Summary
The molecular apparatus that protects us against infection can also injure us by causing autoimmune or autoinflammatory disease. It now seems that at times, defects within the sensing arm of innate immunity contribute to diseases of this type. The initiation of an immune response is often microbe dependent and, in many cases, Toll-like receptor (TLR) dependent. Positive feedback loops triggering immune activation may occur when TLR signaling pathways stimulate host cells in an unchecked manner. Or, immune activation may persist because of failure to eradicate an inciting infection. Or on occasion, endogenous DNA may trigger specific immune responses that beget further responses in a TLR-dependent autoamplification loop. Specific biochemical defects that cause loop-related autoimmunity have been revealed by random germline mutagenesis and by gene targeting. We have also developed some insight into critical points at which feedback loops can be interrupted.
Keywords: monocytes/macrophages, systemic lupus erythematosus, Toll-like receptors/pattern recognition receptors, inflammation, signal transduction, immunotherapies
Introduction
Microbes have exerted a powerful selective pressure throughout the evolution of eukaryotes. Even the earliest eukaryotes must have needed protection against prokaryotic invaders that lived among them. As eukaryotes grew in size and complexity in the wake of the Cambrian explosion, evolving specialized tissues and new internal compartments, their defensive requirements grew. It was not enough to depend upon an integument. Microbes found ways of breaching physical barriers, and if only through occasional accidents, they might gain access to normally sterile interior milieu, plundering the organic molecules of which eukaryotes are composed.
Eukaryotes therefore evolved cellular and humoral systems dedicated to the eradication of microbes. All the while, their cellular constituents retained some level of autonomous resistance to infection (particularly by viruses), utilizing defensive strategies that certainly changed over time. The complex array of defenses we see in vertebrates today, partly redundant and partly non-redundant in character, we have come to know as the ‘immune system’. We traditionally divide it into two branches: innate immunity and adaptive (or acquired) immunity. The former is said to be rapid in its response to microbes and non-specific; the latter is said to be slow, highly specific, and endowed with memory of past infections, which permits a more rapid secondary response. There is some blurring at the edges, because cells of the adaptive immune system are endowed with innate recognition mechanisms in addition to their highly specific recombinatorial receptors. In some sense, cellular elements of the innate immune system can be primed for augmented function by an encounter with microbes and hence display properties of ‘memory’, albeit memory that lacks specificity.
Adaptive immune systems evolved at least twice, around 500 million years ago, each time upon an innate substructure, and one of the greatest mysteries in immunology is precisely how and why this occurred. Adaptive immunity never gained full autonomy and continues to depend upon an innate response to the present day. Moreover, some functions of the innate immune system became superfluous with the new protection afforded by adaptive immunity and were ultimately lost. For this reason, the innate immune response is, by itself, insufficient to provide advanced vertebrates with full protection against microbes. The interdependency of innate and adaptive immunity has introduced complex questions that preoccupy many of us. What are the precise signals elicited from the innate immune system that trigger an adaptive response? What adaptive immune signals feed back and co-activate an innate response? What elements of cell-autonomous immunity are shared by both systems, and just how important are they?
Each of these questions has specific answers, even if we cannot be comprehensive in providing these answers at present. One of the most universal questions to be raised and answered in good measure concerns the initial recognition events that stimulate an immune response. Perhaps more than might have been imagined, the sensors of microbial infection are few in number, specific in what they detect, and non-redundant.
Awareness of infection
The host perceives both what microbes are and what microbes do. In other words, it has evolved to recognize their molecular constituents and also the molecular changes that transpire during infection.
‘Pattern recognition’
The first and most obvious mode of recognition depends upon a system of receptors that engage molecules unique to microbes and different from self. In fact, the direct engagement of molecules from microbes by receptors of the host has been known to occur for a long time. The key receptors themselves proved elusive until recently, however, and some of them are discussed in the course of this review.
Some examples of receptors that directly engage molecules of microbial origin include molecules that can activate complement. For example, the MASP pathway includes soluble mannose binding receptors (ficolins and other collectins) that activate complement via the MASP-2 pathway (1). The properdin pathway (2) includes the protein properdin, which detects microbes, presumably engaging them with thrombospondin-1 (TSP-1) domains, and activates complement. The precise mechanism of recognition remains unknown. In the classical pathway, direct (though yet unknown) contacts between complement precursors themselves and microbes bring about conformational change and activation. In each case, the initial contact may be regarded as a receptor ligand interaction, conserved and specific for the detection of microbes. The existence of a receptor for N-formyl methionine modified peptides has been known for many years as well (3). The existence of a lipopolysaccharide (LPS) receptor was also posited by many, although it remained elusive for a long time (4). We do not yet have a comprehensive list of the microbial molecules ‘seen’ by the mammalian host, but we are developing the impression that the list is relatively short. LPS, peptidoglycans, lipopeptides, and nucleic acids are included, and perhaps account for most of the initial signaling that occurs.
A few microbial proteins are detected as well. Some, like flagellin [a ligand for Toll-like receptor 5 (TLR5)] are expressed across many microbial taxa. But others, like the G-glycoprotein of vesicular stomatitis virus (VSV), and the F protein of respiratory syncytial virus (RSV), are far less broadly expressed. The fact that these latter proteins are perceived by TLR4 raises questions as to how they are detected, and how TLRs can so quickly evolve to engage virally encoded proteins and signal their presence. Conversely, perhaps it is the virus that evolves to favor engagement and triggers an innate response for its own purposes.
It has previously been proposed that the host proteome is restricted in its variation by the innate immune sensing receptors of the host, which are driven to detect all that is foreign though forbidden to detect self (5). Within the structural constraints imposed by the innate sensors, it appears that there is much freedom to evolve. Therefore, most human proteins do not trigger an innate immune response by mouse macrophages and vice versa. Nonetheless, it is possible that more distant species (such as microbes) have proteins that do encroach upon the ligand space of TLRs and other innate immune receptors. The chance finding that certain viral proteins activate mammalian TLRs may simply be illustrative of this phenomenon.
A second means of recognition is perhaps more passive, although formally similar. Modification of host proteins by microbial enzymes may lead to recognition. For example, the coagulases of Staphylococci may initiate a clotting cascade. Fungal proteases lead to activation of a proteolytic cascade that eventuates the production of spaetzle, the ligand for Toll (6). Along the same lines, the action of microbial toxins such as the adenosine diphosphate (ADP)-ribosyltransferase of diphtheria toxin may cause cell death and so produce awareness of infection.
A third mechanism entails the recognition of ‘danger’, nebulous as the term may seem. In a way, this is a variant of the second means of recognition: it is a means of sensing ‘change caused by infection’. We have never actually found a receptor for danger. We do not have a precise molecular definition of danger. Nonetheless, the conclusion that the host responds to sterile injury is inarguable, and it is worth looking for definable molecular events at the apex of this response. Many ligands have been proposed. These include uric acid, heat shock protein 70 (HSP70), high mobility group box 1 (HMGB1), and others. Some of these ligands are clearly inflammatory, while the case has not been convincingly made for others. Host nucleic acids certainly do trigger responses via TLRs and cytoplasmic receptors partly defined and partly obscure.
A fourth mode of recognition occurs via detection of ‘missing self’, the downregulation of major histocompatibility complex (MHC) molecules, for example, and through detection of ‘stress signals’ such as H60 and Rae1, induced by viral infection. These modes of recognition might also be placed in the category of danger. Moreover, insofar as real receptors and real ligands are involved, we can speak about them more tangibly and analyze their overall importance to the immune response.
It is not excluded that other means of recognizing non-self may exist. It is remarkable that the most structurally variable molecules in nature are proteins and that alterations in protein structure most consistently distinguish one species from another. Adaptive immunity is certainly well suited to recognizing such differences. Yet, we do not know of a systematic mechanism used by the innate immune system for seeing protein differences. We can certainly imagine such a system, but it has yet to be found.
TLRs and NLRs and RLHs
In recent years we have come to know something of the principal receptors that detect molecules of microbial origin. Three basic families are well described. These are the ‘outward-looking’ Toll-like receptors (TLRs), and the ‘inward looking’ (cytoplasmic) nucleotide oligomerization domain (NOD)-like receptors (NLRs), and the cytoplasmic retinoic acid-inducible gene I (RIG-I)-like helicases (RLHs). Each seems to occupy a different niche in the perception of infection, and each may correspondingly render different effects in inflammatory disease states.
