Abstract
Mitochondria are involved in many cellular processes; mitochondrial dysfunctions have been associated with apoptosis, aging, and a number of pathological conditions, including osteoarthritis (OA). Mitochondrial proteins are attractive targets for the study of metabolism of the chondrocyte, the unique cell type present in mature cartilage, and its role in tissue degradation. Using a proteomics approach based on two-dimensional DIGE and MALDI-TOF/TOF mass spectrometric identification of mitochondria- enriched protein fractions from human articular chondrocytes, we analyzed mitochondrial protein changes that are characteristic of OA chondrocytes. A total of 73 protein forms were unambiguously identified as significantly altered in OA; 23 of them have been previously described as mitochondrial. An extensive statistical and cluster analysis of the data revealed a mitochondrial protein profile characteristic for OA. This pattern includes alterations in energy production, maintenance of mitochondrial membrane integrity, and free radical detoxification. Real time PCR, Western blot, and immunohistofluorescence assays confirmed a significant decrease of the major mitochondrial antioxidant protein manganese-superoxide dismutase (SOD2) in the superficial layer of OA cartilage. As possible outputs for this antioxidant deficiency, we found an increase of intracellular reactive oxygen species generation in OA chondrocytes and also verified an OA-dependent increase in the mitochondrial tumor necrosis factor-α receptor-associated protein 1 (TRAP1), a chaperone with a reported reactive oxygen species antagonist role. Our results describe the differences between the mitochondrial protein profiles of normal and OA chondrocytes, demonstrating that mitochondrial dysregulation occurs in cartilage cells during OA and highlighting redox imbalance as a key factor in OA pathogenesis.
The mitochondrion is one of the most complex and important organelles found in eukaryotic cells and carries out a wide variety of biochemical processes. Mitochondria are critical subcellular organelles responsible for energy production through the coupling of respiration to the generation of ATP. Mitochondria consist of four components: an outer membrane, an intermembrane space, an inner membrane, and a matrix. These components all function in concert to convert pyruvate and fatty acids to acetyl CoA, which is metabolized by the citric acid cycle to produce NADH. High energy electrons from NADH are then passed to oxygen by means of the respiratory chain in the inner membrane, producing ATP by a chemiosmotic process. Transcription and translation take place in mitochondria, which also actively import proteins and metabolites from the cytosol, influence programmed cell death, and respond to cellular signals such as oxidative stress (1). In addition to their central role in energy metabolism, mitochondria are involved in many cellular processes; mitochondrial dysfunctions have been associated with apoptosis, aging, and a number of pathological conditions, including Parkinson disease, diabetes mellitus, Alzheimer disease, and OA1 (1–3).
OA, the most common age-related cartilage and joint pathology (4), is a slowly progressive degenerative disease characterized by degradation of the matrix and cell death, which result in a gradual loss of articular cartilage integrity (5, 6). The only cell type present in mature cartilage is the chondrocyte, which is responsible for repairing cartilage tissue damaged by OA. Recently the role of mitochondrial dysfunction in OA has been the subject of renewed interest. Some studies have shown that mitochondrial dysfunction mediates several pathways implicated in cartilage degradation (7). These include oxidative stress, inadequacy of chondrocyte biosynthetic and growth responses, up-regulated chondrocyte cytokine-induced inflammation and matrix catabolism, pathologic cartilage matrix calcification, and increased chondrocyte death (necrosis or apoptosis). For example, mitochondrial respiratory chain (MRC) activity in OA chondrocytes showed decreases in complexes I, II, and III compared with normal chondrocytes that caused a reduction in mitochondrial membrane potential (Δψm) and in ATP synthesis. On the other hand, as a compensatory mechanism, the number of mitochondria is increased in OA chondrocytes as demonstrated by a significant increase in mitochondrial mass and in citrate synthase activity (8). Other reports implicate decreased mitochondrial bioenergy reserve as a pathogenic factor in degenerative cartilage disease (9–11).
Taken together, these findings suggest that mitochondrial proteins would be an attractive target for study of the metabolism of chondrocytes and the role they play in cartilage degradation. Most studies analyzing mitochondrial proteins in chondrocytes evaluated single proteins without addressing the mitochondrial proteome. The introduction of proteomics has enabled the simultaneous analysis of changes in multiple proteins. Currently many proteomics studies use two-dimensional gel electrophoresis (2-DE) to separate proteins, and this technology remains one of the key methodologies in proteomics studies. 2-DE gel-based approaches typically resolve hundreds to thousands of intact proteins according to their charge and molecular mass and compare the presence and intensity of protein spots among gel images to allow both qualitative and quantitative analysis. Using this proteomics approach, we have recently described the proteome of normal and OA human chondrocytes as well as their differential protein profile (12, 13). Nevertheless traditional 2-D gel-based strategies have, until recently, lacked the ability of directly quantifying changes in abundance in the same fashion as stable isotope strategies using liquid chromatography coupled with tandem mass spectrometry (14, 15). Therefore, issues such as gel-to-gel variation and normalization of spot intensities across gel sets highly affect both accuracy and sensitivity of quantification. DIGE technology (16) adds an essential quantitative advantage to 2-D gel-based strategies and allows the detection of slight changes in protein abundance with statistical confidence (17). In this approach, samples are labeled with different fluorescent dyes (Cy2, Cy3, and Cy5), not only increasing sensitivity and dynamic range but also allowing sample multiplexing so that two different samples can be run in the same gel together with an internal standard. The use of a pooled sample internal standard permits direct quantitative evaluation of changes and reduces intergel variation and false positives (18, 19), resulting in highly reproducible data with biological significance. Moreover one of the advantages of 2-D DIGE versus non-gel-based quantitative proteomics techniques is that it detects not only changes in protein quantity but also posttranslational modifications of the protein (20).
Because gel-based techniques have a bias toward abundant proteins, proteins in lower quantity are not often detected in the 2-DE analysis of total cellular proteins because of the complexity of these samples. The use of prefractionation methods by subcellular isolation or selective enrichment of a specific group of proteins provides an effective approach to eliminate this drawback. We previously optimized the methodology for isolating mitochondria from human articular chondrocytes and reported their mitochondrial 2-DE reference map (21). In this work, we took advantage of 2-D DIGE technology to study mitochondria-enriched protein fractions and analyze the differential mitochondrial protein profile of osteoarthritic human articular chondrocytes to identify new mitochondrial proteins related with OA pathogenesis.
EXPERIMENTAL PROCEDURES
Reagents, Chemicals, and Antibodies—
Culture media and FCS were from Invitrogen. Culture flasks and plates were purchased from Costar (Cambridge, MA). DIGE materials (IPG buffer and strips and Cy dyes) were from GE Healthcare. Unless indicated, all other chemicals and enzymes were obtained from Sigma-Aldrich. Monoclonal antibodies against human tumor necrosis factor-α (TNFα) receptor-associated protein 1 (TRAP1) and manganese-superoxide dismutase (SOD2) were from BD Biosciences. The corresponding horseradish peroxidase- or phycoerythrin-conjugated secondary antibodies were from Santa Cruz Biotechnologies (Santa Cruz, CA) and DAKO Diagnostics (Glostrup, Denmark), respectively. MitoTracker Green was purchased from Invitrogen.
Cartilage Procurement and Processing—
Macroscopically normal human knee cartilage from adult donors having no history of joint disease was provided by the Tissue Bank and the Autopsy Service at Complejo Hospitalario Universitario A Coruña. Osteoarthritic cartilage was obtained from patients undergoing joint surgery. The study was approved by the institutional Ethics Committee. Once cartilage surfaces were rinsed with saline, scalpels were used to cut parallel vertical sections 5 mm apart from the cartilage surface to the subchondral bone. These cartilage strips were dissected from the bone, and the tissue was incubated with trypsin at 37 °C for 10 min. After removing the trypsin solution, the cartilage slices were treated for 12–16 h with type IV clostridial collagenase in Dulbecco's modified Eagle's medium with 5% FCS to release cartilage cells.
Primary Culture of Chondrocytes—
The isolated chondrocytes were recovered and plated at high density in Dulbecco's modified Eagle's medium supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 1% glutamine, and 10% FCS. The cells were then seeded onto 162-cm2 flasks for proteomics studies or 12-well culture dishes for total RNA extraction. The seeded cells were incubated at 37 °C in a humidified gas mixture containing 5% CO2 balanced with air. The chondrocytes were used at confluency (2–3 weeks in primary culture) after making them quiescent by incubation in a medium containing 0.5% FCS for 48 h. Cell viability was assessed by trypan blue dye exclusion.
Mitochondrial Protein Extraction—
Chondrocytes (20–30 × 106 cells) were recovered from culture flasks by trypsinization and collected by centrifugation at 4 °C. After one wash in 130 mm NaCl, 5 mm KCl, 2.5 mm Tris-HCl (pH 7.5), and 0.7 mm Na2HPO4, the cells were transferred to microcentrifuge tubes and resedimented. For mitochondrial isolation, a differential centrifugation procedure (21) was performed. All centrifugations were carried out at 4 °C. Briefly chondrocytes were resuspended in a swelling buffer containing protease inhibitors and incubated for 20 min on ice. Then 0.4 volumes of an ice-cold 2.5× sucrose buffer was added, and the cells were homogenized by 30 passes through a 25-gauge needle. The resulting homogenates were centrifuged twice for 10 min at 1200 × g to remove nuclei and large cell debris. The supernatants were then centrifuged at 9000 × g for 15 min; each homogenate yielded a mitochondrial pellet and a cytosol-enriched fraction. The mitochondria were thoroughly resuspended in 500 μl of sucrose buffer and centrifuged at 1200 × g to remove further contaminants. The supernatant was finally centrifuged at 9000 × g to obtain the crude mitochondrial pellet. This pellet was solubilized by 1-h incubation with gentle agitation in an isoelectric focusing-compatible urea lysis buffer (21). For protein quantification, 2–4 μl of each protein extract was diluted to 50 μl with water, and protein was quantified in triplicate by the BCA technique (Pierce). Correct quantification was confirmed by loading 5 μg of each sample on a standard SDS-PAGE gel and subsequent Coomassie staining.
DIGE Experimental Design and Protein Labeling—
The proteomics comparison between OA and normal chondrocyte mitochondrial proteins was performed across six DIGE gels using the same pooled sample internal standard to reduce intergel variation. The 12 individual samples were generated from six normal (N) donors (mean age, 65.8 years; age range, 53–83 years) and six OA donors (mean age, 69.8 years; age range, 51–87 years). Proteins in each sample were fluorescently tagged with a set of matched fluorescent dyes according to the manufacturer's protocol for minimal labeling. To eliminate any dye-specific labeling artifacts, three samples of each group (N and OA) were labeled with Cy3, and the other three were labeled with Cy5. The pooled sample internal standard was always Cy2-labeled. In every case, 400 pmol of dye was used for 50 μg of protein. Briefly labeling was performed for 30 min on ice in darkness, and the reaction was quenched with 1 μl of 10 mm l-lysine for 10 min under the same conditions.
2-DE and Imaging of Cy-labeled Proteins—
The six pairs of Cy3- and Cy5-labeled samples (each containing 50 μg of protein) were combined and mixed with a 50-μg aliquot of the Cy2-labeled pooled standard. The mixtures containing 150 μg of protein were diluted 1:1 with rehydration buffer (7 m urea, 2 m thiourea, 4% CHAPS, 4% ampholytes (pH 3–11), and 200 mm DTT). The IPG strips (24 cm, pH 3–11 non-linear) were rehydrated overnight with 450 μl of a rehydration buffer as above but with 2% ampholytes, 0.002% bromphenol blue, and 97 mm DeStreak reagent instead of DTT. The labeled samples were then applied to the strips by cup-loading on a manifold-equipped IPGphor II IEF system (GE Healthcare). Isoelectric focusing was carried out for a total of 70 kV-h using the following conditions: 1 h at 120 V, 1 h at 500 V, 1 h at 1000 V, gradient to 4000 V in 1 h, and finally 12 h at 4000 V. Prior to the second dimension run, the strips were equilibrated first for 15 min in equilibration buffer (100 mm Tris-HCl (pH 8.0), 6 m urea, 30% glycerol, and 2% SDS) with 2% DTT and then for another 15 min in the same buffer supplemented with 2.5% iodoacetamide and 0.002% bromphenol blue. The equilibrated strips were transferred onto 12% homogenous polyacrylamide gels (2.6% C) cast in low fluorescence glass plates using an Ettan-DALT six system (GE Healthcare). Electrophoresis was run at 2 watts/gel for about 17 h at 20 °C.
