Abstract
We recently identified HSulf-1 as a downregulated gene in ovarian carcinomas. Our previous analysis indicated that HSulf-1 inactivation in ovarian cancers is partly mediated by loss of heterozygosity (LOH) and epigenetic silencing. Here we demonstrate that variant hepatic nuclear factor 1 (vHNF1), encoded by transcription factor 2 gene (TCF2, HNF-1β) negatively regulates HSulf-1 expression in ovarian cancer. Immunoblot assay revealed that vHNF1 is highly expressed in HSulf-1 deficient OV207, SKOV3 and TOV-21G cell lines but not in HSulf-1 expressing OSE, OV167 and OV202 cells. By shRNA-mediated downregulation of vHNF1 in TOV21-G cells and transient enhanced vHNF1 expression in OV202 cells, we showed that vHNF1 suppresses HSulf-1 expression in ovarian cancer cell lines. Reporter assay and chromatin immunoprecipitation (ChIP) experiments showed that vHNF1 is specifically recruited to HSulf-1 promoter at two different vHNF1 responsive elements in OV207 and TOV-21G cells. Additionally, downregulation of vHNF1 expression in OV207 and TOV-21G cells increased cisplatin- or paclitaxel-mediated cytotoxicity as determined by both MTT and clonogenic assays and this effect was reversed by downregulation of HSulf-1. Moreover, nude mice bearing TOV-21G cell xenografts with stably downregulated vHNF1 were more sensitive to cisplatin-or paclitaxel-induced cytotoxicity compared to xenografts of TOV-21G clonal lines with nontargeted control shRNA. Finally, immunohistochemical analysis of 501 ovarian tumors including 140 clear cell tumors on tissue microarrays showed that vHNF1 inversely correlates to HSulf-1 expression. Collectively, these results indicate that vHNF1 acts as a repressor of HSulf-1 expression and might be a molecular target for ovarian cancer therapy.
Keywords: vHNF1, HSulf-1, ovarian cancer
Introduction
Our previous studies have shown that HSulf-1, an extracellular sulfatase catalyzing the 6-O desulfation of heparan sulfate (HS) glycosaminoglycans, is downregulated in the majority (75%) of tumor tissues harvested from ovarian cancer patients and is undetectable in clear cell carcinoma (CCC) of the ovary, a particularly aggressive and chemoresistant subtype (1). Loss of HSulf-1 expression augments both autocrine and paracrine proliferation signaling through various EGF-like growth factor family ligands, such as EGF, amphiregulin, heparin-binding EGF-like growth factor (HB-EGF), resulting in an increase tumor cell growth and resistance to drug-induced apoptosis (1, 2). In addition, re-expression of HSulf-1 inhibits both tumor growth and tumor angiogenesis in vivo(3).
Variant hepatic nuclear factor 1 (vHNF1, also called HNF1β, TCF2) protein were initially identified based on its interaction with a sequence essential for liver-specific transcription of several genes postulated to determine the hepatic phenotype (4). Subsequent studies have shown that there is a high level of vHNF1 expression in pancreas, kidney, stomach, lung tissue, and hepatocellular carcinoma (5). Mutations of the human vHNF1 gene are associated with maturity onset diabetes of the young type 5 (MODY5) and renal carcinomas (6).
Despite the fact that it is highly curable if diagnosed early, cancer of the ovaries causes more mortality in American women each year than all other gynecologic malignancies combined (7). Clear cell ovarian cancer represents 2–15% of all epithelial ovarian malignancies and has a number of clinical and histologic characteristics distinguishing it from other types of ovarian cancers (8). For instance, of all of the epithelial ovarian cancers, CCC has the worst prognosis partially because of its low response to standard platinum-based chemotherapy (9–12). Several investigations have indicated that vHNF1 is overexpressed in CCC and can be an excellent molecular marker of this disease (13, 14). Other studies have shown that hypomethylation of vHNF1 CpG island participates in the upregulation of vHNF1 in CCC (15). Furthermore, downregulation of vHNF1 expression by RNA interference induced apoptotic cell death in ovarian CCC cell lines (8). As indicated, our studies of HSulf-1 expression have revealed that there is a much higher frequency of loss of HSulf-1 expression in clear cell tumors (100%) of the ovary compared to serous histological subtype (70%) (1). Therefore, we hypothesized that vHNF1 may negatively regulate HSulf-1 expression in ovarian carcinomas.
In the present study, we report on the expression profile of vHNF1 in ovarian cell lines and primary ovarian cancers by immunoblot and tumor tissue microarray (TMA) analysis. Additionally, chromatin immunoprecipitation (ChIP) and luciferase reporter assays show that vHNF1 is specifically recruited to HSulf-1 promoter at vHNF1 responsive elements in vivo and can markedly reduce HSulf-1 promoter activity in vitro. Using lentiviral-mediated shRNA technology, we also investigated the effect of downregulation of vHNF1 on HSulf-1 expression and sensitivity to chemotherapy of ovarian cancer cells both in vitro and in vivo.
Materials and methods
Cell culture
TOV-21G, SKOV3, OVCAR3, and OVCAR5 cells were purchased from American Type Culture Collection (Manassas, VA, USA). OV167, OV202, and OSE cell lines were established as previously described (16, 17). A2780 cell line was obtained on a MTA from Dr. Thomas Hamilton, Fox Chase Cancer Center, Philadelphia, PA. All cell lines were grown as previously reported (16, 17).