TLRs
TLRs were first identified as receptors for the recognition of discrete molecules of microbial origin when one of them, TLR4, was shown to be absolutely required to detect LPS (7). Gene targeting established the function of most of the other TLRs (8-11). It now appears that each TLR responds to a limited repertoire of ligand molecules (Table 1). In general, these molecules are represented by many microbial taxa.
In some cases, molecules unique to one microbe or at most restricted to a small clade of microbes have been observed to activate TLRs. For example, the F protein of RSV, or the G glycoprotein of VSV, or the Env protein of mouse mammary tumor virus (MMTV) can each activate TLR4 (although in each case in a qualitatively different manner).
Structural and genetic data suggest that TLRs exist as dimeric proteins (either heterodimers or homodimers), and it is widely held that activation entails a conformational change elicited by ligand binding. All of the TLRs are single-spanning type I transmembrane proteins. The ectodomains of TLRs are composed of leucine-rich repeat motifs and adopt an aesthetically pleasing curved solenoid shape, which is believed to be relatively rigid except where interrupted by irregular loops that may act as hinges, permitting some degree of flexibility. The ectodomains of TLRs are variably glycosylated, and glycosylation may restrict the types of interactions that can occur between subunits (12).
On the cytoplasmic side, TLRs have a characteristic protein domain called a TIR [Toll/interleukin-1 (IL-1) receptor] domain. The TIR domain always accounts for the major portion of the cytoplasmic domain and serves two purposes. First, it contains an oligomerization site, maintaining dimeric interactions between TLR subunits. Second, it contains a site that recruits cytoplasmic adapter proteins that also contain TIR domains. The adapter proteins also have oligomerization sites and through ‘face to face’ and ‘back to back’ interactions can presumably form extended chains in the cytoplasm of the cell following activation (13), as described in more detail below. TIR domains are found not only in Toll-like receptors but also in receptors of the IL-1/IL-18/IL-33 family, which do not have leucine-rich repeat motifs in their ectodomains but are built of immunoglobulin-type domain, represented in varying numbers of copies. TIR domains are also represented in plant disease resistance proteins and in some proteins encoded by bacteria and viruses. In microbes, some of the TIR domains almost certainly operate as molecular mimics to disrupt TLR signal transduction.
The IL-1, IL-18, and IL-33 receptors each bind well defined endogenous proteins and elicit inflammatory responses. IL-1, IL-18, and possibly IL-33 as well are each induced in response to TLR signal transduction events, which activate NF-κB. The strategy of cytokine production and subsequent activation of additional TIR domain-mediated signaling may be seen as a device for generalizing the inflammatory response and spreading a warning signal beyond the bounds of the initial inflammatory stimulus. Moreover, at times, signaling via IL-1 is clearly involved in autoinflammatory diseases. These may be seen as the consequence of continuous microbial stimulation or, perhaps at times, sterile inflammatory reactions that result from a failure in normal ‘braking’ mechanisms the terminate TIR domain signaling.
In some cases, it is clear that TLRs act as the membrane-spanning components of receptor complexes. For example, TLR4 is tightly associated with MD-2, a small molecule that binds to a hinge region of the TLR4 ectodomain and physically engages LPS, triggering activation of the complex (14). CD14, a glycosylphosphoinositol-linked leucine-rich repeat protein, also assists in the activation of TLR4, specifically by highly glycosylated forms of LPS. It is not entirely clear how this occurs, but it is reasonable to hypothesize that CD14, too, is part of a multisubunit complex and contributes to receptor activation. Another example may be seen in the case of the TLR2/TLR6 complex, in which CD36 plays an important role in the recognition of some microbial components (15), and CD14 plays an important role in the recognition of others (16). While associated subunits have not been identified for TLR3, TLR5, TLR7, and TLR9, it is possible that these TLRs also behave as parts of complexes that have not yet been fully deciphered.
Some of the TLRs appear to signal chiefly from the cell surface (TLR1/TLR2, TLR2/TLR6, TLR4, and TLR5). Others—those that detect nucleic acids—signal from internal compartments within the cell. These include TLR3, TLR7, and TLR9 (and TLR8 in humans). Because agents that block acidification of the endolysosomal compartment inhibit the perception of TLR3, TLR7, and TLR9 ligands, it is believed that nucleic acid sensing occurs strictly within these compartments. TLRs are expressed by a wide variety of cells, some of which are professional components of the immune system and some of which are not. Although considerable work has gone into the analysis of which cells respond to which TLR ligands, a complete summary of the response patterns would be premature and also beyond the scope of this review. It is interesting, however, to note that the induction of type I IFN responses in vivo seems to depend largely (though not entirely) upon activation of plasmacytoid dendritic cells, also known as interferon (IFN)-producing cells. These cells express TLR9 and other nucleic acid sensing TLRs, and probably most of their response to nucleic acids is TLR mediated (although an alternative pathway for nucleic acid perception exists as described below).
TLR signaling
Very different modes of receptor binding have been proposed for different TLRs based on co-crystallization studies. The lipid A moiety of LPS, for example, is believed to engage MD-2, the small accessory subunit of the TLR complex described above, and to cause a conformational change sensed by the holoprotein complex. Double-stranded RNA is believed to span two subunits of TLR3, uniting them and causing activation (17). Tri-acyl lipopeptides are believed to insert into the interstices of the leucine-rich repeat coils of TLR1 and TLR2, uniting them and causing activation (18). It may be proposed that these essentially extracellular (or intravesicular) events are sensed across the lipid bilayer through torsion and cause a change in the spatial orientation of TIR domains on adjoining TLR subunits. This activity leads to the recruitment and organization of adapter proteins that carry the signal into the cytoplasm.
Four TIR adapter proteins [myeloid differentiation factor 88 (MyD88), MyD88-adapter-like protein (MAL), TIR-domain-containing adapter-inducing IFN-β (TRIF), and translocating chain-associating membrane protein (TRAM)] mediate virtually all TLR signals, and in the absence of two of the adapters (MyD88 and TRIF), no discernable signaling is known to arise from any of the TLRs or from the IL-1 or IL-18 receptors. MyD88 is believed to be capable of signaling by itself (from TLR5, TLR7, TLR8, and TLR9, as well as the IL-1 and IL-18 receptors, none of which is known to depend upon other adapters), but when signaling from the TLR1/TLR2 heterodimer or from the TLR2/6 heterodimer, it depends in part upon the presence of MAL, a second adapter protein, and the one with strongest primary sequence homology with MyD88. MyD88-dependent signaling from the TLR4 complex also depends upon MAL. Hence, MyD88 seems to operate in a functional pair with MAL, at least when signaling is nucleated by some of the TIR receptor proteins.
A number of interpretations may be attached to this observation. According to one basic model, MyD88 never engages the TLR1/TLR2, TLR2/TLR6, or TLR4 receptor TIR domains directly, but instead depends upon MAL to do so. Hence, MyD88 has contact only with MAL, which acts as a ‘bridge’ to the receptor TIR domain. Other possibilities also exist, and the bridge model is contested by specific genetic evidence (13). This evidence comes in the form of receptor-selective mutations of MyD88, including the Pococurante mutation of MyD88: I178N, and the classical P712H mutation first identified in the TLR4 TIR domain in C3H/HeJ mice, and localized to the so-called BB loop of the TIR domain. These mutations behave identically, and both reside on the surface of all TIR domains. Both mutations, when engrafted into MyD88, are capable of interrupting MyD88-dependent signaling from TLR4 and TLR9 but not from TLR2/TLR6. If a simple bridge was provided by MAL in both cases, it is difficult to understand how a mutation of the MyD88 TIR domain could discriminate between the two triggering receptors.
Other hypotheses may be advanced to explain the dependency of MyD88 on MAL for some of the TIR domain receptors but not others. For example, MyD88 may be altered post-translationally (e.g. by phosphorylation) after contact with the TLR1/TLR2 or TLR2/TLR6 heterodimers or the TLR4 homodimer. In this state, it may recruit MAL, and further ‘chaining’ between MyD88 and MAL may be nucleated.
