Abstract
Synaptic vesicles fuse at active zone (AZ) membranes where Ca2+ channels are clustered and that are typically decorated by electron-dense projections. Recently, mutants of the Drosophila melanogaster ERC/CAST family protein Bruchpilot (BRP) were shown to lack dense projections (T-bars) and to suffer from Ca2+ channel–clustering defects. In this study, we used high resolution light microscopy, electron microscopy, and intravital imaging to analyze the function of BRP in AZ assembly. Consistent with truncated BRP variants forming shortened T-bars, we identify BRP as a direct T-bar component at the AZ center with its N terminus closer to the AZ membrane than its C terminus. In contrast, Drosophila Liprin-α, another AZ-organizing protein, precedes BRP during the assembly of newly forming AZs by several hours and surrounds the AZ center in few discrete punctae. BRP seems responsible for effectively clustering Ca2+ channels beneath the T-bar density late in a protracted AZ formation process, potentially through a direct molecular interaction with intracellular Ca2+ channel domains.
Introduction
The arrival of action potentials mediates Ca2+ influx through strategically localized clusters of voltage-operated Ca2+ channels at the synaptic active zone (AZ) membrane. Ca2+ triggers exocytosis of synaptic vesicles, and tight coupling between release-ready vesicles and Ca2+ channels seems important for efficient neurotransmitter release (Neher and Sakaba, 2008). AZs are further characterized by macromolecular cytomatrices named dense bodies (Zhai and Bellen, 2004; Siksou et al., 2007). The role of these electron-dense specializations and of AZ-enriched proteins in the assembly of the AZ and/or the synaptic vesicle exo-endocytosis cycle is under intense investigation (Owald and Sigrist, 2009). Protein architectures constituting and controlling dense bodies remain to be revealed, and their contributions to AZ assembly in general and Ca2+ channel clustering in particular must be defined genetically.
The most straightforward approach would be to use immunolabeling combined with light microscopy. However, individual AZs measure only a few hundred nanometers in diameter, rendering this approach difficult. Recently, stimulated emission depletion (STED) microscopy (Hell, 2007) has proven valuable for high resolution light microscopic studies of synapse architectures (Kittel et al., 2006; Jin and Garner, 2008; Westphal et al., 2008).
The Drosophila melanogaster neuromuscular junction (NMJ) is a leading model for genetic analyses of synapse structure and assembly (Featherstone et al., 2000; Koh et al., 2000; Collins and DiAntonio, 2007). In this preparation, dense body structures termed T-bars may take part in activity-dependent changes of synaptic performance (Prokop and Meinertzhagen, 2006). Furthermore, the Ca2+ channel α1 subunit Cacophony (Cac) was shown to dominate neurotransmitter release at NMJ synapses (Kawasaki et al., 2000).
Proteins of the conserved CAST (CAZ-associated structural protein)/ERC (ELKS–Rab6-interacting protein CAST) family are generic AZ proteins. In mice, CAST/ERC proteins have been shown to localize to AZs of various synapses and to bind other AZ proteins such as RIM (Rab3a-interacting molecule) and Liprin-α (Ohtsuka et al., 2002; Wang et al., 2002; Ko et al., 2003; Deguchi-Tawarada et al., 2004). In Caenorhabditis elegans, the CAST/ERC family member ELKS (glutamine-, leucine-, lysine-, and serine-rich protein) appears to operate genetically downstream of Syd-2/Liprin-α during the assembly of AZs at vulval synapses (Dai et al., 2006; Patel et al., 2006). Recently, the CAST/ERC family member Bruchpilot (BRP), a coiled-coil rich protein of nearly 200 kD, was identified via its localization to Drosophila AZs. Mutants of brp lacked T-bars, and Ca2+ channels were mislocalized at AZs, leading to inefficient vesicle release and changes in synaptic short-term plasticity (Kittel et al., 2006; Wagh et al., 2006).
In this study, we provide evidence that BRP takes up an elongated conformation and is a direct component of the T-bar. The N terminus of BRP is found superimposed on the Ca2+ channel clusters at the AZ center. BRP and Cac arrive at an advanced stage of the protracted synapse assembly process, and both proteins interact in vitro. In contrast, a further AZ-organizing protein, Drosophila Liprin-α (DLiprin-α), localizes to a different subcompartment of the AZ and enters nascent AZs substantially earlier than BRP. Thus, the assembly of the T-bar is instructed by BRP, which seems essential for clustering higher numbers of Ca2+ channels at an advanced stage of AZ maturation.
Results
The AZ protein BRP was recently shown to be crucial for efficient neurotransmission at Drosophila NMJs. Presynaptic AZs missing BRP lacked dense bodies (T-bars), and Ca2+ channel densities were compromised within AZs (Kittel et al., 2006). However, whether BRP performs an essential signaling role in T-bar formation or whether the protein itself is an essential building block of T-bars remained to be clarified. Thus, we entered into a structure-function analysis of BRP.
mAb Nc82 maps toward the C-terminal end of BRP
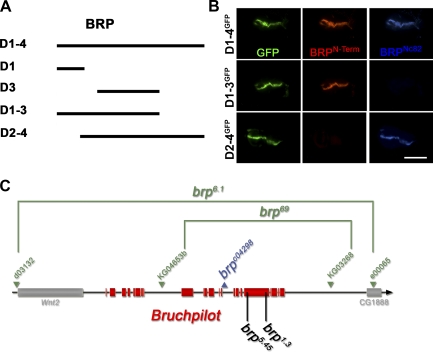
mAb Nc82 is derived from a Drosophila head extract–directed library (Hofbauer et al., 2009) and allowed the first identification of the BRP protein. mAb Nc82 is a widely used marker in Drosophila, both for neuropil in general and for AZs in particular (Wucherpfennig et al., 2003; Kittel et al., 2006; Wagh et al., 2006). Previously, we had loosely mapped the epitope of Nc82 to the region between aa 635 and the end of the 1,740-aa BRP protein (based on cDNA AT09405; Wagh et al., 2006). To define the Nc82 epitope more precisely, various BRP fragments (Fig. 1 A and Table I) were ectopically expressed in wing discs using dpp-Gal4 (Fig. 1 B). In this manner, the mAb Nc82 epitope (hereafter BRPNc82) could be mapped to the region between aa 1,227 and 1,740. Additionally, an antibody directed against an N-terminal peptide (BRPN-Term antibody; aa 62–75; Fig. 1 B) was produced.
Figure 1.
Epitope mapping for mAb Nc82 and genetic analysis of brp. (A) BRP fragments used for transgenic expression experiments. (B) Ectopic expression (in wing imaginal discs) of GFP-tagged BRP fragments missing either N- (D2-4) or C-terminal (D1-3) regions. Wing discs were costained for BRPN-Term (red), BRPNc82 (blue), and GFP (green). The D2-4 construct shows no BRPN-Term reactivity, whereas the D1-3 construct lacks BRPNc82 staining. Bar, 300 µm. (C) Genomic analysis of the brp locus. The deletion mutants (brp69 and brp6.1) and their parental transposon insert lines are shown in green, and the pBac transposon insert line is shown in blue. Position of EMS-induced stop codons of brp1.3 and brp5.45 are shown in black.
Table I.
BRP-reexpressing constructs
| BRP fragments | Start (aa) | End (aa) |
| Domain 1 (D1) | 1 | 320 |
| Domain 2 (D2) | 268 | 617 |
| Domain 3 (D3) | 473 | 1,226 |
| Domain 4 (D4) | 1,152 | 1,740 |
The predicted lengths in aa of the UAS-BRP fragments (relative to full-length BRP [1,740 aa]) are shown.
Structure-function analysis of BRP in T-bar formation
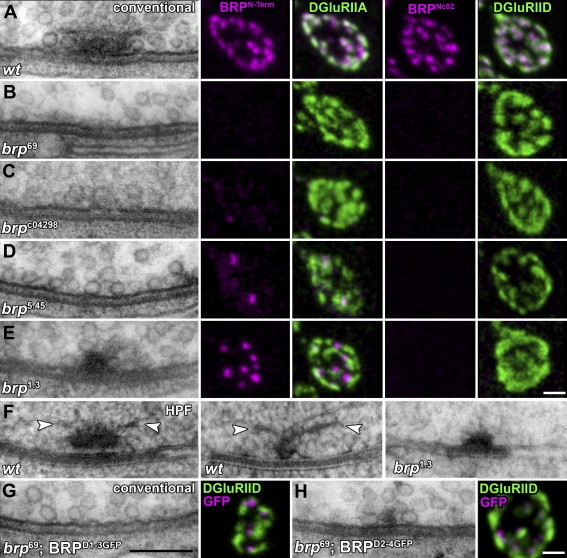
So far, the analysis of BRP function was based on the brp69 allele in which most of the protein-coding sequence (corresponding to aa 283–1,740) is deleted (Fig. 1 C, green). As previously reported, T-bars were missing at brp69 AZs (Fig. 2, compare A and B), and the BRPNc82 label (Fig. 2, A and B) was absent (Kittel et al., 2006). The BRPN-Term label was also completely absent (Fig. 2 B), indicating that the predicted residual protein (corresponding to aa 1–282) is unstable or at least does not localize to the NMJ. To ensure that brp69 reflects a true null phenotype, we produced the deletion mutant brp6.1 (Fig. 1 C, green) in which all genomic sequences of brp were removed (see Materials and methods). This led to a complete loss of BRPNc82/BRPN-Term labels and T-bars, whereas some traces of residual electron-dense material appeared at the same frequency as in brp69 (Kittel et al., 2006; unpublished data). As brp6.1 and brp69 (Kittel et al., 2006) behaved identically in all aspects, our previous analysis based on brp69 reflected a true null situation.
Figure 2.
Combined electron and light microscopic analysis of AZ organization at NMJs of different brp alleles. (A–E, left) Ultrastructure of Drosophila NMJ AZs preserved with conventional room temperature embedding for transmission EM. The following genotypes are depicted: wild-type (wt; A), brp69 (B), brpc04298 (C), brp5.45 (D), and brp1.3 (E). (right) Corresponding confocal images of boutons costained for either BRPN-Term or BRPNc82 (magenta) and DGluRIIA or DGluRIID (green). (F) Wild-type and brp1.3 T-bars preserved using HPF followed by FS. For controls, long (left) and short axis (middle) views of T-bars are depicted; arrowheads indicate filaments emerging from the T-bar pedestal. (G and H, left) Conventionally embedded AZs after expression of BRPD1-3GFP and BRPD2-4GFP in the brp69 mutant background. In BRPD1-3GFP, T-bar formation could not be observed. For BRPD2-4GFP, electron-dense structures much smaller than T-bars were observed. (right) Corresponding confocal images of the reexpression constructs costained for GFP (magenta) and DGluRIID (green). Bars: (G) 200 nm; (E and H) 500 nm.
BRP is a large protein (1,740 aa). To enter into a structure-function analysis of BRP in T-bar assembly, additional brp alleles were looked into. First, a piggyBac-transposon insert (brpc04298; Fig. 1 C, blue; Bellen et al., 2004) located toward the middle of the locus was characterized. At brpc04298 NMJs, the BRPNc82 label was absent, whereas the BRPN-Term label was dramatically reduced (Fig. 2 C). Comparable with our observations for brp69 (Kittel et al., 2006), electron microscopic analysis of brpc04298 showed a complete lack of T-bars (Fig. 2 C), and only traces of electron-dense material remained at AZ membranes. Thus, as this allele is a site-specific insertion but not a deletion (which in principle might eliminate control elements of genes other than brp), this allele provides further proof that BRP is essential for T-bar assembly. However, as the molecular alterations of brpc04298 cannot be predicted easily, we sought to analyze aa point mutations in brp. To do so, a chemical mutagenesis screen (ethyl methyl sulfonate [EMS]) selecting for reduced viability over brp-null alleles was performed.
