Abstract
Formation of the ribbon-like membrane network of the Golgi apparatus depends on GM130 and GRASP65, but the mechanism is unknown. We developed an in vivo organelle tethering assaying in which GRASP65 was targeted to the mitochondrial outer membrane either directly or via binding to GM130. Mitochondria bearing GRASP65 became tethered to one another, and this depended on a GRASP65 PDZ domain that was also required for GRASP65 self-interaction. Point mutation within the predicted binding groove of the GRASP65 PDZ domain blocked both tethering and, in a gene replacement assay, Golgi ribbon formation. Tethering also required proximate membrane anchoring of the PDZ domain, suggesting a mechanism that orientates the PDZ binding groove to favor interactions in trans. Thus, a homotypic PDZ interaction mediates organelle tethering in living cells.
Introduction
Intracellular organelles form membrane networks through fusion and fission events, which must be tightly regulated to preserve organelle identity and morphology (Voeltz and Prinz, 2007). In mammals the Golgi apparatus forms a ribbon-like network comprised of laterally linked stacked cisternae, or ministacks. Each ministack is comprised of subcompartments that carry out ordered processing reactions on cargo passing through the organelle (Puthenveedu and Linstedt, 2005; Pfeffer, 2007). The lateral linkages that connect adjacent ministacks are homotypic and dynamic. That is, analogous cisternal subcompartments are linked with each other and both fusion and fission occur at the sites of contact (Colanzi and Corda, 2007). Disruption of the lateral connections is associated with increased deviation in enzyme distribution among ministacks and processing deficiencies (Puthenveedu et al., 2006). The linkages also appear to act as a control point in cell cycle progression, as a MAP kinase pathway (Acharya et al., 1998; Yoshimura et al., 2005; Shaul and Seger, 2006) triggers unlinking of the Golgi ribbon in late G2 phase of the cell cycle and blockade of this event delays entry into M phase (Feinstein and Linstedt, 2007). Lateral linkage of the Golgi ribbon may also be regulated to allow repositioning of the Golgi apparatus to face the wound edge during the cellular response to a scratch wound (Bisel et al., 2008).
Two of the identified factors required for ribbon formation are the golgin GM130 and its binding partner GRASP65 (Puthenveedu et al., 2006; Marra et al., 2007), which is also required for reassembly of Golgi stacks in an in vitro assay (Barr et al., 1997; Wang et al., 2003). In the absence of GM130 or GRASP65 the Golgi apparatus is fragmented into ministacks that nevertheless mostly retain their juxtanuclear positioning and transport competence (Sutterlin et al., 2005; Puthenveedu et al., 2006; Kodani and Sutterlin, 2008). Significantly, although knockdown of GRASP65 leaves GM130 properly localized on the Golgi (Sutterlin et al., 2005; Puthenveedu et al., 2006), knockdown of GM130 causes loss of GRASP65 (Puthenveedu et al., 2006; Kodani and Sutterlin, 2008). Further, GM130 function requires its ability to bind GRASP65 (Puthenveedu et al., 2006). Based on these findings and the demonstrated ability of the GRASP65 N terminus, which contains a tandem array of PDZ-like domains (Barr et al., 1998; Kuo et al., 2000), to form homo-oligomeric structures (Wang et al., 2003, 2005), we hypothesized that GM130 recruits GRASP65 to the Golgi membrane and that GRASP65 mediates homotypic tethering of adjacent cis cisternae via oligomeric interactions in trans (Puthenveedu et al., 2006).
However, evidence that the GM130–GRASP65 complex plays a direct role in organelle tethering is lacking, as is a detailed understanding of how it might work. The finding that GRASP65 oligomerizes forming complexes in trans was performed with protein purified from bacteria (Wang et al., 2003, 2005). These preparations were not myristoylated, were studied in the absence of membranes, and formed large polydisperse complexes leaving the physiological context of a key aspect of the hypothesis that GRASP65 oligomers bridge membranes an open question. Further, although oligomerization activity mapped to the N-terminal region containing PDZ-like domains, it is not clear whether either or both of these domains mediate the interaction, whether the interaction involves a bona fide PDZ domain interaction, or whether these domains even form PDZ domains at all. Finally, the validity of the model in which GM130–GRASP65 complexes bridge membranes to mediate Golgi ribbon formation is further complicated by recent findings that implicate the proteins in nonclassical secretion occurring outside the Golgi apparatus at specific points of development of two nonvertebrates (Kinseth et al., 2007; Schotman et al., 2008).
Thus, to investigate their sufficiency in organelle tethering in a physiological context, GM130 and GRASP65 constructs were expressed on the mitochondrial outer membrane of mammalian cells containing or lacking endogenous mitochondrial tethering factors. Paralleling its activity at the Golgi, GM130 recruited GRASP65 and GRASP65 was necessary and sufficient for mitochondrial tethering. Tethering depended on the predicted ligand-binding groove of a self-interacting GRASP65 PDZ domain, and mutation of this predicted groove to block tethering also blocked Golgi ribbon formation. The combined results indicate that, after recruitment by GM130, GRASP65 homotypic PDZ-type interactions mediate organelle tethering.
Results
Organelle clustering induced by GRASP65
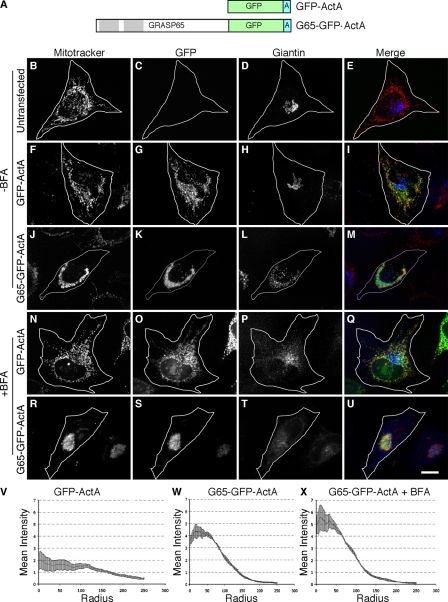
As a test of the hypothesis that GRASP65 directly cross-bridges membranes, we expressed in HeLa cells a modified version of GRASP65 containing a C-terminal membrane anchor sequence specifying targeting to the mitochondrial outer membrane. The mitochondrial-targeting signal, derived from the ActA protein of Listeria monocytogenes (Pistor et al., 1994), was placed after an inserted GFP coding sequence at the C terminus of GRASP65 to yield a cytoplasmically disposed G65-GFP-ActA construct (Fig. 1 A). GFP-ActA, which lacked the GRASP65 sequence but was otherwise identical, served as a control.
Figure 1.
Mitochondrial clustering by GRASP65. Schematic diagram of the constructs (A). Untransfected HeLa cells (B–E) or cells expressing GFP-ActA (F–I) or G65-GFP-ActA (J–M) were analyzed 24 h after transfection using Mitotracker (red) to stain mitochondria, GFP fluorescence (green) to localize the transfected proteins, and giantin (blue) staining to image the Golgi apparatus. An identical analysis was performed after a 30-min BFA treatment on cells expressing GFP-ActA (N–Q) or G65-GFP-ActA (R–U). Bar, 10 µm. Radial profile plots show the spread of mitochondrial fluorescence starting from the centroid and extending to the cell periphery for cells expressing GFP-ActA (V), G65-GFP-ActA (W), or BFA-treated cells expressing G65-GFP-ActA (X). Values are averages corresponding to the fraction of total fluorescence present in each concentric circle drawn from the centroid (n = 3, ±SEM, >15 cells/experiment).
The control GFP-ActA was targeted to mitochondria as indicated by its colocalization with Mitotracker, and it altered neither mitochondrial nor Golgi morphology (Fig. 1, B–I). G65-GFP-ActA also colocalized with Mitotracker staining, indicating mitochondrial targeting but, in striking contrast with GFP-ActA, G65-GFP-ActA expression had a profound effect on mitochondrial location and appearance (Fig. 1, J–M). The mitochondria became clustered in the juxtanuclear region of the cells with little or no mitochondrial staining remaining elsewhere in the cytoplasm. The Golgi apparatus, which appeared intermingled with the clustered mitochondria, was fragmented in these cells. The mitochondrial clusters were strikingly similar to those induced by overexpression of the mitofusin proteins that normally tether mitochondria (Chen et al., 2003; Koshiba et al., 2004), suggesting that G65-GFP-ActA was tethering the membranes to one another.
To exclude the possibility that the juxtanuclear clustering induced by G65-GFP-ActA depended on interaction with Golgi membranes, cells expressing GFP-ActA or G65-GFP-ActA were treated with brefeldin A (BFA). As expected, BFA induced Golgi collapse in control cells (Fig. 1, N–Q) and in cells expressing G65-GFP-ActA (Fig. 1, R–U), and mitochondrial clusters persisted in the latter. In fact, the clusters frequently appeared tighter suggesting that, in the absence of BFA, the Golgi membranes partially constrained, or otherwise limited, interactions between the clustered mitochondria.