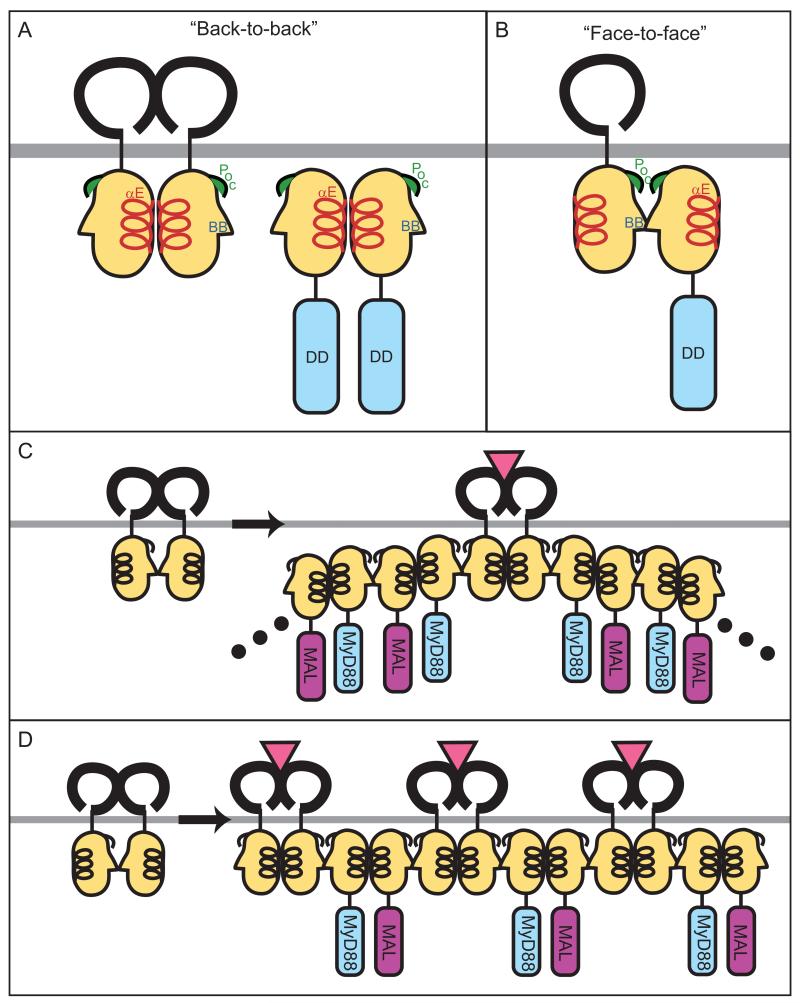
No crystal structure of MyD88 exists, but in the absence of such data, the TIR domains of all of the adapter proteins can be modeled on the existing structures of the TLR1 and TLR2 TIR domains, derived from X-ray crystallography. Docking studies can then be performed to predict how MyD88 is most likely to interact with receptor TIR domains. The most favored mode of interaction between the MyD88 TIR domain and the TLR2 TIR domain (face-to-face interaction) entails reciprocal engagement of surfaces containing both the BB loop and the Poc site, described above. The second most favored mode of interaction (back-to-back interaction) entails the reciprocal engagement of αE helices from both the receptor and adapter TIR domains. This suggests that initial contact between receptor and adapter TIR domains is face-to-face, and back-to-back interaction may follow to allow the recruitment of more adapter molecules (Fig. 1).
Fig. 1. TLR:MyD88 interactions.
The interfaces between two receptors or two adapters or receptor and adapter are predicted from docking studies based on the structures of the TIR domains of TLR1 and TLR2. (A) Binding between two receptors or two adapters may be mediated by ‘back-to-back’ TIR domain (light orange) interactions, which are characterized by homotypic interactions between αE helices (red). The back-to-back interface does not mediate receptor:adapter binding. (B) ‘Face-to-face’ TIR domain interactions likely mediate receptor:adapter binding and involve the BB loop (blue) and Poc site (green) from each molecule. Whether face-to-face interactions can also mediate receptor:receptor or adapter:adapter binding is untested but expected based on molecular docking studies. (C and D) Models for recruitment and ‘chaining’ of adapter molecules upon TLR activation. In (C), ligand binding causes dimerization of two TLRs in a back-to-back fashion, followed by recruitment of adapter (MyD88) to the receptor by a face-to-face interaction. Signal amplification may be achieved by the subsequent recruitment of additional adapters (MyD88 and MAL), which bind each other with alternating face-to-face and back-to-back interactions. Alternatively, as shown in (D), upon ligand binding and back-to-back dimerization of receptors, each receptor complex recruits one adapter by a face-to-face interaction. Adjacent adapters (MyD88 and MAL) then form back-to-back interfaces.
For TLR3 and TLR4, different adapters are also involved in ‘MyD88-independent signaling’ (19-21). One of these adapters, shared by both TLR3 and TLR4, is called TRIF [or TIR-containing adapter molecule-1 (TICAM1)]. TRIF is the sole adapter used by TLR3 and activates both NF-κB and IFN-regulatory factor 3 (IRF3) signal transduction. To activate NF-κB, TRIF interacts with tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6), which is a key signaling intermediate of the MyD88-dependent pathway as well. To activate IRF3, TRIF interacts with one of two protein kinases: TANK-binding kinase 1 (TBK1) or inhibitor of NF-κB kinase i (IKKι), which in turn phosphorylate IRF3, permitting its translocation to the nucleus (22).
TLR4 activates not only TRIF but also TRAM (also called TICAM2), its closest protein homologue, and an adapter protein that exists only to serve TLR4 signaling (19, 21). Like MAL, TRAM has been proposed as a bridging adapter protein. As with MAL, there are few direct data to support this hypothesis, and other possibilities exist.
Some TLR4 stimuli (for example, VSV infection) cause TRAM-dependent (and TRIF-independent) signal transduction, leading to the activation of IRF7 and type I IFN production (23). There is thus considerable flexibility in signal transduction downstream of TLR4. Depending upon the ligand involved, it may be strictly MyD88/MAL dependent, strictly MyD88 independent, or TRAM dependent. In the absence of MyD88 and TRIF (in mice with compound homozygous mutations), no TLR signaling can occur. This would imply that neither MAL nor TRAM is by itself capable of propagating a signal.
MyD88 contains not only a TIR domain but also an N-terminal death domain and is known to recruit protein kinases of the IL-1 receptor-associated kinase (IRAK) family to the activation complex via a death domain interaction. IRAK4 is believed to be activated and to phosphorylate IRAK1 and/or IRAK2, which serve a positive role in signal transduction (24). IRAK-M, the remaining member of this protein family, serves an inhibitory role in signaling (25). The activated IRAK proteins somehow recruit TRAF6, a more distal component of the signaling chain, to the activation complex.
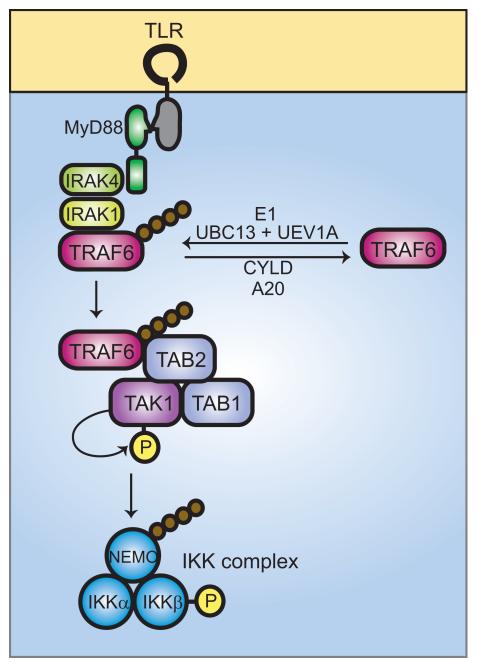
TRAF6, which as noted above is also activated by TRIF (or TRIF/TRAM) recruitment, is a protein with multiple ring-finger motifs that complex zinc and is endowed with catalytic activity, serving as an E3 ubiquitin ligase (26). The intrinsic E3 ligase activity of TRAF6 acts in concert with an E1 ligase as well as two other protein complexes, known as TRAF6-regulated IKK activators (TRIKAs). TRIKA1 is an E2 ubiquitin ligase complex consisting of Ubc13 and a Ubc-like protein Uev1A. Together with TRAF6, TRIKA1 attaches a chain of K63-linked ubiquitin subunits on NF-κB essential modulator (NEMO) (the IKKγ subunit) and on TRAF6 itself. TRIKA2 is a complex of proteins including the transforming growth factor-β-activated kinase 1 (TAK1) and the adapter proteins TAK1-binding protein 1 (TAB1) and TAB2. These proteins act together to permit the recruitment of IKKβ and its activation. TAB2 and TAB3 both bind to the K63-linked ubiquitin chain, which serves as an anchorage site for IKK activation. Once activated, the IKK complex phosphorylates IκB, which is subsequently degraded in K48-ubiquitination dependent process. The role of TRAF6 as a central way-station in signaling via both MyD88-dependent and MyD88-independent pathways makes it the focus of both activating and inactivating signals. The polyubiquitin chains that are attached by TRIKA1 complex proteins can be removed by the action of de-ubiquitination proteins such as CYLD and A20 (Fig. 2).