The brp5.45 allele is characterized by a stop codon at aa position 867 (∼50% protein length), which leads to pupal lethality over brp null with weak escapers (Fig. 1 C). As expected, the BRPNc82 label was absent from brp5.45 NMJs. Although the number of BRPN-Term clusters was reduced over the whole NMJ (Fig. S1 A), those remaining in brp5.45 were slightly smaller, although of comparable intensity as in controls (Fig. 2 D; and Fig. S1, B and C). Despite extensive analysis, T-bars were not detected at brp5.45 NMJs (Fig. 2 D).
The EMS allele brp1.3 delivered paralyzed adult escapers over brp null as the result of a premature stop codon at aa 1,390 (generating a protein 523 aa longer than predicted for brp5.45; Fig. 1 C). Although the number of BRPN-Term clusters was reduced to ∼40% (Fig. S1 A), their sizes and intensities were comparable with controls. At the same time, the BRPNc82 label was absent (Fig. 2 E). T-bar–like structures were observed at brp1.3 NMJs (Fig. 2 E), although at lower frequency than in controls (not depicted). However, upon closer inspection, the T-bar–like structures typically appeared truncated (Fig. 2).
For EM, conventional room temperature embedding procedures, including aldehyde fixation and dehydration of the tissue (Fig. 2, A–E), are prone to shrinkage artifacts. To use an alternative conservation method for the analysis of brp1.3, we introduced high pressure freezing (HPF)/freeze substitution (FS) EM (Gray et al., 2006; Rostaing et al., 2006; Siksou et al., 2007) to larval NMJs. With HPF/FS, NMJ tissue appeared well preserved, as judged by the smooth membrane surfaces of, for example, mitochondria (Fig. S2, A and B) or presynaptic boutons (not depicted). Furthermore, electron-dense structures appeared taller, which was likely caused by a reduced loss of material during HPF/FS embedding (e.g., the synaptic cleft; Fig. 2 F). Unlike T-bars visualized in conventionally embedded tissues, HPF/FS-processed T-bars were characterized by filamentous elements at their distal ends (Fig. 2 F, arrowheads). At brp1.3 NMJs, HPF/FS EM (similarly to our observations obtained with standard EM) typically revealed shortened T-bars (Fig. S2 C, quantification). In conclusion, elimination of aa 1,390–1,740 of BRP did not prevent the formation of T-bar–like assemblies per se. However, these assemblies were significantly smaller than in controls. This result is in line with the assumption that BRP operates as a building block shaping the T-bar.
To both confirm and extend our results, BRP-encoding cDNA fragments (C-terminally GFP tagged for in vivo visualization) were expressed in motoneurons of brp69 larvae (Fig. 2, G and H). As expected (Kittel et al., 2006), full-length BRP localized to AZs and restored T-bar formation (not depicted). The C-terminally truncated fragment D1-3GFP (BRPD1-3GFP; Δ aa 1,227–1,740; Fig. 1 A and Table I) localized to presynaptic sites but was not sufficient for T-bar formation (Fig. 2 G). Thus, expression of BRPD1-3GFP further suggests that the C-terminal region of BRP (distal of aa 1,226; Table I) is important for T-bar formation.
By expressing D2-4GFP (BRPD2-4GFP; Δ aa 1–267; Fig. 1 A and Table I), the role of the N-terminal region of BRP was tested. The expressed protein was found close to individual AZs (Fig. 2 H). Although T-bars were never observed, small electron-dense aggregates (clearly smaller than T-bars) localized to the AZ membrane at high frequency (Fig. 2 H). Thus, the N-terminal region of BRP seems important for the formation of proper, full-sized T-bars, whereas the C-terminal region is required for the assembly of T-bars by itself. Thus, the entire BRP protein appears to take part in configuring the T-bar structure.
BRP epitopes reside at the electron-dense T-bar matrix
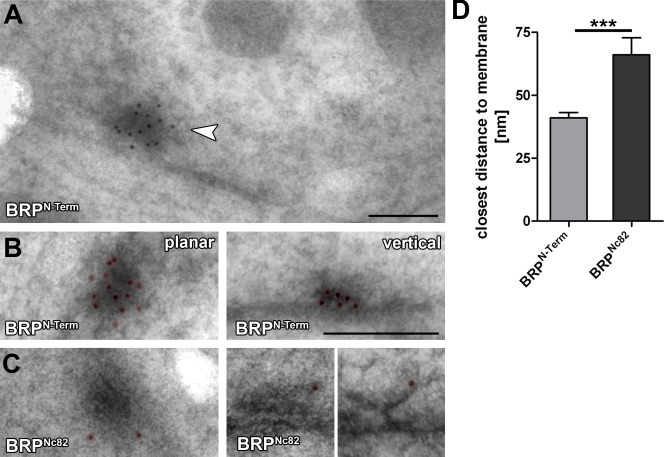
To monitor whether BRP epitopes are associated with the electron-dense T-bar matrix, HPF/FS samples were subjected to immuno-EM. The antibodies to both epitopes, BRPN-Term and BRPNc82, bound to the T-bar matrix (Fig. 3). Notably, the BRPN-Term antibody showed higher labeling efficacy (Fig. 3, compare B with C) and was found throughout cross-sectional views of T-bars in vertical sections (Fig. 3 B, right).
Figure 3.
Immuno-EM versus two BRP epitopes at Drosophila NMJ AZs. (A) High pressure frozen bouton prepared for immunogold labeling, indicating a T-bar labeled for BRPN-Term (arrowhead). (B and C) Magnifications of individual planar (left) and vertically (right) imaged T-bars labeled (after embedding) for either BRPN-Term (B) or BRPNc82 (C). Gold particles are highlighted by red circles. (D) Quantification of BRPN-Term and BRPNc82 signals of the closest distance of individual gold particles to the AZ membrane. Error bars indicate mean ± SEM. ***, P < 0.005. Bars: (A) 150 nm; (B) 200 nm.
In contrast, BRPNc82-conjugated gold particles were typically found at the most distal edge of electron-dense structures in vertical sections (Fig. 3 C, right). From the expression of ectopic BRP fragments (Fig. 1, A and B) and brp1.3 (Fig. 2 E), it is clear that the Nc82 epitope must be located distal to aa 1,390 in the C-terminal quarter of the protein. When comparing BRPN-Term and BRPNc82, the BRPN-Term label appeared significantly closer to the AZ membrane than the BRPNc82 label (Fig. 3 D). Thus, the N-terminal region of BRP seems to reside closer to the AZ membrane than the C-terminal region. However, it should be noted that because of the moderate membrane contrast possible with the immuno-EM technique used, the angle at which AZs are observed cannot be exactly determined. This complicates a quantitative analysis. Collectively, both BRP epitopes are found at the T-bar matrix, and the ultrastructural analysis supports the notion that BRP is a direct component of the T-bar.
BRPNc82 and BRPN-Term epitopes are vertically segregated relative to the AZ membrane
To validate and extend our findings concerning the localization of BRPN-Term and BRPNc82, further light microscopic experiments were performed. Conventional fluorescence microscopy is highly compatible with protein-specific labeling and enables the processing of high sample numbers. Importantly, confocal sectioning of Drosophila NMJ boutons allows for a reliable definition of the orientation of synapses relative to the optical axis because bouton surfaces are nearly spherical. Hereafter, tangentially imaged AZs are called planar AZs, whereas vertically imaged AZs are referred to as vertical AZs.
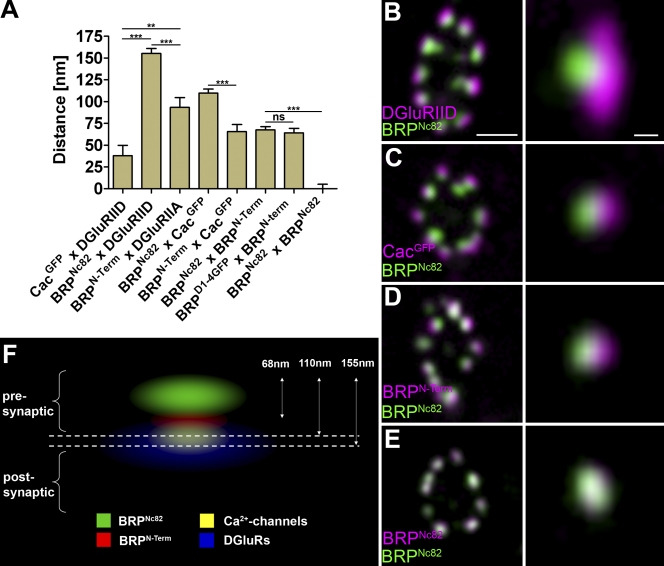
We were interested in visualizing the BRP protein with its distinct epitopes in relation to the remainder of the AZ and the synapse. Therefore, center of mass distances at vertical AZs (along an axis perpendicular to the AZ membrane) were performed with standard confocal microscopy. First, to test this approach, the distance between postsynaptic glutamate receptors (intracellular epitope on Drosophila glutamate receptor subunit IID [DGluRIID]; Qin et al., 2005) and presynaptic Ca2+ channels (CacGFP [GFP at intracellular C terminus of Cac]; Kawasaki et al., 2004) was measured. A value of 40 nm was determined (Fig. 4 A), which is compatible with a distance of ∼35 nm between the cytoplasmic leaflets of pre- and postsynaptic membranes, as measured with EM at NMJ synapses (not depicted).
Figure 4.
Polarized orientation of BRP at AZs. (A) Distances of center to center intensity maxima for different synaptic labels are as follows: CacGFP × DGluRIID, 37.9 ± 11.9 nm (n = 30); BRPNc82 × DGluRIID, 155.2 ± 5.7 nm (n = 30); BRPN-Term × DGluRIIA, 93.4 ± 11.3 nm (n = 30); BRPNc82 × CacGFP, 109.8 ± 4.6 nm (n = 30); BRPN-Term × CacGFP, 65.6 ± 7.9 nm (n = 30); BRPNc82 × BRPN-Term, 67.5 ± 3.8 nm (n = 70); D1-4GFP × BRPN-Term, 64.2 ± 5.1 nm (n = 40); BRPNc82 × BRPNc82, 0.0 ± 5.2 nm (n = 30). Error bars indicate mean ± SEM. **, P < 0.01; ***, P < 0.005. (B–E) Confocal images of midsections through the bouton (left) and single vertically imaged synapses (right) with the bouton lumen facing left. (B) Magenta, DGluRIID; green, BRPNc82. Bars: 500 nm (left) and 100 nm (right). (C) Magenta, CacGFP; green, BRPNc82. (D) Magenta, BRPN-Term; green, BRPNc82. (E) Magenta, BRPNc82; green, BRPNc82. (F) Schematic model of protein epitope distribution at an individual NMJ AZ.
As expected, mAb Nc82 identified diffraction-limited spots opposite the center of postsynaptic densities (PSDs; Fig. 4 B). Notably, BRPNc82 and DGluRIID signals were separated by ∼155 nm (Fig. 4, A and B), and BRPNc82 and CacGFP were separated by ∼110 nm (Fig. 4, A and C). Thus, the epitope recognized by BRPNc82 is clearly localized at a distance away from the presynaptic AZ membrane, which is consistent with our immuno-EM findings (Fig. 3, C and D). Next, we colabeled BRPN-Term and BRPNc82 (Fig. 4 D). The BRPN-Term label was found ∼70 nm closer to the plasma membrane than the C-terminal label (Fig. 4 A).
Our measurements were performed using sandwiches of primary antibodies and labeled secondary antibodies. For distances in the double-digit nanometer range, the size of individual Ig molecules (used for the detection of epitopes) might be relevant. Thus, we sought to independently validate the distance between BRP N and C termini. To do so, BRPD1-4GFP was expressed in the brp69 background, and the distance between the BRP C terminus (endogenous GFP fluorescence) and the N terminus (BRPN-Term antibody) was determined (Fig. 4 A). Again, ∼70 nm was measured. Finally, the center to center distance between BRPN-Term and CacGFP was measured as ∼60 nm (Fig. 4 A). Collectively, we conclude that the BRPNc82 and BRPN-Term epitopes are segregated along an axis perpendicular to the AZ membrane.
