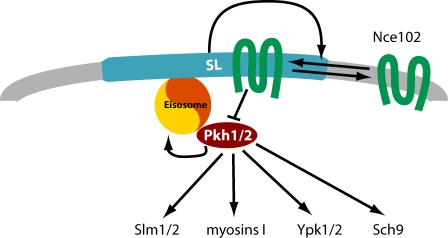
Figure 10.
Model for Nce102 function in sphingolipid sensing. Nce102 (green) senses sphingolipid levels in the plasma membrane by distributing between the thick sphingolipid-rich MCC (blue) and the rest of the plasma membrane (gray) depending on sphingolipid levels. In the MCC, it inhibits Pkh kinases (red) that localize under this domain at eisosomes.
The sensing mechanism of sphingolipids in the plasma membrane by Nce102 suggested in this study is different from a previous model positing that soluble sphingoid bases in the cytoplasm directly control Pkh kinases (Zhang et al., 2004; Liu et al., 2005). It is possible that sphingolipids control the activity of Pkh kinases at many levels. Sphingoid bases could, for example, directly regulate Pkh kinases as in vitro experiments suggest (Zhang et al., 2004), and complex sphingolipids could affect signaling via Nce102. However, the in vivo effect of direct inhibition of Pkh kinase activity toward Pil1 by sphingoid bases is probably minor compared with the Nce102 pathway, as exogenously added PHS was not able to counteract the Δnce102 hyperphosphorylation effect on eisosomes.
Our model of regulation of Pkh kinases by complex sphingolipids via the transmembrane protein Nce102 would also bridge the topological barrier between the cytoplasmic Pkh kinases and complex sphingolipids mainly concentrated in the outer leaflet of the plasma membrane (van Meer et al., 2008).
Downstream of sphingolipid sensing, the Nce102–Pkh pathway regulates phosphorylation of Pil1 that, when increased, always correlated with eisosome disassembly, e.g., in Δnce102 cells. This effect could be blocked by mutating phosphorylated Pil1 residues to alanines that we previously found were required and sufficient for the effect of Pkh kinases on eisosomes (Walther et al., 2007). This is different from findings by Luo et al. (2008), who reported that a mutant form of Pil1 harboring five phosphosites mutated to alanine (S6A, S59A, T233A, S273A, and S299A) cannot assemble properly and argued that phosphorylation of Pil1 is important for assembly of eisosomes rather than disassembly. In this study, even a mutant that lacks these five sites and even two additional sites (S6A, S45A, S59A, S230A, T233A, S273A, and S299A) still assembles into eisosomes correctly (unpublished data). The reason for this difference is unclear but might indicate that additional pathways to the Nce102–Pkh kinase module regulate Pil1 phosphorylation in a complex fashion.
Besides phosphorylation of Pil1, Pkh kinases modify many plasma membrane functions. Therefore, as expected, Δnce102 cells show altered organization of the plasma membrane, as observed for MCCs, MCPs, and the endocytic foci marked by the lipophilic dye FM4-64. Recently, it was also shown that Δnce102 cells have accelerated endocytosis rates of some membrane transporters (Grossmann et al., 2008). Pkh kinases also participate in processes as diverse as cortical actin patch organization, cell integrity signaling, endocytosis, and eisosome organization (Inagaki et al., 1999; Sun et al., 2000; deHart et al., 2002; Roelants et al., 2002; Zhang et al., 2004; Liu et al., 2005; Grosshans et al., 2006; Walther et al., 2007; Luo et al., 2008). Regulation of these diverse processes may help control the composition of the plasma membrane according to need, perhaps constituting part of a negative feedback loop that targets genes involved in sphingolipid synthesis. Indeed, expression of such genes, e.g., LCB2, FEN1, and SUR4, is strongly negatively correlated to the expression of NCE102 when the latter is regulated in response to various physiological conditions, e.g., during diauxic shift, nitrogen depletion, or heat shock (correlations for NCE102/SUR4 are: diauxic shift, −0.94; nitrogen starvation, −0.66; and heat shock, −0.85; Fig. S3, b–d; DeRisi et al., 1997; Gasch et al., 2000). In addition, after prolonged incubation with PHS, we observed Nce102-GFP signal at the vacuole (unpublished data), perhaps reflecting an adaptation to recalibrate the set level of Pkh signaling after long times with high sphingolipids levels.