Fig. 2. Ubiquitin signaling in TLR-mediated activation of NF-κB.
Ligand binding to TLRs leads to the recruitment of MyD88 and, subsequently, IRAK4 and IRAK1. IRAK1 binds to TRAF6. The oligomerization of TRAF6 (not shown) activates its ubiquitin ligase activity, leading to K63-linked polyubiquitination of NEMO and TRAF6 in conjunction with UBC13 and UEV1A. De-ubiquitination can be carried out by CYLD and A20. Ubiquitinated TRAF6 recruits the TRIKA2 complex, consisting of TAB1, TAB2, and TAK1, via binding to the novel zinc finger domain of TAB2. TAK1 kinase becomes activated by autophosphorylation and phosphorylates IKKβ at two activation loop serine residues, resulting in IKK complex activation. The IKK complex then phosphorylates IκB, leading to IκB degradation, and releasing NF-κB for translocation to the nucleus.
The activation of TAK1 (also known as MAP3K7) is a prerequisite not only for activation of IKKβ but also for the activation of mitogen-activated kinase (MAPK) kinases such as MKK6 (MAP2K6), which in turn phosphorylates Jun kinases (JNK), p38 kinase, and extracellular signal regulated kinase 1 (ERK1) and ERK2. Other protein kinases also play a key role in the downstream activation of signaling from all of the TLRs. Among these is Tpl2 (also known as MKK8, MAP3K8, or Cot), first shown to be important for LPS-induced responses not long after the TLRs were identified as signaling receptors (27). Tpl2 is probably also activated by the TRAF6 complex and seems to be essential for the production of TNF, IL-6, and in some cells type I IFNs. Its key function lies in the activation of MAPKs including ERK1, ERK2, and p38.
Other proteins play supporting functions in TLR signaling. For example, UNC93B, a 12-spanning membrane protein located primarily in the endoplasmic reticulum, is required to allow TLR3, TLR7, and TLR9 to transit to the endosomes, where signaling actually occurs (28-30). Similarly, gp96, an endoplasmic reticulum chaperone, is required for TLR4 to reach the cell surface (31).
In the end, hundreds (and in some circumstances thousands) of genes are modulated by TLR activation: some upregulated and some suppressed. These events create the extremely complex changes that are known macroscopically as inflammation. TLR signaling also causes post-transcriptional changes that alter protein synthesis or trafficking within the cell. Most of these lie beyond the scope of discussion in this review, but where individual genes have been studied, it is seen that translational activation can result from TLR activation (in the case of the TNF mRNA, for example), and that degranulation of certain cells (such as mast cells) can be triggered by TLR signaling. While it has been proposed that the TLRs interpret the ‘molecular patterns’ present on microbes as a bar-code reader does and elicit a response finely attuned to the type of infection that is present, direct evidence of strong specificity is lacking. Some TLRs do indeed yield qualitatively different biological responses than others. However, the overlap in signaling is perhaps more striking, and the impression gained is one of a stereotypic response to infection.
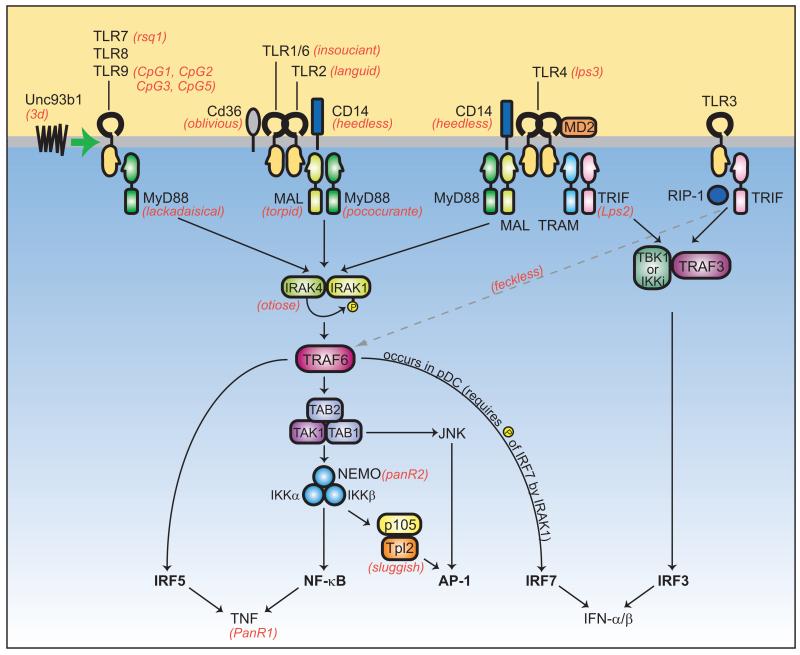
Despite the seeming complexity of the TLR signaling pathways and the potential for redundancy in signaling in general, a number of mutations are capable of interrupting signal transduction completely or almost completely. Indeed, this is how the pathways have become clearly established as they are drawn today, and germline mutations remain the gold standard in pathway analysis. Random germline mutagenesis (forward genetics) and gene targeting (reverse genetics) have both played important roles in the dissection of these pathways. The current illustration based on forward genetic analysis (including both known and unknown genes) is illustrated in Fig. 3.
Fig. 3. Overview of the TLR signaling pathways.
The pathways represent data obtained from both forward and reverse genetic studies. ENU-induced phenotypes are shown in red text. Note that TLR3, TLR7, and T:R9 are endosomal proteins, while TLR1, TLR2, TLR6, TLR4, as well as TLR5 (not shown) are expressed on the plasma membrane. Unc93b1 is an endoplasmic reticulum protein that influences endosome function. TLR activation recruits TRIF, TRAM, MAL, and/or MyD88 and leads to the activation of IRAK1 and IRAK4, or the TBK1 or IKKι family kinases. TRAF6 signaling to the TAB1/TAB2/TAK1 complex results in the activation of NF-κB and AP-1, via the functions of the IKK complex, Tpl2, and JNK. IRF5 is another means by which signaling through MyD88 leads to TNF production. MyD88 and TRAF6 may interact directly with IRF5 in a complex, activating IRF5 and promoting its translocation to the nucleus. Additionally, MyD88, together with TRAF6 and IRAK4, has also been shown to bind IRF7 directly in order to stimulate IFN-α production. This occurs downstream of TLR7, TLR8 and TLR9 in plasmacytoid dendritic cells (pDCs) and requires the phosphorylation of IRF7 by IRAK1. TRAF3 is also involved in TLR7- and TLR9-dependent pDC production of IFN-α; TRAF3 may form a complex with IRAK1 and IRF7 and facilitate phosphorylation of IRF7 (42) (not shown). In the MyD88-independent pathway, TLR3 or TLR4 recruit TRIF and TBK1, the critical kinase required for activation of IRF3. TRAF3 mediates the interaction between TRIF and TBK1. The kinase RIP-1 is required for NF-κB activation downstream of TRIF, and likely requires the direct interaction between TRIF and RIP-1. The point at which RIP-1 signaling impinges on the NF-κB pathway is unknown (dashed gray arrow).