The BRP N terminus displays a confined distribution close to the AZ membrane
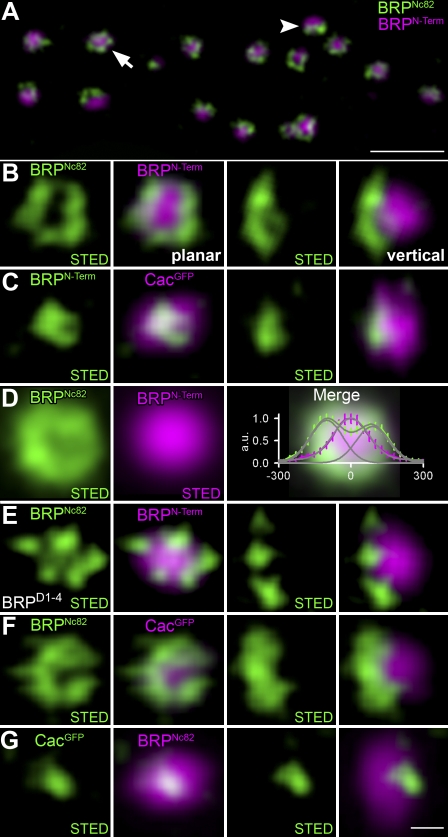
We proceeded to study the molecular organization of AZs at the Drosophila NMJ with STED microscopy to obtain improved optical resolution in xy coordinates. Previously, it was demonstrated that BRPNc82 forms doughnut-shaped structures when visualized at AZs arranged planar to the optical axis (Kittel et al., 2006). BRPNc82 doughnuts were reproduced from planar AZs (Fig. 5, A [arrow] and B) with a resolution displaying an effective point-spread function (PSF) of 80-nm full-width half-maximum. Other than BRPNc82, BRPN-Term did not show a doughnut-shaped distribution when imaged with STED (Fig. 5 C). Instead, the BRPN-Term signal appeared centered within the “doughnut hole” of the BRPNc82 signal at planar AZs, as also apparent after averaging STED signals from individually aligned AZs (Fig. 5 D). The combination of STED resolution (confined to one channel in our experiments) for BRPNc82 and confocal resolution for BRPN-Term revealed a polarized and funnel-like distribution of BRP epitopes (Fig. 5, A–C). Notably, the BRPNc82 signal did not appear fully continuous but instead consisted of discrete foci (Fig. 5, B and F) within an overall circular array. In an additional experiment, we expressed full-length BRPD1-4 (Wagh et al., 2006) in brp69 mutants. The distance between BRPN-Term and BRPNc82 was similar to that observed at control AZs (Fig. 5 E). This suggests that individual BRP molecules can adopt an elongated conformation.
Figure 5.
STED analysis of AZ organization at Drosophila NMJ synapses. (A) Overview of a bouton stained for BRPN-Term (confocal; magenta) and BRPNc82 (STED; green) showing planar (arrow) and vertical (arrowhead) AZs. (B and C) Magnifications of individual planar (left) and vertical (right) AZs stained for BRPNc82 (STED) and BRPN-Term (confocal; B) and BRPN-Term (STED) and CacGFP (confocal; C). (D) Mean normalized planar BRPN-Term (magenta) and BRPNc82 (green) arrangement shown with STED resolution (BRPN-Term, n = 14; BRPNc82, n = 47). (right) The merge superimposed with the intensity profile along one axis through the midpoint for BRPN-Term (magenta) and BRPNc82 (green) is shown. Error bars indicate ± SEM. (E) BRPNc82 (STED) and BRPN-Term (confocal) after expression of full-length BRP cDNA in brp69 background (BRPD1-4). (F and G) Individual planar (left) and vertical (right) AZs stained for BRPNc82 (STED) and CacGFP (confocal; F) and CacGFP (STED) and BRPNc82 (confocal; G). All images were deconvolved using Imspector software. Bars: (A) 1 µm; (G) 100 nm.
The N terminus of BRP overlays the Ca2+ channels at the AZ core
How does the molecular architecture of BRP relate to Ca2+ channels at AZs? Ca2+ channel spots (CacGFP) imaged at standard confocal resolution were found to cocenter with the BRPN-Term label and BRPNc82 doughnuts at planar AZs. At vertical AZs, the CacGFP signal localized toward the AZ membrane relative to both BRPN-Term (Fig. 5 C) and BRPNc82 (Fig. 5, F and G). When imaged with STED resolution, Ca2+ channels consistently localized to small, typically slightly elliptical patches (∼100–150 nm along the longest axis) at the AZ center (Fig. 5 G). Thus, the T-bar organized by BRP overlays the field of Ca2+ channels at the AZ center.
DLiprin-α localizes to discrete compartments surrounding the AZ center
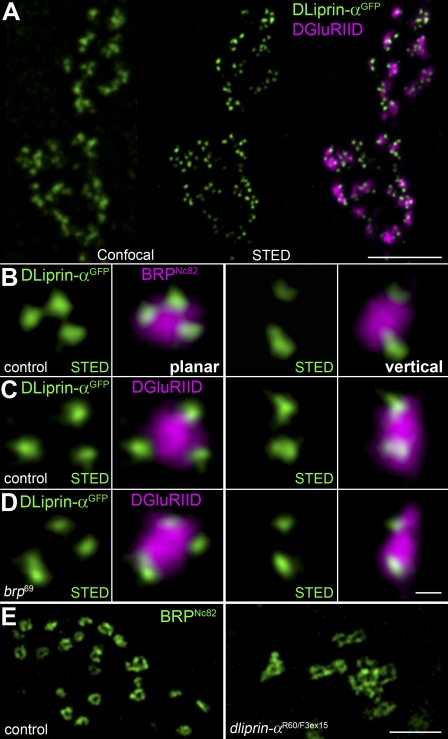
Liprin-α localizes to the AZ and has been shown to be important for the formation of AZs in both Drosophila and C. elegans (Kaufmann et al., 2002; Dai et al., 2006; Patel et al., 2006). To extend our “AZ map,” DLiprin-αGFP was expressed in motoneurons and visualized via αGFP stainings with STED microscopy (Fig. 6 A). DLiprin-α localized to presynaptic AZs opposite DGluRIID-positive PSDs. However, as opposed to BRP, DLiprin-α clustered somewhat lateral from the AZ center. STED resolution revealed that DLiprin-α formed discrete “quantal” clusters at the edge of a single AZ colabeled with BRPNc82 (Fig. 6 B) or DGluRIID (Fig. 6 C).
Figure 6.
STED microscopic analysis of DLiprin-αGFP at AZs. (A) Single confocal sections of NMJs colabeled for DLiprin-αGFP (green, confocal resolution in left image and STED resolution in middle image) and DGluRIID (magenta, confocal overlay in right image). STED images of DLiprin-αGFP reveal substructures beyond the diffraction limit of confocal microscopy. (B–D) STED images of an individual AZ. Discrete dots of DLiprin-αGFP are arranged at the AZ edge (magenta, BRPNc82 [B] and DGluRIID [C and D]). Left, planar AZ; right, vertical AZ. B and C show controls, and D shows brp69. (E) Single confocal slices of control (left) and dliprin-α (right) junctions labeled for BRPNc82 with STED resolution. Atypical clusters of BRP doughnuts are observed at dliprin-α mutant NMJs. Bars: (A) 1.5 µm; (D) 100 nm; (E) 1 µm.
Discrete DLiprin-α clusters were still observable at brp69 NMJs, suggesting that the presence of BRP is not essential for the recruitment of DLiprin-α to the AZ (Fig. 6 D). However, the localization of BRP, as imaged with STED, appeared aberrant at dliprin-α NMJs (Fig. 6 E). Strikingly, individual BRP doughnuts seemed interconnected, which is directly consistent with the previous observation of complex, multi–T-bar AZs at dliprin-α mutant NMJs (Kaufmann et al., 2002).
BRP and Ca2+ channels accumulate late during AZ assembly
So far, we have provided evidence that BRP operates as an essential building block of the T-bar. Notably, fast assembly of T-bars might drive experience-dependent changes of synaptic transmission in the fly central nervous system (Brandstatter et al., 1991; Rybak and Meinertzhagen, 1997). Thus, to learn about the T-bar assembly process in the frame of synapse reorganization, we visualized BRP accumulation in vivo during the developmental formation of individual synapses (Rasse et al., 2005; Fuger et al., 2007; Schmid et al., 2008). Previously, we found that neuromuscular accumulation of glutamate receptors (as DGluRIIAs) in PSDs typically form at a distance from existing PSDs and then grow over several hours before reaching a final mature size (Rasse et al., 2005; Schmid et al., 2008). Thus, for the analysis of AZ assembly in vivo, DGluRIIA was coimaged to serve as a reference point for our temporal analysis (Fig. 7, A, B, and D; and Table II).
Figure 7.
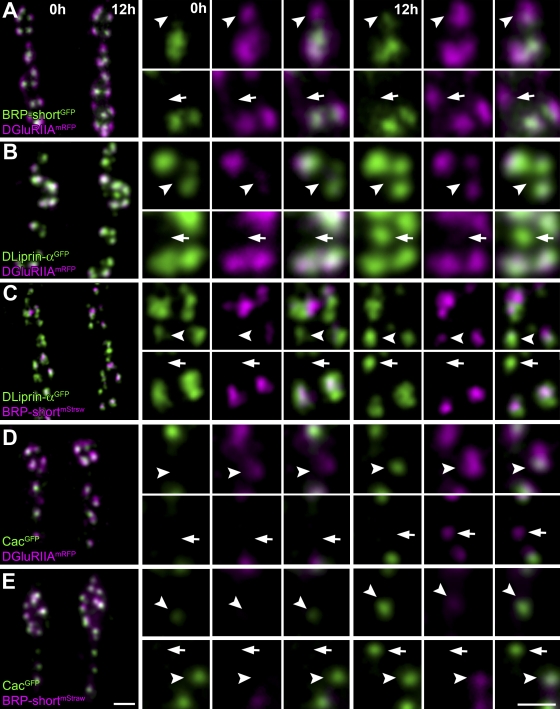
In vivo analysis of synaptic protein accumulation. (A–E) Confocal stacks of sequentially in vivo–imaged NMJs (muscle 26) at Δt = 12 h. NMJs coexpress the indicated labels (green, GFP constructs; magenta, mRFP constructs). (top) Individual in vivo–imaged synapses (arrowheads) positive for only one label at t = 0 h but positive for both labels at t = 12 h are shown. (bottom) A prospective synapse (arrows) positive for only one label at t = 12 h is shown. Bars, 1 µm.
Table II.
Quantification of protein accumulation during AZ assembly using NMJ in vivo imaging
| Coexpressed proteins (A × B) | Both A and B | A before B | B before A |
| BRP × DGluRIIA | BRP+/IIA+ 17/39 (44%) | BRP+/IIA− 0/39 (0%) | BRP−/IIA+ 22/39 (56%) |
| DLiprin-α × DGluRIIA | DLiprin-α+/IIA+ 16/39 (41%) | DLiprin-α+/IIA− 23/39 (59%) | DLiprin-α−/IIA+ 0/39 (0%) |
| BRP × DLiprin-α | BRP+/DLiprin-α+ 8/31 (26%) | BRP+/DLiprin-α− 0/31 (0%) | BRP−/DLiprin-α+ 23/31 (74%) |
| DGluRIIA × Cac | IIA+/Cac+ 36/62 (58%) | IIA+/Cac− 21/62 (34%) | IIA−/Cac+ 5/62 (8%) |
| BRP × Cac | BRP+/Cac+ 42/62 (68%) | BRP+/Cac− 7/62 (11%) | BRP−/Cac+ 13/62 (21%) |
Quantification of the relative accumulation of the indicated synaptic proteins at newly forming AZs (Δt = ∼12 h). A synaptic site was scored positive (+) or negative (−) for a specific protein depending on whether protein fluorescence signals exceeded the mean background level by >2.5-fold. For example, when comparing BRP and DGluRIIA (at synapses forming newly over 12 h), 44% were positive for both proteins, 0% for BRP only, and 56% for DGluRIIA only.