An important role of Nce102 in regulation of sphingolipids is also supported by the ability of its deletion to suppress the growth defect of a mutation in the serine palmitoyl transferase subunit TSC3 (Schuldiner et al., 2005; unpublished data) or the growth inhibition caused by myriocin (Fig. S3 a), both of which reduce the first and rate-limiting step in sphingolipid synthesis.
Salient features of the sphingolipid–Nce102–Pkh kinase signaling network are likely conserved between yeast and other eukaryotes. Nce102 shares homology with the synaptogyrin/cellugyrin protein family (Belfort et al., 2005), but the molecular roles of these proteins are not understood. In contrast, mammalian PDK1 kinases (the homologues of yeast Pkh kinases) are well characterized, and the general architecture of downstream signaling is conserved (Casamayor et al., 1999). In either system, full activation of AGC kinases, such as AKT, Sch9, serum glucocorticoid kinase, or Ypk1/2, requires phosphorylation both by PDK1 (in mammalian cells) or Pkh kinases (in yeast) and in addition by the conserved kinase complex TORC2 (Powers, 2007). For yeast, TORC2 signaling has been implicated in the regulation of sphingolipid biosynthesis (Beeler et al., 1998; Tabuchi et al., 2006; Aronova et al., 2008). This effect of TORC2 is mediated by the Pkh kinase target Ypk2 (Aronova et al., 2008). Additionally, the Slm proteins were identified as targets of both the TORC2 and Pkh kinase signaling pathways and to regulate a late step in the synthesis of complex sphingolipids (Tabuchi et al., 2006). It is therefore likely that the TORC2 and Pkh1/2 pathways collaborate to regulate sphingolipid metabolism in yeast. Given the similarity of the signaling pathway components and its architecture in higher eukaryotes, their output may also be evolutionarily conserved.
Materials and methods
Visual screen
To generate a library of deletion mutants each expressing Pil1-GFP, we performed a modification of the synthetic genetic array screen as described previously (Tong et al., 2001). In short, a MATα strain containing a chromosomal copy of Pil1-GFP marked with a NATR marker, the CAN1 gene disrupted by a construct that encodes the MATa promoter driving expression of the HIS3 gene and the LYP1 gene disrupted by a construct expressing LEU2 from the MFα promoter (KEM108), was crossed to the MATa deletion library BY4741 (each strain marked with KANR). Diploids were selected on G418 and nourseothricin and sporulated. From the sporulation, MATa haploids containing both Pil1-GFP and the deletion gene were selected with successive pinning on –HIS (selection for MATa cells), G418 (selection for the gene deletion from the library), canavanine (selection for haploid cells harboring can1Δ), S-(2-aminoethyl)-l-cysteine hydrochloride (selection for haploid cells harboring lyp1Δ), and nourseothricin (selection for Pil1-GFP)-containing media.
For imaging, cells were grown overnight to saturation in 384-well plates, transferred to 96-well plates, diluted, and grown for 9 h until they reached mid-log phase. Cells were transferred to glass-bottom 96-well plates coated with concanavalin A and imaged with a 40× high NA objective in an ImageXPress Micro (MDS Analytical Technologies). At least six different images were taken for each deletion strain.
The images were visually inspected using the software by the microscope manufacturer and classified using three categories: increased cytosolic Pil1-GFP fluorescence, altered number or eisosomes, and altered pattern of eisosomes. Strains that showed a phenotype in this first round of screening were regrown and imaged by using a confocal microscope (LSM510; Carl Zeiss, Inc.).
