Abstract
Hypusination is a unique posttranslational modification by which lysine is transformed into the atypical amino acid hypusine. eIF5A (eukaryotic initiation factor 5A) is the only known protein to contain hypusine. In this study, we describe the identification and characterization of nero, the Drosophila melanogaster deoxyhypusine hydroxylase (DOHH) homologue. nero mutations affect cell and organ size, bromodeoxyuridine incorporation, and autophagy. Knockdown of the hypusination target eIF5A via RNA interference causes phenotypes similar to nero mutations. However, loss of nero appears to cause milder phenotypes than loss of eIF5A. This is partially explained through a potential compensatory mechanism by which nero mutant cells up-regulate eIF5A levels. The failure of eIF5A up-regulation to rescue nero mutant phenotypes suggests that hypusination is required for eIF5A function. Furthermore, expression of enzymatically impaired forms of DOHH fails to rescue nero clones, indicating that hypusination activity is important for nero function. Our data also indicate that nero and eIF5A are required for cell growth and affect autophagy and protein synthesis.
Introduction
Hypusination is a unique form of posttranslational modification occurring in eukaryotic organisms that transforms the amino acid lysine into the atypical amino acid hypusine (Park et al., 1982, 1997; Gordon et al., 1987). Thus far, a specific lysine residue on eIF5A (eukaryotic initiation factor 5A) is the only known target for this modification (Cooper et al., 1983; Smit-McBride et al., 1989). The formation of hypusine occurs via a two-step process. Each step of this process is catalyzed by a different enzyme (Park et al., 1982). In the first step, deoxyhypusine synthase (DHS) cleaves spermidine and transfers the 4-amino butyl moiety to a specific lysine residue of eIF5A (Wolff et al., 1990). In the second step, deoxyhypusine hydroxylase (DOHH) hydroxylates deoxyhypusine and irreversibly completes the hypusination process (Abbruzzese et al., 1986; Park et al., 2003). The protein responsible for DOHH function has only recently been identified and characterized. DOHH is an atypical hydroxylase consisting of eight HEAT (named after huntingtin, elongation factor 3, protein phosphatase 2A, and target of rapamycin [Tor]) repeat motifs (Joe et al., 1995; Kang et al., 1995; Park et al., 2006). DOHH is a metalloenzyme that requires iron ions for enzymatic activity (Park et al., 1982; Kim et al., 2006). Four conserved histidine–glutamic acid metal-binding motifs have been identified in DOHH (Park et al., 2006). Mutagenesis of these binding motifs impairs the enzyme's ability to bind iron and concomitantly abolishes its enzymatic activity (Kang et al., 2007).
Several lines of evidence suggest that eIF5A plays a role in cell proliferation. Inhibition of eIF5A function through drug-mediated hypusination blockage causes proliferation arrest in mammalian cell lines. Spermidine analogues that inhibit DHS activity as well as various metal ion chelators that inhibit DOHH enzymatic function have been shown to affect proliferation in vitro (Park et al., 1982, 1994; Jakus et al., 1993; Hanauske-Abel et al., 1994). Furthermore, inhibiting hypusination through cellular spermidine depletion also blocks cell proliferation in both mammalian cell lines as well as in yeast (Byers et al., 1994; Chattopadhyay et al., 2003). More direct evidence supporting a role for eIF5A in proliferation comes from genetic studies in yeast. Mutations in TIF51A and TIF51B, which encode eIF5A, or loss of DYS1 (DHS homologue) in yeast cause cell inviability as well as G1–S phase arrest (Schnier et al., 1991; Wöhl et al., 1993; Sasaki et al., 1996; Park et al., 1998; Chatterjee et al., 2006). Interestingly, LIA1 (ligand of eIF5A 1; yeast DOHH) is not essential for viability and has only a mild effect on the proliferative ability of Saccharomyces cerevisiae (Park et al., 2006). Moreover, the function of LIA1 remains unknown as no major phenotypes are associated with its loss.
Lack of evidence supporting a role for yeast DOHH in eIF5A proliferation regulation questions the importance of the second step in the hypusination process. Thus, the only current evidence that argues for a role for DOHH in eIF5A regulation comes from drug studies that block DOHH enzymatic function through metal ion chelators in mammalian cell culture systems (Park et al., 1982; Hanauske-Abel et al., 1994), leading to the argument that the second step in the hypusination process is probably important only in higher eukaryotes. However, the effect of metal ion chelators on proliferation may be nonspecific. Therefore, genetic experiments on DOHH in higher eukaryotic organisms may shed light on the importance of the second step in the hypusination pathway.
This study addresses the function of DOHH in Drosophila melanogaster. Mutations in nero, the Drosophila DOHH homologue, were identified in a genetic screen for genes that regulate bristle number. nero is essential for organismal viability and plays a role in a wide number of important processes such as cell growth, proliferation, and autophagy. These phenotypes are reminiscent of mutations in the Tor pathway, but no clear epistatic relationship was found to exist between these pathways. As eIF5A is the sole known target of hypusination, we analyzed eIF5A function using RNAi. Loss of eIF5A causes phenotypes highly similar to but more severe than nero. The similarities in phenotypes imply that both nero and eIF5A govern the same processes. This also implicates eIF5A in processes besides proliferation, including autophagy and cell growth. Interestingly, we find that eIF5A is highly up-regulated in nero mutants. This up-regulation may partially compensate for the loss of nero and possibly accounts for the differences in the severity of the phenotypes of the loss of either gene. Furthermore, we find that the disruption of DOHH enzymatic activity impairs nero function, suggesting that its hypusination function is critical for its function. Finally, knockdown of either eIF5A or Nero through RNAi in S2 cells affects translation elongation.
Results
nero mutations in Drosophila DOHH affect sensory organ development
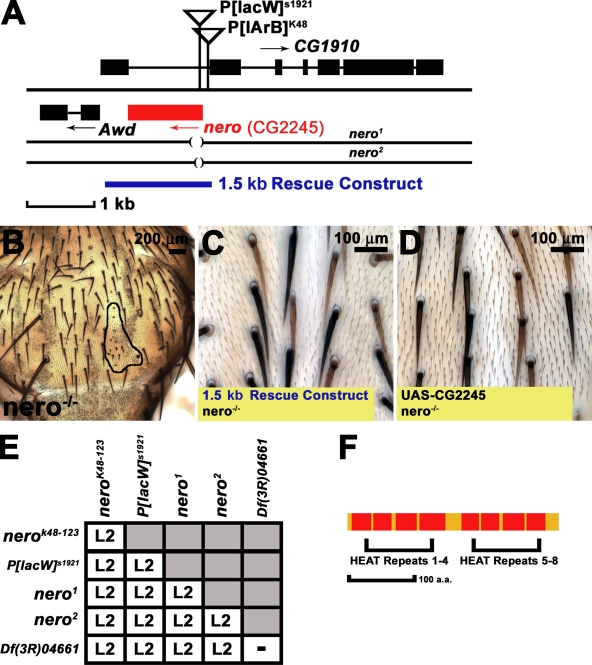
Lyman et al. (1996) performed a P element screen to identify insertions that alter bristle number in fruit flies and identified several insertions within genes previously implicated in bristle development. A similar screen in a genetically well-controlled background confirmed that such screens can be used for the purposes of identifying genes that govern neural development (Norga et al., 2003). One P element strain, P[lArB]K48, which was generated in the initial Lyman et al. (1996) screen, exhibits a subtle bristle loss in homozygous flies. Therefore, we decided to focus on this P element insertion. P[lArB]K48 is inserted in an intron of the CG1910 gene and in close proximity to CG2245, potentially affecting both genes (Fig. 1 A). We identified a second P element, P[lacW]s1921, associated with this locus and inserted within the 5′ untranslatable region of CG2245 (Fig. 1 A; Spradling et al., 1999). We excised both P[lArB]K48 and P[lacW]s1921 and consequently generated three homozygous lethal mutations. To determine whether these mutations affect bristle development, each of the three mutant alleles was recombined onto a flippase (FLP) recombination target (FRT) chromosome for clonal analysis (Xu and Rubin, 1993). Mutant clones generated on the thorax show bristle loss as well as very small bristles (Fig. 1 B). Therefore, we named the gene nero after a nearly bald Flemish comic book character. Two mutations derived from P[lacW]s1921, nero1 and nero2, are associated with small molecular deletions in CG2245 that remove 189 bp and 123 bp, respectively, of the gene's open reading frame (Fig. 1 A). Animals carrying these mutations die as second instar (L2) larvae whether they are homozygous or in heteroallelic combinations (Fig. 1 E). Larvae carrying nero alleles in combination with a deficiency that spans the CG2245 genomic region also die as second instar animals, suggesting that all nero alleles are strong loss of function alleles or null alleles (Fig. 1 E).