The differentially labeled co-resolved proteins within each gel were imaged at a resolution of 100 dots/inch using a Typhoon 9400 laser scanner (GE Healthcare). Cy2-, Cy3-, and Cy5-labeled images of each gel were acquired at excitation/emission values of 488/520, 523/580, and 633/670 nm, respectively. Gels were scanned directly between the glass plates, and the 16-bit image file format images were exported for data analysis. After imaging for Cy dyes, the gels were removed from the plates and subjected to colloidal Coomassie staining.
DIGE Data Analysis—
The DeCyder version 6.5 software (GE healthcare) was used for spot detection and determination of quantity, intergel matching, and statistics. The differential in-gel analysis (DIA) module was used for automatic spot detection and abundance measurements for each individual gel by comparing the normalized volume ratio of each spot from a Cy3- or Cy5-labeled sample to the corresponding Cy2 signal from the pooled sample internal standard. The DIA data sets from each individual gel were collectively analyzed using the biological variation analysis (BVA) module, which allows intergel matching and calculation of average abundance for each protein spot among the six gels of our study. Statistical significance was assessed for each change in abundance using Student's t test and analysis of variance analyses. We considered statistical significance to be at the 95% confidence level when standardized average spot volume ratios exceeded 1.3 in at least four of the six analyzed gels (i.e. 12 of the 18 analyzed images). Calculation of experimental molecular weight and pI for each differential protein spot was carried out using PDQuest 7.3.1 software.
Unsupervised principal component analysis (PCA), hierarchical clustering (HC), and k-means clustering analyses were performed using the DeCyder extended data analysis module on the group of spots identified as significantly changed. These multivariate analyses clustered the individual Cy3- and Cy5-labeled samples based on collective comparison of expression patterns from the set of proteins. The groups of protein expression characteristics are represented by each data point in the PCA plots and by each column in the HC expression matrixes. Mapping of proteins identified by mass spectrometry, biological association network analysis, and database search onto existing pathways and cellular networks was carried out using Pathway Studio 5.0 (Ariadne Genomics, Rockville, MD).
Mass Spectrometry Analysis—
The gel spots of interest were manually excised from the gels and transferred to microcentrifuge tubes. Samples selected for analysis were in-gel reduced, alkylated, and digested with trypsin according to Sechi and Chait (22). Briefly spots were washed twice with water, shrunk with 100% ACN, and dried in a Savant SpeedVac. Then samples were reduced with DTT and subsequently alkylated with iodoacetamide. Samples were digested with 12.5 ng/μl sequencing grade trypsin (Roche Applied Science) for at least 6 h at 37 °C. After digestion, the supernatant was collected, and 1 μl was spotted onto a MALDI target plate (384-spot Teflon®-coated plates) and allowed to air dry at room temperature. Subsequently 0.5 μl of a 3 mg/ml solution of α-cyano-4-hydroxy-trans-cinnamic acid matrix in 0.1% TFA and 50% ACN was added to the dried peptide digest spots and again allowed to air dry. The samples were analyzed using the MALDI-TOF/TOF mass spectrometer 4800 Proteomics Analyzer (Applied Biosystems, Framingham, MA) and 4000 Series Explorer™ software (Applied Biosystems). MALDI-TOF spectra were acquired in reflector positive ion mode using 1000 laser shots per spectrum. Data Explorer version 4.2 (Applied Biosystems) was used for spectra analyses and generating peak picking lists. All mass spectra were internally calibrated using autoproteolytic trypsin fragments and externally calibrated using a standard peptide mixture (Sigma-Aldrich). TOF/TOF fragmentation spectra were acquired by selecting the 10 most abundant ions of each MALDI-TOF peptide mass map (excluding trypsin autolytic peptides and other known background ions) and averaging 2000 laser shots per fragmentation spectrum. The parameters used to analyze the data were a signal to noise threshold of 20, a minimum area of 100, and a resolution higher than 10,000 with a mass accuracy of 20 ppm.
Database Search—
The monoisotopic peptide mass fingerprinting data obtained from MS and the amino acid sequence tag obtained from each peptide fragmentation in MS/MS analyses were used to search for protein candidates using Mascot version 1.9 from Matrix Science. Peak intensity was used to select up to 50 peaks per spot for peptide mass fingerprinting and 50 peaks per precursor for MS/MS identification. Tryptic autolytic fragment-, keratin-, and matrix-derived peaks were removed from the data set used for the database search. The searches for peptide mass fingerprints and tandem MS spectra were performed in the Swiss-Prot release 53.0 and TrEMBL release 37.0 databases without taxonomy restriction, containing 269,293 and 4,672,908 sequence entries, respectively, for each software version and database release. Fixed and variable modifications were considered (Cys as S-carbamidomethyl derivate and Met as oxidized methionine, respectively), allowing one trypsin missed cleavage site and a mass tolerance of 50 ppm. For MS/MS identifications, a precursor tolerance of 50 ppm and MS/MS fragment tolerance of 0.3 Da were used. Identifications were accepted as positive when at least five matching peptides and at least 20% of the peptide coverage of the theoretical sequences matched within a mass accuracy of 50 or 25 ppm with internal calibration. In every case probability scores were significant at p < 0.01. Intracellular localization of the identified proteins was predicted from the amino acid sequence using PSORT II program.
Western Blot Test—
One-dimensional Western blot tests were performed according to standard procedures. Briefly 50 μg of total cellular or mitochondria-enriched proteins were loaded and resolved on standard 10% polyacrylamide SDS-PAGE gels. Separated proteins were then electroblotted onto PVDF membranes (Immobilon P, Millipore, Bedford, MA). Equivalent loadings were verified by Ponceau Red staining after transference. Membranes were blocked in Tris-buffered saline (pH 7.4) containing 0.1% Tween 20 (TBST) and 5% nonfat dried milk for 60 min at room temperature. The blots were then hybridized overnight at 4 °C with antibodies against TRAP1 (1:500) or SOD2 (1:1000) and the housekeeping controls α-ATPase (1:5000, in tests performed on crude mitochondria extracts) or α-Tubulin (1:5000 in tests performed on whole cell extracts). All antibodies were diluted in TBST with 2% nonfat milk. After thorough washing with TBST, immunoreactive bands were detected by chemiluminescence using corresponding horseradish peroxidase-conjugated secondary antibodies and ECL detection reagents (GE Healthcare) and then digitized using an LAS 3000 image analyzer. Quantitative changes in band intensities were evaluated with ImageQuant 5.2 software (GE Healthcare). The densitometric values of the Western blot bands containing the protein of interest (TRAP or SOD2) were normalized against those of α-ATPase or α-Tubulin obtained from the same membranes. Then the relative abundance of SOD2 and TRAP1 was calculated by obtaining the ratio of the normalized densitometric values between normal and OA samples. Statistical p values of the densitometry data were obtained by application of Mann-Whitney U test using SPSS version 15.0 program.
Real Time PCR Assays—
Primers for SOD2, TRAP1, and the housekeeping genes HPRT1, GAPDH, and PBGD were designed using the Universal Probe Library tool from the Roche Applied Science. Primer sequences were as follows: SOD2 forward, 5′-CTGGACAAACCTCAGCCCTA-3′; SOD2 reverse, 5′-TGATGGCTTCCAGCAACTC-3′; TRAP1 forward, 5′-AGACCAATGCCGAGAAAGG-3′; TRAP1 reverse, 5′-TCCTGTGTCATCCCGATACC-3′; HPRT1 forward, 5′-TGACCTTGATTTATTTTGCATACC-3′; HPRT1 reverse, 5′-CGAGCAAGACGTTCAGTCCT-3′; GAPDH forward, 5′-GGAGTCAACGGATTTGGTCGTA-3′; GAPDH reverse, 5′-GGCAACAATATCCACTTTACCAGAGT-3′; PBGD forward, 5′-AGCTATGAAGGATGGGCAAC-3′; and PBGD reverse, 5′-TTGTATGCTATCTGAGCCGTCTA-3′. Total RNA was isolated from cartilage or chondrocytes using TRIzol LS reagent (Invitrogen) following the manufacturer's instructions. Whole RNA was treated with DNase (Invitrogen), and its concentration was determined by spectrophotometry. RNA from each sample (1 μg) was reverse transcribed in a final volume of 20 μl using the Transcriptor First Strand cDNA Synthesis kit (Roche Applied Science). cDNA synthesis was performed at 55 °C for 30 min followed by a final step of 5 min at 85 °C for inactivating the reverse transcriptase. Tubes were finally stored at −20 °C until PCR analyses.
Real time PCR was performed in the LightCycler 480 instrument (Roche Applied Science) with 20-μl reactions containing 10 μl of LightCycler 480 SYBR Green I Master, 7.4 μl of RNase free water, 0.3 μl (0.3 μm) of each primer, and 2 μl of cDNA as PCR template. Cycling parameters were 95 °C for 10 min to activate DNA polymerase followed by 45 cycles of 95 °C for 10 s, 60 °C for 10 s, and a final extension of 72 °C for 10 s. Detection of fluorescence was carried out at the end of each extension step. After amplification, a melting curve was acquired by heating to 95 °C for 5 s, cooling to 70 °C for 1 min, and slowly heating to 95 °C with a continuous fluorescence data collection of 10 acquisitions per °C. PCR data were analyzed using REST (Relative Expression Software Tool) software, which provides statistical information suitable for comparing groups of treated versus untreated samples while taking into account issues of reaction efficiency and reference gene normalization.
Indirect Immunofluorescence—
Normal and OA frozen cartilages were serially sectioned at 4-μm thickness with a cryostat at −30 °C. Slices were washed for 10 min with PBS on glass slides, and mitochondrial staining was performed with 100 μl of 100 nm MitoTracker Green (Invitrogen) for 30 min. The samples were then washed and fixed for 10 min with acetone at 4 °C. The sections were incubated with primary anti-TRAP1 (1:50), anti-SOD2 (1:50) antibodies, or a control antibody for 1 h. After three 10-min washes, a goat anti-mouse phycoerythrin-conjugated antibody (1:20 dilution) was applied for 1 h, and the samples were washed. Finally nuclear staining was performed with 4′,6-dianidino-2-phenylindole dihydrochloride (2 μg/ml) for 30 min at 37 °C. The chambers were stored at 4 °C in darkness until observed by fluorescence microscopy. Quantification of the emitted fluorescence was performed with AnalySIS 5.0 software (Olympus Biosystems, Hamburg, Germany).
Measurement of Reactive Oxygen Species (ROS) Generation in Chondrocytes—
Free radical generation in chondrocytes was assessed by measuring the oxidative conversion of cell-permeable 2′,7′-dichlorofluorescein diacetate to fluorescent dichlorofluorescein (DCF) using fluorescence-activated cell sorting (FACS) (23). Briefly 5 × 105 chondrocytes were seeded in 6-well plates and allowed to reach confluency. Then 2′,7′-dichlorofluorescein diacetate (Fluka, St. Louis, MO) was added to the medium at a final concentration of 10 μm, and cells were incubated for 30 min at 37 °C in the dark. Finally chondrocytes were washed, collected by trypsinization, and suspended in 300 μl of cold PBS. In each sample a minimum of 10,000 cells was subjected to FACS using a FACSCalibur system flow cytometer (BD Biosciences) and CellQuest software (BD Biosciences). Fluorescence intensity was assessed in clusters of several cells identified as regions of interest. The background was identified as an area with minimal cellular fluorescence. Intensity values were reported as the percentage from control values after subtraction of the background.
