Summary
SH3 domains are modules of 50-70 amino acids that promote interactions among proteins, often participating in the assembly of large dynamic complexes. These domains bind to peptide ligands, which usually contain a core Pro-X-X-Pro (PXXP) sequence. Here we identify a class of SH3 domains that binds to ubiquitin. The yeast endocytic protein Sla1, as well as the mammalian proteins CIN85 and amphiphysin, carry ubiquitin-binding SH3 domains. Ubiquitin and peptide ligands bind to the same hydrophobic groove on the SH3 domain surface, and ubiquitin and a PXXP-containing protein fragment compete for binding to SH3 domains. We conclude that a subset of SH3 domains constitutes a distinct type of ubiquitin-binding domain, and that ubiquitin-binding can negatively regulate interaction of SH3 domains with canonical proline-rich ligands.
Introduction
Monoubiquitin and polyubiquitin chains attached to proteins are important regulatory signals in a variety of basic cellular processes (for example, Salghetti et al., 2001; Hoege et al., 2002; Di Fiore et al., 2003; Hicke and Dunn, 2003; Staub and Rotin, 2006). Recently, proteins that are likely to be downstream effectors that recognize and transmit information conferred by ubiquitin signals have been discovered. These proteins generally have small (20–150 amino acid), independently folded ubiquitin-binding domains (UBDs) that can interact directly with monoubiquitin or polyubiquitin chains (reviewed in Hicke et al., 2005). The UBDs characterized to date are structurally disparate and generally bind ubiquitin with low affinity (Kd – 2-500 μM). Many, but not all, UBDs bind to a region on the ubiquitin surface around Ile44. UBDs are found in combination with a large assortment of other modular domains and in proteins of diverse function.
Ubiquitin plays several important roles in the internalization of cell surface proteins into the endocytic pathway (reviewed in Haglund et al., 2003; Hicke and Dunn, 2003). First, ubiquitin is a regulated internalization signal that can be appended to plasma membrane proteins to trigger internalization. Second, ubiquitination regulates the endocytic machinery itself. Key endocytic proteins that are ubiquitinated, primarily monoubiquitinated, include epsins, Eps15, endophilin and CIN85 (Cbl-interacting protein of 85 kDa). In addition to being monoubiquitinated, epsins and Eps15 carry a well-characterized UBD, the ubiquitin-interacting motif (UIM). The epsin UIMs are important for rapid receptor internalization (Shih et al., 2002; Sigismund et al., 2005) and one function of epsin UIMs may be to promote protein-protein interactions among the internalization machinery (M. Dores, J. Schnell and L. Hicke, in preparation).
Numerous protein-protein interactions and protein-protein interaction domains are required for efficient receptor internalization. One model for the action of the internalization machinery is that endocytic proteins combine, not necessarily in a linear sequence, at the cytoplasmic face of the plasma membrane into different complexes that together form a dynamic network to recruit cargo and deform the membrane into a nascent vesicle (for example, Praefcke et al., 2004). Examples of motifs that promote interactions in these complexes are EH (Eps15 homology) domains and LLDLD clathrin-binding sequences. Because epsin UIMs probably also function in formation of endocytic complexes and because multiple endocytic proteins are modified with ubiquitin, we hypothesized that UBD-ubiquitin interactions might generally function in internalization to control assembly and disassembly of the endocytic machinery.
To identify other endocytic proteins that participate in ubiquitin-mediated interactions we screened for yeast proteins in the endocytic machinery that bind to monoubiquitin. In this screen we identified a previously unknown ubiquitin-binding domain in Sla1, the yeast CIN85 homologue. Surprisingly, this unusual UBD is a variant SH3 that binds to ubiquitin and is implicated in the regulation of protein-protein interactions.
Results
Sla1 binds directly to ubiquitin
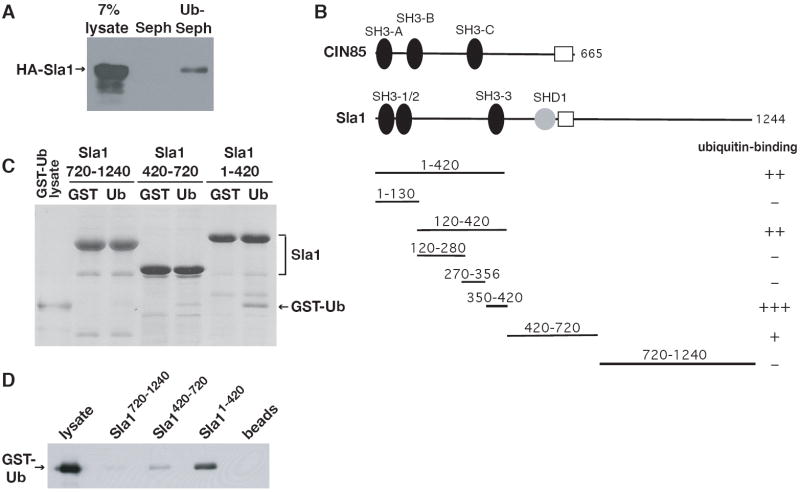
To identify the repertoire of endocytic proteins that binds to monoubiquitin, we tested yeast proteins required for receptor internalization for their ability to bind to monoubiquitin immobilized on Sepharose beads. One protein that reproducibly bound to ubiquitin was Sla1 (Figure 1A). Sla1 regulates the actin cytoskeleton and is required for rapid receptor internalization (Warren et al., 2002). It interacts with numerous other endocytic proteins, including the yeast endophilin (Rvs167), synaptojanin (Sjl2) and Hip1R (Sla2) homologues, and the Rsp5 ubiquitin ligase (Gourlay et al., 2003; Stamenova et al., 2004).
Figure 1. Sla1 binds directly to monoubiquitin.
(A) A lysate prepared from cells expressing HA-tagged Sla1 (LHY4607) was incubated with ubiquitin immobilized on beads (Ub-Seph) or beads alone (Seph). Bound proteins were eluted and Sla1 was detected on an anti-HA immunoblot. (B) Protein-protein interaction domains of Sla1 and CIN85 are labeled. White boxes represent coiled coil domains in the proteins. Fragments of Sla1 tested for binding to ubiquitin are indicated. (C) The indicated His6-tagged Sla1 fragments were immobilized on beads and incubated with lysates from bacteria expressing GST or GST-Ub. Bound proteins were eluted, resolved by SDS-PAGE and stained with Coomassie blue. (D) The proteins eluted from beads incubated with GST-Ub lysates that were visualized by Coomassie staining in (C) were analyzed on an anti-GST immunoblot.
Since Sla1 is a large protein with many binding partners, the observed binding to ubiquitin might be indirect via another protein in the yeast cell lysate. Therefore, we expressed three His6-tagged fragments of Sla1, a.a. 1-420, a.a. 420-720 and a.a. 720-1240 (Figure 1B), which encompass almost the entire protein sequence, in bacteria. The C-terminal fragment lacked the last four amino acids of Sla1 (1241-1244), because these residues seemed to inhibit expression of the fragment in E. coli. The three fragments were immobilized on metal ion affinity beads and incubated with bacterial lysates containing glutathione-S-transferase (GST) or a GST-ubiquitin fusion protein (GST-Ub). The N-terminal Sla1 fragment (a.a. 1-420) bound specifically and robustly to GST-Ub, the central fragment (a.a. 420-720) bound slightly and the C-terminal fragment (a.a. 720-1240) bound little or not at all (Figures 1C and 1D).