Figure 1.
nero mutants disrupt CG2245 and affect bristle size and viability. (A) nero genomic locus. P[lArB]K48 and P[lacW]s1921 are both inserted within CG1910 and in proximity to CG2245 (shown in red). nero1 and nero2 alleles are derived from the excision of P[lacW]s1921 and disrupt the translational start of CG2245 (disrupted black lines). The 1.5-kb genomic rescue construct (shown in blue) rescues nero mutants to viability. (B) nero mutant clones (outlined in black) display a disruption in both the patterning and the size of bristles (genotype: y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS/+; FRT82B nero1/FRT82B hsp70-CD2 y+ tub-GAL80). (C) The 1.5-kb genomic rescue construct rescues bristle defects in nero mutant clones (genotype: y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS/+; P[CaSper4–1.5 KB nero rescue]/+; FRT82B nero1/FRT82B hsp70-CD2 y+ tub-GAL80). (D) Overexpression of the nero cDNA (CG2245) in nero mutant clones rescues bristle size defects (genotype: y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS/+; UAS-nero/+; FRT82B nero1/FRT82B hsp70-CD2 y+ tub-GAL80). (C and D) Rescued mutant bristles are marked by the recessive yellow mutation and appear light brown. (E) The lethal phase of nero mutants as homozygotes, in heteroallelic combinations, or in combination with Df(3R)04661 is L2 lethal. (F) nero encodes a small 302-aa protein composed of two dyads consisting of four HEAT motifs each.
To verify that the nero mutations constitute a loss of CG2245, we generated a 1.5-kb genomic rescue construct containing the CG2245 coding region (Fig. 1 A). This construct is sufficient to rescue the lethality of nero mutant larvae to full viability as well as bristle phenotypes associated with nero clones (Fig. 1 C). Ubiquitous overexpression of CG2245 cDNA under upstream activation sequence (UAS) regulation using the actin-GAL4 driver rescues the lethality associated with nero mutations (Brand and Perrimon, 1993). Furthermore, overexpression of CG2245 in nero mutant clones using mosaic analysis with a repressible cell marker system is able to rescue the bristle phenotypes (Fig. 1 D; Lee and Luo, 1999). Altogether, these experiments demonstrate that nero mutations correspond to a loss of CG2245. Interestingly, nero overexpression in a wide variety of contexts using different GAL4 drivers has no phenotypic effects.
CG2245 encodes a 302-aa protein bearing a high level of homology to eukaryotic DOHH proteins (57% identity with Caenorhabditis elegans and mouse homologues and a 59% identity with its human homologue). Thus, nero likely encodes the Drosophila DOHH homologue. Similar to the yeast and vertebrate homologues, the Drosophila DOHH protein consists of two dyads of four HEAT repeat domains (Fig. 1 F; Park et al., 2006). The human DOHH (hDOHH) protein bears four histidine–glutamic acid motifs important for DOHH enzymatic activity. These motifs are also conserved in the fly homologue, suggesting functional conservation.
Nero localizes to the ER
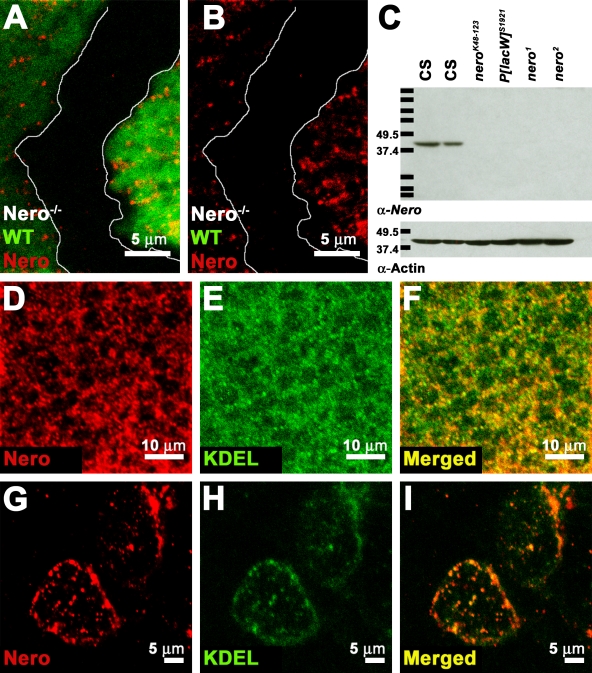
To determine the spatial expression of Nero as well as its subcellular localization, an antibody was raised against the full-length protein. The antibody specifically recognizes the Nero protein in immunohistochemical labeling, as all immunoreactivity is lost in nero mutant clones (Fig. 2, A and B). This antibody also specifically recognizes Nero on Western blots as a single ∼39-kD band in wild-type L2 protein lysates (Fig. 2 C). This band is not detected in nero mutant lysates. Thus, the ∼39-kD band observed in wild-type lysates corresponds to the Nero protein (Fig. 2 C). The inability of the antibody to detect Nero in nero mutants is further evidence that the nero alleles are probably null alleles.
Figure 2.
Nero localizes to the ER. (A–C) Antibodies generated against Nero specifically detect the Nero protein on both immunohistochemical preparations and Western blots. (A and B) Nero antibody fails to recognize the protein in mutant clones marked by the absence of GFP (genotype: y w hs-FLP; FRT82B nero1/FRT82B Ubi-GFP). White lines mark the clonal boundary. WT, wild type. (C) The Nero antibody detects a single ∼39-kD band in Canton-S (CS) L2 larval protein extracts (first and second lanes) but fails to detect the protein in protein extracts from nerok48-123 (third lane), P[lacW]s1921 (fourth lane), nero1 (fifth lane), and nero2 (sixth lane) L2 larvae on Western blots. Protein lysates from 10 larvae were loaded in each lane except the second lane, in which only five larvae were loaded. Actin was probed as a loading control. Protein standards run along with larval lysates are marked with black bars. Their sizes are recorded in kilodaltons on the left. (D–F) Double labeling of Canton-S third instar wing discs using the Nero antibody and the KDEL antibody shows extensive colocalization, demonstrating that Nero is ER associated. (G–I) Double labeling of Canton-S third instar garland cells using Nero and KDEL antibodies.
The Nero protein is expressed in larval imaginal disc cells (unpublished data). The subcellular distribution shows that the protein is excluded from the nucleus. The protein distribution within the cell appears punctate, indicating that Nero is most likely associated with an organelle (Fig. 2 D). Double labeling of imaginal discs with the Nero antibody and a monoclonal antibody that recognizes the ER retention signal KDEL shows extensive colocalization, revealing that the Nero protein localizes at least in part to the ER (Fig. 2, D–F; Pelham, 1990). ER localization was also observed in larval garland cells (Fig. 2, G–I). As the Nero protein does not bear a signal peptide or a C-terminal KDEL sequence tag, Nero most likely associates with the external surface of the ER.
nero is cell autonomously required for cell growth and affects the cell cycle
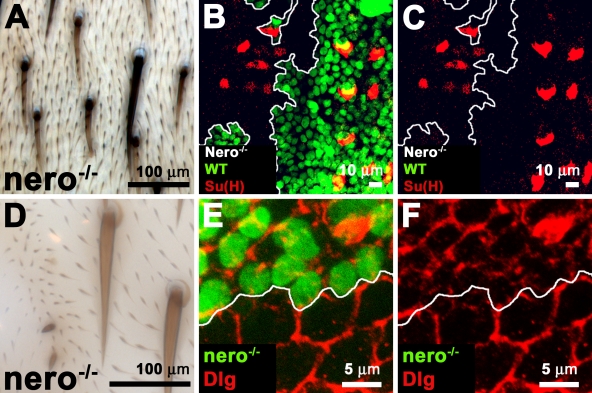
nero mutations cause both mild bristle loss as well as a severe reduction in bristle size (Figs. 1 B and 3 A). Bristles are composed of two external cells, the socket and the shaft. Socket cells dramatically increase in size within a 5-h period between the time points of their birth at 19 h after puparium formation (APF) and 24 h APF (Audibert et al., 2005). To determine whether the specification and the size of socket cells are affected early during the development of these cells, we labeled pupal thoraxes 24 h APF with the suppressor of Hairless (Su(H)) antibody, which is a marker for socket cells. Su(H) is expressed both in the cytoplasm and the nucleus of the socket cell, allowing visualization of the entire cell (Gho et al., 1996). As shown in Fig. 3 (B and C), nero mutant socket cells at this time point are much smaller than wild type. Thus, nero mutant socket cells are specified but fail to reach their mature size.
Figure 3.
nero is required for cell growth. (A) The shaft and socket cells that comprise the external sensory organ on the thorax of the adult fly are typically shorter and smaller in nero mutant clones (genotype: y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS/+; FRT82B nero1/FRT82B hsp70-CD2 y+ tub-Gal80). Mutant bristles are marked by the recessive yellow mutation and appear light brown. (B and C) Socket cells observed 24 h APF labeled with Su(H) are smaller than wild-type (WT) socket cells (genotype: y w hs-FLP/+; UAS-FLP/+; FRT82B nero1/C684-GAL4 FRT82B Ubi-GFP M[3]). The clone is marked by the absence of GFP. (D) Epidermal cells on the adult thorax in the vicinity of nero mutant bristles are often smaller in size, as indicated by smaller trichome size and closer trichome spacing (genotype: y w hs-FLP/+; FRT82B nero1/FRT82B Ubi-GFP). (E and F) nero mutant epidermal cells in the thorax 47 h APF are smaller than adjacent wild-type cells. Thoraxes are labeled with Dlg to reveal cell outlines (genotype: y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS/+; FRT82B nero1/FRT82B hsp70-CD2 y+ tub-GAL80). The mutant clone is marked positively with GFP. (B, C, E, and F) White lines mark the clonal boundary.