RESULTS
2-D DIGE Analysis—
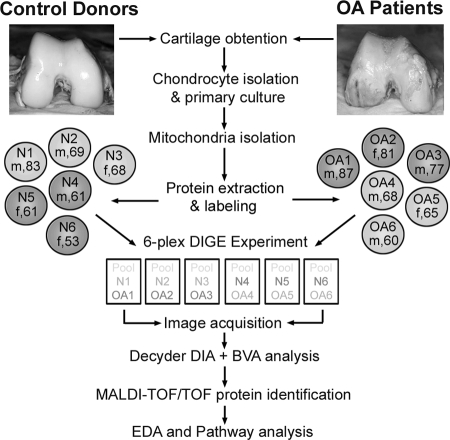
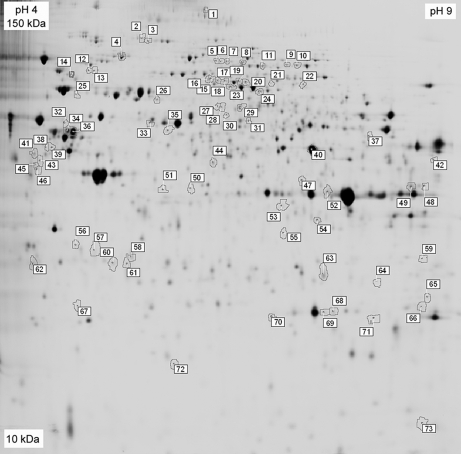
Cartilage obtained from six OA donors and six N controls was used for isolation of chondrocytic mitochondrial proteins and DIGE-MS analysis. Donors were selected according to their clinical diagnoses and similar ages. Fig. 1 describes the experimental procedure followed. Crude mitochondria from chondrocytes were obtained by differential centrifugation according to a protocol set up by our group previously (21) that allowed the obtention of mitochondria-enriched fractions while avoiding high sample losses. After six-plex 2-D DIGE, three individual images were obtained from each gel, corresponding to Cy2-, Cy3-, and Cy5-labeled samples. The 18 gel images were analyzed using DeCyder software. DIA allowed the detection of an average of 2180 protein spots on each image with a 6.98% coefficient of variation between them. Then interimage spot matching was carried out by BVA. In this step, an average of 1691 spots was matched on the gels (17.6% coefficient of variation), and their average abundances among the 18 images of our study were calculated. We considered changes within a 95% confidence interval (p < 0.05) and standardized average spot volume ratios exceeding 1.3 in at least four of the six analyzed gels. This analysis resulted in 28 spots significantly and reproducibly increased in OA and 45 spots that were decreased. These 73 spots are depicted on a DIGE gel image shown in Fig. 2 and listed in Table I.
Fig. 1.
DIGE experimental design. Chondrocytes were obtained from normal (N) and pathological OA knee cartilages and primary cultured. The sex and age of each donor is shown (m, male; f, female). After the first passage, cells were homogenized prior to isolation of mitochondria. Mitochondrial proteins were extracted and labeled with the corresponding Cy dyes. Samples were then mixed and resolved on six independent DIGE gels. Three fluorescence images were obtained from each gel and subjected to image analysis using DeCyder software. After biological variation analysis, spots that exhibited significant quantity modifications in OA were identified by MALDI-TOF/TOF mass spectrometry. Finally extended data analysis (EDA) was carried out to gain further information about protein alterations that are inherent to OA pathology.
Fig. 2.
Representative Cy2-labeled internal standard proteome map indicating the proteins altered in OA. Proteins were resolved in the 3–11 (non-linear) pH range on the first dimension and on 12% acrylamide gels on the second dimension. Proteins that exhibited a significant alteration in expression in OA samples were identified by MALDI-TOF or MALDI-TOF/TOF mass spectrometry and are listed in Table I by the same number as that in the figure.
Table I.
Chondrocytic proteins identified by 2-D DIGE-MS as differentially expressed in osteoarthritis grouped according to their predicted biological function
ECM, extracellular matrix; PM, plasma membrane; Exc, extracellular, secreted; Cyt, cytoplasmic; Nuc, nuclear; ER, endoplasmic reticulum; Mit, mitochondria; Lys, lysosomes; Pex, peroxisomes; Vac, vacuoles; Ves, vesicles; Mem, membrane-associated. The first column (No.) gives the protein spot number according to Fig. 2. TCA, tricarboxylic acid; PI3K, phosphatidylinositol 3-kinase; EGF, epidermal growth factor.
| No. | Protein IDa | Swiss-Prota | Protein name | Av. ratiob | t test | Cellular role | Loc.c | Predictedd
|
Exp.e
|
Scoref | No. pept.g | Cov.h | App.i | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | pI | Molecular weight | pI | ||||||||||||
| % | |||||||||||||||
| Structural and cytoskeleton-related | |||||||||||||||
| 1 | CO1A2 | P08123 | Collagen α-2(I) chain (precursor) | −1.94 | 0.013 | ECM component | Exc | 129.7 | 9.08 | 126.5 | 6.18 | 142 | 20/41 | 24 | 12 |
| 2 | CO6A2 | P12110 | Collagen α-2(VI) chain (precursor) | 1.36 | 0.034 | ECM organization | Exc/Mem | 108.6 | 5.85 | 107.1 | 5.64 | 49 | YGGLHFSDQVEVFSPPGSDRj | 18 | |
| 12 | TGM2 | P21980 | Protein-glutamine γ-glutamyltransferase 2 | 1.44 | 0.011 | Regulation of cell adhesion | ER | 78.4 | 5.11 | 86.2 | 5.15 | 109 | 14/36 | 28 | 18 |
| 13 | ACTB | P60709 | Actin, cytoplasmic 1 | 1.68 | 0.0082 | Cytoskeleton component | Cyt | 42.1 | 5.29 | 83.4 | 5.27 | 181 | 21/29 | 53 | 18 |
| 14 | TGM2 | P21980 | Protein-glutamine γ-glutamyltransferase 2 | 1.66 | 0.028 | Regulation of cell adhesion | ER | 78.4 | 5.11 | 81.1 | 5.02 | 166 | 20/30 | 37 | 12 |
| 18 | EZRI | P15311 | Ezrin (cytovillin) | −1.31 | 0.015 | Cytoskeletal anchoring to PM | Cyt/Mem | 69.0 | 5.94 | 77.3 | 6.34 | 221 | 26/24 | 46 | 18 |
| 19 | EZRI | P15311 | Ezrin (cytovillin) | −1.39 | 0.0051 | Cytoskeletal anchoring to PM | Cyt/Mem | 69.0 | 5.94 | 77.2 | 6.44 | 111 | 12/38 | 20 | 18 |
| 96 | IGFPWSEIRAPDFVFYAPR | ||||||||||||||
| 23 | MOES | P26038 | Moesin | −1.32 | 0.0058 | Cytoskeletal anchoring to PM | Cyt/Mem | 67.8 | 6.08 | 73.8 | 6.38 | 188 | 24/26 | 41 | 18 |
| 26 | PSLT3 | P13797 | Plastin-3 | −1.35 | 0.028 | Actin-bundling protein | Cyt | 70.9 | 5.52 | 67.0 | 5.80 | 86 | 14/36 | 21 | 18 |
| 34 | VIM | P08670 | Vimentin | −1.39 | 0.013 | Cytoskeleton component | Cyt | 53.7 | 5.06 | 54.3 | 5.02 | 194 | 19/31 | 53 | 18 |
| 36 | VIM | P08670 | Vimentin | −1.38 | 0.042 | Cytoskeleton component | Cyt | 53.7 | 5.06 | 53.7 | 5.07 | 258 | 30/20 | 67 | 18 |
| 39 | SCRN1 | Q12765 | Secernin-1 | 1.83 | 0.0025 | Exocytosis regulation | Cyt/Mem | 46.4 | 4.66 | 46.7 | 4.85 | 91 | 10/40 | 42 | 15 |
| 43 | VIM | P08670 | Vimentin | 1.62 | 0.039 | Cytoskeleton component | Cyt | 53.7 | 5.06 | 43.2 | 4.39 | 361 | 32/18 | 65 | 15 |
| 49 | CH3L1 | P36222 | Chitinase-3-like protein 1 (precursor) | −1.63 | 0.0001 | Tissue remodeling | Exc | 43.0 | 8.69 | 40.4 | 8.66 | 100 | 11/39 | 34 | 18 |
| 54 | SDCB1 | O00560 | Syntenin-1 | −1.71 | 0.038 | Cytoskeleton organization, signal transduction | Mem | 32.6 | 7.05 | 34.7 | 7.07 | 96 | 10/40 | 45 | 18 |
| 55 | MYH13 | O95252 | Myosin-13 | 1.59 | 0.024 | Microfilament formation | Cyt | 224.7 | 5.56 | 32.4 | 6.78 | 64 | 13/37 | 9 | 18 |
| 64 | EDIL3 | O43854 | EGF-like repeat and discoidin I-like domain-containing protein 3 (precursor) | −1.78 | 0.0073 | Cell adhesion | Exc | 55.1 | 7.08 | 25.9 | 8.00 | 68 | 12/38 | 20 | 12 |
| Transcription, protein synthesis, and turnover | |||||||||||||||
| 5 | P3H3 | Q8IVL6 | Prolyl 3-hydroxylase 3 (precursor) | 1.46 | 0.0098 | Collagen synthesis | ER | 82.6 | 5.93 | 89.6 | 6.24 | 145 | 16/34 | 34 | 15 |
| 6 | P3H3 | Q8IVL6 | Prolyl 3-hydroxylase 3 (precursor) | 1.37 | 0.0053 | Collagen synthesis | ER | 82.6 | 5.93 | 89.9 | 6.31 | 182 | 20/30 | 38 | 18 |
| 7 | P3H3 | Q8IVL6 | Prolyl 3-hydroxylase 3 (precursor) | 1.37 | 0.04 | Collagen synthesis | ER | 82.6 | 5.93 | 89.9 | 6.35 | 124 | 13/37 | 29 | 18 |
| 9 | EF2 | P13639 | Elongation factor 2 | −1.76 | 0.039 | Protein synthesis | Cyt | 96.2 | 6.41 | 87.3 | 6.80 | 205 | 26/24 | 33 | 15 |
| 10 | EF2 | P13639 | Elongation factor 2 | −1.35 | 0.031 | Protein synthesis | Cyt | 96.2 | 6.41 | 87.1 | 6.88 | 138 | 19/31 | 31 | 18 |
| 11 | IREB1 | P21399 | Iron-responsive element-binding protein 1 | −1.36 | 0.014 | Regulates ferritin transcription | Cyt | 98.9 | 6.23 | 87.1 | 6.62 | 193 | 23/27 | 31 | 18 |
| 11 | PLOD2 | O00469 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 (precursor) | −1.36 | 0.014 | Collagen synthesis | ER/Mem | 85.4 | 6.24 | 87.1 | 6.62 | 128 | 15/35 | 29 | 18 |
| 22 | DDX3X | O00571 | ATP-dependent RNA helicase DDX3X | −1.44 | 0.0033 | Transcription | Nuc/Cyt | 73.6 | 6.73 | 73.6 | 6.95 | 70 | 12/38 | 22 | 18 |
| 58 | CATD | P07339 | Cathepsin D (precursor) | −1.31 | 0.0079 | Acid protease | Lys | 45.0 | 6.10 | 26.9 | 5.55 | 177 | 19/31 | 42 | 18 |
| 66 | PSB1 | P20618 | Proteasome subunit β type 1 (precursor) | −1.43 | 0.015 | Proteolysis | Nuc/Cyt | 26.7 | 8.27 | 23.0 | 8.76 | 108 | 11/39 | 56 | 18 |
| % | |||||||||||||||
| Transport | |||||||||||||||
| 3 | AT1A3 | P13637 | Sodium/potassium-transporting ATPase subunit α-3 | 1.34 | 0.042 | Na+/K+ exchange | Mem | 113.1 | 5.22 | 106.9 | 5.67 | 60 | 7/43 | 9 | 18 |
| 22 | SC23A | Q15436 | Protein transport protein Sec23A | −1.44 | 0.0033 | ER to Golgi protein transport | ER/Mem | 87.0 | 6.64 | 73.6 | 6.95 | 71 | 11/39 | 23 | 18 |
| 33 | VATB2 | P21281 | Vacuolar ATP synthase subunit B, brain isoform | 1.38 | 0.021 | Proton transport | Vac | 56.8 | 5.57 | 56.0 | 5.80 | 181 | 20/30 | 45 | 18 |
| 35 | VATB2 | P21281 | Vacuolar ATP synthase subunit B, brain isoform | −1.3 | 0.0024 | Proton transport | Vac | 56.8 | 5.57 | 53.9 | 5.89 | 123 | 15/35 | 37 | 18 |
| 47 | VDAC2 | P45880 | Voltage-dependent anion-selective channel protein 2 | −1.36 | 0.049 | Anion channel | Mit | 38.6 | 6.32 | 41.2 | 6.91 | 92 | 10/40 | 40 | 18 |