Protein extracts and Western Blot
Protein extraction and Western blot analysis were performed as described previously (2). The antibodies used for immunoblotting were as follows: HSulf-1 (Abcam Cat# 22010), vHNF1 (Santa Cruz Cat# sc7411), HNF1α (Santa Cruz Cat# sc6547).
shRNAs
shRNAs used in this study is Supplementary experiment procedures and Supplementary Table S1.
Plasmid Constructions
The construction of all plasmids used in this study is described in Supplementary experiment procedures. The sequences of primers used for the amplification of constructs are listed in Supplementary Table S1.
Luciferase Reporter Assays
Reporter assay was performed as previously described (18). Also see Supplementary experiment procedures.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed as described (16). Also see Supplementary experiment procedures.
MTT and Clonogenic survival assays
Both assays were performed as previously described (17).
Attenuation of cisplatin-induced cytotoxicity following siRNA mediated downregulation of HSulf-1 in vHNF1 downregulated TOV-21G cells
Following confirmation of upregulation of HSulf-1 in vHNF1 downregulated TOV-21 G cells, upregulated endogenous expression of HSulf-1 was downregulated with siRNA targeting towards 3’-UTR of HSulf-1 (Supplementary Table S1) as previously described (16). Two days following HSulf-1 siRNA transfection, the cells were treated with increasing concentrations of cisplatin ranging from 2.5 to 30 µM for 24 h in a 96-well plate and the percentage of viable cells was determined by MTT assay.
Tumor xenograft experiments
All mice were handled according to the Guide for the Care and Use of Laboratory Animals. Mouse studies were carried out following procedures approved by the Institutional Animal Care and Use Committee at the Mayo Clinic College of Medicine. Female athymic nude mice (NCr nu/nu; Animal Production Area, National Cancer institute- Frederick Cancer Research and Development Center, Frederick, MD), 5 to 6 weeks old, were used for this study. For measurement of tumor growth in vivo, 3 × 106 TOV-21G transfectants stably expressing control shRNA or shvHNF1-1 and /or 2 in 0.1 ml Matrigel (Collaborative Biomedical Products, Bedford, MA, USA) were injected s.c. in the flanks of mice, generating two tumors/mouse (a total of 12 mice for each stable clone). When TOV-21G tumors were palpable, mice were treated intraperitoneally with cisplatin (4mg/kg) or pactlitaxel (20mg/kg) on days 1, 7 and 13 (n=6). On day 14 (24 hours following the last dose for both treatments), 1 mouse per group was sacrificed and the tumor xenografts were monitored for vHNF1 and HSulf-1 expression by immunoblot. The tumor volume was calculated twice a week and by caliper measurements using the following formula: a × b × c, where ‘a’ is the length and ‘b’ is the width and ‘c’ is the height in millimeters. The experiment was terminated 4 weeks following the beginning of treatment.
Tumor tissue microarray
After approval by Institutional Review Boards of Vancouver General Hospital and British Columbia Cancer Agency and Mayo Clinic, archived ovarian epithelial tumor specimens from patients with ovarian cancers obtained prior to chemotherapy were utilized in the analysis. TMA construction and digital imaging system were as previously described (14, 17). Characteristics of patients included in the analysis are shown in Supplementary Table S2. The antibodies used for these studies are for HSulf-1 (Abcam Cat# 31960) and vHNF1 (Santa Cruz Cat # sc7411). The expression level of vHNF1 and HSulf-1 by immunohistochemistry was scored and compared in triplicate specimens by 3 separate observers in a double-blinded fashion. For both proteins, tumors showing no staining (score 0) or weak focal staining (score 1) were considered to be negative, whereas moderate (score 2) or intense staining (score 3) was considered positive.
Statistical analysis
Data are presented as mean ± S.D. of at least three independent experiments. Differences between groups were analyzed using two-tailed Student’s t test. Fisher's exact test was used to determine the strength of the association between the parameters. To correlate the levels of vHNF1 and HSulf-1 with overall patient survival, Kaplan-Meier survival analysis and log-rank test were performed. P<0.05 was considered statistically significant.