The growth defects observed in nero mutant clones are not specific to cells of the bristle lineage. As shown in Fig. 3 D, cells adjacent to nero mutant bristles are smaller in size as indicated by trichome size and spacing. This suggests that nero function is not confined solely to the regulation of bristle size. Indeed, ectodermal cells that lack nero in developing thoraxes 47 h APF are significantly smaller than adjacent wild-type cells (Fig. 3, E and F; Woods and Bryant, 1991). These defects are confined to nero mutant cells, and thus, nero is cell autonomously required for cell growth (Fig. 3, E and F). These nero mutant cells also show a decrease in Discs large (Dlg) expression, which is a basolateral cell membrane marker (Fig. 3, E and F). These effects on cell size are not restricted to the thorax, as rhabdomere size in mutant eye clones is also reduced when compared with adjacent wild-type cells, suggesting a defect in photoreceptor size (unpublished data). Thus, nero is cell autonomously required to achieve correct cell size in many tissues.
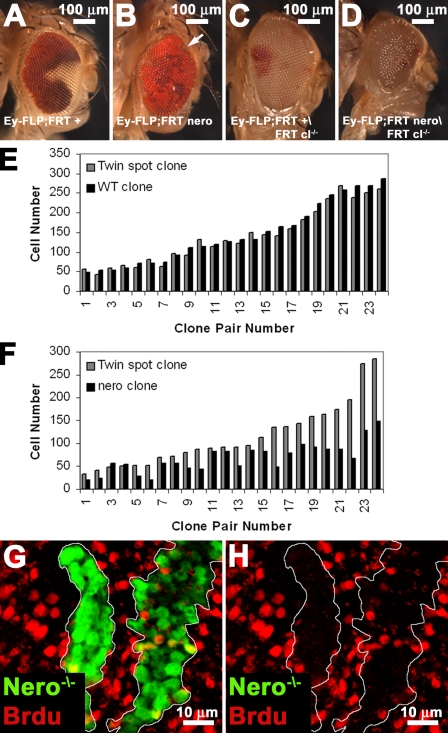
We further analyzed nero function by generating adult somatic clones using the eyeless-FLP system. nero mutant clones generated using the eyeless-FLP technique are generally irrecoverable (Fig. 4, A and B). Interestingly, nero mutant clones are easily obtained using eyeless-FLP when cell competition is ameliorated through the use of an FRT chromosome bearing a cell lethal mutation that effectively removes the mitotic wild-type, sister twin cells (Fig. 4 D). We observe similar results when competition is alleviated using a Minute. This suggests that nero mutant cells are viable but are at a competitive disadvantage. Indeed, nero mutant clones generated using this technique completely overtake the eye (Fig. 4, C and D). However, the resultant nero mutant eyes and heads are much smaller than wild-type heads and resemble the pinhead phenotype associated with mutants in several growth pathways (Fig. 4 D; Oldham et al., 2000). Thus, nero mutations affect cell size and organ size, but nero is not required for cell viability.
Figure 4.
nero regulates organ size, cell number, and proliferation. (A and B) nero clones generated in a wild-type background are poorly competitive. Clones are marked with the absence of white+ and thus appear white. (A) Wild-type clones are easily recovered (genotype: y w eyeless-FLP GMR-lacZ/+; FRT82B+/FRT82B w+). (B) nero clones are usually irrecoverable (genotype: y w eyeless-FLP GMR-lacZ/+; FRT82B nero1/FRT82B w+). The white arrow points to a small nero mutant clone at the edge of the eye. (C and D) To provide nero mutant cells a competitive advantage, clones were made using an FRT-bearing chromosome carrying a recessive cell lethal that effectively eliminates wild-type twin spots. Clones are marked as in A and B. (C) Wild-type clones proliferate and take over most of the eye (genotype: y w eyeless-FLP GMR-lacZ/+; FRT82B+/FRT82B w+ l[3]cl). (D) nero clones generated in a cell lethal background can, like wild type, dominate the entire eye (genotype: y w eyeless-FLP GMR-lacZ/+; FRT82B nero1/FRT82B w+ l[3]cl). nero mutant eyes and heads are smaller than wild-type control heads, suggesting a defect in organ size regulation. Eyes are also rough. (E) Cell number in wild-type (WT) clones is roughly similar to cell number in their twin spots (genotype: y w hs-FLP; FRT82B +/FRT82B Ubi-GFP). Black bars correspond to the clone, and gray bars correspond to the twin spot. (F) nero mutant clones have fewer cells than wild type (genotype: y w hs-FLP; FRT82B nero1/FRT82B Ubi-GFP). Black and gray bars are the same as in E. (G and H) nero mutant clones marked positively with GFP incorporate BrdU more poorly than adjacent wild-type cells (genotype: y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS/+; FRT82B nero1/FRT82B hsp70-CD2 y+ tub-GAL80). White lines mark the clonal boundary.
Quantification of the number of cells in nero mutant clones versus the number of cells in their associated wild-type twin spot in the context of the wing imaginal disc reveals that nero mutant cells are underrepresented. Wild-type clones generated in the wing imaginal disc contain a similar number of cells when compared with the number of cells in their associated twin spot (Fig. 4 E). However, nero clones are smaller and contain fewer cells than their associated wild-type twin spots (Fig. 4 F). The differences in cell number between nero clones and their wild-type twin spots are less obvious in small clones and clearly increase as clones get larger (Fig. 4 F). This is likely caused by differences in Nero protein perdurance in small clones (Garcia-Bellido and Merriam, 1971). nero mutant cells are most likely eliminated because of an inability to compete with wild-type cells.
To determine whether the cell cycle is affected in nero mutant cells, we assayed their ability to incorporate BrdU (Gratzner, 1982). nero mutant clones show a decreased level of BrdU incorporation, indicating a delay in cell cycle progression (Fig. 4, G and H). Thus, the data suggest that nero also plays a role in cellular proliferation.
nero negatively regulates autophagy
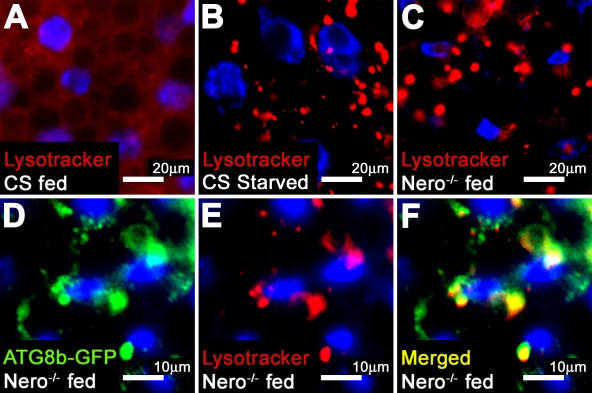
Several growth pathways like the Tor and insulin pathways are linked to the regulation of autophagy (Rusten et al., 2004; Scott et al., 2004). The induction of autophagy in the fat body is a cellular response to amino acid deprivation (Rusten et al., 2004; Scott et al., 2004). Larvae fed with an amino acid source lack autophagosomes in their fat bodies (Fig. 5 A; Rusten et al., 2004; Scott et al., 2004). However, upon amino acid withdrawal, enlarged, acidic intracellular structures are readily visible using the acidic pH reactive dye LysoTracker (Fig. 5 B; Rusten et al., 2004; Scott et al., 2004). These large structures have been shown to correspond to autophagosomes (Rusten et al., 2004; Scott et al., 2004). nero mutant larval fat bodies prominently display enlarged, acidified structures even when the animals are fed, suggesting that nero mutant larvae undergo a constitutive starvation response in the presence of an amino acid source (Fig. 5 C). To rule out a defect in feeding behavior, mutant larvae were fed yeast paste containing food dyes. nero mutant larvae feed properly, as animals show ample consumption of yeast paste (unpublished data).
Figure 5.
nero mutants display autophagic starvation response. (A) Fed second instar Canton-S (CS) larvae lack enlarged, acidic autophagic structures, as marked by LysoTracker fluorescence. (B) Second instar Canton-S animals subjected to a 4-h starvation period show LysoTracker-fluorescent autophagic structures. (C) Fed nero1 larvae display autophagic structures despite nutrient availability. (D–F) LysoTracker-fluorescent structures and ATG8b-GFP fusion proteins colocalize in nero1 mutant larvae, demonstrating that these structures are autophagosomes (genotype: hs-ATG8b/+; FRT82B nero1/FRT82B nero1).
To demonstrate that the acidified structures observed in nero mutant fat bodies are autophagosomes, ATG8b (autophagy-specific gene 8b)-GFP fusion proteins were overexpressed in nero mutants. ATG8b-GFP fusion proteins have been shown to localize to autophagic structures (Scott et al., 2004). LysoTracker-positive structures observed in nero mutant fat body cells colocalize with the ATG8b-GFP fusion proteins, confirming that these structures are indeed autophagosomes (Fig. 5, D–F).