To obtain a quantitative measure of the extent of mitochondrial spread in the transfected cells, a radial profile algorithm was used. For each cell, the algorithm measured the mean signal intensity for a series of concentric circles emanating from the calculated centroid of the fluorescent signal. Average values from many cells over multiple experiments were then used to generate radial profile plots in which the fraction of total mean intensity is expressed as a function of distance from the centroid. The radial profile plot for cells expressing the GFP-ActA control construct was essentially flat, reflecting the uniform spread of the mitochondria throughout the cytoplasm (Fig. 1 V). In contrast, a clearly significant clustering was evident in cells expressing G65-GFP-ActA (Fig. 1 W), and this was slightly accentuated in G65-GFP-ActA–expressing cells treated with BFA (Fig. 1 X). These results indicate that GRASP65, when targeted to the mitochondrial outer membrane, is sufficient to induce organelle clustering.
Nocodazole-induced microtubule disassembly was used to test whether clustering depended on an intact microtubule network. Untreated cells expressing the GFP-ActA control construct exhibited filamentous tubulin staining and filamentous mitochondria and nocodazole converted the tubulin pattern from filamentous to diffuse, and this reduced the filamentous appearance of mitochondria, which, nevertheless, remained dispersed throughout the cytoplasm (Fig. S1, A–D). Significantly, nocodazole had no effect on mitochondrial morphology or distribution in G65-GFP-ActA–expressing cells. That is, mitochondria were found in juxtanuclear clusters in cells lacking microtubules (Fig. S1, E–H). Thus, in contrast to nocodazole-sensitive mitochondrial clustering involving recruitment of motor and/or microtubule binding activity (Hoogenraad et al., 2003; Rios et al., 2004), the microtubule independence GRASP65-mediated clustering argues that it is likely a direct effect of crossbridging the mitochondrial membranes and forming a large structure that, for steric reasons, occupies the juxtanuclear area.
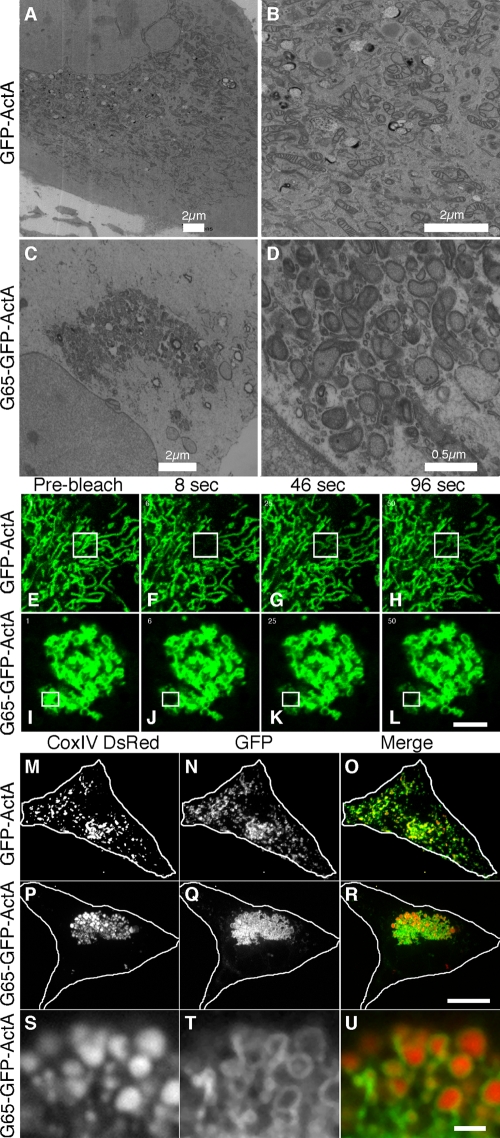
Next, electron microscopy was performed on the BFA-treated transfected cells to assess the ultrastructure of the clustered mitochondria. In untransfected cells, and in cells transfected with the GFP-ActA control plasmid, mitochondria were evident throughout the entire cytoplasm and were frequently well separated from each other (Fig. 2, A and B). As expected, the filamentous aspect apparent using fluorescence microscopy was not evident, presumably due to the low probability of obtaining thin sections with longitudinal profiles of membrane tubules. In contrast, cells transfected with G65-GFP-ActA had prominent clusters of mitochondria in the juxtanuclear region and the remaining cytoplasm was essentially devoid of mitochondria (Fig. 2, C and D). Unlike Golgi stacks, which have extended zones of apposition with uniform gap widths, the mitochondria in the clusters were apposed mostly at discrete sites and at a greater distance. Other membranes may be present within the clusters. Nevertheless, an immunofluorescence assay (not depicted) failed to reveal any accumulation in the clusters of calnexin, an ER marker, or ERGIC53, a marker of the intermediate compartment that accumulates in BFA remnants (Seemann et al., 2000). Interestingly, the outer membranes of individual mitochondria appeared distinct from neighboring outer membranes, suggesting maintenance of mitochondrial integrity within the cluster. Absence of syncytia formation was further supported by FRAP experiments. Cells expressing the control construct, GFP-ActA, exhibited an extended mitochondrial network, and when a small region of the network was bleached, fluorescence was rapidly recovered in the bleached structures (Fig. 2, E–H, movie and quantification in Video 1). This is the expected behavior for a contiguous membrane network established by membrane fusion. In contrast, cells expressing the G65-GFP-ActA construct exhibited a juxtanuclear cluster of mitochondria and there was no recovery of fluorescence observed after photobleaching small portions of the clustered membranes (Fig. 2, I–L, movie and quantification in Video 2).
Figure 2.
Mitochondria are tethered and not fused into syncytia. GFP-ActA (A and B) or G65-GFP-ActA (C and D) transfected HeLa cells were BFA treated, processed for electron microscopy, and shown at 2 magnifications. GFP-ActA (E–H) or G65-GFP-ActA (I–L) transfected cells were imaged live using a scanning laser microscope. A region of interest (marked in figure) was selected and bleached and recovery was monitored in subsequent frames at 2-s intervals. Bar, 2 µm. Mouse embryonic fibroblasts lacking mitofusin-1/2 and expressing the matrix marker COX-IV-DsRed were transfected with GFP-ActA (M–O) or G65-GFP-ActA (P–R), BFA-treated, and processed to reveal mitochondria (red), and the expressed proteins (green). Bar = 10 µm. An enlarged view of a single optical section is also shown (S–U). Bar = 1 µm.
Mitofusins form homotypic interactions in trans, thereby crossbridging mitochondria (Chen and Chan, 2005; Griffin et al., 2006). To test whether mitofusins were involved, GFP-ActA and G65-GFP-ActA were expressed in mfn−/− cells, which are homozygous for deletions in each of the mitofusin genes and contain mitochondria that are incompetent to dock and fuse (Chen et al., 2003; Koshiba et al., 2004). Mitochondria were detected using DsRed fused to presequence of subunit IV of cytochrome c oxidase (COX-IV-DsRed), which is localized to the matrix of mitochondria (Koshiba et al., 2004). The cells were also treated with BFA to disperse Golgi membranes. The GFP-ActA control colocalized with COX-IV-DsRed and the mitochondria appeared as discrete punctate structures distributed throughout the cytoplasm (Fig. 2, M–O). Expression of G65-GFP-ActA caused clustering of the mitochondria in mfn−/− cells (Fig. 2, P–R). Further, because COX-IV-DsRed and G65-GFP-ActA were localized to the matrix and the outer membrane, respectively, it was possible to distinguish individual mitochondria in the clusters after acquiring single optical sections by confocal microscopy. This analysis provided strong evidence that, instead of undergoing fusion to form syncytia, mitochondria remained intact as discrete entities each containing an outer membrane compartment surrounding a matrix compartment (Fig. 2, S–U).
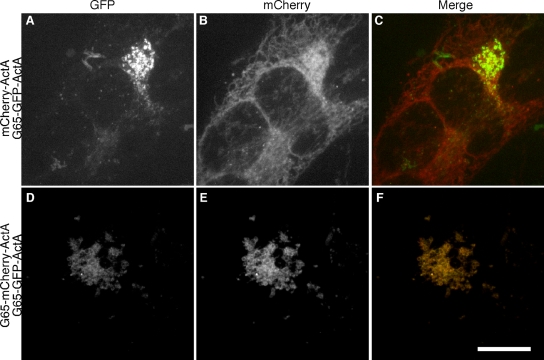
The experiments in this section argue that G65-GFP-ActA tethers adjacent mitochondria to generate juxtanuclear clusters. To verify that interactions in trans mediated mitochondrial clustering, HeLa cells expressing G65-GFP-ActA were fused to HeLa cells expressing either mCherry-ActA or G65-mCherry-ActA. Cycloheximide was used to prevent new protein synthesis and the mitochondria in the resulting heterokaryons were analyzed. Importantly, mitochondria bearing the control construct mCherry-ActA mostly remained strikingly distinct from mitochondria bearing G65-GFP-ActA in that the former retained the filamentous morphology characteristic of control mitochondria, whereas the latter remained clustered (Fig. 3, A–C). Further, the mitochondria bearing G65-mCherry-ActA coalesced with mitochondria bearing G65-GFP-ActA into single clusters (Fig. 3, D–F). Thus, clustering depended on GRASP65 being present in both membranes. Due to a low transfection frequency, we were unable to achieve dual-labeled heterokaryons with mfn−/− cells, thus the apparent mixing of the two markers in HeLa cells could be attributed to mitofusin-dependent membrane fusion (Legros et al., 2002) or membrane transfer (Neuspiel et al., 2008) in the 3 h following cell fusion.