Results
Expression analysis of HSulf-1 and vHNF1 in ovarian cancer cell lines
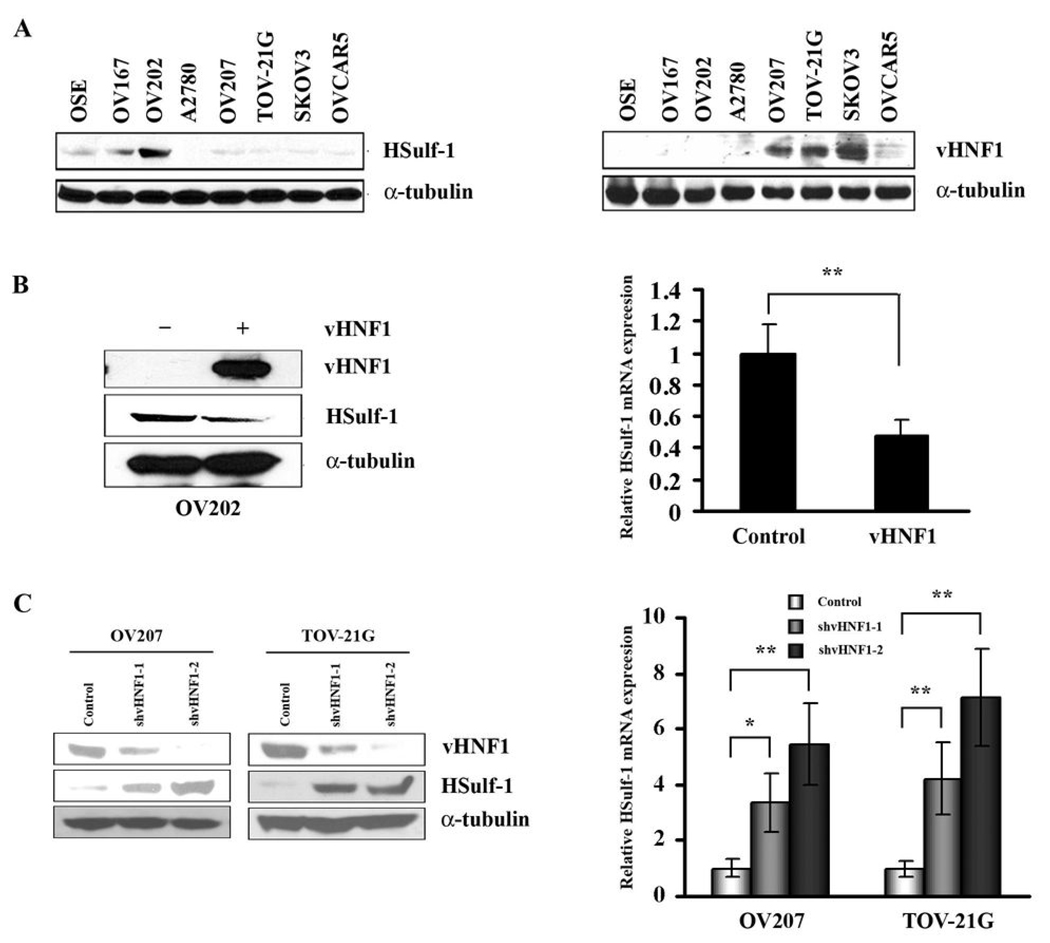
HSulf-1 and vHNF1 expression were evaluated in 7 ovarian cancer cell lines and normal ovarian epithelial cells (OSE) by Western blot. As shown in Fig. 1A, OSE, OV167 and OV202 cells express HSulf-1, while A2780, OV207, TOV-21G, SKOV3, and OVCAR5 have undetectable or very low levels of HSulf-1 expression. However, vHNF1 is highly expressed in HSulf-1 deficient OV207, SKOV3 and TOV-21G cell lines and weakly in A2780, OVCAR5 cells but not in HSulf-1 expressing OSE, OV167 and OV202 cells.
Figure 1. vHNF1 negatively regulates HSulf-1 expression in ovarian cancer cell lines.
A, Immunoblot analysis of HSulf-1 and vHNF1 expression in ovarian cell lines. vHNF1 is highly expressed in HSulf-1 deficient OV207, SKOV3 and TOV-21G cell lines but not in HSulf-1 expressing OSE, OV167 and OV202 cells. B, Enhanced expression of vHNF1 downregulates HSulf-1 expression in OV202 cells. OV202 cells were transfected with vHNF1 expression construct. 48 hours later, endogenous HSulf-1 protein and mRNA level were determined by immunoblot analysis and quantitative real time RT-PCR. C, shRNAs targeting vHNF1 results in up-regulation of HSulf-1 expression at both protein and mRNA level in OV207 and TOV-21G cells determined by immunoblot analysis and quantitative real time RT-PCR. shRNAs targeting two different regions of vHNF1 mRNA was used to generate stable transfectants (shvHNF1-1 and shvHNF1-2) in OV207 and TOV-21G cell lines. The blots were stripped and re-probed with anti-α-tubulin antibody to show equal loading of total protein. Results of real time RT-PCR are shown as mean ± S.D. of triplicates samples. * p<0.05, ** p<0.01.
Forced expression of vHNF1 in OV202 cells downregulated HSufl-1 expression
Lack of vHNF1 expression in HSulf-1 expressing cell lines led us to speculate that vHNF1 could act as a repressor of HSulf-1 expression. In order to test this hypothesis, we transiently transfected vHNF1 expression construct into OV202 cells. 48 h following transfection, HSulf-1 expression levels was determined by immunoblot and real-time quantitative RT-PCR. As shown in Fig. 1B, forced expression of vHNF1 in OV202 cells reduced HSulf-1 expression at both protein and mRNA level.
shRNA mediated downregulation of vHNF1 upregulated HSulf-1 expression
To test if downregulation of endogenous vHNF1 may upregulate HSulf-1 expression, vHNF1 was silenced using lentiviral-mediated shRNAs in OV207 and TOV21-G cells as described in the methods section. The non-target shRNAs served as controls. As shown in Fig. 1C, our immunoblot analysis showed that shRNAs targeting two different regions of vHNF1 open reading frame markedly decreased the expression of vHNF1 compared with the control shRNA in both cell lines. Attenuation of vHNF1 expression resulted in an increase of HSulf-1 protein expression which was more pronounced in TOV21-G cells compared to OV207 cells. Downregulation of vHNF1 also led to increased HSulf-1 expression at mRNA level in both cell lines.