Two optical mid-section and two top-section images were taken for each strain, showing 15–40 cells per image. Images were visually analyzed, and several parameters of the Pil1-GFP signal on the surface of cells were quantitated using an image interpretation script developed in MATLAB that we named EISURAN (both the source code and an executable MATLAB file can be found at http://www.biochem.mpg.de/en/rg/walther/news/index.html). In short, EISURAN first subtracts background from the images and then finds the edge of a cell by image dilation. To facilitate the quantitation of signal from assembled Pil1, the script thresholds the image using Otsu's method (Otsu, 1979). From the resulting binary image, EISURAN calculates the number of positive pixels per cell, a measure for the area covered by eisosomes (percentage of cell surface covered by eisosomes). It then calculates the mean size of the connected clusters (mean pixel number per eisosome cluster on cell surface). To eliminate small background clusters, a minimum of 20 pixels are used for the lower cut off. In some conditions, very large values result caused by connection of eisosomes into very large clusters, which are sometimes string formed. To determine how much Pil1 is in individual eisosomes, EISURAN determines the mean intensity of each cluster by applying the binary image as a mask on the original image, integrating the intensity in each cluster, and averaging this intensity over all clusters in an image (mean integrated intensity per eisosome cluster on cell surface). Quantitation of eisosome number was based on the mean number of eisosomes counted by two independent observers in optical mid sections of mother cells (eisosome number; mean for the wild type = 11.5 ± 1.55).
The cytoplasmic signal was quantitated as the mean intensity of a 50 × 50–pixel area in the cytoplasm (cytoplasmic Pil1-GFP signal) visible in a confocal mid section of the cell using ImageJ (National Institutes of Health). The resulting values were averaged from at least 20 cells, analyzed by hierarchical clustering, and displayed using R software (http://www.r-project.org).
Yeast strains and plasmids
All yeast strains used as well as their genotypes are listed in Table S1. The starter strain used for the screen Pil1-GFP::NATR (KEM 108) was generated in the strain YMS196 (provided by N. Krogan, University of California, San Francisco, San Francisco, CA) by tagging Pil1 with GFP using homologous recombination. Nce102-GFP::HIS and Δnce102::NATR were generated in the W303 wild-type strain by homologous recombination of PCR-generated fragments to yield strains TWY840 and TWY842, respectively (Janke et al., 2004). The Δpil1:KANR NCE102-GFP::HIS was similarly generated by homologous recombination, transforming the NCE102-GFP and marker into the Δpil1 strain TWY226 to yield TWY836. Analogously, Δnce102::NATR Pil1-GFP was generated by transforming a PIL1-GFP fragment with an HIS marker into TWY842 to yield TWY837. The Nce102-TAP:KANR strain was generated by homologous recombination after transforming an Nce102-TAP fragment in the S288C wild-type strain to yield TWY897 (Janke et al., 2004). Strains harboring pkhts mutants and Pil1-GFP were described previously (Walther et al., 2007). Δnce102::NATR was generated by homologous recombination of a PCR fragment in a control strain with the same background as pkhts and used to derive a strain with pkh1ts Δpkh2::LEU Δnce102::NATR Pil1-GFP::URA by crossing, sporulation, and selection of haploid cells to yield strain TWY932. All deletion strains were confirmed by PCR, and strains expressing tagged proteins were confirmed by PCR and Western blot analysis.
A Δpil1::KANR Δnce102::NATR strain was generated by crossing Δpil1 strain TWY226 with the Δnce102 strain TWY841. Sporulation and selection yield TWY898.
pRS306 plasmids containing the PIL1 promoter, the wild type, or mutated ORF fused to GFP used for expression of phosphomimicking and nonphosphorylatable Pil1 were described previously (Walther et al., 2007) and were integrated into the URA locus of TWY898 to yield TWY931 and TWY934.
Pma1-GFP was created by PCR-mediated tagging in wild type or Δnce102::NATR to generate TWY958 and TWY1049, respectively. Similarly, SUR7-GFP strains were generated to yield TWY956 (wild type) and TWY1049 (Δnce102).
The pGalNce102-mars plasmid was created by cloning a fusion PCR product combining the Gal-promoter, the Nce102 ORF, and the mars sequence into the NotI and HindIII sites of pRS306. This plasmid was integrated into the URA3 locus of TWY110 to yield TWY1222.
For SILAC labeling, the lysine auxotroph S288C strain TWY70 was transformed with a Pil1-TAP fragment to get the Pil1-TAP:KANR strain TWY1004 (Janke et al., 2004). To generate a Pil1-TAP:KANR Δnce102:NATR strain, TWY1004 was transformed with a Δnce102 fragment yielding TWY1052 (Janke et al., 2004).