The combination of defects in cell size and autophagy suggests a possible link between nero and the Tor pathway (Oldham et al., 2000; Zhang et al., 2000; Rusten et al., 2004; Scott et al., 2004). Tsc1 (tuberous sclerosis 1) mutations cause constitutive activation of the Tor pathway and have the opposite phenotype of nero mutations. Tsc1 mutations increase cell size, cause tissue overgrowth, increase cell number in clones when compared with their twin spot, and block the induction of autophagy (Gao and Pan, 2001; Potter et al., 2001; Tapon et al., 2001; Rusten et al., 2004; Scott et al., 2004). Therefore, we generated a Tsc1 nero double mutant chromosome to ascertain the possibility of an epistatic relationship between these genes. Unlike either nero or Tsc1 clones, Tsc1 nero double mutant clones cannot be recovered (Fig. S1). Thus, nero and Tsc1 do not exist in a straightforward epistatic relationship. Interestingly, Tsc1 fully suppresses the autophagic defect seen in nero mutant animals (Fig. S1). However, this epistatic relationship is reversed in other contexts such as larval growth and developmental timing. Tsc1 mutant larvae appear to undergo precocious development. We find that this precocious development is suppressed in the Tsc1 nero double mutant larvae (Fig. S1). These results suggest that nero may not play a direct role in Tor signaling and most likely alters cell growth through an independent mechanism.
nero regulates eIF5A
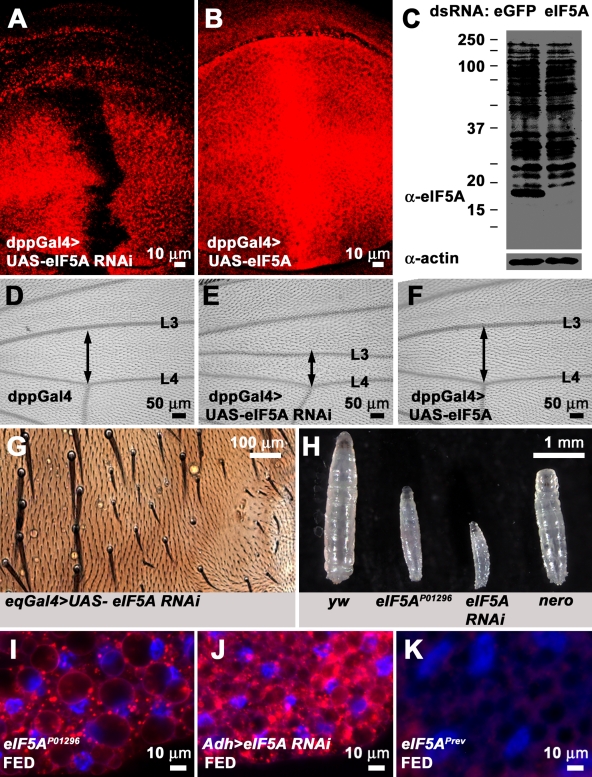
eIF5A is generally believed to be the sole protein that undergoes hypusination in eukaryotic organisms (Cooper et al., 1983; Smit-McBride et al., 1989). Having established a genetic model in which loss of the second step of the hypusination process causes severe phenotypes, we decided to ascertain whether loss of eIF5A causes phenotypes similar to loss of nero, as would be predicted if nero regulated eIF5A function. To test this, we obtained an RNAi line purported to knock down eIF5A (Dietzl et al., 2007). We used a monoclonal antibody raised against the human eIF5A protein to test whether this RNAi line is able to effectively down-regulate eIF5A. Expression of eIF5A double-stranded RNA (dsRNA) under the regulation of dpp-GAL4 strongly reduces the expression of eIF5A in a central stripe that runs down the middle of the wing imaginal disc, proving that the RNAi line is effective and that the antibody is able to specifically recognize the Drosophila eIF5A protein in vivo (Fig. 6 A). Additionally, this antibody is able to recognize the wild-type overexpressed eIF5A protein (Fig. 6 B). Unfortunately, it is difficult to detect the eIF5A protein on Western blots of larval lysates using this antibody. However, it recognizes an ∼17-kD band on Western blots of S2 cell lysates, which corresponds to the expected molecular mass of eIF5A (Fig. 6 C). Furthermore, this 17-kD band is lost in S2 cell lysates treated with eIF5A dsRNA, suggesting that the band specifically corresponds to eIF5A (Fig. 6 C). Altogether, these data suggest that we can efficiently knock down or overexpress eIF5A using UAS-eIF5A RNAi or UAS-eIF5A constructs, respectively. We used UAS-eIF5A RNAi to test whether loss of eIF5A has any phenotypic consequences. Knockdown of eIF5A in the dpp-GAL4 expression domain causes a narrowing of the region between the L3 and L4 wing veins, a phenotype which is often associated with growth mutants in Drosophila (Fig. 6, D and E). However, overexpression of eIF5A appears to have no phenotypic effect on the distance between L3 and L4 veins, and thus, like nero, overexpression of eIF5A has no overt phenotypic consequences (Fig. 6, D and F).
Figure 6.
Loss of eIF5A causes phenotypes reminiscent of nero. (A) Wing imaginal disc in which eIF5A dsRNA has been expressed under the regulation of the dpp-GAL4 driver line and labeled with anti-eIF5A antibody. Anti-eIF5A antibody fails to detect the eIF5A protein in cells expressing eIF5A dsRNA. (B) Wing imaginal disc in which eIF5A under UAS regulation has been overexpressed using dpp-GAL4 and stained with anti-eIF5A antibody. Anti-eIF5A antibody detects the overexpressed eIF5A protein. (C) Anti-eIF5A antibody detects an ∼17-kD band in EGFP dsRNA–treated S2 cells but fails to detect this band in eIF5A dsRNA–treated S2 cells. Actin is used as the loading control. Protein standards run along with S2 cell lysates are marked with black bars. Their sizes are recorded in kilodaltons on the left. (D) Control adult wing (genotype: dpp-GAL/+). (E) Adult wing in which eIF5A dsRNA has been expressed under the regulation of the dpp-Gal4 driver. The distance between the L3 and L4 vein (double-headed arrow) is drastically reduced (genotype: dpp-Gal4/UAS-eIF5A RNAi). (F) Adult wing in which wild-type eIF5A under UAS regulation was overexpressed using dpp-Gal4. The distance between the L3 and L4 vein (double-headed arrow) appears similar to the experimental control (D; genotype: dpp-GAL4/UAS-eIF5A RNAi). (G) RNAi knockdown of eIF5A using eq-GAL4 causes defects in bristle growth similar to nero mutations (genotype: eq-GAL4/+; UAS-eIF5A RNAi). (H) Synchronized larvae 72 h after egg hatching of four different genotypes (from left to right): y w, eIF5AP01296, tub-GAL4/UAS-eIF5A RNAi, and nero1. (I) eIF5AP01296 larval fat bodies undergo constitutive autophagy under fed conditions. (J) RNAi knockdown of eIF5A in the fat body induces constitutive autophagy under fed conditions (genotype: Adh-GAL4/+; UAS-eIF5A RNAi/+). (K) Larval fat bodies of P element revertant eIF5APrev do not undergo autophagy under fed conditions.
We decided to test whether loss of eIF5A causes phenotypes similar to those associated with nero. Flies carrying a transgenic UAS-eIF5A RNAi construct were crossed to the thoracic driver eq-GAL4 to determine whether loss of eIF5A affects bristle size (Pi et al., 2001; Dietzl et al., 2007). We find that eIF5A knockdown results in decreased bristle size on the thorax, which is similar to what we observe in nero mutant clones (compare Fig. 6 G with Fig. 1 B). We also identified a homozygous lethal P element, P[PZ]01296 (eIF5AP01296), inserted within the first intron of eIF5A (Spradling et al., 1999). Excision of this P element reverts the lethality, indicating that the lethality is caused by the insertion. Homozygous mutant eIF5AP01296 larvae exhibit a severe larval growth defect and are much smaller than wild-type larvae, although they live for 8 d (Fig. 6 H). RNAi knockdown of eIF5A using the ubiquitous driver tub-GAL4 severely affects larval growth, and the effect is greater than the phenotype observed in eIF5AP01296 larvae, suggesting that loss of eIF5A using RNAi perhaps constitutes a stronger loss of function condition than eIF5AP01296 (Fig. 6 H). Thus, eIF5AP01296 is most likely a hypomorphic mutation. Interestingly, eIF5A mutations are more severe than nero in terms of larval growth (Fig. 6 H). Finally, we find that the eIF5AP01296 mutation also affects autophagy induction. Similar to nero, eIF5A mutant larval fat bodies undergo constitutive autophagy under fed conditions (Fig. 6 I). The autophagy defect is specific to the P element insertion, as the revertants do not show a defect in autophagy induction (Fig. 6 K). Driving eIF5A RNAi in larval fat bodies using Adh-GAL4 likewise induces autophagy (Fig. 6 J). Thus, the phenotypes associated with loss of eIF5A are very similar to the phenotypes associated with loss of nero, suggesting that Nero and eIF5A function in the same pathway in flies.