Overexpression of vHNF1 decreases HSulf-1 promoter activity via vHREs
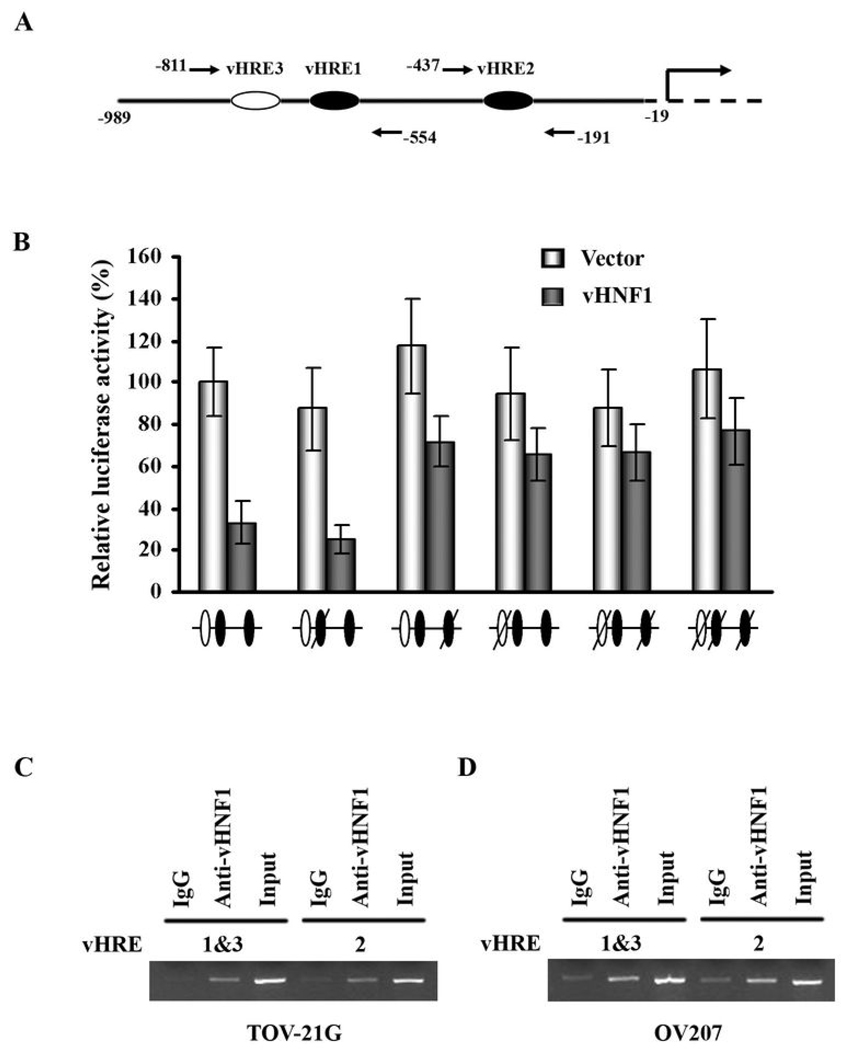
Based on the regulatory effect of HSulf-1 expression by vHNF1, we analyzed the databases TRANFAC and TESS for consensus binding site for vHNF1 transcription factor upstream of transcription initiation site on HSulf-1 promoter. Sequence analysis of HSulf1 promoter using Transcription Element Search System (TESS, http://www.cbil.upenn.edu/cgi-bin/tess/tess) revealed the presence of three vHNF Responsive Elements (vHRE) (5’-ATTAAC-3’) upstream of the transcription initiation site. The diagrammatic representation of these elements in HSulf-1 promoter is shown in Fig. 2A. The 1Kb promoter sequence was cloned into a pGL3-Basic luciferase reporter plasmid. Dual-Luciferase Assays were performed on cell extracts from OV202 cells co-transfected with luciferase reporter plasmid and either pvHNF1 or pcDNA3.1 empty vector. As shown in Fig. 2B, overexpression of vHNF1 significantly reduced HSulf-1 promoter activity compared with controls. Fig. 1B, left panel, lane 2 shows the level of HSulf-1 upon transient transfection of vHNF1 in OV202 cells.
Figure 2. Involvement of vHNF1 in suppression of HSulf-1 promoter activity.
A, Schematic representation of potential vHNF1 binding sites in HSulf-1 promoter and the location of the primers used for ChIP assay. B, Suppression of HSulf-1 transcription by vHNF1 requires functional vHREs. OV202 cells were co-transfected with either wild-type HSulf-1 luciferase reporter construct or plasmids containing mutations in the putative vHNF1 binding sites (slashed oval, white ones indicate vHREs in the opposite orientation) as well as pvHNF1 or pcDNA3.1 empty vector. The relative luciferase activity is presented as a percentage of the luciferase activity of the cells transfected with wild-type promoter construct and pcDNA3.1 empty vector. Results are shown as mean ± S.D. of triplicates samples. The results indicate that both vHRE2 and vHRE3, but not vHRE1 could be functionally relevant to regulate HSulf-1 expression. C&D, vHNF1 is recruited to specific sites within HSulf-1 promoter. ChIP assay was performed to demonstrate in vivo binding of vHNF1 on HSulf-1 promoter. Anti-vHNF1 antibody or goat IgG was used to precipitate sonicated chromatin from TOV-21G (C) and OV207 (D). Input control DNA was diluted 5 fold prior to PCR amplification.