Figure 3.
GRASP65 must be present in opposing mitochondrial membranes. G65-GFP-ActA–transfected HeLa cells were seeded together with either mCherry-ActA (A–C) or G65-mCherry-ActA (D–F) transfected HeLa cells, and the cells were fused with polyethylene glycol. Cycloheximide was added and after 3 h the heterokaryons, without BFA treatment, were fixed and visualized. Bar, 10 µm.
GM130 tethers membranes by recruiting GRASP65
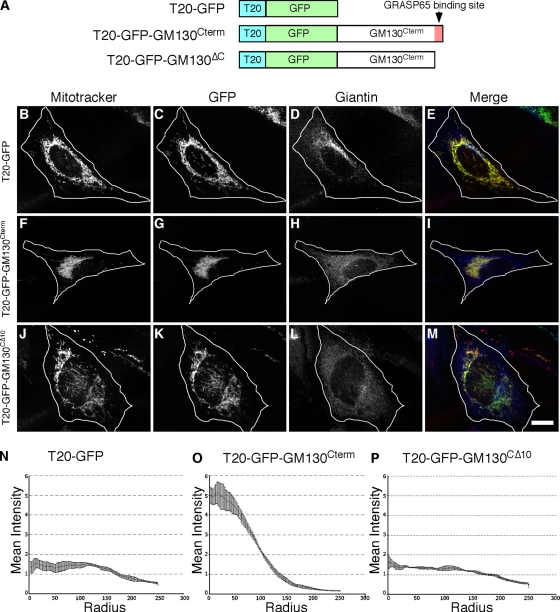
As mentioned above, GM130 is required for Golgi ribbon formation and for targeting of GRASP65 to Golgi membranes. Therefore, a strong prediction is that targeting of GM130 to mitochondria would also induce mitochondrial clustering, but in a manner dependent on its ability to recruit GRASP65. To test this idea, we targeted the GM130 C terminus, which contains the GRASP65 binding site, to mitochondria. One technical challenge was that targeting GM130 to mitochondria required a distinct strategy because GM130 uses its C terminus to interact with GRASP65 (Barr et al., 1998) and the ActA C-terminal membrane anchor was likely to interfere with this interaction. Fortunately, a search for outer membrane targeting sequences that could be used at the N terminus yielded a sequence in TOM20, a component of the outer membrane translocator complex. The N-terminal 40 amino acids of TOM20 contain a membrane-anchoring domain that orients such that the C-terminal hydrophilic sequences are exposed to the cytosol (Waizenegger et al., 2003), and this domain is sufficient for mitochondrial targeting (Kanaji et al., 2000). Thus, we generated the constructs diagrammed (Fig. 4 A) containing the TOM20 signal anchor followed by GFP alone (T20-GFP) or by GFP and the C-terminal 100 amino acids of GM130 (T20-GFP-GM130Cterm). As a further control, we also constructed T20-GFP-GM130CΔ10, which lacked 10 amino acids required for both GRASP65 binding (Barr et al., 1998) and for GM130 function in Golgi linking (Puthenveedu et al., 2006). BFA was used to carry out the assays in the absence of an intact Golgi apparatus.
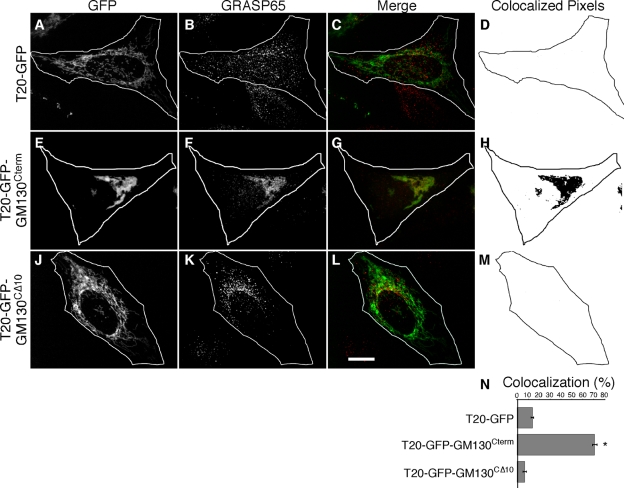
Figure 4.
Clustering by the GRASP65-binding domain of GM130. Schematic diagram of the constructs (A). 24 h after transfection, HeLa cells expressing T20-GFP (B–E), T20-GFP-GM130Cterm (F–I), or T20-GFP-GM130CΔ10 (J–M) were BFA treated for 30 min and analyzed using Mitotracker (red), GFP fluorescence (green), and giantin staining (blue). Bar, 10 µm. Radial profile plots (n = 3, ±SEM, >15 cells/experiment) of T20-GFP (N), T20-GFP-GM130Cterm (O), or T20-GFP-GM130CΔ10 (P).
T20-GFP was targeted to the mitochondria as indicated by colocalization with Mitotracker staining (Fig. 4, B–E). T20-GFP-GM130Cterm was also targeted to mitochondria, but it induced mitochondrial clustering in the juxtanuclear region (Fig. 4, F–I). Mitochondrial clustering was not evident in cells expressing T20-GFP-GM130CΔ10 despite its evident targeting to mitochondrial membranes (Fig. 4, J–M). These results were quantified using the radial profiling analysis (Fig. 4, N–P) and were also observed in cells not treated with BFA (Fig. S2, A–L). Further, T20-GFP-GM130Cterm induced mitochondrial clustering in mfn−/− cells (Fig. S2, M–R) where, similar to the case for G65-GFP-ActA, the integrity of the GFP-labeled outer membranes surrounding their DsRed-labeled matrices appeared intact, suggesting that the mitochondria were clustered by crossbridging rather than fusion (Fig. S2, S–U).
The absence of clustering by T20-GFP-GM130CΔ10 strongly suggests that T20-GFP-GM130Cterm clusters mitochondria by recruiting endogenous GRASP65. As a test, GRASP65 localization was determined. In BFA-treated cells GRASP65 is known to be principally associated with remnant membrane structures localized adjacent to distributed ER exit sites (Seemann et al., 2000; Ward et al., 2001). Consistent with this localization, GRASP65 was present in dispersed punctate structures in cells expressing T20-GFP, which itself was localized to filamentous mitochondria (Fig. 5, A–D). In striking contrast, GRASP65 localization was largely juxtanuclear in cells expressing T20-GFP-GM130Cterm, and it was clearly evident on the clustered mitochondria (Fig. 5, E–H). In the case of cells expressing T20-GFP-GM130CΔ10, GRASP65 retained the control BFA remnant pattern and was distinct from the filamentous mitochondria (Fig. 5, J–M). GRASP65 coincidence with the GFP constructs was analyzed on a pixel-by-pixel basis and the quantified results indicated significant specific recruitment of endogenous GRASP65 to mitochondria by T20-GFP-GM130Cterm (Fig. 5 N). Thus, the GM130 C terminus recruited endogenous GRASP65 to mitochondria and this induced their clustering. Because GM130 is required for GRASP65 localization to Golgi membranes (Puthenveedu et al., 2006; Kodani and Sutterlin, 2008) and because the GRASP65 binding site in GM130 is required for GM130-dependent Golgi linking (Puthenveedu et al., 2006), these results strongly suggest that GM130 links Golgi ribbons by recruiting GRASP65, which in turn is sufficient to link membranes. Additionally, the results show that endogenous levels of GRASP65 are sufficient to induce mitochondrial clustering, ruling out concerns regarding overexpression of exogenous constructs.
Figure 5.
Endogenous GRASP65 is recruited to mitochondria by GM130. HeLa cells expressing T20-GFP (A–D), T20-GFP-GM130Cterm (E–H), or T20-GFP-GM130CΔ10 (J–M) were BFA treated and processed to reveal GFP fluorescence, GRASP65 staining, merged images and, from single optical sections, representations of the colocalized pixels. Bar, 10 µm. GRASP65 recruitment (N) was assayed by determining the fraction of total GFP-positive pixels in single optical sections (chosen to maximize mitochondrial representation) that colocalized with GRASP65 staining (n = 3, ±SEM, >15 cells/experiment; *, P < 0.0001).
Although unlikely, we wished to rule out the possibility that GRASP65 might link membranes by recruiting GM130. Cells were treated with control siRNA or a previously described siRNA targeting GM130 (Puthenveedu et al., 2006) and then transfected with G65-GFP-ActA. In control knockdown cells, G65-GFP-ActA induced mitochondrial clustering and the clusters appeared colabeled with GM130, presumably reflecting binding of GM130 to G65-GFP-ActA (Fig. S3, A–D). Significantly, GM130 recruitment did not appear functionally important. GM130 knockdown cells lacked detectable specific GM130 staining and yet the mitochondria remained clustered (Fig. S3, E–H). These results were also confirmed by radial profile analysis (Fig. S3, I and J). Thus, GRASP65, independent of its binding partner GM130, induces membrane cross-linking, and the role of GM130 in membrane linking appears to be membrane recruitment of GRASP65.