Signal amplification
Soon after Tlr4 was shown to encode the LPS receptor core by positional cloning, its mRNA was found to exist at low abundance: a fact that had undoubtedly frustrated efforts to identify the Lps gene product through expression cDNA cloning. The protein itself also exists at remarkably low copy number. Titration experiments were performed by expressing epitope tagged versions of the wildtype (WT) (Lpsn) and signaling defective, dominant inhibitory (Lpsd) versions of the protein in RAW 264.7 cells: a mouse macrophage line that exhibits robust responses to LPS. Expression levels and LPS signal transduction were then measured in a large number of stable clones. Fewer than 1,000 molecules of the signaling defective isoform suppressed LPS signaling by 50%, suggesting that the endogenous WT protein was expressed at a similar level. Moreover, LPS signaling was dramatically augmented by overexpressing the WT isoform, which suggests that TLR4 itself is the limiting factor in LPS signaling to the level of TNF production (32). This conclusion was later supported by transgenesis experiments performed in vivo: it was shown that increasing the copy number of TLR4 increases sensitivity to the lethal effect of LPS (33).
The lethal effect of LPS in an intact mouse is mediated principally by macrophages (34) and is entirely dependent upon TLR4. One may estimate the quantity of TLR4 protein that delivers the lethal signal based on assumptions about the number of macrophages that exist in normal mice and on the added assumption that each macrophage expresses approximately 1,000 TLR4 monomers, as is the case for RAW 264.7 cells (32). The most generous estimate concerning macrophage numbers (109 macrophages, or about 5% of the body weight of the mouse) leads to the conclusion that less than 0.15 μg of TLR4 mediates a lethal outcome when a large dose of LPS is administered. It may therefore be supposed that each TLR4 complex elicits the production of enormous numbers of cytokine molecules as a result of signal amplification within the TLR4-expressing cell and through secondary inducing effects of the cytokines themselves. Primary signal amplification probably occurs at catalytic steps in the signaling cascade: e.g. IRAK4, TAK1, and Tpl2 mediate phosphorylation of transcription factors, and at the levels of transcription and translation.
NLRs
The NLRs fill a different niche in perception than TLRs, which seem mainly dedicated to sounding an alarm about microbes. Initial understanding of NLRs grew from the observation that similar proteins exist in plants and are involved in disease resistance. Moreover, the importance of NLR family members in immunity was first suggested by the fact that one of them, a trans-activator of major histocompatibility complex (MHC) gene transcription, class II transactivator (CIITA), was mutated in patients with bare lymphocyte syndrome. The prevalence of motifs involved in microbe sensing [such as leucine-rich repeats, caspase recruitment domain (CARD), or the structurally related pyrin domains] suggested that proteins of the NLR superfamily must play a role in host defense. The impression was compounded by strong genetic studies that implicated mutations in NOD2, one member of the superfamily, in Crohn’s disease. It was promptly suggested that NOD2 must serve as an intracellular sensor of LPS (an idea now discarded) and later that NOD2 might detect other molecules of bacterial origin.
It is now known that several classes of NLR proteins exist. One subgroup includes the NOD1 and NOD2 proteins; another includes the Nacht domain-, leucine-rich repeat-, and PYD-containing protein 1 (NALP1) and NALP3 proteins; another includes the IPAF and NAIP proteins. These are regarded as key sensors of ‘something’ caused by infection or injury, though their precise mode of action remains unknown. Many other members of the superfamily exist but have no assigned functions.
Although arguably less capable of driving acute inflammatory responses than TLRs (which can drive a lethal systemic inflammatory response within hours following administration of LPS, for example), the NLRs can definitely produce inflammation of a severe degree over time, in some cases leading to death. The strongest evidence of this conclusion is the fact that activating mutations of NLRs cause severe chronic inflammatory diseases. For example, activating mutations of one family member, NALP3, produce cold-induced autoinflammatory syndrome (CIAS) or Muckle-Wells syndrome, or chronic infantile neurological cutaneous and articular (CINCA) syndrome, also known as neonatal onset multisystem inflammatory disease (NOMID). These three diseases are clinically distinguishable and range from an urticarial disorder induced by cold in the case of CIAS to a lethal disease affecting the skin, joints, and central nervous system in the case of CINCA/NOMID. Indeed, it is interesting to find that rather dramatic and qualitative differences in disease phenotype result from different mutations within this family of proteins (35, 36). Activating mutations of another family member, NOD2, produce Blau’s syndrome, a disease in which uveitis, granulomatous arthritis, and rash are prominent features. There is debate as to whether activating mutations or hypomorphic mutations of NOD2 produce Crohn’s disease. Familial Mediterranean fever, a disease in which recurrent serosal inflammation, arthritis and fever are prominent, results from a mutation in the pyrin gene.
NLR activation and signaling
Some of the NLRs are capable of activating NF-κB. This is so in the case of NOD2, which activates NF-κB through recruitment of receptor-interacting protein 2 (RIP2), a homologue of the RIP that is involved in TNF receptor-mediated signal transduction via TNF receptor 1-associated death domain protein (TRADD) and TRAF2, ultimately to the level of NF-κB. NOD2 activates RIP2, which in turn activates IKK through a ubiquitination-dependent process that is not yet fully understood.
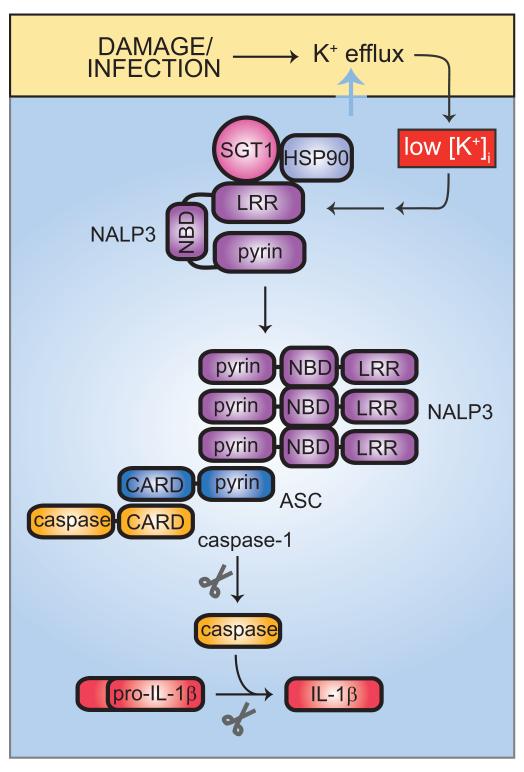
Most of the inflammatory effects of the NLRs seem to be mediated by IL-1, since neutralization of IL-1 strongly mitigates the clinical syndrome seen in CIAS, Muckle-Wells disease, or CINCA/NOMID. However, the ability of NLRs to activate NF-κB may not be the central cause of IL-1 production. Rather, the NLRs activate caspase 1, which is required for the processing and secretion of IL-1. This is exemplified by NALP3, which normally exists in a repressed state, complexed with SGT1 (suppressor of G2 allele of SKP1 homolog) and HSP90. Both proteins are known to be complexed with the structurally related plant disease resistance proteins and to maintain their quiescence prior to infection (Fig. 4).
Fig. 4. IL-1β activation by the NALP3 inflammasome.
Activation of the NALP3 inflammasome is triggered by a wide variety of stimuli, all of which may be united in their ability to cause K+ efflux from the host cell. Low intracellular K+ concentrations lead, through an unknown mechanism, to dissociation of HSP90 and SGT1 from the autorepressed NALP3. NALP3 can then recruit ASC via pyrin-pyrin domain interactions. ASC in turn recruits caspase 1 via CARD-CARD domain binding. Together, NALP3, ASC, and caspase 1 form the core components of the NALP3 inflammasome. Processing of caspase 1 activates it to cleave pro-IL-1β, giving rise to active IL-1β.
Upon damage or infection of the cell, SGT1 and HSP90 dissociate from the LRR region of NALP3, permitting is oligomerization. This in turn leads to the recruitment of the adapter protein ASC (apoptosis-associated speck-like protein containing a C-terminal CARD), which is endowed with a pyrin domain that interacts with the NALP3 pyrin domain, and a CARD domain that interacts with the CARD domain of caspase 1. Caspase 1 is thus activated and processes IL-1β, which can be secreted in its mature form. IL-1β can then activate the IL-1 receptor, leading to TIR-domain signaling via MyD88. As described below, MyD88 signaling can elicit the synthesis of additional IL-1β precursor protein.