| 56 | CLIC1 | O00299 | Chloride intracellular channel protein 1 | −1.5 | 0.023 | Chloride ion channel | Nuc/Mem | 27.2 | 5.09 | 30.9 | 5.10 | 260 | 20/30 | 86 | 18 |
| 57 | CLIC1 | O00299 | Chloride intracellular channel protein 1 | −1.55 | 0.011 | Chloride ion channel | Nuc/Mem | 27.2 | 5.09 | 29.6 | 5.35 | 96 | 9/41 | 50 | 18 |
| 61 | CLIC4 | Q9Y696 | Chloride intracellular channel protein 4 | −1.45 | 0.0043 | Chloride ion channel | Cyt/Ves | 29.0 | 5.45 | 26.6 | 5.55 | 90 | 10/40 | 50 | 15 |
| 72 | FRIL | P02792 | Ferritin light chain | −1.59 | 0.047 | Cellular iron homeostasis | Cyt | 20.1 | 5.51 | 13.8 | 5.95 | 70 | 6/44 | 41 | 18 |
| Signal transduction | |||||||||||||||
| 4 | MVP | Q14764 | Major vault protein | −1.43 | 0.039 | Nucleocytoplasmic transport | Nuc/Cyt | 99.6 | 7.34 | 92.4 | 5.5 | 265 | 27/23 | 47 | 18 |
| 8 | PCD6I | Q8WUM4 | Programmed cell death 6-interacting protein | −1.32 | 0.0058 | Protein concentration and sorting | Cyt/Vac | 96.6 | 6.13 | 89.1 | 6.47 | 127 | 18/32 | 27 | 15 |
| 71 | MVPVSVQQSLAAYNQRFYNELTEILVR | 15 | |||||||||||||
| 15 | IMMT | Q16891 | Mitochondrial inner membrane protein (mitofilin) | 1.5 | 0.038 | Protein binding | Mit | 83.7 | 6.08 | 77.8 | 6.21 | 278 | 27/23 | 44 | 12 |
| 16 | IMMT | Q16891 | Mitochondrial inner membrane protein (mitofilin) | −1.67 | 0.021 | Protein binding | Mit | 83.7 | 6.08 | 77.7 | 6.16 | 233 | 24/26 | 37 | 18 |
| 17 | IMMT | Q16891 | Mitochondrial inner membrane protein (mitofilin) | −1.39 | 0.0066 | Protein binding | Mit | 83.7 | 6.08 | 77.6 | 6.28 | 263 | 26/24 | 41 | 18 |
| 21 | OPA1 | O60313 | Dynamin-like 120-kDa protein, mitochondrial (precursor) | −1.34 | 0.0038 | Mitochondrial membrane organization, antiapoptotic | Mit | 112.2 | 7.88 | 77.3 | 6.73 | 76 | 15/35 | 16 | 15 |
| 28 | DPYL2 | Q16555 | Dihydropyrimidinase-related protein 2 | −1.37 | 0.015 | Nucleic acid metabolism, signal transduction | Cyt | 62.7 | 5.95 | 63.1 | 6.31 | 328 | 29/21 | 67 | 18 |
| 30 | TRXR1 | Q16881 | Thioredoxin reductase 1, cytoplasmic (precursor) | −1.34 | 0.0012 | Signal transduction | Cyt | 55.0 | 6.07 | 57.7 | 6.36 | 70 | 10/40 | 25 | 18 |
| 32 | 2AAA | P30153 | Serine/threonine-protein phosphatase 2A 65-kDa regulatory subunit A α isoform (PP2A, subunit A) | −1.51 | 0.0082 | Assembly of catalytic and regulatory subunits of PP2A | Cyt | 66.0 | 5.00 | 56.2 | 4.99 | 143 | 17/33 | 32 | 18 |
| 38 | DAPP1 | Q9UN19 | Dual adapter for phosphotyrosine, 3-phosphotyrosine, and 3-phosphoinositide | 1.46 | 0.012 | Signal transduction downstream of PI3K | Cyt/Mem | 32.0 | 7.66 | 46.9 | 4.81 | 73 | 7/43 | 21 | 18 |
| 41 | RCN3 | Q96D15 | Reticulocalbin-3 (precursor) | 1.58 | 0.0076 | Protein binding | ER | 37.5 | 4.74 | 46.4 | 4.56 | 102 | 11/39 | 45 | 18 |
| 45 | RCN3 | Q96D15 | Reticulocalbin-3 (precursor) | 1.89 | 0.026 | Protein binding | ER | 37.5 | 4.74 | 42.1 | 4.56 | 144 | 14/36 | 50 | 18 |
| 46 | RCN3 | Q96D15 | Reticulocalbin-3 (precursor) | 1.64 | 0.018 | Protein binding | ER | 37.5 | 4.74 | 41.3 | 4.58 | 102 | 11/39 | 45 | 18 |
| 62 | 1433T | P27348 | 14-3-3 protein θ | −1.37 | 0.0016 | Signaling pathway regulation | Cyt | 28.0 | 4.68 | 28.4 | 4.61 | 130 | 13/37 | 56 | 18 |
| 63 | RAN | P62826 | GTP-binding nuclear protein Ran | 1.34 | 0.032 | Nucleocytoplasmic transport | Nuc/Cyt | 24.6 | 7.01 | 26.8 | 7.17 | 92 | 9/41 | 47 | 18 |
| 73 | NDKB | P22392 | Nucleoside-diphosphate kinase B | 1.36 | 0.012 | Nucleoside triphosphate synthesis | Nuc/Cyt | 17.4 | 8.52 | 9.5 | 8.62 | 96 | 10/40 | 65 | 18 |
| % | |||||||||||||||
| Metabolism | |||||||||||||||
| 20 | PLCD1 | P51178 | 1-Phosphatidylinositol-4,5-bisphosphate phosphodiesterase δ 1 | −1.39 | 0.005 | Phospholipid metabolism | Cyt/Ves | 86.5 | 6.17 | 76.1 | 6.50 | 109 | 14/36 | 25 | 18 |
| 25 | ECHA | P40939 | Trifunctional enzyme subunit α, mitochondrial (precursor) | −1.4 | 0.036 | Fatty acid β oxidation | Mit | 83.7 | 9.16 | 69.2 | 5.11 | 138 | 19/31 | 26 | 18 |
| 42 | VIIVVKDGPGFYTTR | ||||||||||||||
| 27 | CLUS | P10909 | Clusterin (precursor) (apolipoprotein J) | −1.45 | 0.0049 | Lipid metabolism | Exc | 53.0 | 5.89 | 63.7 | 6.28 | 58 | 8/42 | 19 | 15 |
| 31 | GLSK | O94925 | Glutaminase kidney isoform, mitochondrial (precursor) | 1.41 | 0.035 | Glutamine catabolism | Mit | 56.2 | 7.13 | 57.1 | 6.52 | 73 | 12/38 | 25 | 15 |
| 40 | ENOA | P06733 | α-Enolase | −1.33 | 0.0061 | Glycolysis | Cyt | 47.5 | 7.01 | 47.4 | 6.99 | 276 | 27/23 | 67 | 18 |
| 42 | THIM | P42765 | 3-Ketoacyl-CoA thiolase, mitochondrial | 1.61 | 0.0015 | Lipid and fatty acid metabolism | Mit | 42.4 | 8.32 | 45.8 | 8.85 | 359 | 29/21 | 80 | 18 |
| 47 | HIBCH | Q6NVY1 | 3-Hydroxyisobutyryl-CoA hydrolase, mitochondrial (precursor) | −1.36 | 0.049 | Valine catabolism | Mit | 43.8 | 8.38 | 41.2 | 6.91 | 159 | 20/30 | 43 | 18 |
| 48 | G3P | P04406 | Glyceraldehyde-3-phosphate dehydrogenase | 1.59 | 0.041 | Glycolysis | Cyt | 36.2 | 8.57 | 40.4 | 8.74 | 68 | 9/41 | 34 | 18 |
| 47 | LISWYDNEFGYSNR | ||||||||||||||
| 49 | AK1C1 | Q04828 | Aldo-keto reductase family 1 member C1 | −1.63 | 0.0001 | Cholesterol homeostasis | Cyt | 37.2 | 8.02 | 40.4 | 8.66 | 197 | 18/32 | 69 | 18 |
| 52 | AK1C2 | P52895 | Aldo-keto reductase family 1 member C2 | −1.32 | 0.046 | Prostaglandin and steroid metabolic process | Cyt | 37.1 | 7.13 | 39.4 | 7.22 | 225 | 21/29 | 71 | 18 |
| 53 | ESTD | P10768 | S-Formylglutathione hydrolase (esterase D) | −1.58 | 0.0093 | Formaldehyde detoxification | Cyt/Ves | 32.0 | 6.54 | 37.1 | 6.77 | 156 | 14/36 | 69 | 18 |
| Energy production | |||||||||||||||
| 29 | DHSA | P31040 | Succinate dehydrogenase (ubiquinone) flavoprotein subunit, mitochondrial (precursor) | −1.56 | 0.007 | Electron transport, subunit of complex II | Mit | 73.7 | 7.06 | 63.1 | 6.45 | 276 | 28/22 | 54 | 18 |
| 37 | NDUV1 | P49821 | NADH dehydrogenase (ubiquinone) flavoprotein 1, mitochondrial (precursor) | 1.36 | 0.029 | Electron transport, subunit of complex I | Mit | 51.5 | 8.51 | 52.1 | 7.99 | 326 | 27/23 | 60 | 15 |
| 44 | ODPA | P08559 | Pyruvate dehydrogenase E1 component α subunit, somatic form, mitochondrial (precursor) | −1.52 | 0.017 | Oxidative decarboxylation of pyruvate and TCA cycle | Mit | 44.0 | 8.35 | 44.5 | 6.25 | 151 | 18/32 | 40 | 18 |
| 50 | IDH3A | P50213 | Isocitrate dehydrogenase (NAD) subunit α, mitochondrial (precursor) | −1.69 | 0.015 | Oxidative decarboxylation of pyruvate and TCA cycle | Mit | 40.0 | 6.47 | 40.0 | 6.08 | 193 | 19/31 | 41 | 18 |
| 51 | IDH3A | P50213 | Isocitrate dehydrogenase (NAD) subunit α, mitochondrial (precursor) | −1.9 | 0.038 | Oxidative decarboxylation of pyruvate and TCA cycle | Mit | 40.0 | 6.47 | 40.2 | 5.92 | 117 | 14/36 | 38 | 12 |
| 54 | ETFA | P13804 | Electron transfer flavoprotein subunit α, mitochondrial (precursor) | −1.71 | 0.038 | Electron carrier | Mit | 35.4 | 8.62 | 34.7 | 7.07 | 85 | 9/41 | 36 | 18 |
| 59 | KAD4 | P27144 | Adenylate kinase isoenzyme 4, mitochondrial | 1.52 | 0.014 | ATP production | Mit | 25.4 | 8.47 | 27.4 | 8.80 | 177 | 12/38 | 58 | 15 |
| 60 | IDHP | P48735 | Isocitrate dehydrogenase (NADP), mitochondrial (precursor) | −1.53 | 0.0074 | Oxidative decarboxylation of pyruvate and TCA cycle | Mit | 51.3 | 8.88 | 26.7 | 5.48 | 75 | 11/39 | 27 | 18 |
| 67 | NDUS8 | O00217 | NADH dehydrogenase (ubiquinone) iron-sulfur protein 8, mitochondrial (precursor) | 1.32 | 0.031 | Electron transport, subunit of complex I | Mit | 24.2 | 6.00 | 22.1 | 5.09 | 73 | 7/43 | 41 | 18 |
| 71 | ISOC2 | Q96AB3 | Isochorismatase domain-containing protein 2, mitochondrial (precursor) | 1.48 | 0.03 | Pyruvate metabolism | Mit | 22.6 | 7.67 | 19.5 | 8.03 | 168 | 14/36 | 78 | 12 |
| % | |||||||||||||||
| Chaperones and stress | |||||||||||||||
| 24 | TRAP1 | Q12931 | Tumor necrosis factor type 1 receptor-associated protein (Hsp75) | 1.47 | 0.041 | Chaperone, ROS antagonist | Mit | 80.3 | 8.30 | 71.6 | 6.61 | 124 | 19/31 | 35 | 18 |
| 65 | GSTK1 | Q9Y2Q3 | Glutathione S-transferase Kappa 1 | 1.41 | 0.025 | Glutathione conjugation | Pex | 25.6 | 8.50 | 24.0 | 8.81 | 98 | 10/40 | 45 | 18 |
| 68 | SODM | P04179 | Superoxide dismutase (manganese), mitochondrial (precursor) | −1.52 | 0.011 | Superoxide destruction | Mit | 24.9 | 8.35 | 22.4 | 7.32 | 58 | 6/44 | 31 | 18 |
| 69 | SODM | P04179 | Superoxide dismutase (manganese), mitochondrial (precursor) | −1.56 | 0.0024 | Superoxide destruction | Mit | 24.9 | 8.35 | 22.3 | 7.19 | 114 | 10/40 | 55 | 18 |
| 70 | SODM | P04179 | Superoxide dismutase (manganese), mitochondrial (precursor) | −1.65 | 0.018 | Superoxide destruction | Mit | 24.9 | 8.35 | 21.2 | 6.69 | 77 | 7/43 | 40 | 18 |
Protein identity and accession number according to Swiss-Prot and TrEMBL databases.