Yeast culture and drug treatment
Yeast cells were grown according to standard procedure. For microscopy, cells were grown in synthetic complete medium and bound to concanavalin A–treated coverslips. Myriocin (Sigma-Aldrich) and PHS (Sigma-Aldrich) were added in concentrations as indicated and incubated for 1 h or 15 min. Cycloheximide (Sigma-Aldrich) was added in a concentration of 100 µg/ml for 1 h and 15 min before treatment with other drugs.
For SILAC labeling, Pil1-TAP–expressing yeast cells in a wild-type or Δnce102 background were grown in 1 l YNB liquid medium. PIL1-TAP cells were grown in the presence of 20 mg/l normal L-lysine, and PIL1-TAP Δnce102 cells were grown in the presence of 20 mg/l L-lysine-U-13C6,15N2 overnight with at least 10 doublings to OD600 = 0.7.
Microscopy
For fluorescence microscopy, yeast cells were grown to OD = 0.6 in synthetic medium at 30°C unless indicated otherwise. Cells were mounted in synthetic media onto coverslips previously coated with concanavalin A and directly imaged with a spinning-disk confocal microscope (TiLL iMIC CSU22; Andoor) using a back-illuminated EM charge-coupled device camera (iXonEM 897; Andor) and a 100× 1.4 NA oil immersion objective (Olympus). From this setup, 16-bit images were collected using Image iQ (version 1.9; Andor) in the linear range of the camera. For presentation, images were filtered with a smoothening filter averaging 2 pixels, converted to 8-bit images, and cropped using ImageJ software (http://rsbweb.nih.gov/ij/).
FM4-64 assay
To measure the formation of early FM4-64 uptake intermediates, either wild-type or Δnce102 cells were grown in mid-log phase to OD = 0.5. Approximately 5 ml cell culture was harvested by centrifugation and incubated on ice for 5 min. Cells were then labeled for 10 min with 40 µM FM4-64, washed three times in ice-cold medium, and resuspended in RT YPD for 5 min. Cells were killed by addition of 10 mM NaN3 and NaF each and immediately analyzed by fluorescence microscopy.
Isolation of detergent-resistant membranes
Detergent-resistant membranes were isolated essentially as described previously (Bagnat et al., 2000). In brief, 20 ODs Nce102-TAP–expressing yeast cells were harvested by centrifugation. The pellets were washed once with water and lysed in TNE buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 5 mM EDTA) by vortexing with glass beads twice for 5 min each at 4°C. The lysate was cleared of unbroken cells by centrifugation at 500 g for 5 min and incubated with Triton X-100 (1% final) for 30 min on ice. 250 µl lysate was adjusted to 40% Optiprep by adding 500 µl of 60% Optiprep solution and overlaid with 1.2 ml of 30% Optiprep in TXNE (TNE and 0.1% Triton X-100) and 200 µl TXNE. The samples were centrifuged at 55,000 rpm for 2 h in a rotor (S55S; Sorvall), and five fractions of equal volume were collected from the top. Proteins of each fraction were precipitated with 10% trichloroacetic acid for 15 min on ice. Precipitates were resuspended in 50 µl 2× sample buffer (0.24 M Tris, pH 8.0, 8% SDS, 1 mM DTT, 40% glycerol, and 0.4% bromphenol blue) and heated at 65°C. 25-µl aliquots were loaded onto 12% SDS-PAGE gel and analyzed by Western blotting. Nce102-TAP was detected with a rabbit peroxidase antiperoxidase antibody (Sigma-Aldrich), Pma1 with a rabbit anti-Pma1 antibody (Santa Cruz Biotechnology, Inc.), and a horseradish peroxidase–coupled goat anti–rabbit IgG antibody (Santa Cruz Biotechnology, Inc.).