The NALP3 activation pathway has been termed an ‘inflammasome’, although there is no discrete organellar body associated with it. The earlier-described NALP1 protein mediates similar inflammatory effects, in response to cell membrane damage. While there is no question that the NALPs (and other superfamily members) mediate inflammation as described, there is no evidence to support the hypothesis that receptor-like function is responsible for NLR activation or that contact between the NLRs and any molecules of microbial origin actually occurs. Moreover, the NLRs may also mediate adjuvant effects of aluminum hydroxide as well as some microbial adjuvant preparations, responses to uric acid crystals, and to other sterile inflammatory substances. The most important question about the NLRs thus has to do with the mechanism of NLR activation in the first place. How does a small collection of proteins recognize alum, peptidoglycan fragments, uric acid, extracellular adenosine triphosphate, and probably other inflammatory stimuli as well?
It has been proposed that many signals of cellular stress can activate the inflammasome. Among them, low concentrations of cytoplasmic potassium, reactive oxygen species, and possible certain microbial molecules yet unknown have been listed. This begs the question as to the actual identity of primary sensors, but convergence on a single molecule or molecular complex gives at least a good start in tracking the primary sensors down.
RLHs
As already discussed, TLRs can sense nucleic acids. However, there are redundant systems for nucleic acid detection, and at least in part, the RLHs participate in this process. Cytoplasmic sensors for double-stranded RNA (dsRNA) were believed to exist in part because TLR3 or TRIF-deficient cells are still capable of detecting polyinosinic-polycytidylic acid (polyI:C) or dsRNA. In fact, the dominant sensor of polyI:C in vivo appears to be something other than TLR3. Cytoplasmic sensors make sense, because replication often involves the production of unusual ssRNA or dsRNA, and while professional immune cells might detect these atypical molecules by engulfing dying cells that have been infected, a signal from the primary cell (in the form of IFN production, for example) would certainly help to mobilize a response. Perhaps it would do so more quickly than secondary detection would allow.
It is also known that some DNA molecules are not detected via TLR9, but rather, by another sensor, which strongly activates NF-κB. This sensor (or collection of sensors) remains unknown.
The RLHs bear DExD-box RNA helicase domains that can directly engage cytoplasmic RNA. Once engaged, they activate IFN-β promoter stimulator 1 (IPS1), a mitochondria-associated protein also known as mitochondrial antiviral signaling (MAVS), virus-induced signaling adapter (VISA), or CARD adapter inducing IFN-β (CARDIF), through a CARD domain interaction. Acting in conjunction with TRAF3 and Fas-associated death domain protein (FADD), IPS1 signals to activate both IRF3 and IRF7, or to activate NF-κB. The IRFs are activated via TBK1 and IKKι, which also signal in the context of the TLR3 and TLR4 signaling pathways. NF-κB is activated via pro-caspase 8 or pro-caspase 10.
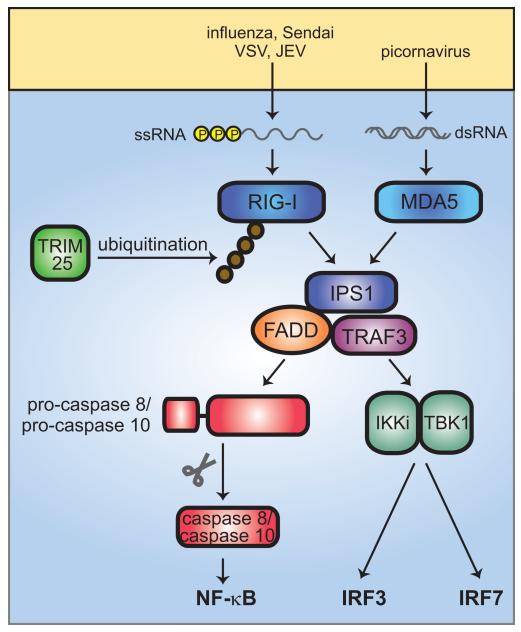
Two principal RLHs drive these events within the cell. They are RIG-I and myeloid differentiation-associated gene 5 (MDA5). RIG-I was originally identified as a retinoic acid-inducible protein and binds 5′-triphosphate single-stranded RNA. Such RNA is present in cells infected by influenza virus, Sendai virus, VSV, Japanese encephalitis virus, and other viruses. RIG-I activation is potentiated by K63 polyubiquitination (much as TRAF6 signaling is enhanced by K63 polyubiquitination as described earlier). Polyubiquitination is accomplished by T-cell receptor interacting molecule 25 (TRIM25), a tripartite motif protein. MDA5 is the dominant sensor of polyI:C in vivo. It is responsive to picornaviruses. Other helicase proteins that may be important sensors include laboratory of genetics and physiology-2 (Lgp2), which lacks a CARD domain but is otherwise homologous to MDA5 and RIG-I, and dsRNA-dependent protein kinase (PKR), an IFN-inducible dsRNA-binding protein kinase (Fig. 5).
Fig. 5. RLH signaling.
The two cytoplasmic nucleic acid sensors, RIG-I and MDA5, respond to different viral infections, recognizing 5′-triphosphate single stranded RNA and dsRNA, respectively. RIG-I also detects dsRNA. RIG-I and MDA5 activate IPS1, which recruits TRAF3 and FADD into a complex. Association of FADD with pro-caspase 8 or pro-caspase 10 results in cleavage to the mature, active caspase 8 or caspase 10, which go on to activate NF-κB. RIG-I and MDA5 signaling also activates IRF3 and IRF7. This occurs through the kinases IKKι and TBK. K63-linked ubiquitination of RIG-I by TRIM25 enhances RIG-I signaling.
On the effector side: what do the signals do?
An immune-sensing apparatus is useless without an effector arm. The host has many ways of countering microbes, but in many cases, the mechanism of the antimicrobial effect remains at least partly obscure. Complement and antimicrobial peptides are capable of directly destroying infectious bacteria and some viruses. Also well studied is the biochemical system for the generation of reactive chemicals such as hypochlorite and nitrogen radicals that can react with microbes and kill them. Some of the effector pathways remain to be fully understood. Even the type I IFNs, which counter the proliferation of most viruses and some bacteria, do so through mechanisms that are only partly deciphered. The induction of 2′,5′ oligoadenylate synthase and its ability to activate nucleases that selectively limit viral proliferation account for at least part of the effect of IFNs but probably not all. Other cell-autonomous immune mechanisms, including, for example, TRIM5α (which protects most mammalian species against human immunodeficiency virus) and the p47 guanosine triphosphatases of mice (which protect against multiple intracellular microbes) are also incompletely understood. The ability of such proteins to offer protection may reflect their recent recruitment as defensive proteins, as they seem species limited in their protective effects.
Positive feedback loops in autoinflammatory disease
One of the key questions to be asked with regard to all of these sensing pathways is related to autoinflammatory disease and, in some measure, to true autoimmune disease. Much research directed toward infectious inflammation was performed on the premise that there were only a limited number of ways to achieve inflammation and, therefore, that the biochemistry of infectious inflammation would ultimately give insight into sterile inflammatory diseases like rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), etc. In some ways, the premise has already been validated. Individual cytokines such as TNF and IL-1 are quite clearly associated with the pathogenesis of RA and were long ago identified because they were produced in response to inflammatory stimuli like LPS. However, we do not yet know what incites chronic inflammatory diseases nor what sustains them, and only such insight will allow treatments aimed at correcting the primary defects.
Increasingly, certain autoinflammatory and autoimmune diseases are seen to be caused by defects within the pathways delineated here. Moreover, we see that diseases once regarded as ‘sterile’ actually are driven by microbes. As might be imagined, defects can affect regulatory steps or effector processes that normally terminate a microbial stimulus by sterilizing the infection. Two examples were revealed by mutations that cause conditional inflammatory diseases. These mutations, induced by ENU mutagenesis, are called Jinx and Spin, and are described in some detail here.
Jinx: a mouse model of hemophagocytic lymphohistiocytosis
Jinx mice were identified by ENU mutagenesis because they were overwhelmed by infection when inoculated with mouse cytomegalovirus (MCMV). It was soon determined that the proximal cause of their immune deficiency was the failure of their natural killer (NK) cells to degranulate. Cytotoxic T lymphocytes also failed to degranulate, although both NK cells and CTLs were capable of producing IFNγ in response to infection.
The mutation was identified by positional cloning in the protein Unc13d: a molecule involved in the exocytosis of toxic granules from lymphoid (but not myeloid) cells. One of four paralogues in mammals, Unc13d, is the only paralogue required for degranulation of lymphoid cells. Unc13a-c are involved in the release of vesicles from neurons, and severe neurological phenotypes result from their targeted deletion. nc13d is also not required for normal pigmentation as are some components of the lymphoid exocytosis machinery.