Average volume ratio OA/N quantified by DeCyder BVA module.
Subcellular localization according to database and PSORT information.
Predicted molecular weight (×103) and pI according to protein sequence and Swiss-2DPAGE database.
Experimental molecular weight (×103) and pI calculated by analysis of the gel images with PDQuest 7.3.1 software.
Mascot MS protein score, or MS/MS ion score, obtained from MALDI-TOF/TOF spectra. In all cases, a probability score <0.01 was obtained.
Number of peptide masses matching/not matching the top hit from MS-Fit PMF. On each spot, the 50 most intense peaks were launched for search.
Amino acid sequence coverage for the identified proteins.
Number of gel images (from a total of 18) where each spot appears.
Amino acid sequence identified by tandem mass spectrometry using MALDI-TOF/TOF MS/MS.
Identification of Differentially Expressed Proteins—
For identification of proteins, spots were selected, digested in-gel, and analyzed by MALDI-PMF-MS. A Mascot database search using the PMF spectra allowed the identification of the proteins present in 70 of the 73 spots obtained from the gels in duplicate. In five cases (spots 11, 22, 47, 49, and 54), two different proteins were identified on the same spot, and we also found 12 redundancies because of posttranslational modifications or proteolysis. The last three proteins from the group of 73 were identified by tandem MS. Altogether 78 different protein isoforms were identified as altered in OA as listed in Table I with detailed information comprising experimental and theoretical molecular weight and pI values, accession number, and identification parameters. A database search was carried out to analyze the predicted subcellular localization of these proteins and to assign them into different functional groups. Most of the identified proteins (30%) were predicted as mitochondrial, and an additional 36% were predicted as either associated with other membrane structures or located in other subcellular organelles, such as the endoplasmic reticulum, vacuoles, or peroxisomes. Most of these proteins are involved in metabolism and energy production processes (27%), and many are structural or cytoskeleton-related (22%).
Multivariate Statistical Analysis—
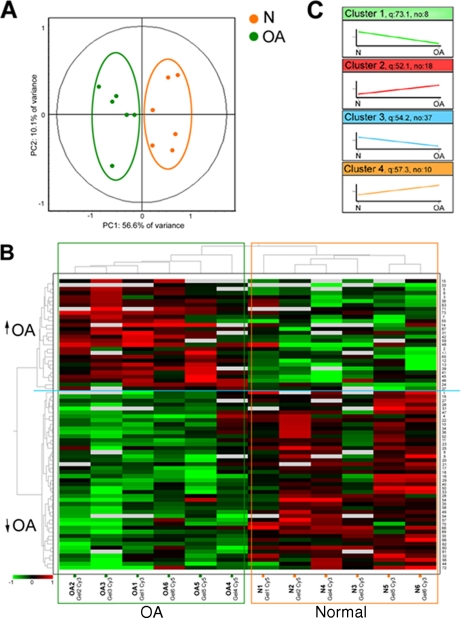
We sought to establish biological significance of the protein changes by performing multivariate statistical tests on the proteins identified by DIGE-MS. As shown in Fig. 3A, PCA indicated how well the OA and N groups are separated. For the 73 proteins altered in the DIGE analysis, the first principal component (PC1) distinguished 56.6% of the variance, and an additional 10.1% variation was distinguished by the second principal component (PC2). Pattern analysis by HC (Fig. 3B) found two different patterns in this group of proteins and clearly differentiated these two diverse profiles, clustering OA (marked with a green square in Fig. 3B) and normal donors (orange square). Protein spot numbering indicated in the heat map correlates with information in Table I. Another type of pattern analysis, the k-means algorithm, was carried out on these 73 protein forms. This analysis clustered them into four groups displaying similar patterns (Fig. 3C). Proteins in each k-means cluster are listed in supplemental Table S1.
Fig. 3.
Unsupervised multivariate analysis of DIGE results. A, principal component analysis clustered the 12 individual Cy3- and Cy5-labeled expression maps into normal (orange) or OA (green) groups differentiated by two principal components that distinguish the variance. B, unsupervised hierarchical clustering of the 12 independent images based on the global expression patterns of the 73 protein spots that were altered in OA and are shown in Table I. Clustering of individual samples is shown on top with the DIGE gel number and type of dye labeling for each sample listed at the bottom. Clustering of individual proteins is shown on the left with relative expression values displayed as an expression matrix (heat map) using a standardized log abundance scale ranging from negative values (green) to positive values (red), and the spot numbers are listed along the right-hand side. These numbers are in agreement with those listed in Table I. C, k-means cluster analysis showing the four different clusters that group the identified proteins. q, cluster quality; no, number of proteins belonging to each cluster. The set of proteins corresponding to each cluster is listed in supplemental Table S1.
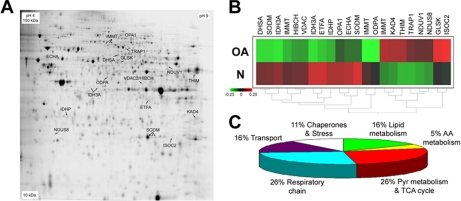
OA-altered Mitochondrial Proteins—
The assignment of predicted subcellular localization to the group of OA-altered proteins led to the identification of 23 protein forms that had been previously characterized as mitochondrial. These proteins are listed in Table II and depicted on the DIGE gel shown in Fig. 4A. An independent HC analysis was carried out on this group of 23 proteins that confirmed the characterization of different mitochondrial protein profiles from the normal and OA samples (Fig. 4B). Most of these proteins (Fig. 4C) are involved with the respiratory chain (26%), carbohydrate (26%) or lipid (16%) metabolism, or mitochondrial transport processes (16%). Other proteins are chaperones or stress-related proteins (11%) or involved in amino acid metabolism (5%).
Table II.
Proteins identified in this work as altered in osteoarthritis that have a previously defined mitochondrial localization
ID, identity; TCA, tricarboxylic acid; E, energy; CH, carbohydrate.
| Ratio | Protein ID | Protein name | Acc. no.a | Biochemical role | Loc.b |
|---|---|---|---|---|---|
| Increased in OA | |||||
| 1.61 | THIM | 3-Ketoacyl-CoA thiolase | P42765 | Lipid metabolism | MIT |
| 1.52 | KAD4 | Adenylate kinase isoenzyme 4, mitochondrial | P27144 | ATP production | MAT |
| 1.50 | IMMT | Mitochondrial inner membrane protein (mitofilin) | Q16891 | Protein binding | MIM |
| 1.48 | ISOC2 | Isochorismatase domain-containing protein 2 | Q96AB3 | Pyruvate metabolism | MIT |
| 1.47 | TRAP1 | TNF receptor-associated protein 1 (Hsp75) | Q12931 | Chaperone, Hsp90 family | MIT |
| 1.41 | GLSK | Glutaminase kidney isoform, mitochondrial precursor | O94925 | Glutamine catabolism | MAT |
| 1.36 | NDUV1 | NADH dehydrogenase (ubiquinone) flavoprotein 1 | P49821 | Complex I subunit, e− to transport chain | MIM |
| 1.32 | NDUS8 | NADH dehydrogenase (ubiquinone) iron-sulfur protein 8, mitochondrial precursor | O00217 | Complex I subunit, e− to ubiquinone | MIM |
| Decreased in OA | |||||
| −1.34 | OPA1 | Dynamin-like 120-kDa protein, mitochondrial | O60313 | Mitochondrial fusion, protection from apoptosis | MIM, IS |
| −1.36 | VDAC2 | Voltage-dependent anion-selective channel protein 2 (outer mitochondrial membrane protein porin 2) | P45880 | Anion transport | MOM |
| −1.36 | HIBCH | 3-Hydroxyisobutyryl-CoA hydrolase, mitochondrial precursor | Q6NVY1 | Valine catabolism | MIT |
| −1.39 | IMMT | Mitochondrial inner membrane protein (mitofilin) | Q16891 | Protein binding | MIM |
| −1.40 | ECHA | Trifunctional enzyme subunit α, mitochondrial (long chain enoyl-CoA hydratase and 3-hydroxyacyl-CoA dehydrogenase) | P40939 | Fatty acid β oxidation | MIT |
| −1.52 | ODPA | Pyruvate dehydrogenase E1 component α subunit | P08559 | TCA cycle | MAT |
| −1.52 | SODM | Manganese-superoxide dismutase | P04179 | Response to ROS stress | MAT |
| −1.53 | IDHP | Isocitrate dehydrogenase (NADP), mitochondrial precursor | P48735 | CH metabolism, E production | MIT |
| −1.56 | SODM | Manganese-superoxide dismutase | P04179 | Response to ROS stress | MAT |
| −1.56 | DHSA | Succinate dehydrogenase (ubiquinone) flavoprotein subunit, mitochondrial precursor | P31040 | Complex II subunit, CH metabolism, TCA cycle | MIM |
| −1.65 | SODM | Manganese-superoxide dismutase | P04179 | Response to ROS stress | MAT |
| −1.67 | IMMT | Mitochondrial inner membrane protein (mitofilin) | Q16891 | Protein binding | MIM |
| −1.69 | IDH3A | Isocitrate dehydrogenase (NAD) subunit α | P50213 | CH metabolism, oxidative decarboxylation of pyruvate, TCA cycle | MIT |
| −1.71 | ETFA | Electron transfer flavoprotein subunit α, mitochondrial precursor | P13804 | Electron transport chain | MAT |
| −1.90 | IDH3A | Isocitrate dehydrogenase (NAD) subunit α | P50213 | CH metabolism, oxidative decarboxylation of pyruvate, TCA cycle | MIT |
Accession number according to Swiss-Prot/TrEMBL database.