Larvae coexpressing two fluorescently tagged synaptic proteins were imaged (Fig. 7), and quantitative data were obtained to analyze the temporal sequence of protein arrival at developing AZs. For a given larval NMJ, two in vivo images were acquired with a time interval of 12 h. Sites were regarded as new synapses if protein labels exceeded the mean background by a factor of 2.5 at the second (t = 12 h) but not at the first time point (t = 0 h). This way, a temporal sequence of molecular AZ assembly was extracted.
We first compared BRP and DGluRIIA accumulation. For visualization of BRP, we used a fragment of the protein (BRP-short), which delivered a label that fully matched the label of endogenous BRP (Schmid et al., 2008). When BRP-shortGFP was examined with STED, doughnuts were detected that resembled those found with BRPNc82 (unpublished data). As previously described (Schmid et al., 2008), the accumulation of DGluRIIA clearly preceded BRP arrival in vivo (Fig. 7 A and Table II). Moreover, all postsynaptic DGluRIIA accumulations eventually incorporated presynaptic BRP, demonstrating that DGluRIIA accumulation reliably indicates the formation of new synapses.
As described in the previous section, presynaptic DLiprin-α localization seems to be largely independent of BRP (Fig. 6 D), which is compatible with DLiprin-α functioning upstream of BRP in AZ assembly. Consistently, DLiprin-α incorporation invariably preceded BRP accumulation (Fig. 7 C and Table II).
BRP is crucial for either the initial formation or the maintenance of Ca2+ channel clusters. Thus, we analyzed Cac localization at individual developing AZs. Cac and BRP appeared highly correlated, and the timing of Cac accumulation at AZs was typically very close to the advent of BRP with a slight tendency of Cac to precede BRP (Fig. 7 E and Table II).
Collectively, we show that newly forming AZs, similar to PSDs (Rasse et al., 2005), are small to begin with and then increase in size over many hours in vivo before accumulating detectable levels of BRP and reaching a final mature size at developing NMJs. This assembly process of individual new synapses is protracted over hours and is characterized by the contribution of pre- and postsynaptic proteins in a defined, overlapping sequence. DLiprin-α appears to be a very early player involved in initializing AZ assembly, whereas BRP, together with Cac, follows only after postsynaptic DGluRIIA incorporation is already clearly detectable (Fig. 7, A and D).
BRP controls Ca2+ channel accumulation at maturing AZs
If Cac slightly precedes BRP during assembly, how can the Cac-clustering defects described in brp69 (Kittel et al., 2006) be explained? To exclude allele-specific effects, we first scored Cac clustering at AZs (opposite DGluRIID receptor fields) in the brp alleles brp6.1, brp69, brpc04298, and brp1.3 (Fig. 8 A). A Cac-clustering defect identical to that found in brp69 was observed in the full-deletion brp6.1. As expected, brpc04298 also showed an identical clustering defect (Fig. 8 A). Previously, we provided evidence that the Cac delocalization in brp69 is responsible for reduced neurotransmitter release. In this study, we took the opportunity to test the influence of BRP on neurotransmission independently of brp69 and recorded from brpc04298 NMJs. All electrophysiological features of brpc04298, including the alterations of short-term plasticity connected to defective Ca2+ channel clustering, were similar to those observed in brp69 (Fig. S3; Kittel et al., 2006). Thus, several independent alleles clearly demonstrate that loss of BRP results in defective clustering of Cac at the AZ, which in turn provokes defects in transmitter release.
Figure 8.
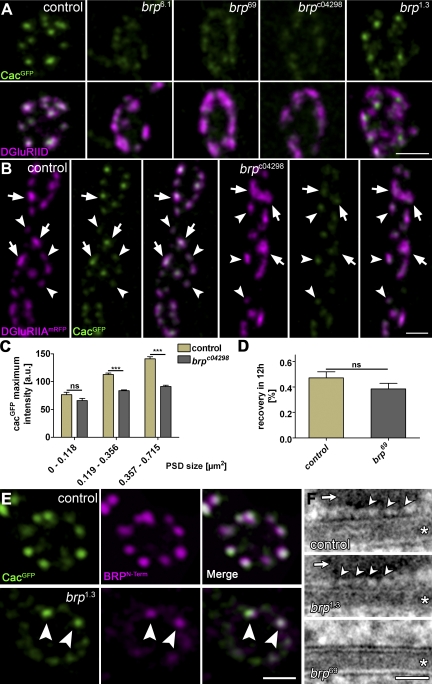
Size-dependent Ca2+ channel–clustering defects in brp. (A) CacGFP (green) clustering at AZs (identified at AZs opposite DGluRIID (magenta)) in control, brp6.1, brp69, brpc04298, and brp1.3 animals. (B) Comparison of Ca2+ channel clustering at control and brpc04298 NMJs expressing CacGFP and DGluRIIAmRFP. Small PSDs in brp mutants opposite CacGFP clusters of comparable intensity to controls (arrowheads). Larger PSDs, indicating a more advanced synaptic maturation state, typically display severely decreased CacGFP intensity values compared with controls (arrows). (C) Statistical analysis shows no significant differences in the maximum CacGFP intensity opposite particularly small PSDs (control: 0–0.118 µm2 = 76.7 ± 4.4 au [n = 66]; 0.119–0.356 µm2 = 113.0 ± 2.3 au [n = 161]; 0.357–0.715 µm2 = 1,41.0 ± 4.0 au [n = 46]; brpc04298: 0–0.118 µm2 = 66.0 ± 3.9 au [n = 34]; 0.119–0.356 µm2 = 83.7 ± 1.6 au [n = 157]; 0.357–0.715 µm2 = 91.4 ± 2.1 au [n = 73]; P = 0.12, P < 0.005, and P < 0.005 by Student’s t test). (D) FRAP experiment for CacGFP with a recovery interval of 12 h. FRAP was comparable between brp69 and control boutons (control: 0.47 ± 0.04 [n = 6]; brp69: 0.38 ± 0.04 [n = 6]; P = 0.3 by Mann-Whitney test). (E) Costaining of BRPN-Term (magenta) and CacGFP (green) at brp1.3 boutons and controls. Those AZs showing restored Cac clustering also show restored BRPN-Term label (arrowheads). (F) Ultrastructure of the area between presynaptic membrane and the T-bar pedestal (arrows) prepared via HPF/FS. Asterisks indicate the synaptic cleft. At control (top) and brp1.3 (middle) AZs, discrete regularly arranged elements emerging from the AZ membrane (∼5–7 nm; arrowheads) are observed within the gap between the AZ membrane and the T-bar pedestal. These elements are not observed at brp69 AZs lacking T-bars (bottom). Error bars indicate mean ± SEM. ***, P < 0.005. Bars: (E) 1 µm; (F) 25 nm.
Notably, the mislocalization of Cac is not absolute, but instead, a certain degree of Cac remained clustered at AZs in brp null. As our in vivo imaging experiments show (Fig. 7), AZs go through a long, protracted assembly process before finally reaching their fully mature size. Thus, we wondered whether BRP may be more important for maturation and potentially less so for initialization of Cac clustering. To address this, we visualized Cac in intact, living brpc04298 mutant larvae together with DGluRIIA as a reference for synapse maturation (Fig. 8 B). In brpc04298 larvae, nascent synapses, identified by their small DGluRIIA accumulations, seemed to accumulate Cac at normal density (Fig. 8, B and C). However, larger, more mature synapses with more substantial DGluRIIA accumulations showed reduced Cac density in the absence of BRP (Fig. 8, B and C). In clear contrast, the recruitment of CacGFP to AZs appears to be independent of presynaptic DLiprin-α, as the localization of Ca2+ channels appears normal in dliprin-α mutants (Fig. S4).
Our data imply the following scenario: BRP, an integral component of the T-bar, is recruited to AZs later than DLiprin-α. BRP seems largely dispensable for the initial Ca2+ channel accumulation. However, the matrix organized by BRP is required for maintaining the continuous clustering of Ca2+ channels beneath the T-bar base during the maturation process, which lasts several hours.
Alternatively, the Ca2+ channel–clustering defect in brp could be caused by impaired Cac transport. To address this, Cac trafficking dynamics were measured using FRAP experiments. Notably, Cac FRAP was slow, with a recovery half-time of ∼12 h (Fig. 8 D), which is similar to other synaptic membrane proteins such as DGluRIIA (Schmid et al., 2008). Importantly, however, Cac FRAP was essentially unaltered at brp boutons, indicating that the long-range transport of Cac to the AZ is not affected by the loss of BRP (Fig. 8 D). Thus, BRP seems to be directly required for Ca2+ channel clustering at AZs but not for its recruitment to the terminal. We aimed to further address the relation between T-bar assembly and Ca2+ channel clustering. Notably, brp1.3 (Fig. 8 A) showed partially restored Cac clustering. Moreover, those AZs positive for BRPN-Term in brp1.3 corresponded to the AZs of restored Cac clustering (Fig. 8 E). Thus, AZ accumulation of the C-terminally truncated but N-terminally intact BRP protein seems to permit the assembly of a distally truncated T-bar, which still functions in clustering Cac.
To further explore this, we prepared larval NMJ AZs via HPF/FS. In this study, a gap between the AZ membrane and the T-bar pedestal (Fig. 8 F, arrows) was observed, where peg-like (Harlow et al., 2001) structures extended from the AZ membrane and distally contacted (or even slightly penetrated) the T-bar pedestal (Fig. 8 F, top, arrowheads). Such pegs were also observed at brp1.3 AZs (Fig. 8 F, middle, arrowheads) but not at AZ membranes of the brp69-null allele (Fig. 8 F, bottom). Thus, these data further support the notion that Cac clustering beneath the T-bar pedestal is restored at brp1.3 AZs. Moreover, the cytoplasmic domains of Ca2+ channels might well extend from the AZ membrane, possibly allowing for a direct interaction with BRP.
Possible direct interaction between the C terminus of Cac and the N terminus of BRP
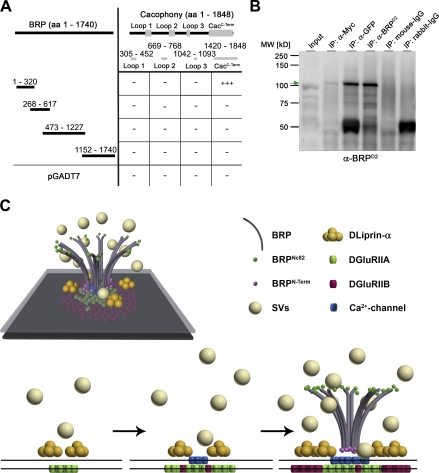
To address a possible interaction between BRP and Ca2+ channels, we tested for protein–protein interactions in vitro. First, all intracellular loops of Cac were tested with BRP constructs in a yeast two-hybrid assay. Only the C-terminal domain of Cac and the N-terminal domain of BRP showed positive for interaction (Fig. 9 A). Thus, in agreement with our hypothesis concerning the orientation of BRP within the T-bar, the BRP N terminus may interact with the Cac C-terminal domain. To further investigate this interaction, we double transfected Schneider S2R+ cells with a GFP-tagged N-terminal construct of BRP (aa 1–617) and Myc-CacC-Term (aa 1,420–1,848). Pull-downs directed against Myc, GFP, and BRP (using a polyclonal antibody directed against aa 268–617 of BRP; see Materials and methods) confirmed a direct interaction (Fig. 9 B).
Figure 9.