GRASP65 PDZ1 is required for homotypic oligomerization and organelle tethering
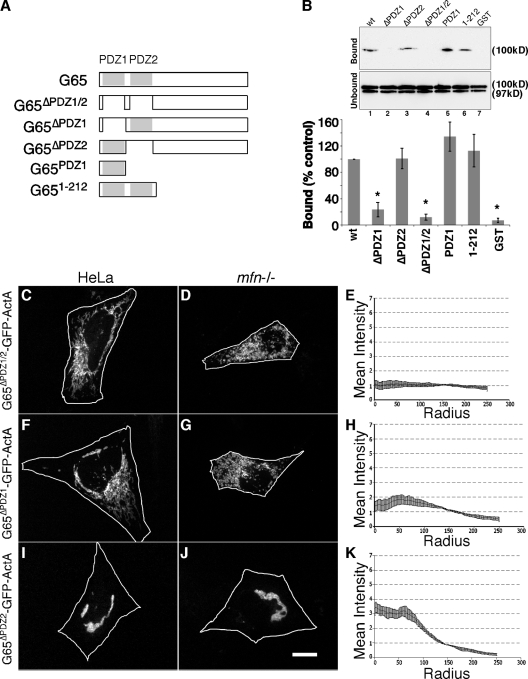
The GRASP65 N terminus contains a tandem array of PDZ-like domains that might mediate organelle tethering. The PDZ domain is a wide-spread protein module involved in protein–protein interactions (Harris and Lim, 2001; Fan and Zhang, 2002; Hung and Sheng, 2002). The canonical structure consists of five to six β-strands (β1–6) and two α-helices (α1, α2) forming a groove such that a ligand inserts between β2 and α2 completing a sheet with β2 and β3 (Doyle et al., 1996; Hung and Sheng, 2002; Im et al., 2003a; Kang et al., 2003). When purified after expression in bacteria, GRASP65 molecules self-interact to form oligomeric complexes, and this activity is mediated by the N terminus (Wang et al., 2005). To test whether one or both PDZ-like domains mediate GRASP65 homotypic interactions, a series of GRASP65 constructs (Fig. 6 A) was purified and attached to beads and incubated in a pull-down assay with HeLa cell extracts containing transfected wild-type GRASP65. GRASP65 specifically bound full-length, bead-attached GRASP65, but it did not bind a version of bead-attached GRASP65 lacking both PDZ1 and PDZ2 (Fig. 6 B). Further, deletion of PDZ1 blocked binding, whereas deletion of PDZ2 did not. Interestingly, in the pull-down assay, the GRASP65 N terminus and even PDZ1 itself was sufficient for the interaction (Fig. 6 B). To confirm that the PDZ1 interaction was direct, GRASP65 with a C-terminal hexahistidine tag was purified from bacteria coexpressing N-myristoyltransferase. The purified protein bound the bead-attached PDZ1 domain in a specific and concentration-dependent manner (Fig. S4).
Figure 6.
PDZ1 mediates homotypic GRASP65 oligomerization and clustering. Schematic diagram of the constructs (A). (B) Immunoblot analysis to detect recovery of G65-myc out of HeLa cell extracts by the indicated purified GST-GRASP65 constructs after incubation with glutathione-agarose beads. 10% of the unbound fraction was loaded for comparison, and for each construct the percentage bound relative to that bound by the wild-type (wt) control is plotted (n = 3, ±SEM; *, P < 0.003). Transfected GRASP65-myc yielded a doublet, which was quantified, although only the upper band bound. Approximately 0.5% of total was bound by the wt control. HeLa or mfn−/− cells expressing G65ΔPDZ1/2-GFP-ActA (C–E), G65ΔPDZ1-GFP-ActA (F–H), and G65ΔPDZ2-GFP-ActA (I–K) were analyzed using GFP fluorescence after BFA treatment (bar, 10 µm) and radial profile plots (n = 3, ±SEM, >15 cells/experiment).
Based on these results, we tested the role of the GRASP65 PDZ-like domains in mitochondrial clustering induced by G65-GFP-ActA. HeLa cells were first transfected with G65ΔPDZ1/2-GFP-ActA in which both PDZ-like domains were excised. The construct appeared stably targeted to mitochondria but the mitochondria remained filamentous and distributed throughout the cytoplasm, indicating that the PDZ-like domains were indeed required for clustering (Fig. 6 C). This finding was confirmed by expressing G65ΔPDZ1/2-GFP-ActA in mfn−/− cells (Fig. 6 D) and by radial profile analysis (Fig. 6 E). Next, we tested G65ΔPDZ1-GFP-ActA and G65ΔPDZ2-GFP-ActA in which the PDZ-like domains were individually deleted. Interestingly, G65ΔPDZ1-GFP-ActA failed to induce mitochondrial clustering in either HeLa or mfn−/− cells (Fig. 6, F–H), but clustering was clearly evident in cells expressing G65ΔPDZ2-GFP-ActA (Fig. 6, I–K). Thus, GRASP65 clustering activity required PDZ1, whereas PDZ2 was dispensable. For an unknown reason constructs containing either PDZ domain in isolation from the rest of the molecule failed to express. In sum, the strong correlation between clustering activity and homotypic binding activity of these constructs argues that organelle clustering by GRASP65 is mediated by homotypic interactions occurring in trans between adjacent mitochondria and that it involves PDZ1.
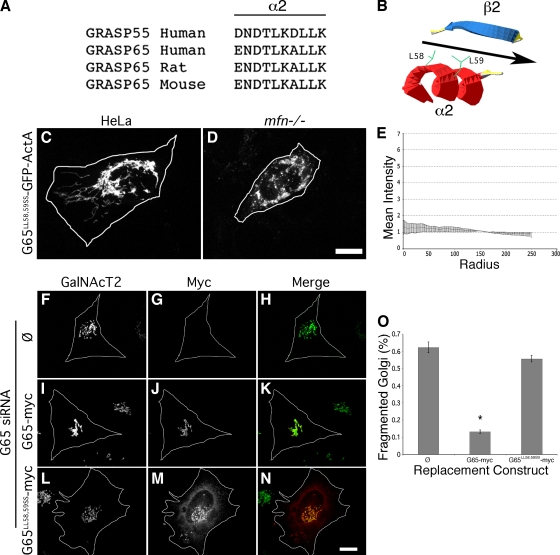
The PDZ-like domains of GRASP65 show low homology with known PDZ domains. However, alignment analysis and computed structural modeling yielded a strong prediction of the sequence comprising the specificity conferring α2 element of the binding groove of PDZ1 (Fig. 7 A). To test whether the binding groove is involved in tethering, we introduced serine substitutions of two leucines in α2 that, in the model, face the binding pocket (Fig. 7 B). The construct was expressed in HeLa or mfn−/− cells. Strikingly, G65LL58,59SS-GFP-ActA failed to cluster mitochondria in both cell types (Fig. 7, C and D). As evidence arguing that this effect was specific, another double amino acid change was made inside α2 (G65LK55,56RR) and several were made outside of α2 (G65LG3,4SS, G65GF16,17RR, and G65EE81,83RR). All constructs exhibited clear localization to mitochondria and the change inside α2 blocked clustering, whereas the changes outside of α2 had no effect on clustering (unpublished data).
Figure 7.
Mutation of the predicted PDZ1 ligand-binding groove blocks clustering. GRASP sequences are shown aligned at the position predicted by the TASSERlite modeling program (Zhang and Skolnick, 2004) to correspond to the second α-helix of the first PDZ domain (A). As illustrated in the diagram (B), this helix is oriented in the model such that two leucines (L58, L59) face the binding pocket formed between the α-helix and a β-strand. HeLa (C) or mfn−/− (D) cells expressing G65LL58,59SS-GFP-ActA were analyzed using GFP fluorescence after BFA treatment (bar = 10 µm) and the radial profile plot for HeLa cells (E) is shown (n = 3, ±SEM, >15 cells/experiment). HeLa cells expressing GalNAcT2-GFP and transfected with GRASP65 siRNA and either no vector (F–H), G65-myc (I–K), or G65LL58,59SS-myc (L–N) were analyzed to assess Golgi morphology (green) and replacement construct expression (red). Bar, 10 µm. Percentage of cells transfected with G65-myc or G65LL58,59SS-myc exhibiting a fragmented Golgi after knockdown with GRASP65 siRNAs (±SEM, n = 3, >50 cells each; *, P < 0.0001).