To determine which of the three vHRE binding sites are required for the inhibitory effect of vHNF1 on HSulf-1 promoter activity, TT→GG (for 1st and 2nd vHRE sites) and TA→GC (for 3rd vHRE in the opposite orientation) mutations that destroy the vHNF1 binding were introduced into the vHNF1 binding sites respectively. Mutation of either vHRE2 or vHRE3 abolished transcriptional repression of HSulf-1 by vHNF1. However, there was no change in vHNF1 promoter activity when vHRE1 was mutated (Fig. 2B). Mutations of both vHRE sites 2 and 3 in the same construct also rescued HSulf-1 promoter activity, although the effects were not additive. Collectively, these results indicate that both vHRE site 2 (close to the transcriptional initiation site) and vHRE site 3, distal to vHRE2 could be functionally relevant to regulate HSulf-1 expression.
vHNF1 is specifically recruited to HSulf-1 promoter
To determine whether vHNF1 is recruited to HSulf-1 promoter, ChIP assays were performed in OV207 and TOV-21G cells. For each cell line, DNA/protein complexes were immunoprecipitated with anti-vHNF1 antibody and then the purified DNA was amplified with primers flanking vHRE1 and 3 sites in one reaction and with primers (Supplementary Table S1) flanking vHRE2 in another. The location of the 3 potential vHNF1 binding sites in HSulf-1 promoter and the primers used for ChIP assay is shown in Fig. 2A. As shown in Fig. 2C and D, vHNF1 is recruited to 2 different regions within the HSulf-1 promoter both in TOV21-G and OV207 cells but not in OV202 cells which do not express vHNF1 (data not shown). The first region spans vHRE1 and vHRE3 which are very close to each other (Fig. 2A), while the second region contains vHRE2. We confirmed that the recruitment was specific because there was undetectable or very little amplification of IgG controls (Lanes 1 and 4 in Figs 2C and D).
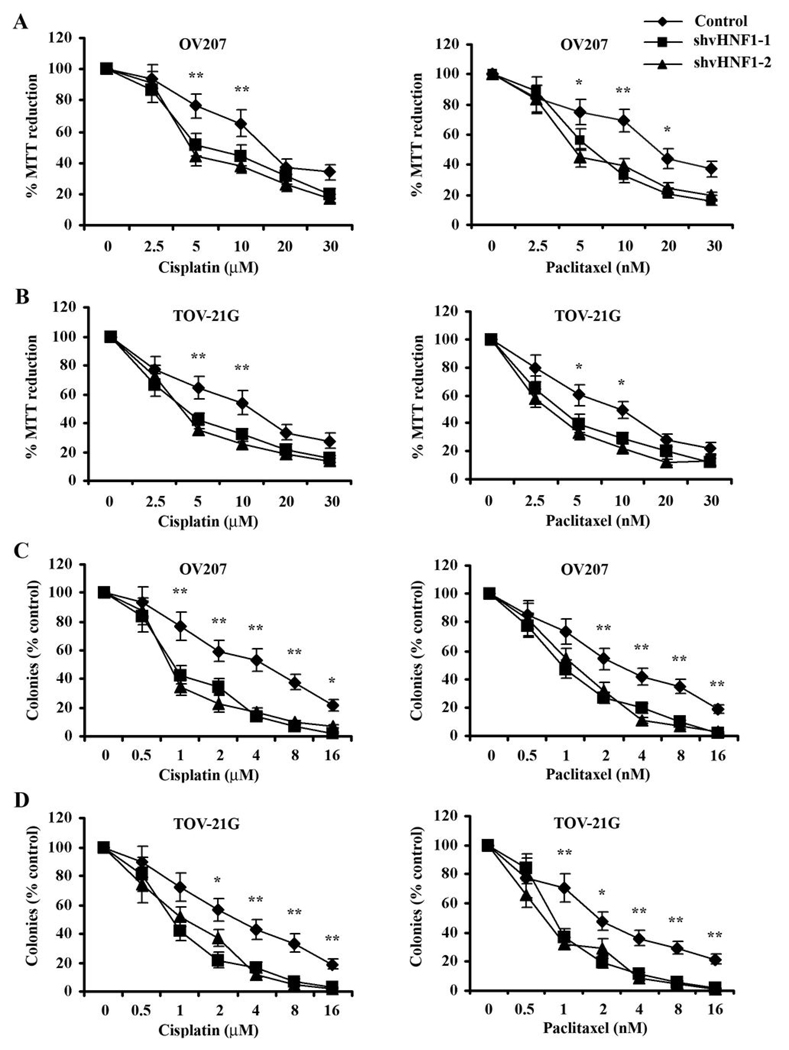
Downregulation of vHNF1 sensitizes ovarian cancer cells to cisplatin- or paclitaxel-mediated cytotoxicity
Our previous data showed that HSulf-1 expression confers chemosensitivity to multiple drugs in ovarian cancer (1). Therefore, we tested the effect of downregulation of vHNF1 on HSulf-1 expression and the sensitivity of cancer cells to cisplatin and paclitaxel. We downregulated vHNF1 expression in the two ovarian clear cell derived cell lines, OV207 and TOV21-G, using lentiviral shRNAs against vHNF1 and generated batch clones. vHNF1 downregulation was confirmed by western blot as shown in Fig. 1C. Both vHNF1 downregulated batch clone and non-target control cells were treated with increasing concentrations of cisplatin or paclitaxel for 24 h and were then incubated in fresh medium for additional 3 days. As shown in Figs. 3A and B, shRNA mediated downregulation of vHNF1 in both OV207 and TOV21-G cells resulted in increased sensitivity to both cisplatin and paclitaxel as determined by MTT reduction assay compared with cells transduced with non-target shRNA. To test whether cisplatin- or paclitaxel-induced antiproliferative effects translated into less clonogenic survival, we tested the clonogenic survival of OV207 and TOV21-G batch clonal lines. Both