The N terminus of BRP physically interacts with the C terminus of Cac in vitro. (A) Scheme of yeast two-hybrid analysis using Cac bait constructs and overlapping BRP prey constructs. Full-length Cac protein comprises three large intracellular loops (aa are indicated) and its intracellular C-terminal region (aa 1,420–1,848). The N terminus of BRP (aa 1–320) interacts with the C-terminal region of Cac (83% of the reported cytoplasmic C terminus, sparing the EF hand and most of its IQ motif; Kawasaki et al., 2002). −, no interaction; +++, interaction of high confidence. (B) Co-IPs from Schneider cell extracts cotransfected with a GFP-tagged N-terminal construct of BRP (D1-2GFP; aa 1–617) and Myc-tagged CacC-Term. Western blotting shows the pull-down of the BRP construct in the anti-Myc IP at ∼100 kD (arrow). The corresponding band is not detected in the control lanes (IgGs). The slightly different migration of the band in the input lane and in the IP lanes is the result of differences in sample buffer. MW, molecular weight. (C) Spatiotemporal model of AZ assembly and organization at Drosophila NMJs. SVs, synaptic vesicles.
We are presently unable to (genetically) test whether this physical interaction of BRP with the Cac C terminus (and not other functions downstream of BRP-mediated T-bar assembly) is essential for Cac clustering. However, we tested whether Cac was important for BRP localization by staining cac-null embryos (Kawasaki et al., 2002). In this study, BRP signals were not reduced compared with controls (Fig. S5). Thus, BRP might be involved in providing slots in the AZ cytomatrix, which define sites of stable Ca2+ channel incorporation, whereas the Ca2+ channels themselves are not involved in clustering BRP. Collectively, our results suggest a model as depicted in Fig. 9 C for the spatiotemporal assembly process of the AZ at the glutamatergic Drosophila NMJ.
Discussion
Efficient neurotransmission is believed to crucially depend on the structural and functional integrity of the presynaptic AZ compartment (Schoch and Gundelfinger, 2006). An ancestral set of AZ components is conserved between Drosophila, C. elegans, and mammals (Stryker and Johnson, 2007; Jin and Garner, 2008). The strong phenotype we observed at Drosophila NMJs in the absence of the ERC/CAST member BRP, lacking T-bar–dense bodies and defective Ca2+ channel clustering (Kittel et al., 2006; Wagh et al., 2006), forms an entry point for studying AZ assembly, which is often complicated by redundant and cooperative interactions between AZ components (Jin and Garner, 2008).
BRP and dense body formation
We addressed whether BRP signals T-bar formation (without being a direct component of the T-bar) or whether the protein itself is an essential building block of this electron-dense structure. In this study, we provide evidence that BRP is a direct T-bar component. Immuno-EM identifies the N terminus of BRP throughout the whole cross section of the T-bar (Fig. 3, A and B), and genetic approaches show that a truncated BRP, lacking the C-terminal 30% of the protein’s sequence, forms truncated T-bars (Fig. 2, E and F). Immuno-EM and light microscopy consistently demonstrate that N- and C-terminal epitopes of BRP are segregated along an axis vertical to the AZ membrane and suggest that BRP is an elongated protein, which directly shapes the T-bar structure (Fig. 9 C).
In brp5.45 (predicted as aa 1–866), T-bars were not detected, whereas brp1.3 (aa 1–1,389) formed T-bar–like structures, although fewer and smaller than normal (Fig. 2, E and F; and Fig. S2 C). Moreover, the BRPD1-3GFP construct (1–1,226) did not rescue T-bar assembly. Thus, domains between aa 1,226 and 1,390 of BRP may also be important for the formation of T-bars. Clearly, however, the assembly scheme for T-bars is expected to be controlled at several levels (e.g., by phosphorylation) and might involve further protein components. Nonetheless, it is highly likely that the C-terminal half of BRP plays a crucial role.
BRP, the dense body, and Ca2+ channel clustering
As BRP represents an essential component of the electron-dense T-bar subcompartment at the AZ center, it might link Ca2+ channel–dependent release sites to the synaptic vesicle cycle (Neher and Sakaba, 2008). Interestingly, light and electron microscopic analysis has located CAST at mammalian synapses both with and without ribbons (tom Dieck et al., 2005; Deguchi-Tawarada et al., 2006; Siksou et al., 2007). Overall, this study is one of the first to genetically identify a component of an electron-dense synaptic specialization and thus paves the way for further genetic analyses of this subcellular structure.
The N terminus of BRP is found significantly closer to the AZ membrane than the C terminus, where it covers a confined area very similar to the area defined by the CacGFP epitope. Electron tomography of frog NMJs suggested that the cytoplasmic domains of Ca2+ channels, reminiscent of pegs, are concentrated directly beneath a component of an electron-dense AZ matrix resembling ribs (Harlow et al., 2001). In addition, freeze-fracture EM identified membrane-associated particles at flesh fly AZs, which, as judged by their dimensions, might well be Ca2+ channels (Feeney et al., 1998). We observed peg-like structures beneath the T-bar pedestal. Similar to fly T-bars, the frog AZ matrix extends up to 75 nm into the presynaptic cytoplasm. Based on the amount of cytoplasmic Ca2+ channel protein (Catterall, 1998), Harlow et al. (2001) concluded that Ca2+ channels are likely to extend into parts of the ribs. Thus, physical interactions between cytoplasmic domains of Ca2+ channels and components of ribs/T-bars might well control the formation of Ca2+ channel clusters at the AZ membrane. However, a short N-terminal fragment of BRP (aa 1–320) expressed in the brp-null background was unable to localize to AZs efficiently and consistently failed to restore Cac clustering (unpublished data).
The mean Ca2+ channel density at AZs is reduced in brp-null alleles. In vitro assays indicate that the N-terminal 20% of BRP can physically interact with the intracellular C terminus of Cac. Notably, we found that the GFP epitope at the very C terminus of CacGFP was closer to the AZ membrane than the N-terminal epitope of BRP (Fig. 4 A). It is conceivable that parts of the Cac C terminus extend into the pedestal region of the T-bar cytomatrix to locally interact with the BRP N terminus. This interaction might play a role in clustering Ca2+ channels beneath the T-bar pedestal.
Clearly, additional work will be needed to identify the contributions of discrete protein interactions in the potentially complex AZ protein interaction scheme. Our study should pave the way for a genetic analysis of spatial relationships and structural linkages within the AZ organization. Moreover, we aim to integrate our findings in the framework of mechanisms for Ca2+ channel trafficking, clustering, and functional modulation (Cao et al., 2004; Evans and Zamponi, 2006; Catterall and Few, 2008).
Timing of AZ assembly, Ca2+ channel accumulation, and synapse maturation
Our imaging assays allowed a temporally resolved analysis of AZ assembly in vivo (Fig. 7). BRP is a late player in AZ assembly, arriving hours after DLiprin-α and also clearly after the postsynaptic accumulation of DGluRIIA. Accumulation of Cac was late as well, although it slightly preceded the arrival of BRP, and impaired Cac clustering at AZs lacking BRP became apparent only from a certain synapse size onwards (Fig. 8, B and C). In this study, we report that new AZs, similar to PSDs (Rasse et al., 2005), form at sites distant from preexisting ones and grow to reach a mature, fixed size. Thus, the late, BRP-dependent formation of the T-bar seems to be required for maintaining high Ca2+ channel levels at maturing AZs but not for initializing Ca2+ channel clustering at newly forming sites. As the dominant fraction of neuromuscular AZs is mature at a given time point, the overall impression is that of a general clustering defect in brp mutants. In reverse, it will be of interest to further differentiate the molecular mechanisms governing early Ca2+ channel clustering. Pre- to postsynaptic communication via neurexin–neuroligin (Missler et al., 2003; Li et al., 2007; Zeng et al., 2007) interactions might well contribute to this process. A further candidate involved in early Ca2+ channel clustering is the Fuseless protein, which was recently shown to be crucial for proper Cac localization at AZs (Long et al., 2008).
In summary, during the developmental formation of Drosophila NMJ synapses, the emergence of a presynaptic dense body, which is involved in accumulating Ca2+ channels, appears to be a central aspect of synapse maturation. This is likely to confer mature release probability to individual AZs (Kittel et al., 2006) and contribute to matching pre- and postsynaptic assembly by regulating glutamate receptor composition (Schmid et al., 2008). Whether similar mechanisms operate during synapse formation and maturation in mammals remains an open question.
Outlook: dense body architectures and synaptic plasticity
In this study, we concentrated on developmental synapse formation and maturation. The question arises whether similar mechanisms to those relevant for AZ maturation might control activity-dependent plasticity as well and whether maturation-dependent changes might be reversible at the level of individual synapses. Notably, experience-dependent, bidirectional changes in the size and number of T-bars (occurring within minutes) were implied at Drosophila photoreceptor synapses by ultrastructural means (Brandstatter et al., 1991; Rybak and Meinertzhagen, 1997). Moreover, at the crayfish NMJ, multiple complex AZs with double-dense body architecture were produced after stimulation and were associated with higher release probability (Wojtowicz et al., 1994). In fact, a recent study has correlated the ribbon size of inner hair cell synapses with Ca2+ microdomain amplitudes (Frank et al., 2009). Thus, a detailed understanding of the AZ architecture might permit a prediction of functional properties of individual AZs.
Materials and methods
Genetics
All fly strains were reared under standard laboratory conditions (Sigrist et al., 2003). Either w1 or w1118 was used as background for transgenesis (BestGene Inc.).
Chemical mutagenesis
The EMS screen was performed according to standard protocols. In brief, isogenic w1118 males were mutagenized with 25 mM EMS solution and crossed to virgins carrying a second chromosomal balancer. For initial mapping, male F1 offspring were crossed with Df(2R)BSC29 virgins, and candidate flies were tested with different brp mutant lines. Genomic DNA was extracted from positive candidate flies, and PCR amplicons containing brp exon clusters were double-strand sequenced to identify the mutations.
Generation of brp deletions
brp chromosomal deletions were constructed using the Flp recombinase system as previously described (Parks et al., 2004). The different parental lines were provided by the Exelixis collection at Harvard Medical School (Boston, MA).
The following genotypes were used for the ectopic expression of the BRP constructs (Fig. 1 A; Fig. 2, G and H; and Fig. 5): for wing disc expression, upstream activator sequence (UAS)–BRPD1-3GFP/+; dpp-Gal4/+. UAS-BRPD2-4GFP/dpp-Gal4. UAS-BRPD1-4GFP/dpp-Gal4. For expression of BRP constructs at NMJs in the brp mutant background, UAS-BRPD1-3GFP/+; Df(2R)BSC29/brp69, ok6-Gal4. Df(2R)BSC29/brp69, ok6-Gal4; UAS-BRPD2-4GFP/+. Df(2R)BSC29/brp69, ok6-Gal4; UAS-BRPD1-4/+. Df(2R)BSC29/brp69, ok6-Gal4. The Gal4 lines and Df(2R)BSC29 were provided by the Bloomington Drosophila Stock Center. For experiments in the dliprin-α mutant background (Fig. 6 D), we used dliprin-αR60/dliprin-αF3ex15 animals (Kaufmann et al., 2002).
Antibodies
For the N-terminal BRP (BRPN-Term) antibody, a rabbit polyclonal antibody was raised (Seqlab) against a synthetic peptide (CREPRDRSRDRSLER). The specificity of the affinity-purified α-BRP antibody was confirmed by immunofluorescence analysis of larval muscle fillet preparations.
The BRPD2 antibody was raised against the BRP domain D2 (aa 268–617; Seqlab) in rabbits. The immunogen was expressed recombinantly as 6×His-tagged fusion protein in Escherichia coli and purified using a protocol including denaturing and refolding of the protein. The antibody containing serum was affinity purified versus the same protein as used for immunization.