Ubiquitin binds to SH3 domains
We focused on the ubiquitin-binding site in the N-terminal Sla1 fragment because this region of the protein bound most effectively to ubiquitin. Sla1 does not carry any of the more than ten UBDs defined to date (Bienko et al., 2005; Hicke et al., 2005; Slagsvold et al., 2005; Bellare et al., 2006; Mattera et al., 2006; Mullally et al., 2006; Penengo et al., 2006). However, in its N-terminal region Sla1 carries three Src homology 3 (SH3) domains, SH3-1, SH3-2 and SH3-3, with the second and third domains separated by a 223 amino acid linker (Figure 1B). We expressed fragments consisting of either SH3-1 and SH3-2 (a.a. 1-130) or the linker region and SH3-3 (a.a. 120-420) in E. coli and observed that all ubiquitin-binding activity was contained in the linker-SH3-3 fragment (Figure 2A, lanes 2 and 3). After further analysis of the linker-SH3-3 fragment, we located the ubiquitin-binding site in the region between amino acids 350 and 420, corresponding to the SH3-3 domain (Figures 2A, lanes 4-6). Precise deletion of the SH3-3 domain from full-length Sla1 expressed in yeast severely diminished ubiquitin binding (Figure 2B), confirming that the SH3-3 domain is responsible for the principal ubiquitin-binding activity of Sla1.
Figure 2. The third SH3 domain of Sla1 binds to ubiquitin and ubiquitinated yeast proteins.
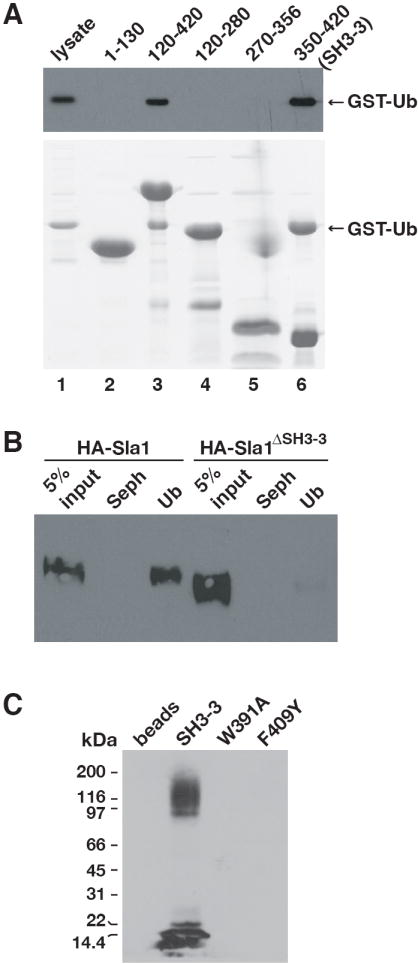
(A) The indicated His6-tagged Sla1 fragments were incubated with lysates from bacteria expressing GST-Ub. Bound proteins were eluted, resolved by SDS-PAGE and detected on an anti-GST immunoblot (top panel) or by staining with Coomassie blue (bottom panel). (B) Lysates were prepared from cells expressing HA-Sla1 (LHY4607) or HA-Sla1ΔSH3-3 (LHY5145) and incubated with ubiquitin immobilized on beads (Ub) or beads alone (Seph). Bound proteins were eluted, resolved by SDS-PAGE and detected on an anti-HA immunoblot. (C) A lysate from wildtype yeast cells (LHY291) was incubated with wildtype or mutant Sla1 SH3-3 domains immobilized on beads, or with beads alone. Bound proteins were eluted, resolved by SDS-PAGE and analyzed on an anti-ubiquitin immunoblot.
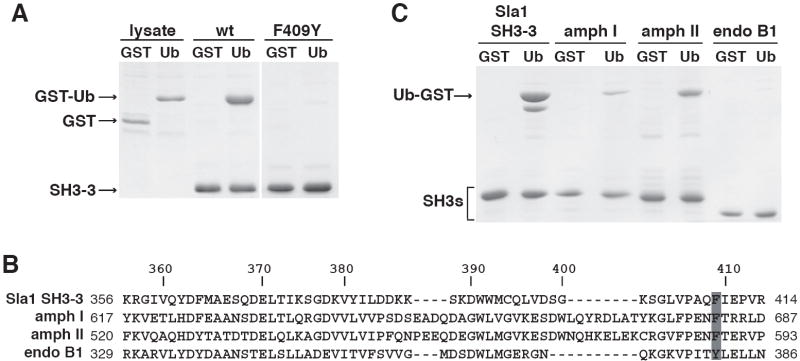
We next tested whether ubiquitinated proteins in the cellular milieu are viable partners for the Sla1 SH3-3 domain. The isolated SH3-3 domain binds to ubiquitin much more efficiently than the full-length Sla1 protein (unpublished data), perhaps because the Sla1-ubiquitin interaction is regulated in the cell. Therefore, we used the isolated SH3-3 domain to detect binding to cellular proteins. Recombinant SH3-3 domain was immobilized and incubated with a lysate prepared from wildtype yeast cells. Ubiquitinated proteins from the lysates bound to the SH3-3 domain (Figure 2C). Furthermore, these interactions were abolished by two different SH3-3 domain mutations, W391A and F409Y, that inhibit binding to free ubiquitin (see below).
Is the ability of the Sla1 SH3-3 domain to bind to ubiquitin unique? Sla1 is similar to mammalian CIN85 (Stamenova et al., 2004), which regulates the internalization of activated growth factor receptors and has been described as a scaffolding protein that binds to endophilin, synaptojanins, Hip1R and the Cbl ubiquitin ligase (Petrelli et al., 2002; Soubeyran et al., 2002; Kowanetz et al., 2004). Like Sla1, CIN85 carries three N-terminal SH3 domains (Figure 1B). We expressed the third SH3 domain of CIN85 (SH3-C) in bacteria and incubated the immobilized domains with bacterial lysates containing GST-Ub, or a fusion protein in which ubiquitin is linked to the N-terminus of GST (Ub-GST). The SH3-C domain bound to GST-Ub and Ub-GST, but binding to Ub-GST was more effective (data not shown). The SH3-C domain and Sla1 SH3-3 domains bound to Ub-GST to a similar extent as analyzed both by immunoblot (Figure 3) and Coomassie staining (data not shown). These observations indicate that ubiquitin-binding is a conserved function of the third SH3 domains of Sla1 and CIN85.
Figure 3. The third SH3 domain of CIN85 binds to ubiquitin.
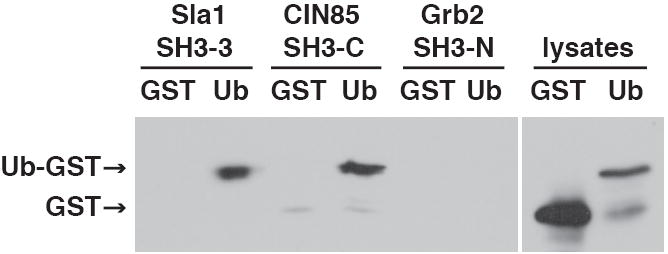
The indicated His6-tagged SH3 domains (described in Experimental Procedures) were incubated with lysates from bacteria expressing GST or Ub-GST. Bound proteins were analyzed on an anti-GST immunoblot. Each binding reaction contained an equivalent amount of SH3 domain.