Cellular localization according to sequence and PSORT II program. MIT, mitochondrion; MAT, mitochondrial matrix; MIM, mitochondrial inner membrane; MOM, mitochondrial outer membrane; IS, intermembrane space.
Fig. 4.
Mitochondrial proteins altered in OA. A database search and subcellular localization analysis allowed the characterization of a group of 22 protein isoforms that exhibit altered expression in OA and are described to be localized in mitochondria. A, Cy2-labeled internal standard proteome map indicating these mitochondrial proteins. B, unsupervised hierarchical clustering of the group of identified mitochondrial proteins. Data are presented as in Fig. 3B but calculating the mean abundance values on each group of samples (N and OA). See Table II (mitochondrial proteins) for protein designations. C, functional distribution of the mitochondrial proteins altered in OA identified in this work. AA, amino acid; Pyr, pyruvate; TCA, tricarboxylic acid.
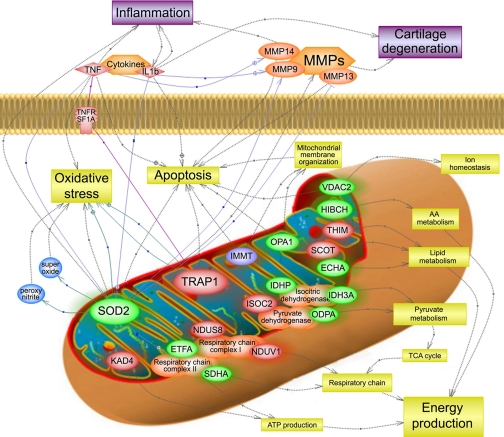
Bioinformatics analysis using Pathway Studio software enabled the characterization of biological association networks related to these differentially expressed mitochondrial proteins. A simplified picture of their interactions is showed in Fig. 5. By this approach, we identified those biochemical pathways that may be altered in osteoarthritic mitochondria. These include production of energy, mitochondrial membrane organization, apoptosis, and oxidative stress response.
Fig. 5.
Pathways and networks associated with mitochondrial chondrocyte proteins identified by DIGE/MS as altered in osteoarthritis. Pathway Studio software was used to map the identified proteins onto characterized human pathways and networks that associate proteins based on known protein-protein interactions, mRNA expression studies, and other biochemical interactions described previously. Abbreviations are shown as in Table II. Increased proteins are shown in red, whereas decreased proteins are depicted in green. IMMT is colored in blue because two isoforms of this protein were decreased, whereas a third was increased. AA, amino acid; TCA, tricarboxylic acid.
Decreased Presence of SOD2 in Osteoarthritic Chondrocytes and Cartilage—
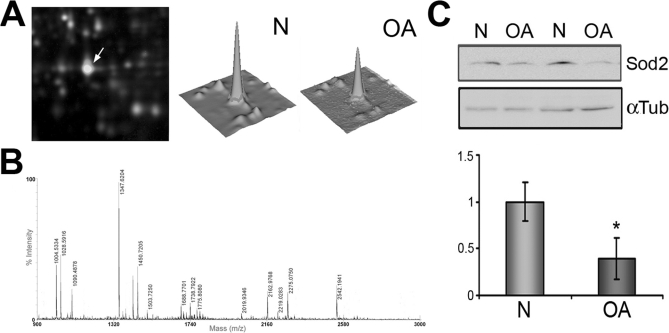
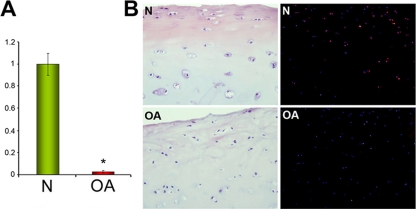
Three protein forms of mitochondrial SOD2 were found to be significantly decreased in OA chondrocytes in our DIGE analysis (one of them is depicted in Fig. 6A) as evidenced after PMF identification (Fig. 6B). Because this protein has been reported to play an essential antioxidant role, being responsible for cellular redox balance maintenance, we sought to confirm its decrease in OA chondrocytes and tissue. First the decrease of SOD2 protein was confirmed on 20 samples from normal and OA chondrocyte protein extracts by Western blot analyses (Fig. 6C). Densitometric analysis revealed a statistically significant (p = 0.043) decrease of 2.56-fold (ratio N/OA = 0.39). To validate these data on cartilage tissue, real time PCR analyses were carried out on 12 RNA samples isolated from age-matched normal (mean age, 54.6 years) and OA (mean age, 58.8 years) cartilages. Results showed a significant (p = 0.031) down-regulation of SOD2 gene expression in OA tissue (Fig. 7A) with a decrease of 34-fold (ratio N/OA = 0.029). Subsequently immunohistofluorescence studies of age-matched N and OA cartilages showed SOD2 (red) to occur mainly in the superficial layer of normal cartilage (Fig. 7B), but this protein was mostly absent in OA tissue.
Fig. 6.
Mitochondrial SOD2 is decreased in OA chondrocytes. A, left, DIGE gel image showing the high abundance of Cy5-labeled protein in spot number 1966; right, three-dimensional view of the fluorescence intensity displayed by this spot where protein from normal chondrocytes is visualized using Cy5 fluorescence and protein from OA chondrocytes is visualized using Cy3 fluorescence. B, tryptic peptide ion spectrum from spot 1966 measured by MALDI-TOF-MS that confirms the identification of SOD2. C, Western blot analysis of SOD2 protein levels in normal and OA chondrocytes showing their decrease in diseased cells. A representative blot is shown along with the numeric data obtained by densitometry analysis of the blots (n = 20). *, p < 0.05. Data are mean values, and error bars indicate standard error of mean.
Fig. 7.
Reduction of SOD2 in OA cartilage. A, decreased values of SOD2 gene expression in OA tissue determined by real time PCR analysis on RNAs extracted from normal and OA cartilages (n = 12). *, p < 0.05. Data are mean values, and error bars indicate standard error of mean. B, histological study on normal and OA cartilages. On the left are shown hematoxylin and eosin stains of the cartilages used for the immunofluorescence study. On the right are indirect immunofluorescence images from frozen N and OA cartilages showing the presence of SOD2 protein primarily localized in the superficial layer of the normal tissue and its significant reduction in OA cartilage. Chondrocyte nuclei (4′,6-dianidino-2-phenylindole dihydrochloride) display a blue color, whereas SOD2 is labeled in red.
Increase of Intracellular ROS Generation in Osteoarthritic Chondrocytes—
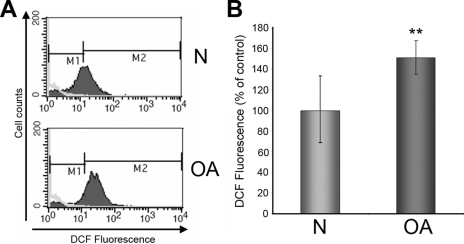
The discovery of the decrease in the important antioxidant protein SOD2 in OA chondrocytes prompted us to investigate possible cellular redox imbalance. We decided to measure the generation of free radicals in cultured N and OA chondrocytes (n = 14) by a flow cytometry probe using DCF as a reporter of ROS production. As shown in Fig. 8, intracellular ROS production in OA chondrocytes increased at an average of 50% when compared with normal cells (n = 14, p = 0.01). These data provide evidence of an oxidative stress dysregulation occurring in osteoarthritic cells.
Fig. 8.
Increase of intracellular ROS production in OA chondrocytes. A, representative flow cytometry graphics of DCF fluorescence emitted by normal (control) and OA chondrocytes that is directly related with intracellular free radical generation levels. B, summary of DCF fluorescence intensities (percent normalized to control) obtained from seven samples of OA chondrocytes and seven normal controls. **, p < 0.01. Error bars indicate standard error of mean. A 50% increase in DCF fluorescence was observed in OA cells.
Increased Abundance of TRAP1 in Osteoarthritic Cartilage—
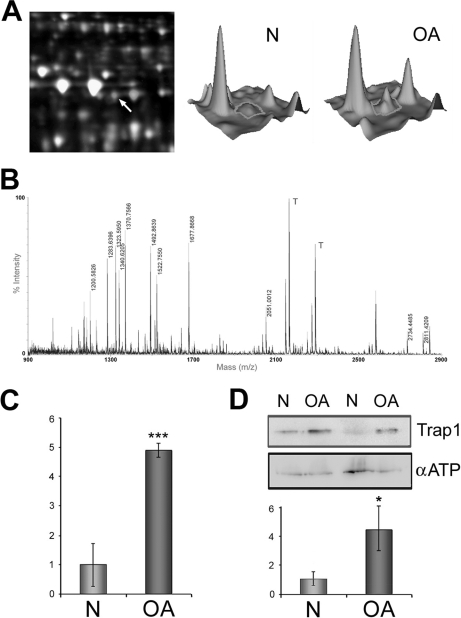
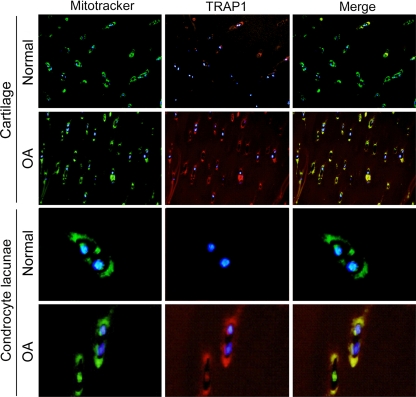
One of the mitochondrial proteins showing an increase in abundance in OA chondrocytes is TRAP1. This protein is the mitochondrial member of the Hsp90 family of protein chaperones and has been reported to antagonize ROS as an attempt to protect cells from oxidative stress-induced apoptosis. Therefore, an increased presence of TRAP1 might also reflect a cellular state of high oxidative stress. Fig. 9A depicts the TRAP1 spot in the DIGE gels and its intensity on normal and OA samples. Unambiguous identification of this protein was carried out by PMF (Fig. 9B). We confirmed this increase at the transcription level by real time PCR analyses on 14 samples from normal and OA freshly isolated chondrocytes. The results showed a significant (p = 0.002) up-regulation of TRAP1 gene expression in osteoarthritic cells with a ratio of expression of 4.89-fold (Fig. 9C). The increased presence of TRAP1 protein in chondrocyte mitochondria was confirmed by Western blot analyses on 20 samples from normal and OA chondrocyte mitochondria-enriched protein extracts (Fig. 9D). Densitometric analysis of the blots showed a significant (p = 0.037) increase of 4.02-fold of TRAP1 in OA chondrocytes. Subsequently immunohistofluorescence assays were performed to evaluate this alteration in cartilage tissue. As shown in Fig. 10, a low level of TRAP1 (red) can be detected in normal cartilage, whereas cartilage mitochondria are easily recognized by MitoTracker green labeling. In OA cartilage, however, a high level of TRAP1 can be noticed. Overlapping images of fluorescence from MitoTracker (green) and TRAP1 (red) establish the mitochondrial localization of TRAP1.
Fig. 9.
The mitochondrial TRAP1 is increased in OA chondrocytes. A, left, DIGE gel image showing the high abundance of Cy5-labeled protein in spot number 606; right, three-dimensional view of the fluorescence intensity displayed by this spot where protein from normal chondrocytes is visualized using Cy3 fluorescence and protein from osteoarthritic chondrocytes is visualized using Cy5 fluorescence. B, tryptic peptide ion spectrum from spot 606 measured by MALDI-TOF-MS that confirms the identification of TRAP1. T, trypsin peaks. C, overexpression values of TRAP1 determined by real time PCR analysis on freshly isolated cells from normal and OA cartilages (n = 14). ***, p < 0.005. D, Western blot analysis of TRAP1 protein levels in normal and OA chondrocyte mitochondria. A representative blot is shown along with the numeric data obtained by densitometry analysis of the blots (n = 20). *, p < 0.05. Error bars indicate standard error of mean.