MS
700 ODs of light-labeled PIL1-TAP and heavy-labeled PIL1-TAP Δnce102 cells were harvested by centrifugation resuspended in 5 ml of buffer (150 mM KOAc, 20 mM Hepes, pH 7.4, 10% glycerol, and complete protease inhibitor cocktail [Roche]) and phosphatase inhibitor cocktail (Sigma-Aldrich) and frozen in liquid nitrogen. Total protein was extracted by bead milling; the thawed lysates were incubated with Triton X-100 (1% final) for 30 min at 4°C and clarified by two consecutive spins of 4 min at 1,000 g. To immunopurify Pil1-TAP, equivalent amounts of the lysates of PIL1-TAP cells and PIL1-TAP Δnce102 cells, according to the protein concentration, were incubated with IgG conjugated to agarose beads (GE Healthcare) for 2 h, washed, and eluted by TEV cleavage and centrifugation. The eluates were mixed, reduced for 20 min at RT in 1 mM DTT, and alkylated for 30 min by 5.5 mM iodoacetamide at RT in the dark. Nu-PAGE sample buffer (Invitrogen) was added, and the sample was loaded onto 4–12% Nu-PAGE Bis-Tris SDS-PAGE gels (Invitrogen). Two Pil1 bands were excised from the gel, and proteins were digested with endoproteinase LysC in gel overnight at RT. The resulting peptides were extracted with 30% acetonitrile and 3% trifluoroacetic acid, reduced in a speed vacuum centrifuge, and desalted and concentrated on a reversed-phase column (C18 StageTip; Rappsilber et al., 2003).
In a different approach, the mixed eluates were precipitated with chloroform/methanol and resuspended in 8 M urea. Proteins were alkylated, reduced, and directly digested in solution overnight at RT with LysC. Peptides were desalted and concentrated on StageTips. Peptides were eluted from the StageTips by passage of 2× 20 µl solvent B (80% acetonitrile and 0.5% acetic acid). The volume was reduced to 4 µl in a speed vacuum centrifuge, and 2 µl solvent A* (2% acetonitrile and 1% trifluoroacetic acid) was added to acidify the sample. Peptides were separated on line to the mass spectrometer by using an HPLC system (1200; Agilent Technologies). 5-µl samples were loaded with constant flow of 500 nl/min onto a 15-cm fused silica emitter with an inner diameter of 75 µm (Proxeon Biosystems) packed in house with reverse-phase 3-µm resin (ReproSil-Pur C18-AQ; provided by Dr. Maisch GmbH). Peptides were eluted with a segmented gradient of 10–60% solvent B over 110 min with a constant flow of 250 nl/min. The HPLC system was coupled to a mass spectrometer (linear trap quadrupole Orbitrap; Thermo Fisher Scientific) via a nanoscale LC interface (Proxeon Biosystems). The spray voltage was set to 2.2 kV, and the temperature of the heated capillary was set to 180°C.
The mass spectrometer was operated in positive-ion mode. Survey full-scan MS spectra (m/z = 300–1,600) were acquired with a resolution of 60,000 at m/z = 400 after accumulation of 1,000,000 ions. The most intense ions (up to five) from the preview survey scan delivered by the Orbitrap were sequenced by collision-induced dissociation (collision energy 35%) in the linear trap quadrupole after accumulation of 5,000 ions. Multistage activation was enabled in all MS/MS events to improve fragmentation of phosphopeptides. Maximal filling times were 1,000 ms for the full scans and 150 ms for the MS/MS. Precursor ion charge state screening was enabled, and all unassigned charge states as well as singly charged peptides were rejected. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 90 s and a relative mass window of 10 ppm. Orbitrap measurements were performed enabling the lock mass option for survey scans to improve mass accuracy (Olsen et al., 2005). Data were acquired using the Xcalibur software (version 2.0.5).
Mass spectra were analyzed using the in house–developed software MaxQuant (version 1.0.1; Cox and Mann, 2008). The data were searched against the yeast database concatenated with reversed copies of all sequences (Moore et al., 2002; Peng et al., 2003) and supplemented with frequently observed contaminants (porcine trypsin, Achromobacter lyticus lysyl-endopeptidase, and human keratins) using Mascot (version 2.2.0; Matrix Science; Perkins et al., 1999).
Carbamidomethylated cysteines were set as fixed, oxidation of methionine, N-terminal acetylation and phosphorylation of serines, threonines, and tyrosines as variable modification. 0.5 D was set as maximum-allowed mass deviation for MS/MS peaks, and a maximum of three missed cleavages were allowed. Maximum false discovery rates were set to 0.01 both on peptide and protein levels. Minimum-required peptide length was six amino acids. Proteins with at least two peptides (thereof one uniquely assignable to the respective sequence) were considered identified.