Outside of SH3 domains in Sla1 fungal homologues, the Sla1 SH3-3 domain is most closely related to the N-terminal SH3 domain in Grb2 and to the SH3 domains in Fyn and FGF. To test the ability of a related SH3 domain to bind to ubiquitin, we expressed the Grb2 SH3-N domain in bacteria and performed a ubiquitin-binding assay. Grb2 SH3-N did not bind to ubiquitin, despite its sequence similarity to the Sla1 SH3-3 domain (Figure 3).
The Sla1 SH3-3 domain binds to the ubiquitin Ile44 surface with moderate affinity
NMR chemical shifts of protein residues are exquisitely sensitive to the local electronic environment, and chemical shift perturbations can be used to determine affinity constants and map intermolecular interfaces of protein complexes. Titrations of 15N-labeled ubiquitin with increasing amounts of purified SH3-3 domain at 35°C caused the selective shift of amide proton and nitrogen resonances of several ubiquitin residues (Figure 4A), indicating a specific association between the two proteins, and a rapidly dissociating and reassociating complex. From the binding isotherms of several non-neighboring ubiquitin residues (Leu8, Lys11, Thr14, Gly47, His68 and Leu73; Figure 4B), we calculated an average apparent Kd of 39 ± 9 μM for the SH3-3 domain–ubiquitin complex.
Figure 4. The Sla1 SH3-3 domain binds with moderate affinity to the ubiquitin Ile44 surface.
(A) Overlays of an expanded region of the 1H-15N correlated spectrum of 15N-labeled ubiquitin showing changes in the amide proton and nitrogen chemical shifts upon addition of the Sla1 SH3-3 domain. (B) Weighted average chemical shift deviations Δav plotted as a function of SH3-3 domain concentration for the six indicated ubiquitin residues. (C) His6-tagged SH3-3 domain was incubated with lysates containing equivalent amounts of GST, GST-Ub or GST-Ub mutants. GST-Ub lysates (total lysates) and proteins eluted from beads (bound to SH3-3) were fractionated by SDS-PAGE and detected by staining with Coomassie blue (top and bottom panels) or on an anti-GST immunoblot (middle panel). (D) Amino acids involved in SH3-3 domain-binding are highlighted on the three-dimensional structure of ubiquitin (PDB accession code 1UBQ). Residues highlighted in purple were identified as important for SH3 domain binding by the experiment shown in (C). Residues highlighted in pink showed significant chemical shifts in the presence of the SH3-3 domain in NMR experiments shown in (A). G47 was identified with both approaches.
Many previously characterized UBDs bind to a hydrophobic surface of ubiquitin including and surrounding Ile44. The resonance shifts of Leu8, Phe45 and His68 observed in the NMR experiments suggested that the SH3-3 domain might bind to the same surface. Therefore, we tested binding of the domain to ubiquitin mutants carrying alanine substitutions for amino acids at or near Ile44. The I44A mutation completely eliminated binding and mutations in several Ile44 neighbors, Val70 and Gly47, reduced binding (Figure 4C). A mutation (F4A) on another surface of the ubiquitin did not affect SH3-3 domain binding. Thus, similar to other UBDs, the SH3-3 domain binds to the Ile44 hydrophobic patch of ubiquitin (Figure 4D).
Location of the ubiquitin-binding site on the Sla1 SH3-3 domain
SH3 domains are independently folded modular domains that typically bind to peptide sequences with a core Pro-X-X-Pro (PXXP) motif to promote protein-protein interactions (reviewed in Mayer, 2001). Ubiquitin does not carry PXXP or a closely related sequence, thus the identification of a ubiquitin-binding site at the Sla1 SH3-3 domain was unexpected.
How does ubiquitin bind to SH3 domains? SH3 domains are formed of two anti-parallel β sheets packed against each other and connected by three variable loops. One side of this globular structure has a relatively shallow, hydrophobic groove in which peptide ligands bind (Mayer, 2001). In addition, several ligands bind to SH3 domains on surfaces other than the hydrophobic groove (Barnett et al., 2000; McGee et al., 2001; Tavares et al., 2001). To determine where ubiquitin binding occurs on the Sla1 SH3-3 domain, we first selected five surface-exposed amino acids in the domain for mutation; two conserved residues in the hydrophobic groove and three non-conserved residues outside the groove (Figure 5A). Mutating the conserved amino acids in the groove, F364A and W391A, abolished ubiquitin binding, whereas mutating the non-conserved residues, L396A, K401A and V413A had no effect (Figure 5B).
Figure 5. Ubiquitin binds to the peptide-binding hydrophobic groove of the Sla1 SH3-3 domain.
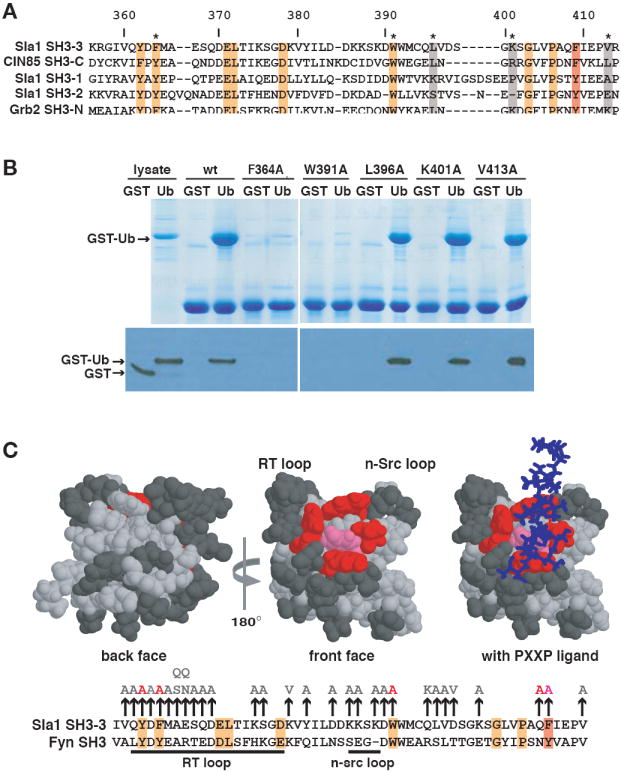
(A) Alignment of Sla1, CIN85 and Grb2 SH3 domains. Numbers above the alignment refer to amino acids of the Sla1 SH3-3 domain. Residues highlighted in orange or red are completely conserved or very similar across the six domains. The position that corresponds to Phe409 of the SH3-3 domain is highlighted with the red box. Non-conserved amino acids that were mutated for the binding analysis shown in (B) are highlighted in grey. (B) The indicated His6-tagged SH3-3 domain mutants immobilized on beads were incubated with lysates containing equivalent amounts of GST or GST-Ub. Lysates or proteins eluted from beads were detected by staining with Coomassie blue (top panel) or on an anti-GST immunoblot (bottom panel). (C) The three-dimensional structures of the Fyn SH3 domain (PDB accession code 1AZG) in the absence and presence of bound PXXP peptide are shown with the residues equivalent to mutated Sla1 SH3-3 domain amino acids highlighted. An alignment of the SH3-3 and Fyn SH3 domain sequences is shown below. Mutation of residues shown in pink (F409) or red (Y362, F364, Q408, W391) abolished ubiquitin-binding. The mutation of residues shown in dark grey had little or no effect on ubiquitin-binding. The position of a PXXP peptide that binds to the Fyn SH3 domain is shown as a stick model in blue. Most of the binding reactions on which these representations are based are shown in (B) or in Supplementary Figure 1. The vertical arrows above the alignment indicate the amino acid substitutions introduced into the SH3-3 domain. Amino acid substitutions in light grey did not affect binding to ubiquitin; the substitutions in red or pink inhibited binding and correspond to residues highlighted on the structural model.