Having mapped residues in GRASP65 that are critical for its tethering activity, we sought to test the role of GRASP65-mediated tethering in Golgi ribbon formation using gene replacement after siRNA-mediated knockdown (Puthenveedu and Linstedt, 2004). GRASP65 expression was inhibited using a siRNA targeting the 3′ untranslated region of the GRASP65 mRNA. As previously reported (Puthenveedu et al., 2006), GRASP65 knockdown induced unlinking of the Golgi ribbon in HeLa cells stably expressing the GFP-tagged Golgi enzyme N-acetylgalactosaminyltransferase 2 (GalNAcT2) (Fig. 7, F–H). The fragmented Golgi phenotype was rescued by expression G65-myc, a version of GRASP65 tagged at the C terminus with the myc epitope and lacking the 3′ untranslated region targeted by the siRNA (Fig. 7, I–K). In marked contrast, failure to rescue was observed for a version of the replacement construct, G65LL58,59SS-myc, containing the same serine substitutions in the predicted binding groove that prevented tethering (Fig. 7, L–N). The replacement constructs were present on Golgi membranes at comparable levels and also showed some cytoplasmic accumulation, which was more evident for the mutated version. The results were quantified confirming a loss of GRASP65 activity in Golgi ribbon formation due to point mutation in the predicted PDZ1 binding groove (Fig. 7 O). Thus, the homotypic PDZ1 interaction revealed by targeting GRASP65 to the mitochondrial outer membrane underlies its ability to maintain the Golgi ribbon.
Another outcome of this analysis was the possible separation of function for the two GRASP65 PDZ-like domains in that tethering activity mapped to PDZ1, whereas previous work had mapped the GM130 binding site to the C-terminal end of PDZ2 (Barr et al., 1998). To test this prediction, we assayed GRASP65-mediated GM130 recruitment to the mitochondria of BFA-treated cells. Consistent with the result for non BFA-treated cells (Fig. S3), G65-GFP-ActA recruited GM130 to mitochondria that were clustered (Fig. S5, A–C). In contrast, GM130 was not recruited to mitochondria by G65ΔPDZ2-GFP-ActA yet, as indicated above (Fig. 6, I–K), the construct exhibited clustering activity (Fig. S5, D–F). Finally, GM130 was recruited to mitochondria bearing G65LL58,59SS-GFP-ActA, which, as expected, failed to cluster mitochondria due to the mutation in PDZ1 (Fig. S5, G–I). These results were confirmed by quantifying colocalization of GM130 with the mitochondrial constructs (Fig. S5 J). Thus, the mitochondrial assay allowed dissection of GRASP65 clustering and GM130 binding activities.
A role for membrane association of GRASP65 N terminus
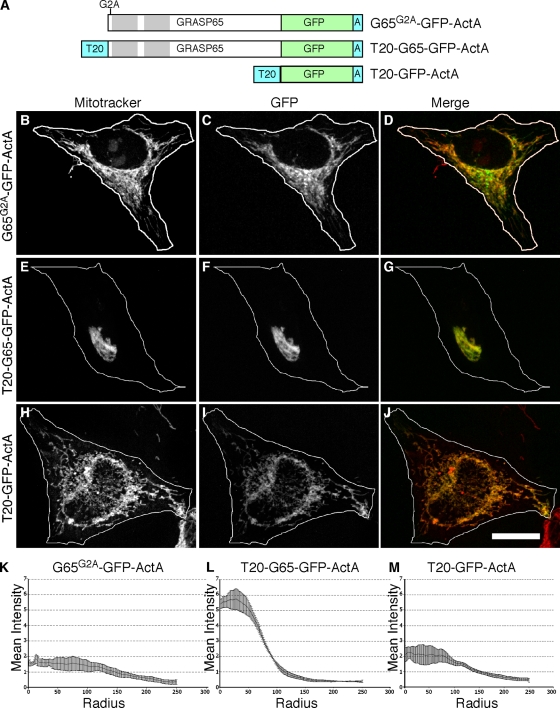
GRASP65 is myristoylated at its N terminus and this modification, together with GM130 binding, mediates GRASP65 localization to the Golgi membrane (Barr et al., 1998; Puthenveedu et al., 2006). Although the G65-GFP-ActA construct is expected to be N-myristoylated, the construct also has a C-terminal transmembrane anchor that would presumably make its membrane integration independent of myristoylation. Indeed, when the construct was modified by alanine substitution of the glycine acceptor site to prevent its myristoylation, the resulting construct, G65G2A-GFP-ActA, was targeted to mitochondria as indicated by colocalization with Mitotracker staining (Fig. 8, A–D). Unexpectedly, however, G65G2A-GFP-ActA failed to cause mitochondrial clustering as verified using radial profile analysis (Fig. 8 K) and also expression in mfn−/− cells (unpublished data). In light of the fact that previous studies of GRASP65 oligomerization-induced crossbridging were performed in vitro using nonmyristoylated protein (Wang et al., 2003, 2005), this result underscores the importance of demonstrating organelle tethering by GRASP65 in vivo.
Figure 8.
Membrane insertion of the GRASP65 N terminus is required. Schematic diagram of the constructs (A). HeLa cells expressing G65G2A-GFP-ActA (B–D and K), T20-G65-GFP-ActA (E–G and L), or T20-GFP-ActA (H–J and M) were BFA treated and analyzed using Mitotracker (red), GFP fluorescence (green), and radial profile plots. Bar, 10 µm.
Interestingly, the N-terminal myristoylation site in the GRASP family of proteins is mostly conserved, and even in cases where it is missing there seems to be an alternative mode of N-terminal membrane association. For example, the Saccharomyces cerevisiae homologue contains an acetylated N-terminal amphipathic helix and Plasmodium falciparum and Plasmodium vivax express splice variants with transmembrane signal anchors in place of the myristoylated N terminus (Behnia et al., 2007; Struck et al., 2008). These observations suggest a critical role for membrane association of the N terminus, whether it is mediated by myristic acid or by other means. If so, the crossbridging activity of the nonmyristoylated G65G2A-GFP-ActA might be rescued by anchoring its N terminus to the mitochondrial outer membrane. As a test, we generated T20-G65-GFP-Acta, which was expected to be N-terminally anchored but not myristoylated because the nonmyristoylated TOM20 signal sequence was introduced at the N terminus of the GRASP65 sequence. As a negative control, T20-GFP-ActA was generated, which contained both N- and C-terminal membrane anchors but lacked the GRASP65 sequence. Remarkably, expression of T20-G65-GFP-ActA induced mitochondrial clustering (Fig. 8, E–G and L), whereas the control construct T20-GFP-ActA did not (Fig. 8, H–J and M). These results demonstrate an additional role for the interaction of the GRASP65 N terminus with membranes. Not only does it contribute to stable membrane targeting (Barr et al., 1998), it is also critical for organelle tethering activity. The finding that the TOM20 signal sequence can substitute for the myristic acid contradicts a possibility in which the myristic acid at the GRASP65 N terminus extends out and inserts in trans into adjacent membranes. Instead, the importance of N-terminal membrane association, taken together with the role of the N-terminal PDZ1 domain, argues that membrane insertion of the N terminus activates and/or orients the PDZ1 binding groove so that it can carry out homotypic interactions in trans. This could obviate two potential problems for the homotypic tethering mechanism: interference by cis interactions in the membrane and interference by interactions with the soluble pool, if any. Indeed, overexpressed G65-myc yielded a clear cytosolic pattern, yet this protein did not appear appreciably targeted to mitochondrial clusters bearing G65-GFP-ActA (Fig. S5, K–P). Thus, membrane association appears to regulate GRASP65 self-association.
Discussion
The ability of GRASP65 to participate directly in organelle tethering was tested using two methods of targeting the protein to the cytoplasmic face of the mitochondrial outer membrane. Each yielded clustering of mitochondria in the absence of the microtubule cytoskeleton or an intact Golgi apparatus, arguing that cluster formation was due to crossbridging connections formed by GRASP65. Indeed, the N-terminal domain of GRASP65, which mediates its homo-oligomerization (Wang et al., 2005), was required for mitochondrial clustering. Of the two PDZ-like domains present in the GRASP65 N terminus, the first was found to be required for both GRASP65 self-interaction and for mitochondrial clustering. Some PDZ domains form oligomers involving a PDZ ligand in one partner binding the groove in the other or, alternatively, a back-to-back association that is independent of the binding groove and therefore not a bona fide PDZ interaction (Xu et al., 1998; Marfatia et al., 2000; Im et al., 2003a,b). To test the involvement of a bona fide PDZ interaction in GRASP65-mediated organelle tethering, the structure of the PDZ1 domain was computationally modeled, allowing us to identify and mutate the putative PDZ1 binding groove. Mutation at this site blocked mitochondrial clustering, confirming the specificity of the assay and strongly suggesting the novel involvement of a homotypic PDZ interaction in homotypic membrane tethering. Further, the same mutation blocked GRASP65-mediated Golgi ribbon formation, arguing that the structure/function analysis performed on the mitochondrial membrane recapitulates key aspects of Golgi membrane network formation and that GRASP65 acts as a homotypic organelle tether.