Online supplemental material
Fig. S1 shows a flowchart of the screening process and the quantitation of Pil1-GFP mutant phenotypes. Fig. S2 shows mass spectrometric analysis of Pil1 phosphorylation in wild-type and Δnce102 or myriocin-treated and -untreated control cells. Fig. S3 shows growth curves of Δnce102 and wild-type cells untreated or treated with myriocin as well as microarray profiles of NCE102 versus sphingolipid biosynthesis gene expression. Fig. S4 shows Nce102-GFP localization depending on sphingolipids, phosphomimicking Pil1 mutant being enhanced by deletion of NCE102, and epistasis between Δnce102 and Pkh kinase mutants. Fig. S5 shows colocalization of Pkh2 and Nce102 depending on sphingolipid synthesis. Table S1 contains yeast strains used. Table S2 shows the phenotypes and classification of Pil1-GFP visual screen hits. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200811081/DC1.
Acknowledgments
We thank Nevan Krogan and Dijana Avdic for help generating the PIL1-GFP deletion library, Ron Vale for use of his high throughput microscope, and Nico Stuurman for his invaluable expertise to image the Pil1-GFP deletion library. We also thank Stefan Jentsch, Robert Farese Jr., Lena Karotki, Natalie Krahmer, and Doris Berchtold for discussions, Howard Riezman (University of Geneva, Geneva, Switzerland) for providing reagents, and Andrew Kruchinsky for sharing unpublished data.
This work was supported by the Max Planck Society, the German Research Association (DFG; grant WA 1669/2-1), the German-Israeli Foundation (grant G-2193-1838.9), and the Human Frontiers Science Program (to T.C. Walther). K. Moreira is a recipient of an aging training grant from National Institutes of Health/National Institute on Aging. P. Walter is a Howard Hughes Medical Institute Investigator. P.S. Aguilar is supported by the ANII (DCI–ALA/2007/19.040).
Footnotes
Abbreviations used in this paper: LC, liquid chromatography; MCC, membrane compartment occupied by Can1; MCP, membrane compartment occupied by Pma1; MS, mass spectrometry; PHS, phytosphingosine; SILAC, stable isotope labeling by amino acids in cell culture; TAP, tandem affinity purification.
References
- Anderson R.G., Jacobson K. 2002. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains.Science. 296:1821–1825 [DOI] [PubMed] [Google Scholar]
- Aronova S., Wedaman K., Anderson S., Yates J., III, Powers T. 2007. Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae.Mol. Biol. Cell. 18:2779–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronova S., Wedaman K., Aronov P.A., Fontes K., Ramos K., Hammock B.D., Powers T. 2008. Regulation of ceramide biosynthesis by TOR complex 2.Cell Metab. 7:148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat M., Keranen S., Shevchenko A., Shevchenko A., Simons K. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast.Proc. Natl. Acad. Sci. USA. 97:3254–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler T., Bacikova D., Gable K., Hopkins L., Johnson C., Slife H., Dunn T. 1998. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Δ mutant.J. Biol. Chem. 273:30688–30694 [DOI] [PubMed] [Google Scholar]
- Belfort G.M., Bakirtzi K., Kandror K.V. 2005. Cellugyrin induces biogenesis of synaptic-like microvesicles in PC12 cells.J. Biol. Chem. 280:7262–7272 [DOI] [PubMed] [Google Scholar]
- Berchtold D., Walther T.C. 2009. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain.Mol. Biol. Cell. 20:1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice S.E., Alford C.W., Cowart L.A. 2009. Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae.J. Biol. Chem. 284:7588–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A., Torrance P.D., Kobayashi T., Thorner J., Alessi D.R. 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast.Curr. Biol. 9:186–197 [DOI] [PubMed] [Google Scholar]
- Cleves A.E., Cooper D.N., Barondes S.H., Kelly R.B. 1996. A new pathway for protein export in Saccharomyces cerevisiae.J. Cell Biol. 133:1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification.Nat. Biotechnol. 26:1367–1372 [DOI] [PubMed] [Google Scholar]