The results of this experiment suggested that the ubiquitin-binding site in the SH3-3 domain overlaps the surface that binds PXXP ligands in other SH3 domains. Although many SH3 domains bind to PXXP motifs, different domains exhibit specificity for particular PXXP-containing peptides. Often residues in the variable loops flanking the hydrophobic groove, known as the RT and n-Src loops (see Figure 5C), influence binding specificity (for example, Noble et al., 1993; Yu et al., 1994; Wu et al., 1995). To ascertain whether residues in the RT and n-Src loops participate in determining ubiquitin-binding specificity, we individually mutated many of the SH3-3 domain surface residues in these loops. We also mutated residues on other surfaces of the SH3-3 domain to further localize the ubiquitin-binding site. Most of the mutations had no effect on ubiquitin binding. Three mutations (Y362A, Q408A, F409A) in addition to the two described above, eliminated ubiquitin-binding (Supplementary Figure 1). These mutations were all in residues residing in the hydrophobic groove (Figure 5C). Several mutations, E367N and D390A, in the RT and n-Src loops respectively, had a minor, but reproducible effect on ubiquitin-binding (Supplementary Figure 1). Mapping of the mutated residues onto the three-dimensional structure of the closely related Fyn SH3 domain bound to a peptide ligand indicates that the Sla1 SH3-3 domain ubiquitin-binding site coincides with the PXXP peptide-binding site on other SH3 domains (Figure 5C).
An alignment of the ubiquitin-binding Sla1 SH3-3 and CIN85 SH3-C domains with the non-binding Sla1 and Grb2 SH3 domains indicates that only one surface residue is identical in the SH3-3 and SH3-C domains, and different in the other SH3 domains (Figure 5A; Val360 and Val380 are buried residues). This amino acid is the Phe at position 409 in the Sla1 SH3-3 domain, which resides in the hydrophobic groove. CIN85 SH3-C also carries a Phe at this position, whereas the two Sla1 SH3 domains and the two Grb2 SH3 domains that do not bind ubiquitin have a Tyr. Mutation of Phe409 to Tyr in the SH3-3 domain abolished ubiquitin-binding as measured by GST-Ub binding experiments (Figure 6A) and by NMR (data not shown), suggesting that the amino acid present in this position is specifically important for ubiquitin-binding. Phe409 corresponds to the 75th position in the consensus alignment of SH3 domains (Figure 6B; SMART database, http://smart.embl-heidelberg.de), thus we will refer to this as position 75 in the SH3 domain sequences discussed below.
Figure 6. Ubiquitin binds to several SH3 domains carrying Phe at position 75.
(A) Binding of wildtype and F409Y SH3-3 domains to GST and GST-Ub was assayed as described for Figure 4B. (B) Alignment of the Sla1 SH3-3 domain sequence with sequences of the SH3 domains from the amphiphysins and endophilin B1. The numbering before and after the sequences refers to the protein amino acids that are the beginning and end of the indicated SH3 domains. The numbering above refers to the amino acids of Sla1. The residue highlighted in dark grey corresponds to Phe409 (Phe75) of the Sla1 SH3-3 domain. (C) Binding of amphiphysin and endophilin SH3 domains to ubiquitin. The indicated SH3 domains were expressed in E. coli and binding to GST and Ub-GST was assayed as described for Figure 2.
In addition to Sla1 and CIN85, other proteins important for receptor internalization, amphiphysin I and amphiphysin II, carry an SH3 domain with Phe at position 75. Endophilins, which are related to the amphiphysins and also function at the internalization step of endocytosis, have a Tyr at this position (Figure 6B). We expressed the human amphiphysin and endophilin SH3 domains in bacteria and found that the amphiphysin I and amphiphysin II SH3 domains bound to ubiquitin, whereas the endophilin BI SH3 domain did not (Figure 6C). By contrast to the Sla1 SH3-3 domain, the amphiphysin SH3 domains bound significantly better to Ub-GST as compared to GST-Ub (data not shown), suggesting that perhaps different SH3 domains interact with slightly different ubiquitin surfaces. Our observations suggest that Phe 75 is one determinant of SH3 domain ubiquitin-binding and indicate that SH3 domains in several different endocytic proteins bind to ubiquitin.
Ubiquitin and PXXP ligands compete for interaction with SH3 domains
During assembly of primary endocytic vesicles at the plasma membrane, amphiphysins bind through their SH3 domains to a PXRPXR sequence in the proline-rich region (PRD, a.a. 751-838) of the dynamin GTPase (Grabs et al., 1997). The Sla1 SH3-3 domain has no known interacting partners besides ubiquitin, and does not bind to several PXXP-containing sequences predicted by phage display screens to be ligands for this SH3 domain (Tong et al., 2002, data not shown). However, we serendipitously found that the SH3-3 domain, like the amphiphysin SH3 domains, bound to the dynamin PRD. Immobilized SH3-3 domain was incubated with lysates from bacteria expressing GST, Ub-GST or a GST-PRD fusion protein. Analysis of the bound proteins by Coomassie staining indicated that GST-PRD and Ub-GST bound to the SH3-3 domain to a similar extent (Figure 7A).
Figure 7. Ubiquitin and a PXXP-containing ligand compete for binding to the Sla1 SH3-3 domain.