When maintained in the absence of infection, mice of the Jinx stock develop no overt disease. In humans, however, positional cloning had established that the orthologue of Unc13d (Munc13-4) is mutated in type III hemophagocytic lymphohistiocytosis (HLH). Type III HLH is a severe inflammatory disease in which myeloid cells (particularly macrophages) invade the marrow and lymphoid organs, and anemia, thrombocytopenia, and fever develop, leading to a fatal outcome within a few months of diagnosis. Observational data had suggested an infectious etiology in humans, although no specific agent has been solidly linked to disease pathogenesis.
When mice are infected with MCMV at low doses, they survive but do not develop HLH. However, when infected with normally survivable doses of the arenavirus lymphocytic choriomeningitis virus (LCMV), mice develop an HLH-like disease. In this disease, overproduction of IFNγ is observed coupled with activation of both myeloid and lymphoid compartments and a failure to control the proliferation of LCMV. One hypothesis concerning the pathogenesis of HLH, then, would be as follows. (i) Mice are infected with LCMV, and present antigen to CD8+ T cells, which in turn produce IFNγ. (ii) Lymphoid cells fail to degranulate, and therefore fail to kill antigen-expressing infected cells, which continue to shed virus and infect other cells in the vicinity. (iii) IFNγ drives the expansion of myeloid cells, and their activation. Among these are dendritic cells, capable of presenting antigen. (iv) The lymphoid and myeloid compartments each stimulate the other, and the viral pathogen, which ultimately drives the process, is never cleared.
In support of this model, mutational inactivation of the IFNγ gene prevents HLH (Crozat and Beutler, unpublished data). So too, in another model of HLH, does antibody against IFNγ (37). The initial sensing mechanism for LCMV is uncertain. However, the existence of an interruptible loop would seem likely (Fig. 6).
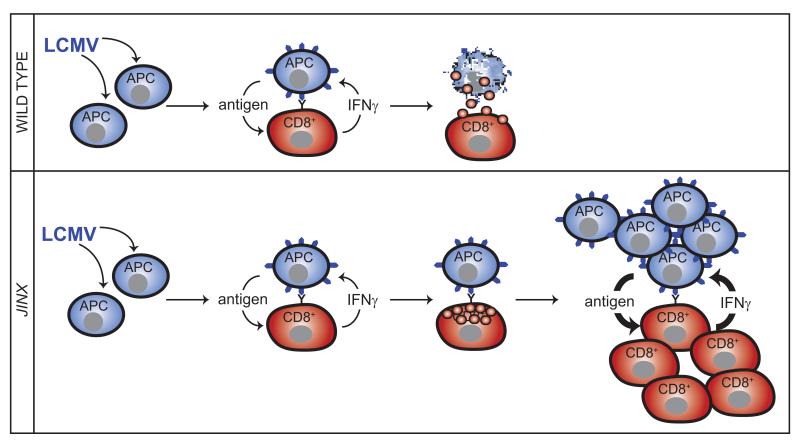
Fig. 6. HLH-like disease in Unc13djinx mice is caused by an uninhibited cytokine production loop.
In WT animals, LCMV-infected antigen-presenting cells (APCs) present antigen to CD8+ T cells, stimulating them to produce IFNγ, which may further promote antigen presentation. Degranulation by CD8+ T cells halts this cycle by killing the target APCs. In Unc13djinx mice, however, lymphoid cells fail to degranulate and kill LCMV-infected targets, which continue to shed virus and infect other cells. Persistent production of IFNγ drives the expansion of myeloid cells (not shown), which present antigen and further stimulate lymphoid cell proliferation and activation. The viral pathogen is never cleared, and mice develop clinical features of HLH, including thrombocytopenia, neutrophilia, splenomegaly, and elevated serum IFN-γ.
Spin: a mouse with severe autoinflammatory and autoimmune disease dependent on microbes TIR domain signaling via the IL-1 receptor
The Spin mutation (38) was induced by random germline mutagenesis and was marked by spontaneous inflammation of the footpads, inflammatory lesions in the lungs, salivary glands, and bone marrow. In addition, Spin mice develop anti-chromatin antibodies (both IgM and IgG). All of these abnormalities are fully suppressed when newborn mice are derived into a germfree environment. However, when conventionalized, the mice promptly develop inflammatory lesions and autoimmunity.
The requirement for microbial flora suggested that TLR-dependent signal transduction might contribute to the development of autoimmunity. Indeed, by combining the Spin mutation with Poc (a markedly hypomorphic allele of MyD88) or with Otiose (a null allele of Irak4) or with a knockout allele of the Il1r1 gene, the phenotype was also fully suppressed. Null alleles of Tnf or Stat1 failed to suppress disease.
The Spin mutation was positionally cloned and found to be a hypomorphic allele of Ptpn6, encoding the protein tyrosine phosphatase Shp1. The classical alleles of Shp1, motheaten and motheaten viable, are more severe than Spin (which is compatible with a normal lifespan). While humans with PTPN6 mutations have not yet been reported, an SLE-like disease would be anticipated in our own species as in mice. A B-cell-specific knockout mutation of Ptpn6 causes autoimmune disease (39), suggesting that the myeloid compartment is not essential for autoimmunity per se. A motheaten allele causes autoinflammatory disease, even on a recombination-activating gene-1 (Rag-1)-deficient background, indicating that the lymphoid compartment is not essential for inflammation to occur.
It would now appear that disease in Ptpn6-deficient mice must be initiated by microbes, and without microbes, inflammation and autoimmunity never result. The first signals may be propagated via any of the microbe sensors described earlier, although it is perhaps most likely that sensing occurs through activation of TLRs. TLR activation induces expression of the IL-1β gene, and NLR activation leads to processing of the IL-1β precursor. Mature IL-1β interacts with its own receptor and causes TIR domain signaling, which, in the absence of sufficient Ptpn6 activity, leads to the production of still more IL-1β (Fig. 7).
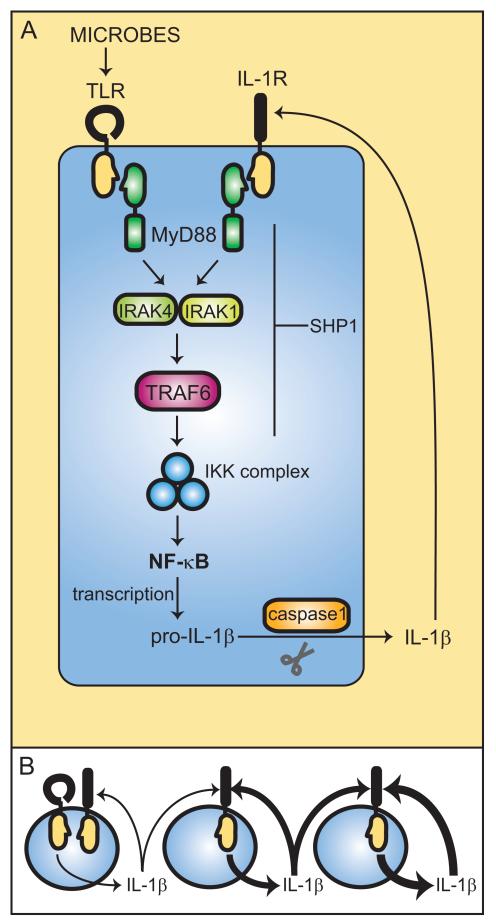
Fig. 7. Unchecked IL-1 signaling causes inflammation and autoimmunity in Ptpn6spin mice.
(A) The IL-1 signaling loop is initiated by microbial stimulation of TLRs, which through canonical TLR signaling via IRAK1, IRAK4, TRAF6, IKK complex, and NF-κB activation, leads to IL-1β production. IL-1β activates its own receptor, which also signals via the same TLR pathway components to produce IL-1β. Through an unknown mechanism, impaired SHP1 function in Ptpn6spin cells results in a failure to downregulate IL-1 signaling, which continues to stimulate its own production. This uninhibited IL-1 signaling loop (B) ultimately causes chronic inflammation and autoimmunity in Ptpn6spin mice. Derivation into a germfree environment and inactivating mutations in MyD88, IRAK4, and IL-1R suppress disease in Ptpn6spin mutants.