- Daquinag A., Fadri M., Jung S.Y., Qin J., Kunz J. 2007. The yeast PH domain proteins Slm1 and Slm2 are targets of sphingolipid signaling during the response to heat stress.Mol. Cell. Biol. 27:633–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- deHart A.K., Schnell J.D., Allen D.A., Hicke L. 2002. The conserved Pkh–Ypk kinase cascade is required for endocytosis in yeast.J. Cell Biol. 156:241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Xiong X., Krutchinsky A.N. 2009. Unifying fluorescence microscopy and mass spectrometry for studying protein complexes in cells.Mol. Cell. Proteomics. 8:1413–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J.L., Iyer V.R., Brown P.O. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale.Science. 278:680–686 [DOI] [PubMed] [Google Scholar]
- Fischer M., Haase I., Simmeth E., Gerisch G., Muller-Taubenberger A. 2004. A brilliant monomeric red fluorescent protein to visualize cytoskeleton dynamics in Dictyostelium.FEBS Lett. 577:227–232 [DOI] [PubMed] [Google Scholar]
- Friant S., Lombardi R., Schmelzle T., Hall M.N., Riezman H. 2001. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast.EMBO J. 20:6783–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. 2000. Genomic expression programs in the response of yeast cells to environmental changes.Mol. Biol. Cell. 11:4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans B.L., Grotsch H., Mukhopadhyay D., Fernandez I.M., Pfannstiel J., Idrissi F.Z., Lechner J., Riezman H., Geli M.I. 2006. TEDS site phosphorylation of the yeast myosins I is required for ligand-induced but not for constitutive endocytosis of the G protein-coupled receptor Ste2p.J. Biol. Chem. 281:11104–11114 [DOI] [PubMed] [Google Scholar]
- Grossmann G., Opekarova M., Malinsky J., Weig-Meckl I., Tanner W. 2007. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast.EMBO J. 26:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann G., Malinsky J., Stahlschmidt W., Loibl M., Weig-Meckl I., Frommer W.B., Opekarova M., Tanner W. 2008. Plasma membrane microdomains regulate turnover of transport proteins in yeast.J. Cell Biol. 183:1075–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G., Gable K., Kohlwein S.D., Beaudoin F., Napier J.A., Dunn T.M. 2002. The Saccharomyces cerevisiae YBR159w gene encodes the 3-ketoreductase of the microsomal fatty acid elongase.J. Biol. Chem. 277:35440–35449 [DOI] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O'Shea E.K. 2003. Global analysis of protein localization in budding yeast.Nature. 425:686–691 [DOI] [PubMed] [Google Scholar]
- Inagaki M., Schmelzle T., Yamaguchi K., Irie K., Hall M.N., Matsumoto K. 1999. PDK1 homologs activate the Pkc1-mitogen-activated protein kinase pathway in yeast.Mol. Cell. Biol. 19:8344–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes.Yeast. 21:947–962 [DOI] [PubMed] [Google Scholar]
- Jin H., McCaffery J.M., Grote E. 2008. Ergosterol promotes pheromone signaling and plasma membrane fusion in mating yeast.J. Cell Biol. 180:813–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.C., Hamamoto S., Schekman R. 2002. Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum.J. Biol. Chem. 277:22395–22401 [DOI] [PubMed] [Google Scholar]
- Liu K., Zhang X., Lester R.L., Dickson R.C. 2005. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9.J. Biol. Chem. 280:22679–22687 [DOI] [PubMed] [Google Scholar]
- Luo G., Gruhler A., Liu Y., Jensen O.N., Dickson R.C. 2008. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover.J. Biol. Chem. 283:10433–10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinska K., Malinsky J., Opekarova M., Tanner W. 2003. Visualization of protein compartmentation within the plasma membrane of living yeast cells.Mol. Biol. Cell. 14:4427–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinska K., Malinsky J., Opekarova M., Tanner W. 2004. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells.J. Cell Sci. 117:6031–6041 [DOI] [PubMed] [Google Scholar]
- Moore R.E., Young M.K., Lee T.D. 2002. Qscore: an algorithm for evaluating SEQUEST database search results.J. Am. Soc. Mass Spectrom. 13:378–386 [DOI] [PubMed] [Google Scholar]