Transmission EM
For HPF, about one to three Drosophila late second/early third instar larvae were placed in aluminum specimen carrier of 200-µm depth (type A; Bal-Tec), filled with yeast paste, and covered with a lid. The samples were frozen immediately in an HPM machine (HPM010; Bal-Tec) and rapidly transferred to liquid nitrogen for storage.
FS and embedding were performed in acetone in either an EM AFS (automatic freeze substitution; for morphology; Leica) or an EM AFS2 (for immunocytochemistry; Leica). Two separate protocols were used for morphological and immunocytochemical analysis (Rostaing et al., 2006; Siksou et al., 2007). For immunocytochemistry, the substitution was performed in pure acetone without uranyl acetate.
For morphology, 55–60-nm (gray silver) sections, and for immunocytochemistry, 85-nm (silver gold) sections were cut using an EM Ultracut 6 (Leica). Sections were collected on formvar-coated 100 mesh grids. For morphological experiments, sections were dried and poststained with uranyl acetate and lead citrate as described previously (Schmid et al., 2006). For immunocytochemistry, grids were placed in droplets of PBS, pH 7.2, until labeling procedure started. Immunocytochemistry was performed as described previously (Rostaing et al., 2006; Siksou et al., 2007). Rbα-BRPN-Term (1:500) and Mα-BRPNc82 (Nc82; 1:2; provided by E. Buchner, Universität Würzburg, Würzburg, Germany) antibodies were used. Micrographs were taken with a 1,024 × 1,024 charge-coupled device detector (HSS 512/1024; Proscan Electronic Systems) in an electron microscope (EM 902A; Carl Zeiss, Inc.) operated in bright field mode.
Conventional room temperature embedding was essentially performed as described previously (Wagh et al., 2006). Images were obtained from dissected preparations of third instar larvae (NMJ 6/7; segments A2/A3). Instead of 1-h fixation in 1% osmium tetroxide, the fixation was performed in 1% osmium tetroxide and 0.8% KFeCn in 0.1 M cacodylate buffer. After infiltration in epon resin, muscles were cut out (six animals for each genotype) and embedded in a single block.
For T-bar size quantification, T-bars or residual T-bars were taken from vertical AZs. The electron density of the T-bar was measured from the AZ membrane to the T-bar platform if present (height) or along the AZ membrane (width).
Molecular cloning
For expression constructs of DLiprin-αGFP, a 3.6-kb fragment of pOT2 LD27334 was subcloned into pBluescript KS(+) (Agilent Technologies) using the SalI and EcoRI restriction sites introduced by PCR primers and subsequently double-strand sequenced. The insert was excised and inserted into pENTR4 (Invitrogen) via SalI and NotI sites. The final expression construct of DLiprin-αGFP was obtained using the Gateway system (Invitrogen). In brief, pENTR4 DLiprin-α was recombined with pTWG (a Drosophila pUASt Gateway vector developed in the laboratory of T. Murphy, The Carnegie Institution of Washington, Baltimore, MD).
The BRP constructs used for expression in Drosophila flies and Drosophila Schneider cell culture were obtained by PCR using the corresponding cDNA as template (brp cDNA; Wagh et al., 2006) and cloned into pENTER, a modified version of pENTR4 using the SpeI and Asp718I restriction sites.
The pUbiP Gateway destination vectors, used for coexpression experiments in S2R+ cell culture, were obtained from A. Herzig (Max Planck Institute, Göttingen, Germany). These contained a ubiquitin promoter and either an N- or C-terminal tag.
Yeast two-hybrid constructs for cac and brp were obtained by PCR using the corresponding cDNA as template (cac cDNA was provided by R.W. Ordway, The Pennsylvania State University, Philadelphia, PA; brp cDNA; Wagh et al., 2006) and cloned into the bait vector pGBKT7 (Clontech Laboratories, Inc.) or the prey vector pGADT7 (Clontech Laboratories, Inc.) using the restriction sites introduced with the PCR primers.
Vectors used for fusion protein expression in Drosophila Schneider cell culture were made from respective modified pENTR4 clones containing truncated brp or truncated cac using the Gateway system.
Molecular cloning in detail
pENTER.
The multiple cloning site of pENTR4 (Invitrogen) was modified by oligonucleotide annealing using two primers with a 5′-phosphate modification (MWG-Biotech AG): 5′-CATGGGAACTAGTCCCGGGCGCGCCGCGGCCGCGGTACCAGC-3′ and 5′-TCGAGCTGGTACCGCGGCCGCGGCGCGCCCGGGACTAGTTCC-3′. The annealed oligonucleotides were ligated into a previously cut pENTR4 vector using the NcoI and XhoI restriction sites. This modification resulted in a loss of the ccdB gene and a new multiple cloning site.
pTWmStrawberry.
In short, the Drosophila Gateway vector pTWG containing an EGFP tag placed downstream of the Gateway cassette was used as a template to replace the EGFP by mStrawberry (mStraw; pRSETB mStraw was provided by R.Y. Tsien, University of California, San Diego, La Jolla, CA). For general information about the Drosophila Gateway vector collection, please visit http://www.ciwemb.edu/labs/murphy/Gateway%20vectors.html#_Copyright_2003,_Carnegie. The following fragments were amplified by PCR using either pTWG (α- and γ-mStraw) or pRSETB mStraw (β-mStraw) as templates: α-mStraw, 5′-CTTCCATGTCGGCAGAATGCT-3′ and 5′-GTTATTCTCCTCGCCCTTGCTCACCAT-3′; β-mStraw, 5′-ACCGGCGGCATGGACGAGCTGTACAAG-3′ and 5′-TGGATCCGATCCAGACATGA-3′; and γ-mStraw, 5′-ATGGTGAGCAAGGGCGAGGA-3′ and 5′-CTTGTACAGCTCGTCCATGC-3′.
In two independent PCRs using the Elongase Enzyme Mix (Invitrogen), the three fragments were fused and subsequently ligated into a previously cut pTWG backbone using XbaI–HpaI. After transformation into E. coli strain DB3.1, the cells were plated on selective media containing ampicillin and chloramphenicol. Digestion and double-strand sequencing confirmed the successful replacement of the fluorophore.
pENTR4 and pENTER cloning.
All final plasmids of pENTR4 or pENTER constructs were double-strand sequenced before any Gateway recombination with destination vectors.
pENTR4 DLiprin-α.
5′-GAGCGTCGACATGTGGAACATGATGTGCGACGTA-3′ and 5′-GGAATTCGCGGCCGCGAAGCACTGCGCTGCTCA-3′; recombined with pTWG (C-terminal EGFP tag).
pENTER BRP D1-2 (aa 1– 617).
5′-GAGTACTAGTATGGGCAGTCCATACTACCGC-3′ and 5′-GTCTGGTACCTGCTCTTTCCGCATCCGAC-3′; recombined with pUbiP-rfA-EGFP (C-terminal EGFP).
pENTER BRP D1-3 (aa 1–1,226).
5′-GAGTACTAGTATGGGCAGTCCATACTACCGC-3′ and 5′-GTCTGGTACCCATTTGCGCCTTCTCCAGTTC-3′; recombined with pTWG (C-terminal EGFP tag).
pENTER BRP D2-4 (aa 269–1,740).
5′-GAGTACTAGTGAGGAGGAGCGTCAGATGTTCC-3′ and 5′-GTCTGGTACCGAAAAAGCTCTTCAAGAAGCCAGC-3′; recombined with pTWG (C-terminal EGFP tag).
pENTER BRP D1-4 (aa 1–1,740).
5′-GAGTACTAGTATGGGCAGTCCATACTACCGC-3′ and 5′-GTCTGGTACCGAAAAAGCTCTTCAAGAAGCCAGC-3′; recombined with pTWG (C-terminal EGFP tag).
pENTER BRP D2 (aa 269–617).
5′-GAGTACTAGTGAGGAGGAGCGTCAGATGTTCC-3′ and 5′-GTCTGGTACCTGCTCTTTCCGCATCCGAC-3′; recombined with pDEST17 (N-terminal 6×His tag; Invitrogen) for antibody production.
pENTER BRP D3 (BRP short; aa 473–1,226).
5′-GAGTACTAGTATGGGCAGTCCATACTACCGC-3′ and 5′-GTCTGGTACCCATTTGCGCCTTCTCCAGTTC-3′; recombined with pTWG (C-terminal EGFP tag) and pTWmStraw.
pENTER Cac C terminus (aa 1,420–1,848).
pGADT7 Cac C terminus was NcoI–XhoI digested, and the 2.2-kb insert was ligated into pENTER using the same restriction sites. The double-strand–sequenced plasmid was recombined with pUbiP-10xmyc-rfA (N-terminal 10×Myc tag).
pGBKT7 Cac Loop 1 (aa 305–452).
5′-GATGCCATGGTGCTCAACTTAGTTCTTGGTGTC-3′ and 5′-GATGCTCGAGGAAAGACGAGGACGATCACG-3′ were used. The PCR product was cut using NcoI–XhoI and ligated into pGBKT7 using NcoI–SalI.
pGBKT7 Cac Loop 2 (aa 669–768).
5′-GATGCCATGGATAATTTGGCGAATGCCCAAGAA-3′ and 5′-GATGGAATTCCAAAATAACCACCCAATGGGC-3′ were used. The PCR product was cut and ligated into pGBKT7 using NcoI–EcoRI.
pGBKT7 Cac Loop 3 (aa 1,042–1,093).
5′-GATGCCATGGTTACGTTTCAAGAGCAAGGCGAA-3′ and 5′-GATGCTCGAGACACCACAATTCGCCACACTTTATA-3′ were used. The PCR product was cut using NcoI–XhoI and ligated into pGBKT7 using NcoI–SalI.
pGBKT7 Cac C terminus (aa 1,420–1,848).
5′-GATGCCATGGCGTTATTCGCTTTGATTCGTGA-3′ and 5′-GATGCTCGAGAGCACCAATCCTCCTCATCCGAA-3′ were used. The PCR product was cut using NcoI–XhoI and ligated into pGBKT7 using NcoI–SalI and into pGADT7 using NcoI–XhoI.
pGADT7 BRP D1 (aa 1–320).
5′-GAGTCATATGGGCAGTCCATACTACCGC-3′ and 5′-GTCTATCGATGTGCCGCTGGTAGTCCTG-3′ were used. The PCR product was cut using ClaI–NdeI and ligated into pGADT7.
pGADT7 BRP D2 (aa 268–617):.
5′-GAGTCATATGGAGGAGGAGCGTCAGATGTTCC-3′ and 5′-GTCTATCGATTGCTCTTTCCGCATCCGAC-3′ were used. The PCR product was cut using ClaI–NdeI and ligated into pGADT7.
pGADT7 BRP D3 (aa 473–1,226).
pENTER BRP D3 was digested with SpeI–Asp718I, and the insert was subcloned into pSL1180fa. The insert was released again using NcoI–NdeI and ligated into a modified version of pGADT7 (pGADT7 IIB). Within the pGADT7 IIB vector, a point mutation deleting the first NcoI restriction site was introduced.
pGADT7 BRP D4 (aa 1,152–1,740).
5′-GAGTCATATGCATGAGAAGCTACTGAAGAAGGTCG-3′ and 5′-GTCTATCGATGAAAAAGCTCTTCAAGAAGCCAGC-3′ were used. The PCR product was cut using ClaI–NdeI and ligated into pGADT7.
Yeast two hybrid
In principle, all cotransformation experiments were conducted according to the yeast two-hybrid protocols of Clontech Laboratories, Inc. using the strain AH109. To ensure the presence of both cotransformed plasmids, the yeast was plated on minimal synthetic-defined/−Leu/−Trp medium plates. After growing for 2–3 d, at least 10 clones each were analyzed on synthetic-defined/−Ade/−His/−Leu/−Trp/X–α- galactosidase medium plates to select for positive interaction. If >90% of the clones grew and turned blue in color, this was regarded as a positive interaction of high confidence (+++). As a positive control, pGBKT7-p53 was cotransformed with pGADT7 containing the SV40 large T antigen. Negative controls consisted either of laminin as bait together with the prey to be tested or the corresponding bait together with the empty prey vector.