Homotypic interactions underlie homotypic tethering
Golgi membranes form a network that is subcompartmentalized such that there is extensive continuity within a subcompartment and little continuity between subcompartments. The model that GRASP65 is the tether for fusion of cis Golgi membranes is appealing because a homotypic interaction mediates crossbridging of like membranes. This is conceptually analogous to homotypic mitofusin interactions in homotypic fusion of mitochondria (Koshiba et al., 2004). Because the GRASP65 interaction is a PDZ-ligand type interaction it raises the possibility that crossbridging is stabilized by reciprocal interactions in which each PDZ domain contributes a ligand that associates with the binding groove of the partner. Alternatively, a higher order set of interactions may take place in which a PDZ domain contributes a ligand to one partner and a binding groove to another. In either case, because it is homotypic, the interaction is likely to involve twofold symmetry. Our data also suggest that the interaction involves an internal PDZ ligand rather than a more typical C-terminally positioned ligand. Whereas the terminal carboxylic acid in a C-terminally positioned ligand is coordinated by a conserved GLGF motif in the PDZ domain (Harris and Lim, 2001; Hung and Sheng, 2002), the C terminus is replaced by a sharp β-turn or loop structure in an internal ligand (Brenman et al., 1996; Gee et al., 1998; Christopherson et al., 1999; Harris et al., 2001). This key structural feature can be highly degenerate in sequence, making identification difficult. The mechanism of homotypic membrane fusion at sites of contact formed by GRASP65 crossbridging remains to be determined, but it is noteworthy that GM130, which stabilizes GRASP65 on the Golgi membrane, interacts with the SNARE protein syntaxin-5 (Diao et al., 2008), suggesting that this interaction may coordinate tethering with fusion.
Membrane contact may regulate GRASP65 interactions
Membrane recruitment and tethering activity of GRASP65 requires two contact points with the organellar membrane. The first contact is binding to the GM130 C terminus. GM130 is required for GRASP65 Golgi localization (Puthenveedu et al., 2006; Kodani and Sutterlin, 2008). Recruitment of endogenous GRASP65 by a mitochondrial GM130 C terminus was sufficient for mitochondrial clustering, and the same deletion of ten amino acids that disrupts GM130 function in Golgi ribbon formation (Puthenveedu et al., 2006) blocked clustering activity. These amino acids are required for GM130 to bind GRASP65 (Barr et al., 1998) and, as expected, in their absence GM130 failed to recruit GRASP65 to the mitochondrial outer membrane. Thus, the results support a model in which GM130 recruits GRASP65 to the Golgi membrane and the recruited GRASP65 is the active factor in tethering.
The second point of membrane contact is the myristoylated GRASP65 N terminus. Under normal circumstances, membrane targeting of GRASP65 requires myristoylation in conjunction with GM130 binding (Barr et al., 1998). The N-terminal myristic acid is immediately adjacent to the PDZ1 module. Mutation of the glycine residue that becomes myristoylated blocked G65-GFP-ActA–mediated mitochondrial clustering, but this construct has its own membrane anchor and the myristoylation site was not needed for its stable membrane association. Thus, the requirement for myristoylation suggests that the N terminus must be imbedded in the membrane and, indeed, substituting a transmembrane domain restored tethering. It can be further argued that the myristic acid is not needed for proper folding or even for the homotypic interaction, per se, because nonmyristoylated GRASP65, purified after expression in bacteria, retains oligomerization activity (Wang et al., 2005). In the cellular context, however, membrane association may facilitate the interaction. One idea is that N-terminal anchoring positions the binding groove such that it faces the cytosol favoring trans-pairing over cis-pairing. Another possibility, not mutually exclusive, is that membrane association triggers a conformational change that activates GRASP65 for binding, thereby preventing soluble GRASP65 from inadvertently inhibiting the trans pairing interactions. In sum, these findings uncover a direct functional role, beyond membrane targeting, for anchoring the GRASP65 N terminus.
Functional divergence of GRASP isoforms may maintain subcompartment identity
Mammals and other vertebrates that form Golgi ribbon networks express two GRASP proteins. GRASP65 is principally localized to the cis Golgi, whereas GRASP55 is localized to medial and trans cisternae (Shorter et al., 1999). In common with GRASP65, GRASP55 has a tandem arrangement of PDZ-like domains at its N terminus (Shorter et al., 1999) and GRASP55 is required for Golgi ribbon formation (Feinstein and Linstedt, 2008). Each protein interacts with itself but does not interact with the other GRASP (unpublished data), suggesting specificity-conferring differences in the PDZ1 domain of each molecule. Thus, it is likely that GRASP65 and GRASP55 act in parallel reactions with GRASP65 supporting membrane fusion to laterally link and elongate cis cisternae and GRASP55 doing the same for medial cisternae. From an evolutionary perspective, functional divergence of the GRASP PDZ domains after gene duplication may have been a necessary step to maintain Golgi subcompartments once microtubule-based motility brought Golgi ministacks into close proximity in the region of the microtubule organizing center. PDZ modules are well suited as divergence in the PDZ1 binding groove and ligand, and also in the interactions determining compartmental localization, would confer specificity such that lateral fusion within the membrane network is specific between cis and medial subcompartments.
GRASP65 and GRASP55 were initially characterized as requirements for stacking Golgi cisternae in an in vitro assay (Barr et al., 1997; Shorter et al., 1999). Subsequently, it was observed that Golgi stacks persist after effective GRASP knockdown (Sutterlin et al., 2005; Puthenveedu et al., 2006; Feinstein and Linstedt, 2008). Although the mechanism elucidated here could also underlie cisternal stacking, this would place a homotypic interaction in a heterotypic membrane linkage. Further, we noted that the ultrastructure of the juxtaposed mitochondrial membranes linked by G65-GFP-ActA was distinct from the parallel less than 15 nm (Mollenhauer and Morre, 1991) spacing present in Golgi stacks. Even so, given its ability to bridge membranes by self-association and the inherent slop in protein targeting, it is arguable that to some extent GRASP65 participates in linking cisternae, not just laterally, but also in a stacked configuration. A possible explanation for the predominant role of the GRASP proteins in lateral homotypic Golgi connections is that membrane insertion and orientation of PDZ1 might render its binding curvature sensitive such that ligand binding by PDZ1 is favored at the highly curved rim regions and disfavored at the relatively flat intra-cisternal contacts within a ministack. The enrichment of the GRASP65 binding partner GM130 at the rim regions of cis cisternae is consistent with this (Marra et al., 2001). Intriguingly, the clustered mitochondria that we observed using electron microscopy exhibited regions of high and low curvature, and the closest points of contact were frequently present at zones of highest curvature.
In lower eukaryotes a single GRASP gene is present and Golgi membranes, even when present as stacked cisternae, are neither confined to a central position nor fused laterally to form a ribbon-like membrane network. Nevertheless, the yeast GRASP homologue possesses an acetylated N-terminal amphipathic helix adjacent to a PDZ-like domain, indicating conservation, via an alternative mechanism, of membrane anchoring, and suggesting a commonality in mechanism (Behnia et al., 2007). Possibly, ribbon formation in vertebrates is a more extreme form of cisternal elongation performed by lower eukaryotes. If so, homotypic membrane tethering mediated by membrane-anchored PDZ1 could represent the fundamental mechanism of GRASP65 action. The physical distance separating cisternae or ministacks may contribute to prevention of lateral fusion which, given the presence of only a single GRASP, might otherwise impair maintenance of subcompartment identity. Recent observations indicate that the GRASP present in Dictyostelium and Drosophila is required in developmentally specific steps involving nonconventional secretion (Kinseth et al., 2007; Schotman et al., 2008). It is not yet clear whether this activity involves PDZ1-mediated membrane tethering.
In summary, our results indicate a direct role for GRASP65 in Golgi membrane network formation and, for the first time, identify a PDZ interaction as a membrane-tethering mechanism. Homotypic PDZ-mediated membrane crossbridging provides a compelling view of how subcompartments are maintained in the Golgi network and suggests modes of regulation that uncouple Golgi ministacks to promote mitotic entry and subsequent Golgi partitioning.
Materials and methods
Constructs
For GFP-ActA, residues 599–624 of the ActA protein from Listeria monocytogens were cloned into the HindIII and BamHI sites of pEGFP-C1 (Clontech Laboratories, Inc.). GRASP65 was then inserted upstream of EGFP using an NheI site to yield G65-GFP-ActA. mCherry was substituted for GFP using Age1 and Sac1 sites to yield mCherry-ActA and G65-mCherry-ActA, respectively. PDZ1 (residues 6–74) and/or PDZ2 (residues 85–167) were deleted using a PCR-based loop-out modification of the QuikChange protocol (Stratagene). Point mutations were introduced using QuikChange. For TOM20-GFP, four rounds of loop-in PCR yielded TOM20 (residues 1–47) upstream of EGFP in pEGFP-C1. GM130 (residues 788–888) was inserted into BamHI and HindIII sites to yield TOM20-GFP-GM130Cterm. A stop codon (position 878) by QuikChange yielded TOM20-GFP-GM130CΔ10. For GST-G65, GRASP65 was inserted into the EcoRI site of pGEX-4T-1 (GE Healthcare). QuikChange introduced a stop codon for GST-G651-212. Deletion of the PDZ domains was as above. For GST-G65PDZ1, GRASP65 (residues 6–74) was inserted into pGEX-2T using the BamHI and EcoRI sites. For G65-myc, GRASP65 was inserted into pCS2-MT (Turner and Weintraub, 1994). For G65-His, GRASP65 was inserted into the EcoR1 site of pRSET-B (Invitrogen) followed by removal and reinsertion of the N-terminal hexahistidine tag to achieve a C-terminal position.