Fig. 10.
High abundance of TRAP1 in OA cartilage. Representative indirect immunofluorescence of TRAP1 (red) and simultaneous mitochondria staining (green) on N and OA cartilages (n = 6) shows the increased abundance of TRAP1 in OA tissue and its colocalization with mitochondria. In the bottom rows, a magnification of OA chondrocyte lacunae is shown to depict the mitochondrial distribution of TRAP1.
DISCUSSION
OA is the most frequently occurring arthropathy, and its prevalence increases with age. Because specific mitochondrial alterations had been described previously in OA chondrocytes, we extracted cartilage cells from age-matched donors to carry out a proteomics approach with the aim of identifying possible mitochondrial pathway alterations in the disease. The 2-D DIGE-MS approach enabled a higher confidence differential protein profile than that obtained by classical 2-DE strategies. At the conclusion of the present study, we had identified 73 protein forms with altered abundance or posttranslational modifications in mitochondria-enriched fractions from OA human articular chondrocytes compared with normal. Hierarchical clustering of these differential spots enabled the definition of a protein profile characteristic for OA (Fig. 3B) that is branded by alterations in structural components and proteins related with signal transduction or metabolism and by a decrease in transport proteins (Table I). All these changes should be studied more deeply as they might be useful for the development of new early diagnosis tools for OA.
Of the identified proteins, 30% were predicted to be mitochondrial after database searches. An additional 21% of the proteins were located in other subcellular organelles such as endoplasmic reticulum, vacuoles, or peroxisomes, a common characteristic of differential centrifugation enrichment processes (24, 25). Another 15% of identified proteins were membrane-associated proteins. Many reported intracellular localizations of these proteins are exclusively based on sequence information, lacking any further assays. Therefore, some of the proteins classified as non-mitochondrial may either be associated to this organelle or have a transitional mitochondrial localization. Some of the identified proteins were characterized previously in an analysis on N and OA whole chondrocyte extracts (13). Interestingly the reported increase in OA chondrocytes of an extracellular structural protein (collagen type IV) and decrease of a cytoplasmic transport protein (chloride intracellular channel protein 1) were confirmed by the present study.
We focused the analysis on the proteins previously characterized as mitochondrial to identify functions that are distorted in OA chondrocytes (Table II). Pathway Studio software was used to identify biological association and interaction networks of the altered mitochondrial proteins, which are shown in Fig. 5. Most of these proteins (73%) are involved in metabolism and energy production (Fig. 4C), either participating in lipid, amino acid, or carbohydrate metabolism or as essential components of MRC. These data demonstrate the mitochondrial dysregulation of energy production that takes place in OA that was first described by our group from the detection of decreased activity of complexes II and III of MRC (8). Using our proteomics approach we found a decrease in a subunit of complex II, a succinate dehydrogenase flavoprotein, which may play a role in this complex II dysfunction. The increased abundance of two subunits of complex I, NDUV1 and NDUS8, may also affect MRC efficiency.
Mitochondrial membrane organization is essential for maintaining correct mitochondrial function. The inner membrane protein mitofilin (IMMT) has recently been reported to control cristae morphology (26) to facilitate proper mitochondrial function. In our proteomics screening, we found two mitofilin isoforms decreased in OA, whereas a third was increased. These data suggest an OA-dependent alteration of mitofilin posttranslational processing that should be further studied. Another protein involved in cristae remodeling, OPA1, was shown in this study to be altered in OA. OPA1 is a dynamin-like GTPase that was first identified as a cause for optic atrophy (27, 28). Subsequently a role for OPA1 as a major organizer of the mitochondrial inner membrane was reported, suggesting its involvement in cytochrome c sequestration (29). An essential participation in mitochondria fusion processes was described for OPA1 (30), although it was reported to happen independently from the control of apoptotic cristae remodeling (31). Most recently, the necessary role of OPA1 in mitochondrial dynamics has been extensively discussed (32, 33). Altogether these data support a hypothesis for altered mitochondrial function in OA chondrocytes in which dysregulation of cristae remodeling stemming from defects in IMMT and OPA1 contribute to increased apoptosis levels in OA chondrocytes.
Finally we focused on two mitochondrial proteins whose predicted role is related to oxidative stress. Mitochondrial dysregulation and oxidative stress balance have been reported to play an essential role in OA development and progression (34, 35). Oxidation-related cartilage changes during the aging process have been studied, highlighting a possible relationship between aging, chronic inflammation, and cartilage degradation in OA (36). The activity of ROS, essential for cartilage degradation, is balanced by enzymatic and non-enzymatic antioxidants, which act by inhibiting oxidative enzymes or scavenging free radicals. Modifications of the antioxidative system in OA remain unknown at this time, and little information is available on the antioxidative status of chondrocytes (37). Therefore, our finding of the deficiency of a major intracellular antioxidant protein, SOD2, in chondrocyte mitochondria may be a key to understanding OA pathophysiology. In an earlier study (38), lowered antioxidative capacity was observed in the degenerating regions of OA cartilage. These investigators showed that the resulting oxidative stress induced telomere genomic instability, replicative senescence, and dysfunction of chondrocytes, possibly leading to the development and progression of OA. Nevertheless only extracellular SOD (EC-SOD, SOD3) was found to be decreased in OA cartilage extracellular spaces (39) and recently also in OA synovial fluid (40). Therefore, the finding of decreased mitochondrial SOD2 within OA chondrocytes reinforces a hypothesis that inadequate control of ROS, which affects chondrocyte intracellular metabolism, is an essential factor in OA pathophysiology (41). These data are also supported by our finding of higher ROS generation occurring in OA chondrocytes. Moreover a very recent work based on microarray gene expression profiling of osteoarthritic bone finds a 6-fold down-regulation of SOD2 in OA tissue (42). Despite the lack of real time PCR validation of this result, an alteration in the antioxidant system of OA subchondral bone is indicated. Combined with our results on cartilage tissue and cells, these data suggest a global alteration of redox balance in the bone, cartilage, and synovial fluid of the OA joint that could be due to a decrease of antioxidant proteins such as SOD2. Although there is no consistent evidence at this time that an additional antioxidant supply is effective for relief of OA symptoms and prevention of structural damage in OA cartilage (43), our results point to the potential utility of SODs and SOD mimetics as antioxidant treatments in OA.
TRAP1 is a member of the Hsp90 family of molecular chaperones that was first identified by its ability to bind the type 1 tumor necrosis factor receptor (44). Although it is generally homologous to cytoplasmic Hsp90 and both proteins are inhibited by compounds such as geldanamycin, TRAP1 is reported to be located in the mitochondria (45) and to have functional properties different from its homologue (46). Subsequent studies have suggested a role for TRAP1 in protecting cells from apoptosis caused by formation of ROS (47). Several recent reports describe how this protein regulates ROS production, thus defending the cells from oxidative stress-mediated apoptosis (48–50). As mentioned earlier, increases of ROS production and chondrocyte apoptosis are characteristic features of OA pathophysiology (6). Therefore, we more thoroughly investigated differences of TRAP1 abundance in OA chondrocyte mitochondria. Both gene expression and protein abundance analyses confirmed the increase of TRAP1 in OA chondrocytes, whereas fluorescence microscopy assays demonstrated its high amount in mitochondria from OA cartilage cells. Considering the reported ROS antagonist role of TRAP1, we hypothesize that the increased presence of TRAP1 in OA tissue might be a compensatory output displayed by chondrocyte mitochondria to hold out a high oxidative stress environment.
Altogether our work provides some insight into the diverse pathways that are altered in OA mitochondria. Defects in the production of energy, mitochondrial membrane organization, and apoptosis have been pointed out, exemplifying the consequences of mitochondrial dysregulation in cartilage cells during the progression of the disease. The decrease of mitochondrial SOD we found is a particularly relevant finding relative to maintenance of a normal intracellular redox balance in chondrocytes. Extensive studies on all these protein distortions are needed to amplify the study of OA pathogenesis. The goal of these studies will be to identify new diagnostic and therapeutic targets that will ultimately enable development of novel therapeutic strategies for OA.
Supplementary Material
Acknowledgments
We are thankful to Purificación Filgueira for technical assistance in fluorescence microscopy and to Pilar Cal for expert secretarial assistance. We express appreciation to the Pathology Service and to Lourdes Sanjurjo and Dolores Velo from the Orthopaedics Department of the Complejo Hospitalario Universitario A Coruña for providing cartilage samples.
Footnotes
Published, MCP Papers in Press, September 9, 2008, DOI 10.1074/mcp.M800292-MCP200
The abbreviations used are: OA, osteoarthritis; 2-D, two-dimensional; 2-DE, two-dimensional gel electrophoresis; BVA, biological variation analysis; Cy, cyanine; DCF, dichlorofluorescein; DIA, differential in-gel analysis; MRC, mitochondrial respiratory chain; PMF, peptide mass fingerprinting; ROS, reactive oxygen species; SOD2, manganese-superoxide dismutase; TNFα, tumor necrosis factor-α; TRAP1, TNFα receptor-associated protein 1; N, normal; PCA, principal component analysis; HC, hierarchical clustering; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PBGD, porphobilinogen deaminase; FACS, fluorescence-activated cell sorting; IMMT, inner membrane protein mitofilin; SOD, superoxide dismutase.