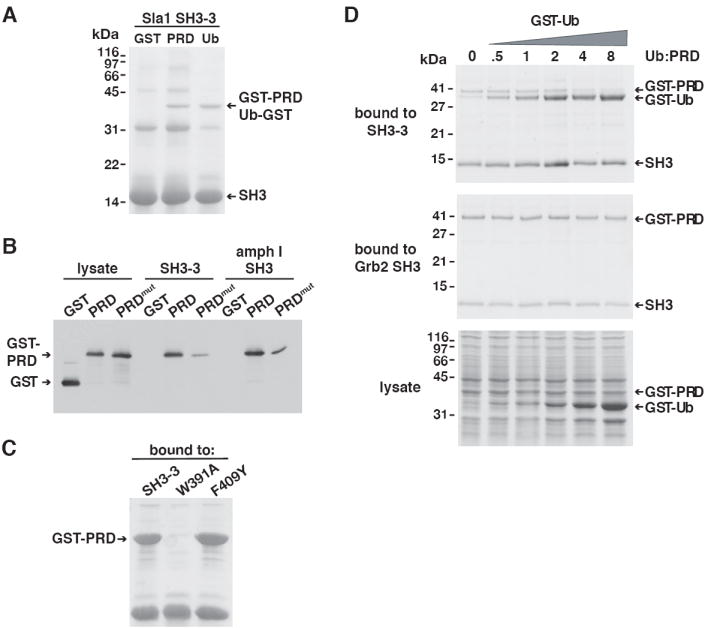
(A) The Sla1 SH3-3 domain was immobilized on beads and incubated with lysates containing GST, GST-PRD or Ub-GST. Bound proteins were analyzed by Coomassie staining. GST-PRD and Ub-GST migrated with a similar mobility. (B) Bacterial lysates containing GST, GST-PRD or GST-PRDmut were incubated with the Sla1 SH3-3 and the amphiphysin I SH3 domains immobilized on beads. Proteins bound to each SH3 domain were eluted, resolved by SDS-PAGE and analyzed on an anti-GST immunoblot. (C) A lysate prepared from cells expressing GST-PRD was incubated with wildtype, W391A or F409Y Sla1 SH3-3 domains immobilized on beads. Proteins bound to each SH3-3 domain were eluted, resolved by SDS-PAGE and stained with Coomassie blue. (D) A lysate containing GST-PRD was mixed with increasing concentrations of GST-Ub and then incubated with immobilized Sla1 SH3-3 or N-terminal Grb2 SH3 domains. GST-Ub rather than Ub-GST was used in this experiment because GST-Ub migrates with a different mobility than GST-PRD. To enhance detection of proteins in the binding assay, SH3-3 levels were raised to a concentration in which GST-PRD did not saturate the SH3-3 domain binding sites, hence competition by GST-Ub only became evident after saturation of the binding site by GST-Ub (at ~2x Ub:PRD). Under conditions in which SH3-3 domain binding sites were saturated by GST-PRD, competition by GST-Ub was observed at equimolar Ub and PRD concentrations (unpublished data). The lysates (bottom panel) or eluted proteins bound to the SH3 domains (SH3-3 domain – top panel; Grb2 SH3 domain – middle panel) were resolved by SDS-PAGE and stained with Coomassie blue. Bound proteins were separated on Tris-Tricine gels to resolve GST-PRD and GST-Ub.
To determine whether the SH3-3 domain, like the amphiphysin SH3 domain, binds to a PXXPXR sequence in the dynamin PRD we tested binding of the SH3-3 domain to a mutant PRD (PRDmut). PRDmut carries a PSRPNR→PSDANR mutation that disrupts its PXXPXR sequences and severely diminishes binding to the amphiphysin SH3 domain (Grabs et al., 1997). Incubation of bacterial lysates containing GST-PRD and GST-PRDmut with the SH3-3 or amphiphysin SH3 domains indicated that the mutation disrupted binding to both domains to a similar extent (Figure 7B).
We next tested how mutations in the SH3-3 domain that inhibit ubiquitin-binding affect binding to PXXP sequence in the dynamin PRD. Mutation of W391, a conserved amino acid that is generally required for binding to PXXP ligands in other SH3 domains, inhibited GST-PRD binding, whereas the F409Y mutation had no effect (Figure 7C). Although binding of the dynamin PRD to the SH3-3 domain may not be a physiologically relevant interaction because there is no known dynamin-like protein that is part of the yeast endocytic machinery, these data indicate that the Sla1 SH3-3 domain can bind to both ubiquitin and to PXXP-containing ligands. Furthermore, the SH3-3 domain F409Y mutation specifically inhibited ubiquitin-binding but did not affect interaction with a PXXP ligand.
Since the Sla1 SH3-3 and amphiphysin SH3 domains bind to ubiquitin at a site that overlaps the PXXP-binding site, ubiquitin and PXXP-containing ligands may compete for binding to the same SH3 domain. To test this idea, we analyzed the PRD-SH3-3 domain interaction in the presence of increasing amounts of ubiquitin. The analysis was simplified by using GST-Ub, which, unlike Ub-GST, migrated with a slightly different mobility than GST-PRD. We titrated GST-Ub into bacterial lysates containing a fixed amount of GST-PRD and incubated these lysates with immobilized SH3-3 domain. As a control we also performed this competition experiment with the N-terminal Grb2 SH3 domain, an SH3 domain that binds to the dynamin PRD (data not shown and Figure 7D), but does not bind to ubiquitin (Figure 3). Binding of GST-PRD to the SH3-3 domain decreased with increasing concentrations of GST-Ub. Concomitant with the decrease in GST-PRD binding, GST-Ub-binding increased (Figure 7D). GST-Ub and GST-PRD also compete for binding to the amphiphysin I SH3 domain, although at significantly higher Ub:PRD ratios, consistent with the inefficient binding of GST-Ub to the amphiphysin I domain (data not shown). By contrast, GST-Ub did not compete for binding of GST-PRD to the Grb2 SH3 domain (Figure 7D). Together, these observations indicate that monoubiquitin and a PXXP-containing ligand compete for binding to both the Sla1 SH3-3 and amphiphysin SH3 domains.
Discussion
The UBDs characterized to date come in many shapes and sizes, ranging from the UIM (ubiquitin-interacting motif), a 20 a.a. α-helix, to the ~150 a.a. UEV (ubiquitin E2 variant) that is comprised of a mix of β-strands and α-helices (reviewed in Hicke et al., 2005). Here we identify and characterize a previously unknown UBD found in a familiar structure, the SH3 domain. Ubiquitin has little in common with previously identified SH3 domain ligands. Ubiquitin does not carry a PXXP-like sequence. Furthermore, ubiquitin is unlikely to bind SH3 domains as a helical peptide because a ubiquitin β-sheet is the site of SH3 domain-binding and a large conformational change in ubiquitin induced upon SH3 domain binding is implausible (Khorasanizadeh et al., 1993). Nevertheless, ubiquitin binds to the same hydrophobic groove on SH3 domains that is the binding site of canonical PXXP peptide ligands. This conclusion is supported by our observation that only SH3 domain residues that reside on this hydrophobic surface were required for interaction with ubiquitin; mutations of amino acids on other SH3 domain surfaces had no effect. Ubiquitin-binding SH3 domains contact ubiquitin at Ile44 and neighboring amino acids, the site of interaction of ubiquitin with all UBDs described to date.
Ubiquitin as a unconventional SH3 domain ligand
Binding of specific PXXP peptides to individual SH3 domains is determined primarily by residues in the RT and n-Src loops, however mutations in the Sla1 SH3-3 domain loops had little or no effect on binding to ubiquitin. Instead, other features of the SH3-3 domain influence ubiquitin interaction, one being the identity of an amino acid at consensus position 75 on the SH3 domain hydrophobic groove. Although a Phe at position 75 of the SH3-3 domain is important for ubiquitin-binding, another feature(s) of the SH3 domain must also contribute to the ability to interact with ubiquitin because two of nine other Phe75 SH3 domains we tested did not bind ubiquitin detectably (unpublished data). Furthermore, mutation of Tyr75 to Phe in the first Sla1 SH3 domain did not confer ubiquitin-binding ability to this domain (unpublished data).