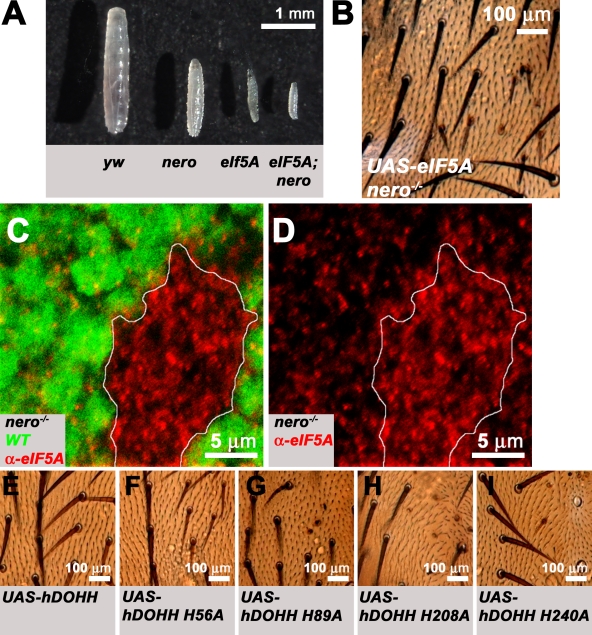
If the target of nero function is eIF5A, loss of eIF5A ought to be epistatic to nero. Thus, eIF5A; nero double mutant larvae should resemble eIF5A mutant larvae. We find that eIF5A; nero double mutant larvae display a slightly more severe larval growth defect than eIF5A mutants alone (Fig. 7 A). As eIF5AP01296 is probably not a null allele, the more severe phenotype is presumably a consequence of increased inactivation of residual eIF5A function in these animals.
Figure 7.
nero regulates eIF5A levels in vivo and Nero's DOHH activity is required for nero function. (A) Synchronized larvae 72 h after egg hatching of four different genotypes (from left to right): y w, nero1, eIF5AP01296, and eIF5AP01296; nero1. The growth of eIF5AP01296; nero1 double mutant larvae appears to be more impaired than either eIF5AP01296 or nero1 homozygous mutant larvae. (B) Overexpression of eIF5A in nero mutant clones fails to rescue bristle growth defects associated with nero mutant clones (genotype: y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS/+; UAS-eIF5A/+; FRT82B nero1/FRT82B hsp70-CD2 y+ tub-GAL80). (C and D) eIF5A levels are dramatically up-regulated in nero mutant clones in the wing imaginal disc. nero mutant clones are marked by the absence of GFP (green; genotype: y w hs-FLP; FRT82B nero1/FRT82B Ubi-GFP). White lines mark the clonal boundary. WT, wild type. (E) Overexpression of hDOHH in nero mutant clones rescues bristle size defects (genotype: y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS/+; UAS-hDOHH/+; FRT82B nero1/FRT82B hsp70-CD2 y+ tub-GAL80). (F–I) Overexpression of mutated forms of hDOHH in nero mutant clones fails to rescue bristle size defects. Genotypes are essentially identical to E except mutated forms of hDOHH are expressed under the UAS regulation. (F) Genotype: y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS/+; UAS-DOHH H56A/+; FRT82B nero1/FRT82B hsp70-CD2 y+ tub-GAL80. (G) Genotype: y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS/+; UAS-hDOHH H89A/+; FRT82B nero1/FRT82B hsp70-CD2 y+ tub-GAL80. (H) Genotype: y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS/+; UAS-hDOHH H208A/+; FRT82B nero1/FRT82B hsp70-CD2 y+ tub-GAL80. (I) Genotype: y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS/+; UAS-hDOHH H240A/+; FRT82B nero1/FRT82B hsp70-CD2 y+ tub-GAL80. (B and E–I) Mutant bristles are marked by the recessive yellow mutation and appear light brown.
Interestingly, the larval growth defect caused by the loss of eIF5A is more severe than loss of nero (Figs. 6 H and 7 A). This may be the result of several causes. One possibility is that eIF5A has both hypusination-dependent and -independent functions. Another possibility is that nero is required only for optimal eIF5A function, and thus, eIF5A maintains some modicum of activity in nero mutants. If eIF5A is still partially active even in the absence of hypusination, overexpression of eIF5A ought to ameliorate nero mutant phenotypes. To test this, we overexpressed eIF5A in nero mutant clones. We found that overexpression of eIF5A is not sufficient to rescue bristle size defects associated with nero mutant clones (Fig. 7 B). Moreover, we were surprised to find that eIF5A is strongly up-regulated in nero mutant clones (Fig. 7, C and D). This strong up-regulation possibly accounts for the weaker phenotype observed in nero mutant clones and may represent an adaptive response of the cells to the inefficiently functioning eIF5A protein.
Finally, to directly assess the requirement for DOHH activity, we overexpressed wild-type and enzymatically inactive versions of hDOHH in nero mutant clones. Alanine replacement of histidines 56, 89, 207, and 240 of hDOHH, all of which are conserved in the Drosophila protein, has been shown to abolish hypusine enzymatic function and iron-binding ability without affecting eIF5A substrate binding (Kang et al., 2007). These residues were mutated separately in the hDOHH cDNA and introduced into flies to determine whether these proteins could complement the loss of nero. Overexpression of the wild-type hDOHH protein fully complements defects in bristle size associated with nero mutant clones on the thorax and does not cause any other obvious phenotype (Fig. 7 E). This result demonstrates that nero is the DOHH homologue in flies. However, overexpression of the four different enzymatically inactive mutant proteins in nero mutant clones fails to rescue defects in bristle size, revealing the importance of the hypusination activity for nero function (Fig. 7, F–I).
Both Nero and eIF5A regulate protein synthesis
Recent evidence supports the idea that eIF5A regulates translation elongation. Gregio et al. (2009) have shown that the polysome (number of ribosomes per mRNA) profile of a yeast eIF5A mutant resembles that of a translation elongation mutant.
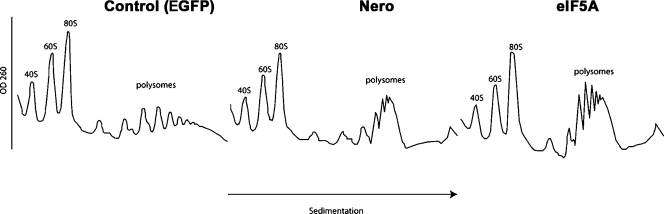
As eIF5A is thought to be the only target for Nero, we predicted that reduction of either Nero or eIF5A should have a similar effect on protein synthesis. To test this hypothesis, we used polysome profiling. Polysome sedimentation in sucrose gradients is one of the most powerful techniques for studying translational control. In this technique, the mRNAs are separated by ultracentrifugation as a function of the number of ribosomes to which they are associated. Conditions that reduce translation elongation result in the binding of mRNAs to multiple ribosomes, thus causing an increase in sedimentation toward the polysomal fraction. We established conditions to decrease levels of either Nero or eIF5A using dsRNA in Drosophila S2 cells (unpublished data). Similar to the yeast eIF5A mutant, RNAi-mediated knockdown of either Nero or eIF5A increases polysome size, indicating a defect in translation elongation (Fig. 8; Gregio et al., 2009). These data suggest that Nero-mediated eIF5A hypusination regulates translation rates.
Figure 8.
RNA knockdown of either Nero or eIF5A blocks translation elongation in Drosophila S2 cells. Polysome analysis of EGFP, nero, and eIF5A dsRNA–treated S2 cells. Cells were harvested, lysed, and fractionated by centrifugation on a 10–50% sucrose gradient. Polysomes were analyzed as described in Materials and methods. EGFP (left), nero (middle), and eIF5A dsRNA–treated (right) images are shown. The positions of the polysomes and ribosomes are indicated.
Discussion
Hypusination, the transformation of the amino acid lysine into the atypical amino acid hypusine, is a process that has been documented in numerous eukaryotic organisms from yeast to humans (Park et al., 1982, 1997; Gordon et al., 1987). The precise function of eIF5A and the significance of hypusination with regard to eIF5A function remain obscure. This study presents the functional characterization of the Drosophila DOHH homologue nero and looks at nero function in relation to eIF5A.
nero is an essential gene and encodes a highly conserved homologue of hDOHH
nero is required for organismal viability in Drosophila. This is in stark contrast to yeast, in which yeast DOHH (LIA1) is not an essential gene (Park et al., 2006). This has lead to the hypothesis that the second step in the hypusination process may not be essential in regard to eIF5A function in lower eukaryotes (Park et al., 2006). We find that DOHH activity is important in higher eukaryotes. The generation of null mutations of nero and the rescue of these mutants using the Drosophila DOHH cDNA/genomic rescue constructs formally confirm that DOHH function is required in higher eukaryotic organisms such as Drosophila.