This work was supported in part by grants from the Ministerio de Educación y Ciencia, Spain (Grant SAF2005-06211), Fondo Investigación Sanitaria, Spain (Grant CIBER-CB06/01/0040), and Secretaria I+D+i (Xunta de Galicia) (Grant PGIDIT06PXIB916358PR). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Green, D. R., and Reed, J. C. ( 1998) Mitochondria and apoptosis. Science 281, 1309–1312 [DOI] [PubMed] [Google Scholar]
- 2.Finkel, T., and Holbrook, N. J. ( 2000) Oxidants, oxidative stress and the biology of ageing. Nature 408, 239–247 [DOI] [PubMed] [Google Scholar]
- 3.Blanco, F. J., Lopez-Armada, M. J., and Maneiro, E. ( 2004) Mitochondrial dysfunction in osteoarthritis. Mitochondrion 4, 715–728 [DOI] [PubMed] [Google Scholar]
- 4.Heinegard, D., Bayliss, M., and Lorenzo, P. ( 1998) Biochemistry and metabolism of normal and osteoarthritic cartilage, in Osteoarthritis (Brandt, K. D., Doherty, M., and Lohmander, L. S., eds) pp. 74–84, Oxford University Press, New York
- 5.Pritzker, K. ( 1998) Pathology of osteoarthritis, in Osteoarthritis (Brandt, K. D., Doherty, M., and Lohmander, L. S., eds) pp. 50–61, Oxford University Press, New York
- 6.Kim, H. A., and Blanco, F. J. ( 2007) Cell death and apoptosis in osteoarthritic cartilage. Curr. Drug Targets 8, 333–345 [DOI] [PubMed] [Google Scholar]
- 7.Blanco, F., López-Armada, M., and Rego, I. ( 2007) Mitochondria and chondrocytes: role in osteoarthritis, in Osteoarthritis, Inflammation and Degradation: A Continuum (Buckwalter, J. A., Lotz, M., and Stoltz, J.-F., eds) pp. 192–205, IOS Press, Amsterdam
- 8.Maneiro, E., Martin, M. A., de Andres, M. C., Lopez-Armada, M. J., Fernandez-Sueiro, J. L., del Hoyo, P., Galdo, F., Arenas, J., and Blanco, F. J. ( 2003) Mitochondrial respiratory activity is altered in osteoarthritic human articular chondrocytes. Arthritis Rheum. 48, 700–708 [DOI] [PubMed] [Google Scholar]
- 9.Johnson, K., Jung, A., Murphy, A., Andreyev, A., Dykens, J., and Terkeltaub, R. ( 2000) Mitochondrial oxidative phosphorylation is a downstream regulator of nitric oxide effects on chondrocyte matrix synthesis and mineralization. Arthritis Rheum. 43, 1560–1570 [DOI] [PubMed] [Google Scholar]
- 10.Tomita, M., Sato, E. F., Nishikawa, M., Yamano, Y., and Inoue, M. ( 2001) Nitric oxide regulates mitochondrial respiration and functions of articular chondrocytes. Arthritis Rheum. 44, 96–104 [DOI] [PubMed] [Google Scholar]
- 11.Carlo, M. D., Jr., and Loeser, R. F. ( 2003) Increased oxidative stress with aging reduces chondrocyte survival: correlation with intracellular glutathione levels. Arthritis Rheum. 48, 3419–3430 [DOI] [PubMed] [Google Scholar]
- 12.Ruiz-Romero, C., Lopez-Armada, M. J., and Blanco, F. J. ( 2005) Proteomic characterization of human normal articular chondrocytes: a novel tool for the study of osteoarthritis and other rheumatic diseases. Proteomics 5, 3048–3059 [DOI] [PubMed] [Google Scholar]
- 13.Ruiz-Romero, C., Carreira, V., Rego, I., Remeseiro, S., Lopez-Armada, M. J., and Blanco, F. J. ( 2008) Proteomic analysis of human osteoarthritic chondrocytes reveals protein changes in stress and glycolysis. Proteomics 8, 495–507 [DOI] [PubMed] [Google Scholar]
- 14.Gygi, S. P., Rist, B., Gerber, S. A., Turecek, F., Gelb, M. H., and Aebersold, R. ( 1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 15.Ross, P. L., Huang, Y. N., Marchese, J. N., Williamson, B., Parker, K., Hattan, S., Khainovski, N., Pillai, S., Dey, S., Daniels, S., Purkayastha, S., Juhasz, P., Martin, S., Bartlet-Jones, M., He, F., Jacobson, A., and Pappin, D. J. ( 2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 16.Unlu, M., Morgan, M. E., and Minden, J. S. ( 1997) Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 18, 2071–2077 [DOI] [PubMed] [Google Scholar]
- 17.Tonge, R., Shaw, J., Middleton, B., Rowlinson, R., Rayner, S., Young, J., Pognan, F., Hawkins, E., Currie, I., and Davison, M. ( 2001) Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics 1, 377–396 [DOI] [PubMed] [Google Scholar]
- 18.Lilley, K. S., and Friedman, D. B. ( 2004) All about DIGE: quantification technology for differential-display 2D-gel proteomics. Expert Rev. Proteomics 1, 401–409 [DOI] [PubMed] [Google Scholar]
- 19.Marouga, R., David, S., and Hawkins, E. ( 2005) The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal. Bioanal. Chem. 382, 669–678 [DOI] [PubMed] [Google Scholar]
- 20.Kolkman, A., Dirksen, E. H., Slijper, M., and Heck, A. J. ( 2005) Double standards in quantitative proteomics: direct comparative assessment of difference in gel electrophoresis and metabolic stable isotope labeling. Mol. Cell. Proteomics 4, 255–266 [DOI] [PubMed] [Google Scholar]
- 21.Ruiz-Romero, C., Lopez-Armada, M. J., and Blanco, F. J. ( 2006) Mitochondrial proteomic characterization of human normal articular chondrocytes. Osteoarthritis Cartilage 14, 507–518 [DOI] [PubMed] [Google Scholar]
- 22.Sechi, S., and Chait, B. T. ( 1998) Modification of cysteine residues by alkylation. A tool in peptide mapping and protein identification. Anal. Chem. 70, 5150–5158 [DOI] [PubMed] [Google Scholar]
- 23.Mancini, M., Sedghinasab, M., Knowlton, K., Tam, A., Hockenbery, D., and Anderson, B. O. ( 1998) Flow cytometric measurement of mitochondrial mass and function: a novel method for assessing chemoresistance. Ann. Surg. Oncol. 5, 287–295 [DOI] [PubMed] [Google Scholar]
- 24.Brunet, S., Thibault, P., Gagnon, E., Kearney, P., Bergeron, J. J., and Desjardins, M. ( 2003) Organelle proteomics: looking at less to see more. Trends Cell Biol. 13, 629–638 [DOI] [PubMed] [Google Scholar]
- 25.Andersen, J. S., and Mann, M. ( 2006) Organellar proteomics: turning inventories into insights. EMBO Rep. 7, 874–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.John, G. B., Shang, Y., Li, L., Renken, C., Mannella, C. A., Selker, J. M., Rangell, L., Bennett, M. J., and Zha, J. ( 2005) The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell 16, 1543–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eiberg, H., Kjer, B., Kjer, P., and Rosenberg, T. ( 1994) Dominant optic atrophy (OPA1) mapped to chromosome 3q region. I. Linkage analysis. Hum. Mol. Genet. 3, 977–980 [DOI] [PubMed] [Google Scholar]
- 28.Delettre, C., Lenaers, G., Griffoin, J. M., Gigarel, N., Lorenzo, C., Belenguer, P., Pelloquin, L., Grosgeorge, J., Turc-Carel, C., Perret, E., Astarie-Dequeker, C., Lasquellec, L., Arnaud, B., Ducommun, B., Kaplan, J., and Hamel, C. P. ( 2000) Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26, 207–210 [DOI] [PubMed] [Google Scholar]
- 29.Olichon, A., Baricault, L., Gas, N., Guillou, E., Valette, A., Belenguer, P., and Lenaers, G. ( 2003) Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 278, 7743–7746 [DOI] [PubMed] [Google Scholar]
- 30.Arnoult, D., Grodet, A., Lee, Y. J., Estaquier, J., and Blackstone, C. ( 2005) Release of OPA1 during apoptosis participates in the rapid and complete release of cytochrome c and subsequent mitochondrial fragmentation. J. Biol. Chem. 280, 35742–35750 [DOI] [PubMed] [Google Scholar]
- 31.Frezza, C., Cipolat, S., Martins de Brito, O., Micaroni, M., Beznoussenko, G. V., Rudka, T., Bartoli, D., Polishuck, R. S., Danial, N. N., De Strooper, B., and Scorrano, L. ( 2006) OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126, 177–189 [DOI] [PubMed] [Google Scholar]
- 32.Merkwirth, C., Dargazanli, S., Tatsuta, T., Geimer, S., Lower, B., Wunderlich, F. T., von Kleist-Retzow, J. C., Waisman, A., Westermann, B., and Langer, T. ( 2008) Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 22, 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, Y., Ouyang, Y., Yang, L., Beal, M. F., McQuibban, A., Vogel, H., and Lu, B. ( 2008) Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl. Acad. Sci. U. S. A 105, 7070–7075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson, K., Svensson, C. I., Van Etten, D., Ghosh, S. S., Murphy, A. N., Powell, H. C., and Terkeltaub, R. ( 2004) Mediation of spontaneous knee osteoarthritis by progressive chondrocyte ATP depletion in Hartley guinea pigs. Arthritis Rheum. 50, 1216–1225 [DOI] [PubMed] [Google Scholar]
- 35.Henrotin, Y. E., Bruckner, P., and Pujol, J. P. ( 2003) The role of reactive oxygen species in homeostasis and degradation of cartilage. Osteoarthritis Cartilage 11, 747–755 [DOI] [PubMed] [Google Scholar]
- 36.Henrotin, Y., Blanco, F., Aigner, T., and Bodo, K. ( 2007) The significance of oxidative stress in articular cartilage ageing and degradation. Curr. Rheumatol. Rev. 3, 261–274 [Google Scholar]
- 37.Henrotin, Y., Kurz, B., and Aigner, T. ( 2005) Oxygen and reactive oxygen species in cartilage degradation: friends or foes? Osteoarthritis Cartilage 13, 643–654 [DOI] [PubMed] [Google Scholar]
- 38.Yudoh, K., Nguyen, T., Nakamura, H., Hongo-Masuko, K., Kato, T., and Nishioka, K. ( 2005) Potential involvement of oxidative stress in cartilage senescence and development of osteoarthritis: oxidative stress induces chondrocyte telomere instability and downregulation of chondrocyte function. Arthritis Res. Ther. 7, R380–R391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Regan, E., Flannelly, J., Bowler, R., Tran, K., Nicks, M., Carbone, B. D., Glueck, D., Heijnen, H., Mason, R., and Crapo, J. ( 2005) Extracellular superoxide dismutase and oxidant damage in osteoarthritis. Arthritis Rheum. 52, 3479–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Regan, E. A., Bowler, R. P., and Crapo, J. D. ( 2008) Joint fluid antioxidants are decreased in osteoarthritic joints compared to joints with macroscopically intact cartilage and subacute injury. Osteoarthritis Cartilage 16, 515–521 [DOI] [PubMed] [Google Scholar]
- 41.Afonso, V., Champy, R., Mitrovic, D., Collin, P., and Lomri, A. ( 2007) Reactive oxygen species and superoxide dismutases: role in joint diseases. Joint Bone Spine 74, 324–329 [DOI] [PubMed] [Google Scholar]
- 42.Hopwood, B., Tsykin, A., Findlay, D. M., and Fazzalari, N. L. ( 2007) Microarray gene expression profiling of osteoarthritic bone suggests altered bone remodelling, WNT and transforming growth factor-beta/bone morphogenic protein signalling. Arthritis Res. Ther. 9, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henrotin, Y., and Kurz, B. ( 2007) Antioxidant to treat osteoarthritis: dream or reality? Curr. Drug Targets 8, 347–357 [DOI] [PubMed] [Google Scholar]
- 44.Song, H. Y., Dunbar, J. D., Zhang, Y. X., Guo, D., and Donner, D. B. ( 1995) Identification of a protein with homology to hsp90 that binds the type 1 tumor necrosis factor receptor. J. Biol. Chem. 270, 3574–3581 [PubMed] [Google Scholar]
- 45.Cechetto, J. D., and Gupta, R. S. ( 2000) Immunoelectron microscopy provides evidence that tumor necrosis factor receptor-associated protein 1 (TRAP-1) is a mitochondrial protein which also localizes at specific extramitochondrial sites. Exp. Cell Res. 260, 30–39 [DOI] [PubMed] [Google Scholar]
- 46.Felts, S. J., Owen, B. A., Nguyen, P., Trepel, J., Donner, D. B., and Toft, D. O. ( 2000) The hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J. Biol. Chem. 275, 3305–3312 [DOI] [PubMed] [Google Scholar]
- 47.Masuda, Y., Shima, G., Aiuchi, T., Horie, M., Hori, K., Nakajo, S., Kajimoto, S., Shibayama-Imazu, T., and Nakaya, K. ( 2004) Involvement of tumor necrosis factor receptor-associated protein 1 (TRAP1) in apoptosis induced by β-hydroxyisovalerylshikonin. J. Biol. Chem. 279, 42503–42515 [DOI] [PubMed] [Google Scholar]
- 48.Im, C. N., Lee, J. S., Zheng, Y., and Seo, J. S. ( 2007) Iron chelation study in a normal human hepatocyte cell line suggests that tumor necrosis factor receptor-associated protein 1 (TRAP1) regulates production of reactive oxygen species. J. Cell. Biochem. 100, 474–486 [DOI] [PubMed] [Google Scholar]
- 49.Hua, G., Zhang, Q., and Fan, Z. ( 2007) Heat shock protein 75 (TRAP1) antagonizes reactive oxygen species generation and protects cells from granzyme M-mediated apoptosis. J. Biol. Chem. 282, 20553–20560 [DOI] [PubMed] [Google Scholar]
- 50.Montesano Gesualdi, N., Chirico, G., Pirozzi, G., Costantino, E., Landriscina, M., and Esposito, F. ( 2007) Tumor necrosis factor-associated protein 1 (TRAP-1) protects cells from oxidative stress and apoptosis. Stress 10, 342–350 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.