Besides ubiquitin, there are other variations on the common PXXP SH3 domain-binding ligands. Motifs similar to PXXP include the PXXXPR sequence recognized by SH3 domains in CIN85 (Kowanetz et al., 2003) and the PXXDY sequence that binds the SH3 domain in Eps8 (Mongiovi et al., 1999). There are also SH3 domain-binding motifs that do not include prolines, but that bind to the hydrophobic groove, specifically the RKxxYxxY motif that binds to the Fyb, Fyn and Lck SH3 domains (Kang et al., 2000), and the RXXK motif that binds as a helix to SH3 domains of Grb2, GADS and Hrs (Kato et al., 2000; Berry et al., 2002). Ubiquitin does not carry any of these motifs and has a compact globular structure — it appears to be a unique SH3 domain ligand that does not bind as a helical peptide.
Competition of ubiquitin and PXXP ligands for binding to SH3 domains
In addition to binding ubiquitin, the Sla1 SH3-3 domain binds to a PXXP-containing peptide from human dynamin, indicating that the SH3-3 domain can bind to PXXP ligands. The Sla1 SH3-3 domain was also reported to select class I RXXPXXP-containing peptides in a phage display screen (Tong et al., 2002), but the interaction of the SH3-3 domain with proteins carrying these peptides was not detected by a yeast two-hybrid analysis and, in our experiments, the Sla1 SH3-3 domain did not bind to RXXPXXP or PXXP fragments from several yeast proteins involved in endocytosis or actin regulation (data not shown). Natural ligands of the SH3-3 domain, other than ubiquitin, remain to be identified.
The amphiphysin SH3 domains also bind to ubiquitin and to PXXPXR sequences in dynamin, indicating that multiple SH3 domains can bind both ubiquitin and peptide ligands. There are several other examples of promiscuous SH3 domains that bind several ligands (Mayer, 2001). The overlapping binding sites for both PXXP peptides and ubiquitin on SH3 domains, and direct competition binding experiments, indicate that these ligands bind to SH3 domains in a mutually exclusive manner. The affinity of ubiquitin for the Sla1 SH3-3 domain (Kd = 39 μM) is similar to that of low-affinity SH3 domain-PXXP ligand interactions. Furthermore, the SH3-3 domain can bind to ubiquitinated proteins in the context of a yeast cell lysate. Thus, competition between ubiquitin and PXXP ligands for binding to SH3 domains in vivo is feasible. We suggest that competition for binding to SH3 domains by ubiquitin and peptide ligands acts to regulate assembly of different protein complexes in a spatial and temporal manner. For instance, amphiphysins and CIN85 homologues may bind to ubiquitin and proline-containing ligands at different times in the assembly of primary endocytic vesicles or at different stages in the endocytic pathway.
Role of ubiquitin-binding SH3 domains in endocytosis
The sorting and internalization of plasma membrane proteins into primary endocytic vesicles is controlled by proteins, such as Sla1/CIN85, that carry multiple protein-protein and protein-lipid interaction domains, and participate in numerous protein-protein interactions. SH3 domain proteins are prevalent in the endocytic pathway, where they promote protein-protein interactions required to sort cargo, regulate actin assembly and participate in the formation of primary endocytic vesicles at the plasma membrane (reviewed in Marsh and McMahon, 1999; Slepnev and De Camilli, 2000; Conner and Schmid, 2003; Engqvist-Goldstein and Drubin, 2003). The assembly of the dynamic complexes that include SH3 domain-containing proteins and carry out cargo sorting and vesicle budding is mediated by high specificity, low affinity interactions. These interactions are probably transient and often at least partially redundant. Because key endocytic proteins are likely to have different binding partners at different stages in the internalization process, the low affinity of the interactions is probably important for the creation of complexes that are only transiently stable and can undergo rapid assembly and disassembly.
Interactions between characterized UBDs and ubiquitin are typically of low affinity (Kd ~ 2-500 μM, Hicke et al., 2005) and between SH3 domains and peptides of moderate affinity (Kd ~ 1-100 μM, Kuriyan and Cowburn, 1997; Landgraf et al., 2004). SH3 domains in many proteins are used to regulate protein complex assembly (Mayer, 2001) and they have been used as examples of protein-protein interactions that act as switches in the assembly of large multimeric complexes (Lim, 2002). We propose that the SH3-3–ubiquitin partnership is one switch in the network of interactions that control assembly and disassembly of the endocytic and actin-regulating machinery.
Other UBDs in endocytic proteins, specifically the epsin UIMs, are also likely to function to promote protein-protein interactions among the endocytic machinery (M. Dores, J. Schnell and L. Hicke, in preparation). Furthermore, multiple components of the endocytic machinery are monoubiquitinated, including epsins, Eps15/Ede1 and endophilin/Rvs167. We propose that ubiquitin–UBD interactions generally regulate receptor internalization by acting as switches that regulate the spatial and temporal assembly of dynamic protein complexes at the plasma membrane. Ubiquitin-binding to SH3 domains of proteins involved in other cellular processes may function in a similar manner.
Experimental Procedures
Strains, media and reagents
Genetic manipulations and cell transformations were performed by standard techniques (Sherman, 1991). Yeast strains were grown in synthetic minimal media (YNB; US Biological, Swampscott, MA) or rich YPUAD medium (2% bacto-peptone, 1% yeast extract, 2% glucose supplemented with 20 mg/l adenine, uracil, and tryptophan).
GST antibodies were obtained from Amersham Biosciences (Piscataway, NJ) and ubiquitin antibodies were from Upstate (Charlottesville, VA). Anti-HA (hemagglutinin) antiserum was prepared by the Northwestern Monoclonal Antibodies Research Facility from the 12CA5 hybridoma cell line. α-Sla1 polyclonal antiserum was raised against an N-terminal Sla1 fragment (a.a. 1-420). GST-Sla11-420 was expressed in E. coli, immobilized on glutathione-Sepharose beads (Genscript Corporation, Piscataway, NJ), and Sla11-420 was cleaved with Prescission™ Protease according to the manufacturer’s instructions (Amersham Biosciences, Piscataway, NJ). Purified Sla11-420 was used to inject two New Zealand White rabbits (Covance Research Products Inc., Denver, PA). The antiserum specifically recognized a large protein present in lysates prepared from wildtype but not sla1Δ cells (unpublished data).
Plasmids
A triple HA epitope was introduced at the N-terminus of Sla1 by a two-step protocol: a NotI site was introduced after the first codon of SLA1 by QuikChange™ mutagenesis (Stratagene, La Jolla, CA), followed by ligation of DNA encoding a triple HA epitope into the NotI site to generate the centromere-based plasmid pHA-SLA1 (LHP2016). pHA-SLA1 completely rescued the growth and internalization defects of Δsla1 cells. The ΔSH3-3 (deletion of a.a. 356-413; LHP2202) mutation was introduced into pHA-SLA1 by QuikChange™ mutagenesis.
Plasmids encoding hexahistidine (His6)-tagged Sla1 fragments were described previously (Stamenova et al., 2004) or were generated by ligation-independent cloning of the relevant PCR-amplified fragments into the pET-30 bacterial expression vector (Novagen, Madison, WI). Plasmids encoding SH3 domains from Grb2 (SH3-N – a.a. 1-57, LHP2408), CIN85 (SH3-C – a.a. 265-330, LHP2191), amphiphysin I (a.a. 625-695, LHP2548), amphiphysin II (a.a. 523-593, LHP2549) or endophilin B1 (a.a. 309-365, LHP2550) were constructed with DNA PCR-amplified from plasmids obtained from the American Type Culture Collection (Manassas, VA; Grb2), Ivan Dikic (Goethe University School of Medicine, Frankfurt, Germany; CIN85) or Pietro De Camilli (Yale University, New Haven, CT; amphiphysins/endophilin).