Cell culture and immunofluorescence
HeLa cells were grown in MEM and mitofusin null cells were grown in DMEM containing 5 µg/ml of uridine (Sigma-Aldrich). The media also contained 10% fetal bovine serum (Atlanta Biological) and 100 IU/ml of penicillin and streptomycin (Sigma-Aldrich), and the cells were maintained at 37°C in a 5% CO2 incubator. Transient transfection of HeLa was performed with Transfectol (GeneChoice) according to the manufacturer's specifications; after 24 h the cells were labeled by adding Mitotracker (Invitrogen) to 15 nM for 30 min and fixed. Transient transfection of mitofusin-null cells was with LIPO2000 (Invitrogen) according to the manufacturer's specifications, and fixation was after 24 h. Paraformaldehyde fixation and immunofluorescence staining were as described previously (Jesch and Linstedt, 1998). Antibodies were: rabbit anti-GM130 (Puthenveedu and Linstedt, 2001), mouse anti-giantin (Linstedt and Hauri, 1993), and Alexa 568– or Cy5-conjugated anti-secondary antibodies (Invitrogen). Knockdown of GM130 by RNA interference was as described previously (Puthenveedu et al., 2006). The GRASP65 sequence targeted by siRNA was AAAAGAGATCACTGTTTAAGT. For gene replacement, transfection was performed with 60 nM chemically synthesized siRNA (Ambion) using Oligofectamine (Invitrogen). After 24 h, the cells were transfected with plasmids using Transfectol (GeneChoice). After another 24 h the cells were retransfected with siRNA and, after another 24 h, Golgi fragmentation was analyzed. For cell fusion, HeLa cells were transfected using JetPIE (Polyplus-transfection), and after 24 h, trypsinized and seeded in 1:1 ratios. After 16–24 h, the cells were treated for 30 min with 20 µg/ml cycloheximide (Sigma-Aldrich) and then 30 s with 45% polyethylene glycol (Roche) in DMEM. After five washes with DMEM the cells were cultured for 3 h in the continued presence of cycloheximide and then fixed and analyzed.
Image capture and analysis
Microscopy was performed using a spinning-disk confocal scan head equipped with three-line laser and independent excitation and emission filter wheels (PerkinElmer) and a 12-bit Orca ER digital camera (Hamamatsu Photonics) mounted on an Axiovert 200 microscope with a 100x, 1.4 NA oil-immersion objective (Carl Zeiss, Inc.). Sections at 0.3-µm spacing were acquired using Imaging Suite software (PerkinElmer). The “Radial profile analysis” plug-in of ImageJ (http://rsbweb.nih.gov/ij) was used after background subtraction and selecting the region of interest using the wand function. The “Co-localization” plug-in of ImageJ used single optical sections chosen to maximize mitochondrial representation (Guo et al., 2008). FRAP was performed using a Meta/UV DuoScan Inverted Spectral Confocal Microscope system (LSM 510; Carl Zeiss, Inc.). Fluorescence was bleached to 20% of its initial value and recovery was monitored for 120 s at intervals of 10 s
Protein purification and binding assays
Proteins were purified and eluted from glutathione agarose as described previously (Guo et al., 2008). For the binding assays, HeLa cells were transiently transfected with G65-myc and harvested 24–48 h after transfection in lysis buffer (10 mM Hepes, pH 7.2, 100 mM KCl, 1% Triton X-100, and 1 mM DTT) in the presence of protease inhibitors. Lysates were precleared with glutathione-agarose beads and then rotated for 2 h at 4°C with 50 µg of purified protein. Complexes were collected by addition of 10 µl glutathione-agarose beads, rotation for 1 h at 4°C, and centrifugation in a microfuge. The isolated beads were washed four times with 1 ml PBS containing 1 mM DTT and 0.1% Tween 20, and analyzed by immunoblot and enhanced chemiluminescence using a digital camera (model LAS-3000; Fujifilm) and ImageGauge software (Fujifilm). G65-His was purified using Ni-NTA beads (Invitrogen) and the manufacturer's protocol from BL21(DE3)-pLysS cells cotransformed with pBB131 encoding N-myristoyltransferase (Duronio et al., 1990) that were IPTG induced in the presence of 200 µM myrstic acid. After elution and dialysis against binding buffer (20 mM Hepes, 200 mM KCl, 1 mM EDTA, 0.01% Triton X-100, and 1.4 mM β-mercaptoethanol), the purified G65-His was incubated for 4 h at 4°C in a 200-µl volume with GST or GST-PDZ1 immobilized on 5 µl of glutathione beads in the presence of 10 µg bovine serum albumin and protease inhibitors. Recovery of G65-His on the beads after washing was analyzed by immunoblot using an anti-His antibody (Bethyl Laboratories).
Online supplemental material
Clustering is shown in nocodazole-treated cells (Fig. S1), in cells expressing T20-GFP-GM130cterm (Fig. S2), and in cells lacking GM130 (Fig. S3). Also shown is self-interaction of purified GRASP65 (Fig. S4) and recruitment by G65-GFP-ActA of GM130, but not GRASP65 (Fig. S5). Videos show fluorescence recovery after bleaching for GFP-ActA (Video 1) and G65-GFP-ActA (Video 2). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200902110/DC1.
Acknowledgments
We thank Tim Feinstein for critical assistance and also thank Jeanne Morin-Leisk, Alex Ritter, Sapna Puri, Tina Lee, Fred Lanni, James Fitzpatrick, and Sashi Pandit. We thank David Chan (Cal Tech) and Julie Theriot (Stanford) for sharing reagents.
Funding was provided by grants RSG-03-148-01-CSM and GM-56779 to A.D. Linstedt.
Footnotes
Abbreviation used in this paper: BFA, brefeldin A.
References
- Acharya U., Mallabiabarrena A., Acharya J.K., Malhotra V. 1998. Signaling via mitogen-activated protein kinase kinase (MEK1) is required for Golgi fragmentation during mitosis.Cell. 92:183–192 [DOI] [PubMed] [Google Scholar]
- Barr F.A., Puype M., Vandekerckhove J., Warren G. 1997. GRASP65, a protein involved in the stacking of Golgi cisternae.Cell. 91:253–262 [DOI] [PubMed] [Google Scholar]
- Barr F.A., Nakamura N., Warren G. 1998. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae.EMBO J. 17:3258–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnia R., Barr F.A., Flanagan J.J., Barlowe C., Munro S. 2007. The yeast orthologue of GRASP65 forms a complex with a coiled-coil protein that contributes to ER to Golgi traffic.J. Cell Biol. 176:255–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisel B., Wang Y., Wei J.H., Xiang Y., Tang D., Miron-Mendoza M., Yoshimura S., Nakamura N., Seemann J. 2008. ERK regulates Golgi and centrosome orientation towards the leading edge through GRASP65.J. Cell Biol. 182:837–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenman J.E., Christopherson K.S., Craven S.E., McGee A.W., Bredt D.S. 1996. Cloning and characterization of postsynaptic density 93, a nitric oxide synthase interacting protein.J. Neurosci. 16:7407–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Chan D.C. 2005. Emerging functions of mammalian mitochondrial fusion and fission.Hum. Mol. Genet. 14:R283–R289 [DOI] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., Chan D.C. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development.J. Cell Biol. 160:189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson K.S., Hillier B.J., Lim W.A., Bredt D.S. 1999. PSD-95 assembles a ternary complex with the N-methyl-D-aspartic acid receptor and a bivalent neuronal NO synthase PDZ domain.J. Biol. Chem. 274:27467–27473 [DOI] [PubMed] [Google Scholar]
- Colanzi A., Corda D. 2007. Mitosis controls the Golgi and the Golgi controls mitosis.Curr. Opin. Cell Biol. 19:386–393 [DOI] [PubMed] [Google Scholar]
- Diao A., Frost L., Morohashi Y., Lowe M. 2008. Coordination of golgin tethering and SNARE assembly: GM130 binds syntaxin 5 in a p115-regulated manner.J. Biol. Chem. 283:6957–6967 [DOI] [PubMed] [Google Scholar]
- Doyle D.A., Lee A., Lewis J., Kim E., Sheng M., MacKinnon R. 1996. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.Cell. 85:1067–1076 [DOI] [PubMed] [Google Scholar]
- Duronio R.J., Jackson-Machelski E., Heuckeroth R.O., Olins P.O., Devine C.S., Yonemoto W., Slice L.W., Taylor S.S., Gordon J.I. 1990. Protein N-myristoylation in Escherichia coli: reconstitution of a eukaryotic protein modification in bacteria.Proc. Natl. Acad. Sci. USA. 87:1506–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J.S., Zhang M. 2002. Signaling complex organization by PDZ domain proteins.Neurosignals. 11:315–321 [DOI] [PubMed] [Google Scholar]
- Feinstein T.N., Linstedt A.D. 2007. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition.Mol. Biol. Cell. 18:594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein T.N., Linstedt A.D. 2008. GRASP55 regulates Golgi ribbon formation.Mol. Biol. Cell. 19:2696–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee S.H., Sekely S.A., Lombardo C., Kurakin A., Froehner S.C., Kay B.K. 1998. Cyclic peptides as non-carboxyl-terminal ligands of syntrophin PDZ domains.J. Biol. Chem. 273:21980–21987 [DOI] [PubMed] [Google Scholar]