control and vHNF1 downregulated batch clones were treated with various concentrations of cisplatin or paclitaxel for 24 hours, allowed to grow for additional ten days in drug-free medium, and then counted for surviving colonies after Coomassie staining. Consistent with the previous results, downregulation of vHNF1 attenuated clonogenic survival of cells transduced with shRNAs targeting vHNF1 compared to the control cells (Figs. 3C and D). However, shRNAs targeting vHNF1 have no effect on both HSulf-1 protein expression and cisplatin-induced cytotoxicity in vHNF1 deficient OV202 (Supplementary Fig. S1). These results suggest that downregulation of vHNF1 promotes chemotherapy-induced cytotoxicity in ovarian cancer cells.
Figure 3. shRNAs targeting vHNF1 promotes cisplatin/paclitaxel toxicity.
A&B, OV207 and TOV-21G batch clonal cells stably expressing shRNAs to vHNF1 were exposed to varying concentrations of cisplatin or paclitaxel for 24 h and tested by MTT reduction assay. Suppression of vHNF1 expression sensitized cancer cells to drug-induced antiproliferative effects in both cell lines. C&D, OV207 and TOV-21G batch clonal cells expressing shRNAs targeting vHNF1 or non-targeting shRNA were treated with the indicated concentrations of cisplatin or paclitaxel for 24h, washed and allowed to form colonies. Stable inhibition of vHNF1 led to decreased clonogenic survival compared with the controls. Results are expressed as mean ± S.D. of triplicates samples. * p<0.05, ** p<0.01.
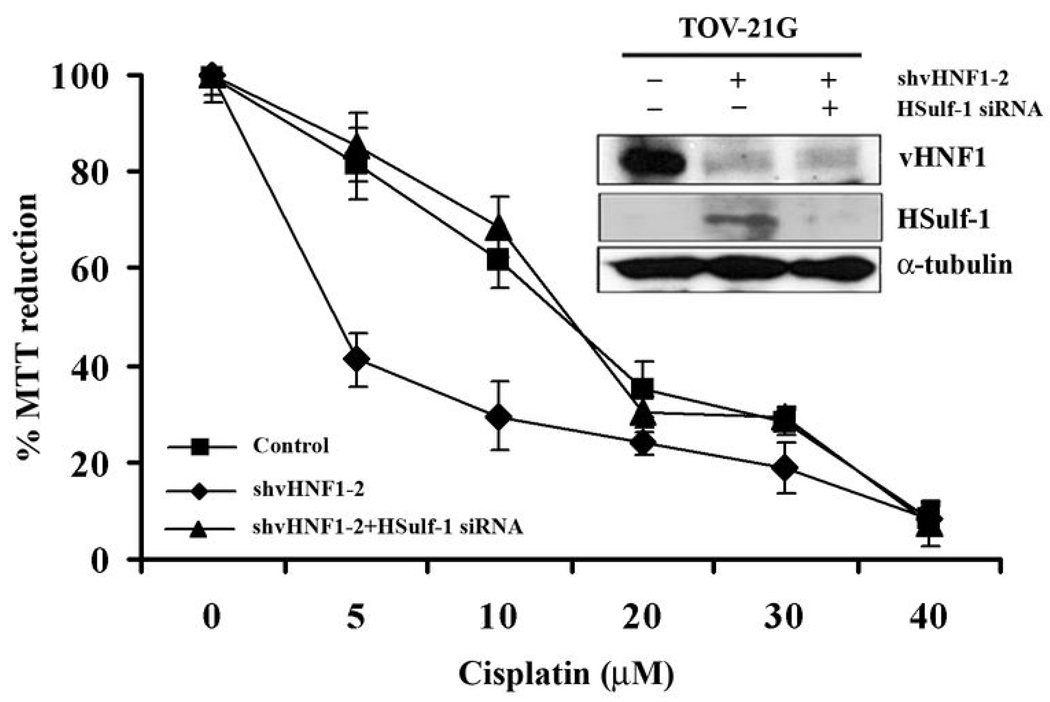
Specific contribution of HSulf-1 in conferring cisplatin-induced cytotoxicity in TOV21-G cells following vHNF1 downregulation
Our results indicate that downregulation of vHNF1 in OV207 and TOV21-G cells results in the upregulation of HSulf-1 expression. Additional studies indicate that under these conditions, the cells become sensitive to cisplatin-induced cytotoxicity. However, downregulation of vHNF1 could result in changes of several vHNF1 target gene expression (19). To determine the specific contribution of HSulf-1 in cisplatin-induced cytotoxicity, we transiently downregulated HSulf-1 expression with siRNA in TOV-21G cells with downregulated vHNF1 expression. Downregulation of both vHNF1 and HSulf-1 was determined by immunoblot analysis (Fig. 4, inset). As seen in Figure 4, there is loss of both vHNF1 and HSulf-1 expression in cells transfected with HSulf-1 siRNA (Fig. 4, inset lane3), whereas the Scrambled siRNA transduced controls express HSulf-1 but not vHNF1 (Fig. 4, inset lane2). We used shvHNF1-2 batch clones in this experiment as it showed higher efficacy of vHNF1 knockdown than the other transfectant shvHNF1-1 in TOV-21G cells (Fig. 1C). Two days following HSulf-1 siRNA transfection, the cells were treated with increasing concentrations of cisplatin for 24 h and the percentage of viable cells was determined by MTT assay. Results, shown in Fig. 4, indicate that there was an increase in cell survival following HSulf-1 knockdown in vHNF1 downregulated TOV21-G cells compared to those with HSulf-1 expression. These results suggest a specific contribution of HSulf-1 in cisplatin-induced cytotoxicity.