There is uncertainty as to which cells are of key importance in each of these events. However, it would seem that an unchecked loop is involved in the pathogenesis of autoimmunity and inflammatory disease in Spin mice much as it is in Jinx mice, albeit a rather different kind of loop. The point at which Ptpn6 normally exerts a braking effect in this loop also remains uncertain. However, many of the molecules already discussed in the description of TLR signaling pathways may be regarded as candidates.
TLR7/TLR9 signaling and SLE
Another ‘loop pathogenesis’ model of autoimmunity has been reported by Marshak-Rothstein, who envisioned a circumstance in which B cells with receptors specific for anti-chromatin antibodies might internalize DNA and/or ribonucleoproteins and be driven to proliferate through activation of TLR7 (in the case RNA or ribonucleoprotein complexes were targeted) or TLR9 (in the case DNA or nucleoprotein complexes were targeted). This would lead to the production of more antibody against an internalizable TLR ligand and a vicious cycle of stimulation by immune complexes. In some mouse models of SLE, the existence of such a loop has been validated by the finding that TLR7 (40) or MyD88 (41) mutations inhibit the development of disease. It remains to be seen whether autoantibody production is truly driven by nucleoprotein immune complexes as envisioned in the model, and importantly, the initial origin of autoantibodies remains mysterious.
From genetics to biochemistry to therapy
For decades inflammation biology research has been built on the premise that sterile inflammation utilizes the same biochemical pathways as inflammation triggered by infectious agents. Some therapeutic successes grew from this assumption, as individual mediators were targeted for the treatment of RA and other diseases. The primary cause remains elusive for many such diseases to this day, but when we do know the causes based on genetic analysis, we are often able to link them to inflammation in a rational manner, develop strategies for intervention, and in some cases, apply these strategies.
It is not usually the case that the mutated proteins are themselves suitable targets for intervention. Kinases, proteases, and other enzymes are likely to present much better targets. As enzymes permit signal amplification, they often represent the key ‘choke points’ at which an inhibitor can abrogate signaling. Moreover, they are subject to selective inhibition by mimetic drugs that can be created by design or by screening.
To the extent that we now know much more about the biochemistry of signaling leading to inflammatory cytokine production, we are in a much better position to block cytokine synthesis than before. More commonly than we know, diseases that we call autoinflammatory or autoimmune may depend upon microbes after all. Also, if we understand that certain inflammatory processes depend upon forward feedback loops, the least redundant part of the loop may productively be targeted.
References
- 1.Runza VL, Schwaeble W, Mannel DN. Ficolins: novel pattern recognition molecules of the innate immune response. Immunobiology. 2008;213:297–306. doi: 10.1016/j.imbio.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Schwaeble WJ, Reid KB. Does properdin crosslink the cellular and the humoral immune response? Immunol Today. 1999;20:17–21. doi: 10.1016/s0167-5699(98)01376-0. [DOI] [PubMed] [Google Scholar]
- 3.Boswell RN, Austen KF, Goetzl EJ. A chemotactic receptor for val(ala)-gly-ser-glu on human. Immunol Commun. 1976;5:469–479. doi: 10.3109/08820137609033861. [DOI] [PubMed] [Google Scholar]
- 4.Sultzer BM. Genetic control of leucocyte responses to endotoxin. Nature. 1968;219:1253–1254. doi: 10.1038/2191253a0. [DOI] [PubMed] [Google Scholar]
- 5.Beutler B. Neo-ligands for innate immune receptors and the etiology of sterile inflammatory disease. Immunol Rev. 2007;220:113–128. doi: 10.1111/j.1600-065X.2007.00577.x. [DOI] [PubMed] [Google Scholar]
- 6.Chamy LE, Leclerc V, Caldelari I, Reichhart JM. Sensing of ‘danger signals’ and pathogen-associated molecular patterns defines binary signaling pathways ‘upstream’ of Toll. Nat Immunol. 2008;9:1165–1170. doi: 10.1038/ni.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi O, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 9.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi O, et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 11.Hemmi H, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88- dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 12.Choe J, Kelker MS, Wilson IA. Crystal structure of human Toll-like receptor 3 (TLR3) ectodomain. Science. 2005;309:581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 13.Jiang Z, et al. Details of Toll-like receptor:adapter interaction revealed by germ-line mutagenesis. Proc Natl Acad Sci USA. 2006;103:10961–10966. doi: 10.1073/pnas.0603804103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HM, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Hoebe K, et al. CD36 is a sensor of diacylglycerides. Nature. 2005;433:523–527. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 16.Jiang Z, et al. CD14 is required for MyD88-independent LPS signaling. Nat Immunol. 2005;6:565–570. doi: 10.1038/ni1207. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin MS, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Hoebe K, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signaling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto M, et al. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 22.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 23.Georgel P, et al. Vesicular stomatitis virus glycoprotein G activates a specific antiviral Toll-like receptor 4-dependent pathway. Virology. 2007;362:304–313. doi: 10.1016/j.virol.2006.12.032. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc Natl Acad Sci USA. 2002;99:5567–5572. doi: 10.1073/pnas.082100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kobayashi K, Hernandez LD, Galan JE, Janeway CA, Jr., Medzhitov R, Flavell RA. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell. 2002;110:191–202. doi: 10.1016/s0092-8674(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 26.Chen ZJ. Ubiquitin signalling in the NF-κB pathway. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dumitru CD, et al. TNF-α induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell. 2000;103:1071–1083. doi: 10.1016/s0092-8674(00)00210-5. [DOI] [PubMed] [Google Scholar]
- 28.Tabeta K, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 29.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J.Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 31.Randow F, Seed B. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat Cell Biol. 2001;3:891–896. doi: 10.1038/ncb1001-891. [DOI] [PubMed] [Google Scholar]
- 32.Du X, Poltorak A, Silva M, Beutler B. Analysis of Tlr4-mediated LPS signal transduction in macrophages by mutational modification of the receptor. Blood Cells Mol Dis. 1999;25:328–338. doi: 10.1006/bcmd.1999.0262. [DOI] [PubMed] [Google Scholar]
- 33.Kalis C, Kanzler B, Lembo A, Poltorak A, Galanos C, Freudenberg MA. Toll-like receptor 4 expression levels determine the degree of LPS-susceptibility in mice. Eur J Immunol. 2003;33:798–805. doi: 10.1002/eji.200323431. [DOI] [PubMed] [Google Scholar]
- 34.Galanos C, Freudenberg MA. Mechanisms of endotoxin shock and endotoxin hypersensitivity. Immunobiology. 1993;187:346–356. doi: 10.1016/S0171-2985(11)80349-9. [DOI] [PubMed] [Google Scholar]
- 35.Ryan JG, Kastner DL. Fevers, genes, and innate immunity. Curr Top Microbiol Immunol. 2008;321:169–184. doi: 10.1007/978-3-540-75203-5_8. [DOI] [PubMed] [Google Scholar]
- 36.Ting JP, Kastner DL, Hoffman HM. CATERPILLERs, pyrin and hereditary immunological disorders. Nat Rev Immunol. 2006;6:183–195. doi: 10.1038/nri1788. [DOI] [PubMed] [Google Scholar]
- 37.Jordan MB, Hildeman D, Kappler J, Marrack P. An animal model of hemophagocytic lymphohistiocytosis (HLH): CD8+ T cells and interferon gamma are essential for the disorder. Blood. 2004;104:735–743. doi: 10.1182/blood-2003-10-3413. [DOI] [PubMed] [Google Scholar]
- 38.Croker BA, et al. Inflammation and autoimmunity caused by a SHP1 mutation depend on IL-1, MyD88, and a microbial trigger. Proc Natl Acad Sci USA. 2008;105:15028–15033. doi: 10.1073/pnas.0806619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pao LI, et al. B cell-specific deletion of protein-tyrosine phosphatase Shp1 promotes B-1a cell development and causes systemic autoimmunity. Immunity. 2007;27:35–48. doi: 10.1016/j.immuni.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 40.Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Sadanaga A, et al. Protection against autoimmune nephritis in MyD88-deficient MRL/lpr mice. Arthritis Rheum. 2007;56:1618–1628. doi: 10.1002/art.22571. [DOI] [PubMed] [Google Scholar]
- 42.Oganesyan G, et al. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]