Biochemistry
Drosophila Schneider S2R+ cells were provided by A. Herzig and cultured at 25°C in an ambient atmosphere in Schneider’s Drosophila medium (BioWest) supplemented with 10% FCS + 2 mM L-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen). Medium was exchanged every 3–4 d. Cell cultures were split every 10–14 d. Cell cotransfection was conducted using the Effectene transfection reagent kit (QIAGEN). Cell lysis was performed with lysis buffer containing 25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 10% glycerol (vol/vol), 1% NP-40 (vol/vol), and complete protease inhibitor (Roche) for 45 min at 0°C. Total protein concentrations were determined by bicinchoninic acid protein assay (Thermo Fisher Scientific).
For coimmunoprecipitations (co-IPs), 350 µg total protein extract from whole cell lysates was mixed with 20 µl protein A agarose bead suspension (Affi-prep protein A support; Bio-Rad Laboratories) precoupled with either monoclonal Mα-Myc antibody (9E10; Santa Cruz Biotechnology, Inc.), polyclonal Rb-αGFP (A11122; Invitrogen), Rbα-BRPD2, or the respective IgG control from mouse or rabbit (Dianova). After incubation at room temperature, the coupled beads were thoroughly washed repeatedly and eluted by boiling in 40 µl of Laemmli buffer.
10 µl IP eluates and 30 µg whole cell lysates were subjected to denaturing SDS-PAGE using Tris-HCl NuPAGE 4–12% gradient gels and then transferred to a nitrocellulose membrane (iBlot; Invitrogen). The membrane was probed with Rbα-BRPD2 (1:500).
Image acquisition
Conventional confocal images were acquired at room temperature with a 63× 1.4 NA oil objective suited in a confocal microscope (TCS-SP5; Leica). Images taken from fixed samples were from third instar larval NMJs 6/7 (segments A2 and A3). NMJs depicted in live experiments derive from muscles 26 and 27 in segments A2 and A3. The fluorescence detection was set with the acousto optical beam splitter between 500 and 530 nm for GFP and between 575 and 620 nm for mRFP and mStraw. Photomultiplier gain was set to 1,250 V. GFP was excited using the 488-nm ArKr laser line, whereas mRFP and mStraw were excited with a 561-nm diode-pumped solid-state laser. For STED images, the STED setup (TCS; Leica) was used in combination with a 100× 1.4 NA oil objective at 20°C (Leica). Dye (Atto647N; Atto-Tec) was excited with a pulsed laser at 635 nm and depleted with a laser adjusted to 760 nm (Mai Tai Ti:Sapphire; Newport Spectra-Physics). Detection of the Atto-647N was performed with avalanche photodiodes and optical filters permeable for light of wavelengths between 650 and 710 nm. Diode gain was continuously set to 310 V. Excitation laser power varied according to the sample but always ranged between 5.0 and 5.6 V.
Immunostainings
Immunostainings and dissections were performed as described previously (Qin et al., 2005). Larvae were incubated with antibody solutions overnight at 4°C. Larvae were mounted either in Vectashield (Vector Laboratories) or Mowiol. The following antibody dilutions were used: Mα-Nc82 (provided by E. Buchner), 1:200; Mα-DGluRIIA (8B4D2; Developmental Studies Hybridoma Bank), 1:100; Rbα-BRPN-Term, 1:250; Rbα-DGluRIID (Qin et al., 2005), 1:500; Mα-GFP (Invitrogen), 1:500; Rbα-GFP (Invitrogen), 1:500; and HRP-Cy5 (Dianova), 1:250. For standard immunostainings, secondary antibodies were diluted 1:500. For STED stainings, dye (Atto647N) conjugation to secondary antibodies giving sheep α-M-Atto647N and sheep α-Rb-Atto647N were performed according to producer protocols (http://www.atto-tec.com) and used 1:100. Embryos (w1118 and elav-Gal4, l(1)L13HC129; Kawasaki et al., 2002) were staged by time (22–24 h after egg laying) and morphology, dissected, and stained as described for larvae.
Live imaging
The following strains were used for in vivo imaging experiments: for Ca2+ channels, ok6-Gal4/+; CacGFP, DGluRIIAmRFP/+ (control); Df(2R)BSC29, ok6-Gal4/brp; CacGFP, DGluRIIAmRFP/+ (brp mutant background). For temporal analysis of AZ assembly, ok6-Gal4/+; BRP-shortGFP/DGluRIIAmRFP. ok6-Gal4/+; DLiprin-αGFP/DGluRIIAmRFP. ok6-Gal4, BRP-shortmStraw/+; DLiprin-αGFP/+. ok6-Gal4/+; CacGFP, DGluRIIAmRFP/+. ok6-Gal4, BRP-shortmStraw/+; CacGFP/+.
Imaging of intact Drosophila larvae along with larval anaesthetization was performed as described previously (Rasse et al., 2005; Fuger et al., 2007; Schmid et al., 2008). For FRAP experiments, intense 488-nm laser light was applied inside a region of interest of ∼10 µm of edge length (zoom, 25) bleaching both green and red fluorescent dyes. After an incubation of 12 h at 25°C, the junctions were imaged and compared with the prebleached pictures. Control regions were conserved at the junctions for internal control of intensity levels.
Image processing
Images were acquired using Application Suite Advanced Fluorescence (LAS-AF; Leica) software.
Confocal imaging
Confocal stacks were processed with ImageJ software (National Institutes of Health). Single slices and confocal stacks were deconvolved using the ImageJ plug-ins iterative deconvolution and iterative deconvolution 3D, respectively (provided by B. Dougherty, OptiNav, Inc., Redmond, WA). To generate the PSF for deconvolution, the ImageJ plug-in diffraction PSF 3D (provided by B. Dougherty) was used. The PSF settings were adjusted according to our hardware parameters, emission wavelengths, and image dimensions.
For analysis of FRAP data, intensity ratios between bleached areas and control nonbleached regions were retrieved. To compare several experiments, prebleached ratios were set to 1, and postbleached images were normalized accordingly. Recoveries were calculated by subtracting ratios generated immediately after the bleaching from ratios acquired 12 h after bleaching.
For quantification of areas and intensities, maximum projections, acquired with comparable confocal settings, were thresholded at 30 arbitrary units (au), and remaining areas were measured via the analyze particle function of ImageJ.
To obtain unbiased mean BRPNc82 and BRPN-Term distributions with STED resolution (Fig. 5 D), we used STED images of BRPNc82–Atto647N (or BRPN-Term–Atto647N), which were simultaneously recorded with confocal images of the BRPN-Term–Alexa Fluor 488 (or BRPNc82–Alexa Fluor 488). From these images, AZs were selected, which appeared planar to the optical slice. The confocal channel of each image of a planar AZ was automatically fitted with a 2D Gaussian function with Mathematica (version 5.0; Wolfram Research). The peaks of the Gaussian functions were used to automatically align and subsequently average the corresponding STED signal. The averaged STED images (BRPNc82 and BRPN-Term) were finally aligned according to the corresponding confocal counterpart. The BRPN-Term signal was fitted with a single Gaussian (standard deviation = 52 nm), and the BRPNc82 signal was fitted with the sum of two Gaussians (standard deviation = 48 nm and peak distance Δx = 189 nm).
STED imaging
STED images were processed using a linear deconvolution software integrated into the Imspector Software package (Max-Planck Innovations GmbH). Regularization parameters ranged from 1e−10 to 1e−12. The PSF was generated by using a 2D Lorentzian function with its half-width and half-length fitted to the half-width and half-length obtained by images of 25-nm crimson beads conjugated to Atto647N.
Electrophysiology
Two electrode voltage clamp recordings were essentially conducted as previously described (Kittel et al., 2006). All experiments were performed on male third instar larval NMJs (muscle 6; segments A2 and A3) in HL3 (70 mM NaCl, 5 mM KCl, 20 mM MgCl2, 10 mM NaHCO3, 5 mM trehalose, 115 mM sucrose, 5 mM Hepes, and 1 mM CaCl2, pH 7.2). Muscle cells had an input resistance ≥4 MΩ. Intracellular electrodes were filled with 3 M KCl, and resistances ranged from 10–25 MΩ. Stimulation artifacts of evoked excitatory junctional currents were removed for clarity. Genotypes used were brpc04298/Df(2R)BSC29 and w1118 as controls.
Statistics
Data were analyzed using the Mann-Whitney rank sum test for linear independent data groups (Prism; GraphPad Software, Inc.). Means are annotated ± SEM. Asterisks are used to denote significance (*, P < 0.05; **, P < 0.01; ***, P < 0.005; not significant, P > 0.05).
Online supplemental material
Fig. S1 shows a quantification of BRPN-Term expression levels in several brp mutants. Fig. S2 shows a comparison of mitochondrion ultrastructure either preserved using conventional room temperature embedding with aldehyde fixation and dehydration or HPF/FS along with T-bar size quantification of conventional and HPF/FS-prepared control and brp1.3 larvae. Fig. S3 shows amplitude and rise time of evoked excitatory junctional current recorded from brpc04298 NMJs. Fig. S4 shows that dliprin-α AZs do not suffer from Cac-clustering defects. Fig. S5 shows that BRP clustering is not disturbed at cac AZs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200812150/DC1.
Acknowledgments
We thank Christine Quentin, Claudia Wirth, and Franziska Zehe for excellent technical assistance. Furthermore, we thank all colleagues who provided us with reagents.
This work was supported by grants from the Deutsche Forschungsgemeinschaft to S.J. Sigrist (grants Exc 257, SI849/2-1 and 2-2, TP A16/SFB 551, TP B23/SFB581), by funds of the Max Planck Society to D. Owald, and by formel.1 grants to S. Hallermann and R.J. Kittel from the Medical Faculty of the University of Leipzig.
Footnotes
Abbreviations used in this paper: au, arbitrary units; AZ, active zone; BRP, Bruchpilot; Cac, Cacophony; DGluR, Drosophila glutamate receptor subunit; DLiprin-α, Drosophila Liprin-α; EMS, ethyl methyl sulfonate; FS, freeze substitution; HPF, high pressure freezing; IP, immunoprecipitation; mStraw, mStrawberry; NMJ, neuromuscular junction; PSD, postsynaptic density; PSF, point spread function; STED, stimulated emission depletion; UAS, upstream activator sequence.