Plasmids encoding GST fused to the PRD region of human dynamin (GST-PRD) or to the PRD carrying a PSRPNR→PSDANR mutation (GST-PRDmut) were kindly provided by Pietro De Camilli (Grabs et al., 1997). Plasmids encoding Ub-GST, GST-Ub, or GST-Ub mutants have been described (Shih et al., 2002; Kang et al., 2003; Swanson et al., 2003) or were generated by QuikChange™ mutagenesis. All SH3-3 domain point mutations were introduced into pET30-SH3-3 (LHP2167) by QuikChange™ mutagenesis.
Introduction of mutations, the in-frame fusion of tag-encoding sequences and the absence of unintentional PCR-introduced mutations in amplified DNA fragments were verified by automated sequencing.
Immunoblots
After fractionation by SDS-PAGE, lysates and bound proteins were transferred to nitrocellulose or PVDF membranes. Immunoblotting of membranes with antibodies was performed as previously described (Shih et al., 2002). Prior to incubation with ubiquitin antiserum, membranes were incubated in 6 M guanidine-HCl to thoroughly denature the transferred proteins.
Preparation of bacterial and yeast lysates
Yeast and bacterial lysates used for binding to Ub-Sepharose or to SH3 domains were prepared as described previously (Shih et al., 2002).
Ubiquitin-binding assays
E. coli cells were induced to express His6-tagged Sla1 fragments or SH3 domains proteins with 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 3-4 h at 18-37°C after overnight growth at 37°C. Purification and immobilization of the His6-tagged proteins and subsequent binding to GST, GST-Ub, Ub-GST or GST-PRD was also performed as described (Shih et al., 2002; Stamenova et al., 2004). Total lysates and bound proteins were resolved by SDS-PAGE and stained with Coomassie Brilliant Blue G-250 (Bio-Rad Laboratories, Hercules, CA) or analyzed by immunoblotting.
Binding of proteins from yeast lysates to ubiquitin-Sepharose or to immobilized His6-tagged SH3 domains was performed as described previously (Shih et al., 2002) with the following modification: total yeast lysate proteins (2.0 mg) were incubated with immobilized SH3 domains, TALON metal affinity beads (Clontech, Palo Alto, CA), 40 μl 25% ubiquitin-Sepharose slurry (Boston Biochem, Cambridge, MA) or Sepharose CL-4B (Sigma-Aldrich, Inc., St. Louis, MO) in 1 ml MES buffer at 4°C for 1 h.
Ubiquitin-PRD competition binding assays
For competition experiments, the Sla1 SH3-3 or Grb2 N-terminal SH3 domains were immobilized on TALON metal affinity beads as previously described. The SH3 domains (1.6 μM SH3-3, 0.8 μM Grb2 SH3) were incubated with E. coli lysate that contained a constant amount of GST-PRD (5 μM) and increasing amounts of GST-Ub (0-40 μM). Total lysates and bound proteins were analyzed by SDS-PAGE and Coomassie staining.
NMR titration experiments
As described above, E. coli cells were grown and induced to express His6-tagged Sla1 SH3-3 domain, except that 1 mM IPTG was used for induction. Cells were harvested and cell pellets were suspended in phosphate buffer (20 mM Na2PO4, pH 7.2, 0.3 M NaCl, 1 mM phenyl-methylsulfonyl fluoride, 1 mM leupeptin, 1 mM pepstatin, 0.1% Triton X-100), lysed by sonication and centrifuged. The supernatant was incubated with a TALON metal affinity resin for 1 h, centrifuged, and the beads were washed with 50 mM Tris, pH 8.0 containing 100 mM NaCl and 5 mM CaCl2. The SH3-3 domain was removed from the beads by cleavage with Factor Xa following the vendor’s protocol (Novagen). The domain was further purified to homogeneity using reverse-phase HPLC and its identity was confirmed by electrospray ionization mass spectrometry (ESI-MS). Uniformly 15N-labeled ubiquitin was produced as described previously (Beal et al., 1996; Kang et al., 2003). The extent of isotope incorporation in the sample determined by ESI-MS was >97% for 15N.
A 0.134 mM 15N-ubiquitin sample was titrated with increasing amounts of unlabeled wild-type Sla1 SH3-3 domain in pH 6.0 buffer at 35°C. The progress of the titration was monitored by recording one-dimensional (1D) 1H and two-dimensional (2D) 1H-15N correlated spectra. All samples contained phosphate buffer, pH 6.0, 10% D2O, 0.2% (w/v) NaN3, and 2 mM DTT-d11. Chemical shift deviations were computed using the formula Δav = √{[(ΔHN)2 + (ΔN/5)2]/2}. Analysis of the binding isotherms were performed as described (French et al., 2005).
Supplementary Material
Acknowledgments
We are grateful to Pietro De Camilli, Ivan Dikic, David Drubin and Ishwar Radhakrishnan for reagents, and we thank Amy deHart for yeast strain construction. The manuscript was improved by the critical comments of Olke Uhlenbeck, Andreas Matouschek and Ingrid Jordens. The work described was supported by National Institutes of Health Grants DK53257 and DK61299. We acknowledge the use of instruments in the Keck Biophysics Facility at Northwestern University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barnett P, Bottger G, Klein AT, Tabak HF, Distel B. The peroxisomal membrane protein Pex13p shows a novel mode of SH3 interaction. EMBO J. 2000;19:6382–6391. doi: 10.1093/emboj/19.23.6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc Natl Acad Sci USA. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellare P, Kutach AK, Rines AK, Guthrie C, Sontheimer EJ. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA. 2006;12:292–302. doi: 10.1261/rna.2152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DM, Nash P, Liu SK, Pawson T, McGlade CJ. A high-affinity Arg-X-X-Lys SH3 binding motif confers specificity for the interaction between Gads and SLP-76 in T cell signaling. Curr Biol. 2002;12:1336–1341. doi: 10.1016/s0960-9822(02)01038-2. [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Polo S, Hofmann K. When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol. 2003;4:491–497. doi: 10.1038/nrm1124. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- French M, Swanson K, Shih SC, Radhakrishnan I, Hicke L. Identification and characterization of modular domains that bind ubiquitin. Methods Enzymol. 2005;399:135–157. doi: 10.1016/S0076-6879(05)99009-5. [DOI] [PubMed] [Google Scholar]
- Gourlay CW, Dewar H, Warren DT, Costa R, Satish N, Ayscough KR. An interaction between Sla1p and Sla2p plays a role in regulating actin dynamics and endocytosis in budding yeast. J Cell Sci. 2003;116:2551–2564. doi: 10.1242/jcs.00454. [DOI] [PubMed] [Google Scholar]
- Grabs D, Slepnev VI, Songyang Z, David C, Lynch M, Cantley LC, De Camilli P. The SH3 domain of amphiphysin binds the proline-rich domain of dynamin at a single site that defines a new SH3 binding consensus sequence. J Biol Chem. 1997;272:13419–13425. doi: 10.1074/jbc.272.20.13419. [DOI] [PubMed] [Google Scholar]