- Nagiec M.M., Nagiec E.E., Baltisberger J.A., Wells G.B., Lester R.L., Dickson R.C. 1997. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene.J. Biol. Chem. 272:9809–9817 [DOI] [PubMed] [Google Scholar]
- Olsen J.V., de Godoy L.M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. 2005. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap.Mol. Cell. Proteomics. 4:2010–2021 [DOI] [PubMed] [Google Scholar]
- Ong S.E., Blagoev B., Kratchmarova I., Kristensen D.B., Steen H., Pandey A., Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics.Mol. Cell. Proteomics. 1:376–386 [DOI] [PubMed] [Google Scholar]
- Otsu N. 1979. A threshold selection method from gray-level histograms.IEEE Trans. Sys. 9:62–66 [Google Scholar]
- Paul S., Gable K., Beaudoin F., Cahoon E., Jaworski J., Napier J.A., Dunn T.M. 2006. Members of the Arabidopsis FAE1-like 3-ketoacyl-CoA synthase gene family substitute for the Elop proteins of Saccharomyces cerevisiae.J. Biol. Chem. 281:9018–9029 [DOI] [PubMed] [Google Scholar]
- Peng J., Elias J.E., Thoreen C.C., Licklider L.J., Gygi S.P. 2003. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome.J. Proteome Res. 2:43–50 [DOI] [PubMed] [Google Scholar]
- Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data.Electrophoresis. 20:3551–3567 [DOI] [PubMed] [Google Scholar]
- Powers T. 2007. TOR signaling and S6 kinase 1: yeast catches up.Cell Metab. 6:1–2 [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics.Anal. Chem. 75:663–670 [DOI] [PubMed] [Google Scholar]
- Roelants F.M., Torrance P.D., Bezman N., Thorner J. 2002. Pkh1 and pkh2 differentially phosphorylate and activate ypk1 and ykr2 and define protein kinase modules required for maintenance of cell wall integrity.Mol. Biol. Cell. 13:3005–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai H., Okamoto Y., Luberto C., Mao C., Bielawska A., Domae N., Hannun Y.A. 2000. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae.J. Biol. Chem. 275:39793–39798 [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Collins S.R., Thompson N.J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J.F., et al. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile.Cell. 123:507–519 [DOI] [PubMed] [Google Scholar]
- Simons K., Ikonen E. 1997. Functional rafts in cell membranes.Nature. 387:569–572 [DOI] [PubMed] [Google Scholar]
- Simons K., Toomre D. 2000. Lipid rafts and signal transduction.Nat. Rev. Mol. Cell Biol. 1:31–39 [DOI] [PubMed] [Google Scholar]
- Simons K., Vaz W.L. 2004. Model systems, lipid rafts, and cell membranes.Annu. Rev. Biophys. Biomol. Struct. 33:269–295 [DOI] [PubMed] [Google Scholar]
- Sun Y., Taniguchi R., Tanoue D., Yamaji T., Takematsu H., Mori K., Fujita T., Kawasaki T., Kozutsumi Y. 2000. Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast.Mol. Cell. Biol. 20:4411–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi M., Audhya A., Parsons A.B., Boone C., Emr S.D. 2006. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation.Mol. Cell. Biol. 26:5861–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Pagé N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants.Science. 294:2364–2368 [DOI] [PubMed] [Google Scholar]
- van Meer G., Voelker D.R., Feigenson G.W. 2008. Membrane lipids: where they are and how they behave.Nat. Rev. Mol. Cell Biol. 9:112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T.A., Emr S.D. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast.J. Cell Biol. 128:779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Brickner J.H., Aguilar P.S., Bernales S., Pantoja C., Walter P. 2006. Eisosomes mark static sites of endocytosis.Nature. 439:998–1003 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Aguilar P.S., Frohlich F., Chu F., Moreira K., Burlingame A.L., Walter P. 2007. Pkh-kinases control eisosome assembly and organization.EMBO J. 26:4946–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Lester R.L., Dickson R.C. 2004. Pil1p and Lsp1p negatively regulate the 3-phosphoinositide-dependent protein kinase-like kinase Pkh1p and downstream signaling pathways Pkc1p and Ypk1p.J. Biol. Chem. 279:22030–22038 [DOI] [PubMed] [Google Scholar]