The Nero protein appears to be highly conserved in Drosophila. Nero shares 59% amino acid identity with its hDOHH homologue, and the hDOHH protein can substitute for the Drosophila protein in vivo. Overexpression of hDOHH in nero mutant clones is able to fully rescue the short bristle defect observed in nero mutant clones. Thus, nero constitutes a true homologue of hDOHH. Analysis of hDOHH function has shown that DOHH is a metalloenzyme that requires iron ions for enzymatic activity (Park et al., 1982; Kim et al., 2006). Four histidine–glutamic acid metal-binding motifs have been identified in hDOHH, and these motifs have been shown to be required for DOHH enzymatic function (Park et al., 2006; Kang et al., 2007). These four motifs are fully conserved in the Drosophila homologue. Interestingly, overexpression of hDOHH carrying mutations in these metal ion–binding motifs fails to complement nero mutations, arguing that these residues are also important for DOHH function in Drosophila. All together, Nero appears to be highly conserved both in terms of amino acid homology as well as important residues critical for its function.
nero is required for cell growth, protein synthesis, and autophagy regulation
Clonal analyses of nero mutations during imaginal disc development implicate nero in cell growth. Loss of nero causes small bristles, epidermal cells, and photoreceptors. Furthermore, removing nero function in the developing Drosophila eye-antennal disc causes decreased head size, suggesting that nero function is required for organ size. Mutations that negatively affect cell growth share an array of common phenotypes such as impaired cell competitive ability and altered cell cycle. Such combined defects in cell growth, impaired competitive ability, and altered cell cycle are often attributed to impaired or reduced translation (Neufeld et al., 1998; Moreno et al., 2002). We find that loss of nero also affects both competitive ability and altered cell cycle phasing, as indicated by the fact that nero mutant cells are poorly recovered and that they poorly incorporate BrdU. All together, these phenotypes suggest that nero plays a role in the regulation of translation.
Consistent with the notion that Nero regulates eIF5A activity, inhibition of Nero or eIF5A by RNAi causes a similar impairment in translation elongation. This finding is consistent with data generated in other experimental systems. eIF5A has been shown to bind to components of the translational machinery in a hypusine-dependent manner (Zanelli et al., 2006). Furthermore, depletion of yeast eIF5A has been shown to block protein synthesis as well as alter polysome profiles (Kang and Hershey, 1994; Gregio et al., 2009). Our data support a function for DOHH in eIF5A-mediated translational control.
Interestingly, nero also regulates the induction of autophagy. Unlike effects on translation, defects in the induction of autophagy are generally not common to all cell growth pathways. In Drosophila, defects in autophagy regulation have been associated with mutations that affect the insulin and Tor growth signaling pathways (Rusten et al., 2004; Scott et al., 2004). Amino acid nutrient availability has been proposed as an upstream regulator of Tor activity (Dann and Thomas, 2006). This is in line with the observation that starvation induces autophagy in Drosophila (Rusten et al., 2004; Scott et al., 2004). This prompted us to test nero for a potential role in the Tor pathway. However, we were unable to generate any conclusive genetic evidence to link the two pathways, allowing us to exclude at the very least an integral role for nero in the Tor growth pathway.
Phenotypes associated with loss of eIF5A resemble nero, and nero regulates eIF5A levels
Inhibition of eIF5A using RNAi-mediated knockdown causes phenotypes similar to the ones observed in nero mutants. Knockdown of eIF5A, the target of hypusination, recapitulates nero mutant phenotypes, including decreased bristle size, autophagy, and defects in larval growth/development. These observations suggest that both nero and eIF5A regulate similar processes in vivo and argue that nero is linked to eIF5A function.
Interestingly, we find that eIF5A is up-regulated in nero mutants. This effect on eIF5A levels in DOHH mutants has not previously been reported. It is likely that the up-regulation of eIF5A constitutes an adaptive response by the cell to the loss of nero. Because the eIF5A mutant phenotype appears to be more severe than the defect observed in nero mutant animals, the up-regulation of eIF5A in nero mutants may explain the observed differences in larval growth associated with eIF5A and nero double mutants. Thus, incompletely hypusinated forms of eIF5A may still be partially functional and able to mildly ameliorate the nero mutant phenotype. This model is also compatible with the idea that the loss of nero causes only a partial loss of eIF5A activity. In conclusion, our work highlights the importance of nero/DOHH function in Drosophila, implicates eIF5A and nero in cell growth and autophagy regulation, and provides genetic evidence that links eIF5A function with nero/DOHH regulation.
Materials and methods
Drosophila stocks and genetics
We used the following stocks: y w (control), Canton-S (control), P[lArB]K48 (provided by T.F. MacKay, North Carolina State University, Raleigh, NC; Lyman et al., 1996), FRT82B Tsc1Q87X (provided by P.H. Patel [Fred Hutchinson Cancer Research Center, Seattle, WA] and F. Tamanoi, [University of California, Los Angeles, Los Angeles, CA]; Tapon et al., 2001), hs-ATG8b-GFP (provided by T.P. Neufeld, University of Minnesota, Minneapolis, MN; Scott et al., 2004), eq-GAL4 (Pi et al., 2001), UAS-CG3186 RNAi (Dietzl et al., 2007), Adh-GAL4/CyO (Fischer et al., 1988), Ubx-FLP; FRT82B Ubi-GFP M(3), y w hs-FLP tub-GAL4 UAS-GFP-6xMYC-NLS; FRT82B hsp70-CD2 y+ tub-GAL80/TM6 (provided by G. Struhl, Columbia University, New York, NY), y w; D/TM3 Kr-GAL4 UAS-GFP, and FRT82B Ubi-GFP (provided by G. Halder, The University of Texas M.D. Anderson Cancer Center, Houston, TX). y w; Act-GAL4/CyO, Df(3R)04661, y w eyeless-FLP GMR-lacZ; FRT82B w+ l(3)cl, FRT82B w+, P[PZ]01296 (Spradling et al., 1999), tub-Gal4/TM3 (Lee and Luo, 1999), and P[lacW]s1921 (Spradling et al., 1999) were obtained from the Bloomington Drosophila Stock Center.
nero mutants were generated through imprecise excision of P[lArB]K48 and P[lacW]s1921. nerok48-123 was generated from P[lArB]K48. nero1 and nero2 alleles were generated from the mobilization of P[lacW]s1921. Genomic lesions in nero1 and nero2 mutants were determined using PCR. eIF5APrev was generated through the excision of P[PZ]01296.
Transgenic flies
To make a genomic rescue construct for nero, Drosophila genomic sequences corresponding to the nero locus were isolated from the DS00235 P1 clone (Berkeley Drosophila Genome Project). The P1 clone was digested with SacI, and this fragment was subcloned into the SacI site of the pBluescript cloning vector (Agilent Technologies). A 1.5-kb fragment was recovered from this construct by digesting with SwaI and MluI and was subcloned into the HpaI site of P[CaSper4]. To make UAS-nero, the CG2245 coding region was isolated from LD09536 (Berkeley Drosophila Genome Project Drosophila Gene Collection) and cloned into the EcoRI and XhoI sites of the P[UAST] vector. To make UAS-eIF5A, the CG3186 coding region was PCR amplified from RE47768 (Berkeley Drosophila Genome Project Drosophila Gold Collection) with adapters bearing EcoRI and XhoI sites and cloned into these sites of the P[UAST] vector. To make UAS-hDOHH and its enzymatically inactive versions, human cDNA (National Institutes of Health Mammalian Gene Collection) for DOHH was obtained from Invitrogen, PCR amplified, and cloned in pBluescript. H56A, H89A, H208A, and H240A point mutations in hDOHH were generated using a GeneTailor Site-Directed Mutagenesis system (Invitrogen). The wild-type and mutant versions were subsequently cloned into P[UAST]. All of these constructs were subsequently introduced into flies.
Antibody generation, Western blotting, immunohistochemistry, and BrdU assay
A full-length Nero–His tag fusion protein was generated by cloning the full-length Nero cDNA into the EcoRI and XhoI sites of the pET28a cloning vector (EMD). Coding sequences were amplified from LD09536 using primers that introduced EcoRI and XhoI restriction sites. The protein was induced in BL21 DE3 bacteria using IPTG and purified using His-Bind resin (EMD). Purified fusion proteins were injected into guinea pigs for antibody production (Cocalico Biologicals, Inc.).
We used standard protocols for Western blotting. Western Lightning Western Blot Chemiluminescence Reagent (PerkinElmer) was used to detect signal. Western blots were incubated in primary antibody using the following dilutions: 1:2,000 guinea pig anti–Nero GP25, 1:1,500 rabbit anti-eIF5A [EP526Y] (Abcam), and 1:10,000 mouse anti–actin C4 (MP Biomedicals).
For immunohistochemistry, larvae were dissected in PBS and fixed for 20 min in 4% formaldehyde in PBS. Standard protocols were used to label larval tissues with antibodies. Incubations in primary antibodies were performed overnight using the following dilutions: 1:100 mouse anti–Dlg 4F3 (Developmental Studies Hybridoma Bank), 1:400 rabbit anti-eIF5A [EP526Y] (Abcam), 1:400 mouse anti-KDEL (Assay Designs), 1:1,000 guinea pig anti–Nero GP26, and 1:2,000 rat anti-Su(H) (provided by F. Schweisguth, Institut Pasteur, Paris, France; Gho et al., 1996). Fluorescent Cy3- and Alexa Fluor 488–conjugated secondary antibodies (provided by Jackson ImmunoResearch Laboratories and Invitrogen, respectively) were applied for 2 h at 1:200.