- Haglund K, Di Fiore PP, Dikic I. Distinct monoubiquitin signals in receptor endocytosis. Trends Biochem Sci. 2003;28:598–604. doi: 10.1016/j.tibs.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Ann Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Kang H, Freund C, Duke-Cohan JS, Musacchio A, Wagner G, Rudd CE. SH3 domain recognition of a proline-independent tyrosine-based RKxxYxxY motif in immune cell adaptor SKAP55. EMBO J. 2000;19:2889–2899. doi: 10.1093/emboj/19.12.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang RS, Daniels CM, Francis SA, Shih SC, Salerno WJ, Hicke L, Radhakrishnan I. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin-binding. Cell. 2003;113:621–630. doi: 10.1016/s0092-8674(03)00362-3. [DOI] [PubMed] [Google Scholar]
- Kato M, Miyazawa K, Kitamura N. A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J Biol Chem. 2000;275:37481–37487. doi: 10.1074/jbc.M007251200. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S, Peters ID, B TR, Roder H. Folding and stability of a tryptophan-containing mutant of ubiquitin. Biochem. 1993;32:7054–7063. doi: 10.1021/bi00078a034. [DOI] [PubMed] [Google Scholar]
- Kowanetz K, Husnjak K, Holler D, Kowanetz M, Soubeyran P, Hirsch D, Schmidt MH, Pavelic K, De Camilli P, Randazzo PA, Dikic I. CIN85 associates with multiple effectors controlling intracellular trafficking of epidermal growth factor receptors. Mol Biol Cell. 2004;15:3155–3166. doi: 10.1091/mbc.E03-09-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowanetz K, Szymkiewicz I, Haglund K, Kowanetz M, Husnjak K, Taylor JD, Soubeyran P, Engstrom U, Ladbury JE, Dikic I. Identification of a novel proline-arginine motif involved in CIN85-dependent clustering of Cbl and downregulation of epidermal growth factor receptors. J Biol Chem. 2003;278:39735–39746. doi: 10.1074/jbc.M304541200. [DOI] [PubMed] [Google Scholar]
- Kuriyan J, Cowburn D. Modular peptide recognition domains in eukaryotic signaling. Annu Rev Biophys Biomol Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- Landgraf C, Panni S, Montecchi-Palazzi L, Castagnoli L, Schneider-Mergener J, Volkmer-Engert R, Cesareni G. Protein interaction networks by proteome peptide scanning. PLoS Biol. 2004;2:E14. doi: 10.1371/journal.pbio.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA. The modular logic of signaling proteins: building allosteric switches from simple binding domains. Curr Opin Struct Biol. 2002;12:61–68. doi: 10.1016/s0959-440x(02)00290-7. [DOI] [PubMed] [Google Scholar]
- Marsh M, McMahon HT. The structural era of endocytosis. Science. 1999;285:215–220. doi: 10.1126/science.285.5425.215. [DOI] [PubMed] [Google Scholar]
- Mattera R, Tsai YC, Weissman AM, Bonifacino JS. The Rab5 guanine nucleotide exchange factor Rabex-5 binds ubiquitin and functions as a ubiquitin ligase through an atypical UIM and a zinc finger domain. J Biol Chem. 2006;281:6874–6883. doi: 10.1074/jbc.M509939200. [DOI] [PubMed] [Google Scholar]
- Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- McGee AW, Dakoji SR, Olsen O, Bredt DS, Lim WA, Prehoda KE. Structure of the SH3-guanylate kinase module from PSD-95 suggests a mechanism for regulated assembly of MAGUK scaffolding proteins. Mol Cell. 2001;8:1291–1301. doi: 10.1016/s1097-2765(01)00411-7. [DOI] [PubMed] [Google Scholar]
- Mongiovi AM, Romano PR, Panni S, Mendoza M, Wong WT, Musacchio A, Cesareni G, Di Fiore PP. A novel peptide-SH3 interaction. EMBO J. 1999;18:5300–5309. doi: 10.1093/emboj/18.19.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullally JE, Chernova T, Wilkinson KD. Doa1 is a Cdc48 adapter that possesses a novel ubiquitin-binding domain. Mol Cell Biol. 2006;26:822–830. doi: 10.1128/MCB.26.3.822-830.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble ME, Musacchio A, Saraste M, Courtneidge SA, Wierenga RK. Crystal structure of the SH3 domain in human Fyn; comparison of the three-dimensional structures of SH3 domains in tyrosine kinases and spectrin. EMBO J. 1993;12:2617–2624. doi: 10.2210/pdb1shf/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S. The endophilin-CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met. Nature. 2002;416:187–190. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- Praefcke GJ, Ford MG, Schmid EM, Olesen LE, Gallop JL, Peak-Chew SY, Vallis Y, Babu MM, Mills IG, McMahon HT. Evolving nature of the AP2 alpha-appendage hub during clathrin-coated vesicle endocytosis. EMBO J. 2004;23:4371–4383. doi: 10.1038/sj.emboj.7600445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti SE, Caudy AA, Chenoweth JG, Tansey WP. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shih SC, Katzmann KJ, Schnell JD, Sutanto M, Emr SC, Hicke LH. Epsins and Vps27/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol. 2002;4:389–393. doi: 10.1038/ncb790. [DOI] [PubMed] [Google Scholar]
- Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, Di Fiore PP, Polo S. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci U S A. 2005;102:2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, Trambaiano D, Wakatsuki S, Stenmark H. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem. 2005;280:19600–19606. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- Slepnev VI, De Camilli P. Accessory factors in clathrin-dependent synaptic vesicle endocytosis. Nat Rev Neurosci. 2000;1:161–172. doi: 10.1038/35044540. [DOI] [PubMed] [Google Scholar]
- Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I. Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature. 2002;416:183–187. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- Stamenova SD, Dunn R, Adler AS, Hicke L. The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85-endophilin complex, Sla1-Rvs167. J Biol Chem. 2004;279:16017–16025. doi: 10.1074/jbc.M313479200. [DOI] [PubMed] [Google Scholar]
- Staub O, Rotin D. Role of ubiquitylation in cellular membrane transport. Physiol Rev. 2006;86:669–707. doi: 10.1152/physrev.00020.2005. [DOI] [PubMed] [Google Scholar]
- Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 2003;22:4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares GA, Panepucci EH, Brunger AT. Structural characterization of the intramolecular interaction between the SH3 and guanylate kinase domains of PSD-95. Mol Cell. 2001;8:1313–1325. doi: 10.1016/s1097-2765(01)00416-6. [DOI] [PubMed] [Google Scholar]
- Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, et al. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295:321–324. doi: 10.1126/science.1064987. [DOI] [PubMed] [Google Scholar]
- Warren DT, Andrews PD, Gourlay CW, Ayscough KR. Sla1p couples the yeast endocytic machinery to proteins regulating actin dynamics. J Cell Sci. 2002;115:1703–1715. doi: 10.1242/jcs.115.8.1703. [DOI] [PubMed] [Google Scholar]
- Wu X, Knudsen B, Feller SM, Zheng J, Sali A, Cowburn D, Hanafusa H, Kuriyan J. Structural basis for the specific interaction of lysine-containing proline-rich peptides with the N-terminal SH3 domain of c-Crk. Structure. 1995;3:215–226. doi: 10.1016/s0969-2126(01)00151-4. [DOI] [PubMed] [Google Scholar]
- Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.