References
- Bellen H.J., Levis R.W., Liao G., He Y., Carlson J.W., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M., et al. 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes.Genetics. 167:761–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstatter J.H., Shaw S.R., Meinertzhagen I.A. 1991. Terminal degeneration and synaptic disassembly following receptor photoablation in the retina of the fly’s compound eye.J. Neurosci. 11:1930–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.Q., Piedras-Renteria E.S., Smith G.B., Chen G., Harata N.C., Tsien R.W. 2004. Presynaptic Ca2+ channels compete for channel type-preferring slots in altered neurotransmission arising from Ca2+ channelopathy.Neuron. 43:387–400 [DOI] [PubMed] [Google Scholar]
- Catterall W.A. 1998. Structure and function of neuronal Ca2+ channels and their role in neurotransmitter release.Cell Calcium. 24:307–323 [DOI] [PubMed] [Google Scholar]
- Catterall W.A., Few A.P. 2008. Calcium channel regulation and presynaptic plasticity.Neuron. 59:882–901 [DOI] [PubMed] [Google Scholar]
- Collins C.A., DiAntonio A. 2007. Synaptic development: insights from Drosophila.Curr. Opin. Neurobiol. 17:35–42 [DOI] [PubMed] [Google Scholar]
- Dai Y., Taru H., Deken S.L., Grill B., Ackley B., Nonet M.L., Jin Y. 2006. SYD-2 Liprin-alpha organizes presynaptic active zone formation through ELKS.Nat. Neurosci. 9:1479–1487 [DOI] [PubMed] [Google Scholar]
- Deguchi-Tawarada M., Inoue E., Takao-Rikitsu E., Inoue M., Ohtsuka T., Takai Y. 2004. CAST2: identification and characterization of a protein structurally related to the presynaptic cytomatrix protein CAST.Genes Cells. 9:15–23 [DOI] [PubMed] [Google Scholar]
- Deguchi-Tawarada M., Inoue E., Takao-Rikitsu E., Inoue M., Kitajima I., Ohtsuka T., Takai Y. 2006. Active zone protein CAST is a component of conventional and ribbon synapses in mouse retina.J. Comp. Neurol. 495:480–496 [DOI] [PubMed] [Google Scholar]
- Evans R.M., Zamponi G.W. 2006. Presynaptic Ca2+ channels–integration centers for neuronal signaling pathways.Trends Neurosci. 29:617–624 [DOI] [PubMed] [Google Scholar]
- Featherstone D.E., Rushton E.M., Hilderbrand-Chae M., Phillips A.M., Jackson F.R., Broadie K. 2000. Presynaptic glutamic acid decarboxylase is required for induction of the postsynaptic receptor field at a glutamatergic synapse.Neuron. 27:71–84 [DOI] [PubMed] [Google Scholar]
- Feeney C.J., Karunanithi S., Pearce J., Govind C.K., Atwood H.L. 1998. Motor nerve terminals on abdominal muscles in larval flesh flies, Sarcophaga bullata: comparisons with Drosophila.J. Comp. Neurol. 402:197–209 [PubMed] [Google Scholar]
- Frank T., Khimich D., Neef A., Moser T. 2009. Mechanisms contributing to synaptic Ca2+ signals and their heterogeneity in hair cells.Proc. Natl. Acad. Sci. USA. 106:4483–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuger P., Behrends L.B., Mertel S., Sigrist S.J., Rasse T.M. 2007. Live imaging of synapse development and measuring protein dynamics using two-color fluorescence recovery after photo-bleaching at Drosophila synapses.Nat. Protoc. 2:3285–3298 [DOI] [PubMed] [Google Scholar]
- Gray N.W., Weimer R.M., Bureau I., Svoboda K. 2006. Rapid redistribution of synaptic PSD-95 in the neocortex in vivo.PLoS Biol. 4:e370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow M.L., Ress D., Stoschek A., Marshall R.M., McMahan U.J. 2001. The architecture of active zone material at the frog’s neuromuscular junction.Nature. 409:479–484 [DOI] [PubMed] [Google Scholar]
- Hell S.W. 2007. Far-field optical nanoscopy.Science. 316:1153–1158 [DOI] [PubMed] [Google Scholar]
- Hofbauer A., Ebel T., Waltenspiel B., Oswald P., Chen Y.C., Halder P., Biskup S., Lewandrowski U., Winkler C., Sickmann A., et al. 2009. The Wuerzburg hybridoma library against Drosophila brain.J. Neurogenet. 23:78–91 [DOI] [PubMed] [Google Scholar]
- Jin Y., Garner C.C. 2008. Molecular mechanisms of presynaptic differentiation.Annu. Rev. Cell Dev. Biol. 24:237–262 [DOI] [PubMed] [Google Scholar]
- Kaufmann N., DeProto J., Ranjan R., Wan H., Van Vactor D. 2002. Drosophila liprin-alpha and the receptor phosphatase Dlar control synapse morphogenesis.Neuron. 34:27–38 [DOI] [PubMed] [Google Scholar]
- Kawasaki F., Felling R., Ordway R.W. 2000. A temperature-sensitive paralytic mutant defines a primary synaptic calcium channel in Drosophila.J. Neurosci. 20:4885–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki F., Collins S.C., Ordway R.W. 2002. Synaptic calcium-channel function in Drosophila: analysis and transformation rescue of temperature-sensitive paralytic and lethal mutations of cacophony.J. Neurosci. 22:5856–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki F., Zou B., Xu X., Ordway R.W. 2004. Active zone localization of presynaptic calcium channels encoded by the cacophony locus of Drosophila.J. Neurosci. 24:282–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel R.J., Wichmann C., Rasse T.M., Fouquet W., Schmidt M., Schmid A., Wagh D.A., Pawlu C., Kellner R.R., Willig K.I., et al. 2006. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release.Science. 312:1051–1054 [DOI] [PubMed] [Google Scholar]
- Ko J., Na M., Kim S., Lee J.R., Kim E. 2003. Interaction of the ERC family of RIM-binding proteins with the liprin-alpha family of multidomain proteins.J. Biol. Chem. 278:42377–42385 [DOI] [PubMed] [Google Scholar]
- Koh Y.H., Gramates L.S., Budnik V. 2000. Drosophila larval neuromuscular junction: molecular components and mechanisms underlying synaptic plasticity.Microsc. Res. Tech. 49:14–25 [DOI] [PubMed] [Google Scholar]
- Li J., Ashley J., Budnik V., Bhat M.A. 2007. Crucial role of Drosophila neurexin in proper active zone apposition to postsynaptic densities, synaptic growth, and synaptic transmission.Neuron. 55:741–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A.A., Kim E., Leung H.T., Woodruff E., III, An L., Doerge R.W., Pak W.L., Broadie K. 2008. Presynaptic calcium channel localization and calcium-dependent synaptic vesicle exocytosis regulated by the Fuseless protein.J. Neurosci. 28:3668–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M., Zhang W., Rohlmann A., Kattenstroth G., Hammer R.E., Gottmann K., Sudhof T.C. 2003. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis.Nature. 423:939–948 [DOI] [PubMed] [Google Scholar]
- Neher E., Sakaba T. 2008. Multiple roles of calcium ions in the regulation of neurotransmitter release.Neuron. 59:861–872 [DOI] [PubMed] [Google Scholar]
- Ohtsuka T., Takao-Rikitsu E., Inoue E., Inoue M., Takeuchi M., Matsubara K., Deguchi-Tawarada M., Satoh K., Morimoto K., Nakanishi H., Takai Y. 2002. Cast: a novel protein of the cytomatrix at the active zone of synapses that forms a ternary complex with RIM1 and munc13-1.J. Cell Biol. 158:577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owald D., Sigrist S.J. 2009. Assembling the presynaptic active zone.Curr. Opin. Neurobiol. doi:10.1016/j.conb.2009.03.003 [DOI] [PubMed] [Google Scholar]
- Parks A.L., Cook K.R., Belvin M., Dompe N.A., Fawcett R., Huppert K., Tan L.R., Winter C.G., Bogart K.P., Deal J.E., et al. 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome.Nat. Genet. 36:288–292 [DOI] [PubMed] [Google Scholar]
- Patel M.R., Lehrman E.K., Poon V.Y., Crump J.G., Zhen M., Bargmann C.I., Shen K. 2006. Hierarchical assembly of presynaptic components in defined C. elegans synapses.Nat. Neurosci. 9:1488–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop A., Meinertzhagen I.A. 2006. Development and structure of synaptic contacts in Drosophila.Semin. Cell Dev. Biol. 17:20–30 [DOI] [PubMed] [Google Scholar]
- Qin G., Schwarz T., Kittel R.J., Schmid A., Rasse T.M., Kappei D., Ponimaskin E., Heckmann M., Sigrist S.J. 2005. Four different subunits are essential for expressing the synaptic glutamate receptor at neuromuscular junctions of Drosophila.J. Neurosci. 25:3209–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasse T.M., Fouquet W., Schmid A., Kittel R.J., Mertel S., Sigrist C.B., Schmidt M., Guzman A., Merino C., Qin G., et al. 2005. Glutamate receptor dynamics organizing synapse formation in vivo.Nat. Neurosci. 8:898–905 [DOI] [PubMed] [Google Scholar]
- Rostaing P., Real E., Siksou L., Lechaire J.P., Boudier T., Boeckers T.M., Gertler F., Gundelfinger E.D., Triller A., Marty S. 2006. Analysis of synaptic ultrastructure without fixative using high-pressure freezing and tomography.Eur. J. Neurosci. 24:3463–3474 [DOI] [PubMed] [Google Scholar]
- Rybak J., Meinertzhagen I.A. 1997. The effects of light reversals on photoreceptor synaptogenesis in the fly Musca domestica.Eur. J. Neurosci. 9:319–333 [DOI] [PubMed] [Google Scholar]
- Schmid A., Qin G., Wichmann C., Kittel R.J., Mertel S., Fouquet W., Schmidt M., Heckmann M., Sigrist S.J. 2006. Non-NMDA-type glutamate receptors are essential for maturation but not for initial assembly of synapses at Drosophila neuromuscular junctions.J. Neurosci. 26:11267–11277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A., Hallermann S., Kittel R.J., Khorramshahi O., Frolich A.M., Quentin C., Rasse T.M., Mertel S., Heckmann M., Sigrist S.J. 2008. Activity-dependent site-specific changes of glutamate receptor composition in vivo.Nat. Neurosci. 11:659–666 [DOI] [PubMed] [Google Scholar]
- Schoch S., Gundelfinger E.D. 2006. Molecular organization of the presynaptic active zone.Cell Tissue Res. 326:379–391 [DOI] [PubMed] [Google Scholar]
- Sigrist S.J., Reiff D.F., Thiel P.R., Steinert J.R., Schuster C.M. 2003. Experience-dependent strengthening of Drosophila neuromuscular junctions.J. Neurosci. 23:6546–6556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siksou L., Rostaing P., Lechaire J.P., Boudier T., Ohtsuka T., Fejtova A., Kao H.T., Greengard P., Gundelfinger E.D., Triller A., Marty S. 2007. Three-dimensional architecture of presynaptic terminal cytomatrix.J. Neurosci. 27:6868–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryker E., Johnson K.G. 2007. LAR, liprin alpha and the regulation of active zone morphogenesis.J. Cell Sci. 120:3723–3728 [DOI] [PubMed] [Google Scholar]
- tom Dieck S., Altrock W.D., Kessels M.M., Qualmann B., Regus H., Brauner D., Fejtova A., Bracko O., Gundelfinger E.D., Brandstatter J.H. 2005. Molecular dissection of the photoreceptor ribbon synapse: physical interaction of Bassoon and RIBEYE is essential for the assembly of the ribbon complex.J. Cell Biol. 168:825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh D.A., Rasse T.M., Asan E., Hofbauer A., Schwenkert I., Durrbeck H., Buchner S., Dabauvalle M.C., Schmidt M., Qin G., et al. 2006. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila.Neuron. 49:833–844 [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu X., Biederer T., Sudhof T.C. 2002. A family of RIM-binding proteins regulated by alternative splicing: implications for the genesis of synaptic active zones.Proc. Natl. Acad. Sci. USA. 99:14464–14469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal V., Rizzoli S.O., Lauterbach M.A., Kamin D., Jahn R., Hell S.W. 2008. Video-rate far-field optical nanoscopy dissects synaptic vesicle movement.Science. 320:246–249 [DOI] [PubMed] [Google Scholar]
- Wojtowicz J.M., Marin L., Atwood H.L. 1994. Activity-induced changes in synaptic release sites at the crayfish neuromuscular junction.J. Neurosci. 14:3688–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucherpfennig T., Wilsch-Brauninger M., Gonzalez-Gaitan M. 2003. Role of Drosophila Rab5 during endosomal trafficking at the synapse and evoked neurotransmitter release.J. Cell Biol. 161:609–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Sun M., Liu L., Chen F., Wei L., Xie W. 2007. Neurexin-1 is required for synapse formation and larvae associative learning in Drosophila.FEBS Lett. 581:2509–2516 [DOI] [PubMed] [Google Scholar]
- Zhai R.G., Bellen H.J. 2004. The architecture of the active zone in the presynaptic nerve terminal.Physiology (Bethesda). 19:262–270 [DOI] [PubMed] [Google Scholar]