BrdU incorporation was conducted as previously described (Lee et al., 2005). However, discs were fixed in 4% formaldehyde for 1 h. 1:200 mouse anti–BrdU G3G4 (Developmental Studies Hybridoma Bank) was used to detect BrdU (Sigma-Aldrich) followed by subsequent detection with a fluorescent secondary antibody as described in the previous paragraph.
Autophagy assay
Autophagy assays were conducted as previously described (Scott et al., 2004). Larvae were dissected and incubated for 10 min in a 1:1,000 dilution of LysoTracker red DND-99 (Invitrogen) in PBS with DAPI to a final concentration of 0.4 µg/ml. LysoTracker was replaced with PBS before mounting and visualization.
Cell counting experiments
Newly hatched larvae were collected and placed in vials with standard food and allowed to develop at a density of 100 larvae per vial. 48 h after being selected, larvae were heat shocked at 37°C for 15 min. Larvae were allowed to develop to third instars and then dissected and fixed in 4% formaldehyde for 20 min. Dissected discs were treated with 400 µg/ml RNase A in PBS for 30 min at 37°C and then stained with propidium iodide (Invitrogen) at 1 µg/ml for 20 min before mounting.
Cell culture, RNAi, and polysome profile analysis
dsRNAs to eIF5A, nero, and EGFP were generated using a T7 RiboMAX Express RNAi system (Promega). Cells were treated as previously described (Clemens et al., 2000). 5 × 107 S2 cells were incubated with 100 µg/ml cycloheximide for 10 min, washed twice with cold PBS + 100 µg/ml cycloheximide, resuspended in lysis buffer (10 mM Hepes-KOH, pH 7.4, 5 mM MgCl2, 150 mM KCl, 0.5% NP-40, 0.5 mM DTT, 100 µg/ml cycloheximide, 100 U/ml RNase inhibitor [Promega], and protease inhibitor [Roche]), homogenized with 10 strokes in a 3-ml dounce tissue grinder (Wheaton) on ice, and then centrifuged for 10 min at 14,000 g. The supernatants were loaded onto 10–50% sucrose gradients, which were prepared in 10 mM Hepes-KOH, pH 7.4, 5 mM MgCl2, and 150 mM KCl, and centrifuged in a rotor (SW40; Beckman Coulter) at 35,000 rpm for 2 h. Gradients were analyzed by piercing the tube with a tube piercer (Brandel), passing 60% sucrose through the bottom of the tube using a syringe pump (Brandel) at a constant flow rate (0.75 ml/min), and monitoring the absorbance of the material eluting from the tube using a UV detector (UA-6; ISCO, Inc.) as previously described (Costa-Mattioli et al., 2005, 2007).
Microscopy
Samples were mounted in Vectashield mounting medium (Vector Laboratories) for confocal imaging. Confocal images were acquired using a confocal microscope (LSM510; Carl Zeiss, Inc.) with its accompanying software using Plan-Apochromat 40× NA 1.4 and Plan-Apochromat 63× NA 1.4 objectives (Carl Zeiss, Inc.). Images of fly adult thoraces (treated by boiling in 10% KOH) mounted in 70% glycerol were obtained using an LSM510 confocal microscope, camera (AxioCam HRc; Carl Zeiss, Inc.), Plan-Apochromat 10× NA 0.45 objective, and AxioVision 3.1 software (Carl Zeiss, Inc.). Images of fly eyes were captured with a camera (MicroFire; Olympus) mounted to a stereomicroscope (MZ16; Leica) using ImagePro Plus 5.0 acquisition software (Media Cybernetics). Fat bodies mounted in PBS for autophagy analysis were visualized using a microscope (Imager.Z1; Carl Zeiss, Inc.), camera (AxioCam MRm; Carl Zeiss, Inc.), AxioVision release 4.3 software (Carl Zeiss, Inc.), and either the Plan-Apochromat 10× NA 0.45 lens or the Plan-Apochromat 20× NA 0.75 lens. All images were later processed using ImageJ (National Institutes of Health) and Photoshop 7.0 (Adobe). All images were captured at room temperature.
Online supplemental material
Fig. S1 shows that no clear epistatic relationship can be established between nero and the Tor pathway. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200904161/DC1.
Acknowledgments
We thank Koen Norga for plasmid rescue. We are grateful to T.F. MacKay, T.P. Neufeld, G. Struhl, G. Halder, P.H. Patel, F. Tamanoi, and the Bloomington Drosophila Stock Center for fly stocks. We are also grateful to F. Schweisguth and the Developmental Studies Hybridoma Bank for providing antibodies. We would also like to thank H. Pan and Y. He for technical assistance.
Confocal microscopy was supported by the Baylor College of Medicine Intellectual and Developmental Disabilities Research Center. This publication was made possible by a grant from the National Institute of Environmental Health Sciences, National Institutes of Health (T32 ES07332) to P.H. Patel. H.J. Bellen is a Howard Hughes Medical Institute Investigator.
Note added in proof. After this manuscript was accepted for publication, a paper appeared in Nature online (Saini et al. 2009. Nature. doi:10.1038/nature08034) showing that the depletion or inactivation of eIF5A in yeast (S. cerevisiae) results in the accumulation of polysomes, indicating that eIF5A promotes translation elongation. These data are in agreement with our observation that RNAi-mediated knockdown of either Nero or eIF5A blocks translation elongation and thus increases the size of polysomes in Drosophila S2 cells.
Footnotes
Abbreviations used in this paper: APF, after puparium formation; DHS, deoxyhypusine synthase; Dlg, Discs large; DOHH, deoxyhypusine hydroxylase; dsRNA, double-stranded RNA; FLP, flippase; FRT, FLP recombination target; hDOHH, human DOHH; Su(H), suppressor of Hairless; Tor, target of rapamycin; UAS, upstream activation sequence.
References
- Abbruzzese A., Park M.H., Folk J.E. 1986. Deoxyhypusine hydroxylase from rat testis. Partial purification and characterization.J. Biol. Chem. 261:3085–3089 [PubMed] [Google Scholar]
- Audibert A., Simon F., Gho M. 2005. Cell cycle diversity involves differential regulation of Cyclin E activity in the Drosophila bristle cell lineage.Development. 132:2287–2297 [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes.Development. 118:401–415 [DOI] [PubMed] [Google Scholar]
- Byers T.L., Lakanen J.R., Coward J.K., Pegg A.E. 1994. The role of hypusine depletion in cytostasis induced by S-adenosyl-L-methionine decarboxylase inhibition: new evidence provided by 1-methylspermidine and 1,12-dimethylspermine.Biochem. J. 303:363–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee I., Gross S.R., Kinzy T.G., Chen K.Y. 2006. Rapid depletion of mutant eukaryotic initiation factor 5A at restrictive temperature reveals connections to actin cytoskeleton and cell cycle progression.Mol. Genet. Genomics. 275:264–276 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay M.K., Tabor C.W., Tabor H. 2003. Spermidine but not spermine is essential for hypusine biosynthesis and growth in Saccharomyces cerevisiae: spermine is converted to spermidine in vivo by the FMS1-amine oxidase.Proc. Natl. Acad. Sci. USA. 100:13869–13874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens J.C., Worby C.A., Simonson-Leff N., Muda M., Maehama T., Hemmings B.A., Dixon J.E. 2000. Use of double stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways.Proc. Natl. Acad. Sci. USA. 97:6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper H.L., Park M.H., Folk J.E., Safer B., Braverman R. 1983. Identification of the hypusine-containing protein hy+ as translation initiation factor eIF-4D.Proc. Natl. Acad. Sci. USA. 80:1854–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M., Gobert D., Harding H., Herdy B., Azzi M., Bruno M., Bidinosti M., Ben Mamou C., Marcinkiewicz E., Yoshida M., et al. 2005. Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2.Nature. 436:1166–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M., Gobert D., Stern E., Gamache K., Colina R., Cuello C., Sossin W., Kaufman R., Pelletier J., Rosenblum K., et al. 2007. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory.Cell. 129:195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann S.G., Thomas G. 2006. The amino acid sensitive TOR pathway from yeast to mammals.FEBS Lett. 580:2821–2829 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila.Nature. 448:151–156 [DOI] [PubMed] [Google Scholar]
- Fischer J.A., Giniger E., Maniatis T., Ptashne M. 1988. GAL4 activates transcription in Drosophila.Nature. 332:853–856 [DOI] [PubMed] [Google Scholar]
- Gao X., Pan D. 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth.Genes Dev. 15:1383–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A., Merriam J.R. 1971. Genetic analysis of cell heredity in imaginal discs of Drosophila melanogaster.Proc. Natl. Acad. Sci. USA. 68:2222–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gho M., Lecourtois M., Geraud G., Posakony J.W., Schweisguth F. 1996. Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling.Development. 122:1673–1682 [DOI] [PubMed] [Google Scholar]