Figure 4. Specific contribution of HSulf-1 in cisplatin-induced cytotoxicity in vHNF1 downregulated TOV21-G cells.
TOV-21G batch clonal cells stably expressing shvHNF1-2 were treated with vehicle (DMSO) or varying concentrations of cisplatin for 24 h and assayed by MTT reduction assay. vHNF1 downregulated clone (♦) were more sensitive to cisplatin-induced cytotoxicity compared to nontargeted shRNA control clone (■). Downregulation of vHNF1 (Lane 2, top panel in the inset) leads to upregulation of HSulf-1 (lane 2 middle panel, inset). However, siRNA mediated downregulation of HSulf-1 expression in these cells (Lane 3, middle panel, inset) resulted in resistance to cisplatin-induced cytotoxicity (▲). The blots were stripped and re-probed with anti-α-tubulin antibody to show equal loading of total protein (bottom panel, inset).
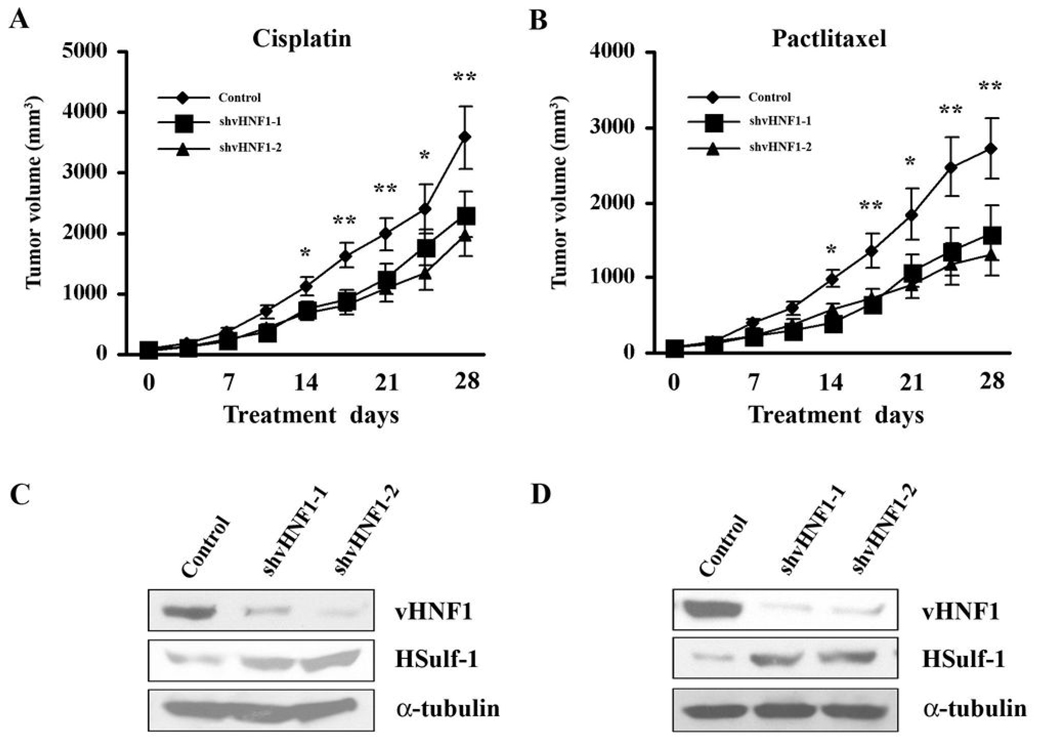
vHNF1 inhibition enhances the antiproliferative activity of cisplatin or paclitaxel in vivo
Xenografts of TOV-21G transfectants stably expressing control shRNA or shvHNF1-1/2 were generated and treated as described in the methods section. Measurements of tumors volume are graphically displayed in Figs. 5A and B. Downregulation of vHNF1 significantly increased the in vivo sensitivity of TOV-21G cells to both drugs used in this study, which is consistent with our in vitro experiments. Sustained downregulation of vHNF1 and HSulf-1 upregulation in the xenografts are confirmed by immunoblot (Figs. 5C and D). This is the first demonstration that vHNF1 gene silencing can induce HSulf-1 expression and potentiate the antiproliferative effect of cisplatin and /or paclitaxel in vivo.