- Griffin E.E., Detmer S.A., Chan D.C. 2006. Molecular mechanism of mitochondrial membrane fusion.Biochim. Biophys. Acta. 1763:482–489 [DOI] [PubMed] [Google Scholar]
- Guo Y., Punj V., Sengupta D., Linstedt A.D. 2008. Coat-tether interaction in Golgi organization.Mol. Biol. Cell. 19:2830–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris B.Z., Lim W.A. 2001. Mechanism and role of PDZ domains in signaling complex assembly.J. Cell Sci. 114:3219–3231 [DOI] [PubMed] [Google Scholar]
- Harris B.Z., Hillier B.J., Lim W.A. 2001. Energetic determinants of internal motif recognition by PDZ domains.Biochemistry. 40:5921–5930 [DOI] [PubMed] [Google Scholar]
- Hoogenraad C.C., Wulf P., Schiefermeier N., Stepanova T., Galjart N., Small J.V., Grosveld F., de Zeeuw C.I., Akhmanova A. 2003. Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport.EMBO J. 22:6004–6015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung A.Y., Sheng M. 2002. PDZ domains: structural modules for protein complex assembly.J. Biol. Chem. 277:5699–5702 [DOI] [PubMed] [Google Scholar]
- Im Y.J., Lee J.H., Park S.H., Park S.J., Rho S.H., Kang G.B., Kim E., Eom S.H. 2003a. Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization.J. Biol. Chem. 278:48099–48104 [DOI] [PubMed] [Google Scholar]
- Im Y.J., Park S.H., Rho S.H., Lee J.H., Kang G.B., Sheng M., Kim E., Eom S.H. 2003b. Crystal structure of GRIP1 PDZ6-peptide complex reveals the structural basis for class II PDZ target recognition and PDZ domain-mediated multimerization.J. Biol. Chem. 278:8501–8507 [DOI] [PubMed] [Google Scholar]
- Jesch S.A., Linstedt A.D. 1998. The Golgi and endoplasmic reticulum remain independent during mitosis in HeLa cells.Mol. Biol. Cell. 9:623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaji S., Iwahashi J., Kida Y., Sakaguchi M., Mihara K. 2000. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane.J. Cell Biol. 151:277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang B.S., Cooper D.R., Devedjiev Y., Derewenda U., Derewenda Z.S. 2003. Molecular roots of degenerate specificity in syntenin's PDZ2 domain: reassessment of the PDZ recognition paradigm.Structure. 11:845–853 [DOI] [PubMed] [Google Scholar]
- Kinseth M.A., Anjard C., Fuller D., Guizzunti G., Loomis W.F., Malhotra V. 2007. The Golgi-associated protein GRASP is required for unconventional protein secretion during development.Cell. 130:524–534 [DOI] [PubMed] [Google Scholar]
- Kodani A., Sutterlin C. 2008. The Golgi protein GM130 regulates centrosome morphology and function.Mol. Biol. Cell. 19:745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T., Detmer S.A., Kaiser J.T., Chen H., McCaffery J.M., Chan D.C. 2004. Structural basis of mitochondrial tethering by mitofusin complexes.Science. 305:858–862 [DOI] [PubMed] [Google Scholar]
- Kuo A., Zhong C., Lane W.S., Derynck R. 2000. Transmembrane transforming growth factor-alpha tethers to the PDZ domain-containing, Golgi membrane-associated protein p59/GRASP55.EMBO J. 19:6427–6439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F., Lombes A., Frachon P., Rojo M. 2002. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins.Mol. Biol. Cell. 13:4343–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt A.D., Hauri H.P. 1993. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa.Mol. Biol. Cell. 4:679–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfatia S.M., Byron O., Campbell G., Liu S.C., Chishti A.H. 2000. Human homologue of the Drosophila discs large tumor suppressor protein forms an oligomer in solution. Identification of the self-association site.J. Biol. Chem. 275:13759–13770 [DOI] [PubMed] [Google Scholar]
- Marra P., Maffucci T., Daniele T., Tullio G.D., Ikehara Y., Chan E.K., Luini A., Beznoussenko G., Mironov A., De Matteis M.A. 2001. The GM130 and GRASP65 Golgi proteins cycle through and define a subdomain of the intermediate compartment.Nat. Cell Biol. 3:1101–1113 [DOI] [PubMed] [Google Scholar]
- Marra P., Salvatore L., Mironov A., Jr., Di Campli A., Di Tullio G., Trucco A., Beznoussenko G., Mironov A., De Matteis M.A. 2007. The biogenesis of the Golgi ribbon: the roles of membrane input from the ER and of GM130.Mol. Biol. Cell. 18:1595–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer H.H., Morre D.J. 1991. Perspectives on Golgi apparatus form and function.J. Electron Microsc. Tech. 17:2–14 [DOI] [PubMed] [Google Scholar]
- Neuspiel M., Schauss A.C., Braschi E., Zunino R., Rippstein P., Rachubinski R.A., Andrade-Navarro M.A., McBride H.M. 2008. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers.Curr. Biol. 18:102–108 [DOI] [PubMed] [Google Scholar]
- Pfeffer S.R. 2007. Unsolved mysteries in membrane traffic.Annu. Rev. Biochem. 76:629–645 [DOI] [PubMed] [Google Scholar]
- Pistor S., Chakraborty T., Niebuhr K., Domann E., Wehland J. 1994. The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton.EMBO J. 13:758–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M.A., Linstedt A.D. 2001. Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130.J. Cell Biol. 155:227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M.A., Linstedt A.D. 2004. Gene replacement reveals that p115/SNARE interactions are essential for Golgi biogenesis.Proc. Natl. Acad. Sci. USA. 101:1253–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M.A., Linstedt A.D. 2005. Subcompartmentalizing the Golgi apparatus.Curr. Opin. Cell Biol. 17:369–375 [DOI] [PubMed] [Google Scholar]
- Puthenveedu M.A., Bachert C., Puri S., Lanni F., Linstedt A.D. 2006. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution.Nat. Cell Biol. 8:238–248 [DOI] [PubMed] [Google Scholar]
- Rios R.M., Sanchis A., Tassin A.M., Fedriani C., Bornens M. 2004. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation.Cell. 118:323–335 [DOI] [PubMed] [Google Scholar]
- Schotman H., Karhinen L., Rabouille C. 2008. dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling.Dev. Cell. 14:171–182 [DOI] [PubMed] [Google Scholar]
- Seemann J., Jokitalo E., Pypaert M., Warren G. 2000. Matrix proteins can generate the higher order architecture of the Golgi apparatus.Nature. 407:1022–1026 [DOI] [PubMed] [Google Scholar]
- Shaul Y.D., Seger R. 2006. ERK1c regulates Golgi fragmentation during mitosis.J. Cell Biol. 172:885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Watson R., Giannakou M.E., Clarke M., Warren G., Barr F.A. 1999. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system.EMBO J. 18:4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck N.S., Herrmann S., Langer C., Krueger A., Foth B.J., Engelberg K., Cabrera A.L., Haase S., Treeck M., Marti M., et al. 2008. Plasmodium falciparum possesses two GRASP proteins that are differentially targeted to the Golgi complex via a higher- and lower-eukaryote-like mechanism.J. Cell Sci. 121:2123–2129 [DOI] [PubMed] [Google Scholar]
- Sutterlin C., Polishchuk R., Pecot M., Malhotra V. 2005. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division.Mol. Biol. Cell. 16:3211–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D.L., Weintraub H. 1994. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate.Genes Dev. 8:1434–1447 [DOI] [PubMed] [Google Scholar]
- Voeltz G.K., Prinz W.A. 2007. Sheets, ribbons and tubules — how organelles get their shape.Nat. Rev. Mol. Cell Biol. 8:258–264 [DOI] [PubMed] [Google Scholar]
- Waizenegger T., Stan T., Neupert W., Rapaport D. 2003. Signal-anchor domains of proteins of the outer membrane of mitochondria: structural and functional characteristics.J. Biol. Chem. 278:42064–42071 [DOI] [PubMed] [Google Scholar]
- Wang Y., Seemann J., Pypaert M., Shorter J., Warren G. 2003. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor.EMBO J. 22:3279–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Satoh A., Warren G. 2005. Mapping the functional domains of the Golgi stacking factor GRASP65.J. Biol. Chem. 280:4921–4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward T.H., Polishchuk R.S., Caplan S., Hirschberg K., Lippincott-Schwartz J. 2001. Maintenance of Golgi structure and function depends on the integrity of ER export.J. Cell Biol. 155:557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.Z., Choudhury A., Li X., Montell C. 1998. Coordination of an array of signaling proteins through homo- and heteromeric interactions between PDZ domains and target proteins.J. Cell Biol. 142:545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Yoshioka K., Barr F.A., Lowe M., Nakayama K., Ohkuma S., Nakamura N. 2005. Convergence of cell cycle regulation and growth factor signals on GRASP65.J. Biol. Chem. 280:23048–23056 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Skolnick J. 2004. Automated structure prediction of weakly homologous proteins on a genomic scale.Proc. Natl. Acad. Sci. USA. 101:7594–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]