- Gordon E.D., Mora R., Meredith S.C., Lee C., Lindquist S.L. 1987. Eukaryotic initiation factor 4D, the hypusine-containing protein, is conserved amongst eukaryotes.J. Biol. Chem. 262:16585–16589 [PubMed] [Google Scholar]
- Gratzner H.G. 1982. Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: a new reagent for detection of DNA replication.Science. 218:474–475 [DOI] [PubMed] [Google Scholar]
- Gregio A.P., Cano V.P., Avaca J.S., Valentini S.R., Zanelli C.F. 2009. eIF5A has a function in the elongation step of translation in yeast.Biochem. Biophys. Res. Commun. 380:785–790 [DOI] [PubMed] [Google Scholar]
- Hanauske-Abel H.M., Park M.H., Hanauske A.R., Popowicz A.M., Lalande M., Folk J.E. 1994. Inhibition of the G1-S transition of the cell cycle by inhibitors of deoxyhypusine hydroxylation.Biochim. Biophys. Acta. 1221:115–124 [DOI] [PubMed] [Google Scholar]
- Jakus J., Wolff E.C., Park M.H., Folk J.E. 1993. Features of the spermidine-binding site of deoxyhypusine synthase as derived from inhibition studies. Effective inhibition by bis- and mono-guanylated diamines and polyamines.J. Biol. Chem. 268:13151–13159 [PubMed] [Google Scholar]
- Joe Y.A., Wolff E.C., Park M.H. 1995. Cloning and expression of human deoxyhypusine synthase cDNA. Structure-function studies with the recombinant enzyme and mutant proteins.J. Biol. Chem. 270:22386–22392 [DOI] [PubMed] [Google Scholar]
- Kang H.A., Hershey J.W. 1994. Effect of initiation factor eIF-5A depletion on protein synthesis and proliferation of Saccharomyces cerevisiae.J. Biol. Chem. 269:3934–3940 [PubMed] [Google Scholar]
- Kang K.R., Wolff E.C., Park M.H., Folk J.E., Chung S.I. 1995. Identification of YHR068w in Saccharomyces cerevisiae chromosome VIII as a gene for deoxyhypusine synthase. Expression and characterization of the enzyme.J. Biol. Chem. 270:18408–18412 [DOI] [PubMed] [Google Scholar]
- Kang K.R., Kim Y.S., Wolff E.C., Park M.H. 2007. Specificity of the deoxyhypusine hydroxylase-eukaryotic translation initiation factor (eIF5A) interaction: identification of amino acid residues of the enzyme required for binding of its substrate, deoxyhypusine-containing eIF5A.J. Biol. Chem. 282:8300–8308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Kang K.R., Wolff E.C., Bell J.K., McPhie P., Park M.H. 2006. Deoxyhypusine hydroxylase is a Fe(II)-dependent, HEAT-repeat enzyme. Identification of amino acid residues critical for Fe(II) binding and catalysis.J. Biol. Chem. 281:13217–13225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.B., Park J., Jung J.U., Chung J. 2005. Nef induces apoptosis by activating JNK signaling pathway and inhibits NF-kappaB-dependent immune responses in Drosophila.J. Cell Sci. 118:1851–1859 [DOI] [PubMed] [Google Scholar]
- Lee T., Luo L. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis.Neuron. 22:451–461 [DOI] [PubMed] [Google Scholar]
- Lyman R.F., Lawrence F., Nuzhdin S.V., Mackay T.F. 1996. Effects of single P-element insertions on bristle number and viability in Drosophila melanogaster.Genetics. 143:277–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E., Basler K., Morata G. 2002. Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development.Nature. 416:755–759 [DOI] [PubMed] [Google Scholar]
- Neufeld T.P., de la Cruz A.F., Johnston L.A., Edgar B.A. 1998. Coordination of growth and cell division in the Drosophila wing.Cell. 93:1183–1193 [DOI] [PubMed] [Google Scholar]
- Norga K.K., Gurganus M.C., Dilda C.L., Yamamoto A., Lyman R.F., Patel P.H., Rubin G.M., Hoskins R.A., Mackay T.F., Bellen H.J. 2003. Quantitative analysis of bristle number in Drosophila mutants identifies genes involved in neural development.Curr. Biol. 13:1388–1396 [DOI] [PubMed] [Google Scholar]
- Oldham S., Montagne J., Radimerski T., Thomas G., Hafen E. 2000. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin.Genes Dev. 14:2689–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.H., Cooper H.L., Folk J.E. 1982. The biosynthesis of protein-bound hypusine (N epsilon -(4-amino-2-hydroxybutyl)lysine). Lysine as the amino acid precursor and the intermediate role of deoxyhypusine (N epsilon -(4-aminobutyl)lysine).J. Biol. Chem. 257:7217–7222 [PubMed] [Google Scholar]
- Park M.H., Wolff E.C., Lee Y.B., Folk J.E. 1994. Antiproliferative effects of inhibitors of deoxyhypusine synthase. Inhibition of growth of Chinese hamster ovary cells by guanyl diamines.J. Biol. Chem. 269:27827–27832 [PubMed] [Google Scholar]
- Park M.H., Lee Y.B., Joe Y.A. 1997. Hypusine is essential for eukaryotic cell proliferation.Biol. Signals. 6:115–123 [DOI] [PubMed] [Google Scholar]
- Park M.H., Joe Y.A., Kang K.R. 1998. Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae.J. Biol. Chem. 273:1677–1683 [DOI] [PubMed] [Google Scholar]
- Park J.H., Wolff E.C., Folk J.E., Park M.H. 2003. Reversal of the deoxyhypusine synthesis reaction. Generation of spermidine or homospermidine from deoxyhypusine by deoxyhypusine synthase.J. Biol. Chem. 278:32683–32691 [DOI] [PubMed] [Google Scholar]
- Park J.H., Aravind L., Wolff E.C., Kaevel J., Kim Y.S., Park M.H. 2006. Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme.Proc. Natl. Acad. Sci. USA. 103:51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H.R. 1990. The retention signal for soluble proteins of the endoplasmic reticulum.Trends Biochem. Sci. 15:483–486 [DOI] [PubMed] [Google Scholar]
- Pi H., Wu H.J., Chien C.T. 2001. A dual function of phyllopod in Drosophila external sensory organ development: cell fate specification of sensory organ precursor and its progeny.Development. 128:2699–2710 [DOI] [PubMed] [Google Scholar]
- Potter C.J., Huang H., Xu T. 2001. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size.Cell. 105:357–368 [DOI] [PubMed] [Google Scholar]
- Rusten T.E., Lindmo K., Juhasz G., Sass M., Seglen P.O., Brech A., Stenmark H. 2004. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway.Dev. Cell. 7:179–192 [DOI] [PubMed] [Google Scholar]
- Sasaki K., Abid M.R., Miyazaki M. 1996. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae.FEBS Lett. 384:151–154 [DOI] [PubMed] [Google Scholar]
- Schnier J., Schwelberger H.G., Smit-McBride Z., Kang H.A., Hershey J.W. 1991. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae.Mol. Cell. Biol. 11:3105–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.C., Schuldiner O., Neufeld T.P. 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body.Dev. Cell. 7:167–178 [DOI] [PubMed] [Google Scholar]
- Smit-McBride Z., Dever T.E., Hershey J.W., Merrick W.C. 1989. Sequence determination and cDNA cloning of eukaryotic initiation factor 4D, the hypusine-containing protein.J. Biol. Chem. 264:1578–1583 [PubMed] [Google Scholar]
- Spradling A.C., Stern D., Beaton A., Rhem E.J., Laverty T., Mozden N., Misra S., Rubin G.M. 1999. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes.Genetics. 153:135–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N., Ito N., Dickson B.J., Treisman J.E., Hariharan I.K. 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation.Cell. 105:345–355 [DOI] [PubMed] [Google Scholar]
- Wöhl T., Klier H., Ammer H., Lottspeich F., Magdolen V. 1993. The HYP2 gene of Saccharomyces cerevisiae is essential for aerobic growth: characterization of different isoforms of the hypusine-containing protein Hyp2p and analysis of gene disruption mutants.Mol. Gen. Genet. 241:305–311 [DOI] [PubMed] [Google Scholar]
- Wolff E.C., Park M.H., Folk J.E. 1990. Cleavage of spermidine as the first step in deoxyhypusine synthesis. The role of NAD.J. Biol. Chem. 265:4793–4799 [PubMed] [Google Scholar]
- Woods D.F., Bryant P.J. 1991. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions.Cell. 66:451–464 [DOI] [PubMed] [Google Scholar]
- Xu T., Rubin G.M. 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues.Development. 117:1223–1237 [DOI] [PubMed] [Google Scholar]
- Zanelli C.F., Maragno A.L., Gregio A.P., Komili S., Pandolfi J.R., Mestriner C.A., Lustri W.R., Valentini S.R. 2006. eIF5A binds to translational machinery components and affects translation in yeast.Biochem. Biophys. Res. Commun. 348:1358–1366 [DOI] [PubMed] [Google Scholar]
- Zhang H., Stallock J.P., Ng J.C., Reinhard C., Neufeld T.P. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR.Genes Dev. 14:2712–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]