Figure 5. Effect of modulating vHNF1 expression on the antiproliferative activity of cisplatin or paclitaxel in vivo.
A and B, Female immune deficient BALB/c nude mice at 4 weeks of age were bilaterally, s.c. injected with 3 × 106 TOV-21G transfectants stably expressing control shRNA or shvHNF1-1/2, generating two tumors/mouse. When TOV-21G tumors were palpable, mice were treated intraperitoneally with cisplatin (4mg/kg) (A) or paclitaxel (20mg/kg) (B) on days 1, 7 and 13 (n=6). The experiment was terminated 4 weeks following the beginning of treatment. Each point represents the means ± S.D. of 10 tumors. *P < 0.05, **P < 0.01. C&D: Immunoblot analysis of vHNF1 and HSulf-1 expression in xenograft tumor homogenates from animals treated with cisplatin (C) or paclitaxel (D) at day 14. The blots were stripped and re-probed with anti-α-tubulin antibody to show equal loading of total protein.
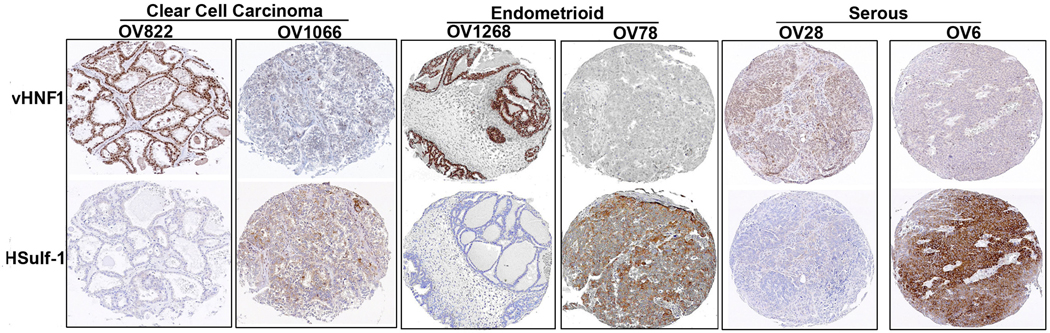
vHNF1 inversely correlates to HSulf1 expression in primary ovarian cancers
To evaluate the potential association between vHNF1 and HSulf-1 expression and clinicopathological parameters in ovarian cancer, a TMA containing a total of 501 primary ovarian tumor samples was analyzed (Fig. 6). vHNF1 immunostaining was predominantly found in the nucleus. 20.6% (103/501) of ovarian tumors had positive staining of vHNF1, of which 63.1% (65/103) of cases showed negative staining for HSulf-1. Conversely, higher level of HSulf-1 was detected in 61.8% (246/398) of samples which were negative for vHNF1 staining. Thus, vHNF1 was inversely correlated with HSulf-1 expression level (p<0.0001). There was no relationship between vHNF1 and the disease stage and grade (p>0.05). Additionally, serous tumors with moderate to high levels of HSulf-1 (127/186, 68.3%) showed a trend toward improved survival as assessed by Kaplan-Meier survival analysis and log-rank test (p=0.0465) (Supplementary Fig. 2) However, we did not observe this trend in either the endometrioid or clear cell tumors potentially due to the limited numbers of HSulf-1 positive tumors with these two subtypes (p>0.05; data not shown).
Figure 6. Inverse correlation between vHNF1 and HSulf-1 expression in primary ovarian Cancers.
Immunohistochemistry analysis of representative tumors (Two each of CCCs, endometrioid and serous tumors) on the TMA showing expression of vHNF1 (top panel) and HSulf-1 (Bottom Panel).
Discussion
We have previously shown that mRNA encoding HSulf-1 is downregulated in ovarian cancer (1). HSulf-1 downregulation modulates heparin binding growth factor signaling and epigenetic modifications contribute in part to its downregulation in human cancers (16, 20, 21). We have also reported that HSulf-1 is a key regulator of VEGF-mediated proliferation and that loss of HSulf-1 leads to increased tumorigenesis and angiogenesis in vivo (3). In particular, patients with ovarian cancers expressing higher levels of HSulf-1 showed a 94.4% response rate (17/18) compared to a 58% (7/12) response rate among those with weak or moderate levels of HSulf-1 expression (16). Accordingly, HSulf-1 downregulation appears to represent a mechanism by which ovarian cancer cells enhance growth factor signaling and diminish apoptosis.
Clear cell ovarian cancers represents 2–15% of all epithelial ovarian malignancies (22) and is associated with a poorer outcome compared to serous carcinoma (12, 23). Several investigations indicated that vHNF1 could be an excellent marker and a possible molecular target for therapy for ovarian CCC (8, 24, 25). Recent studies by Kobel et al (14) showed that, vHNF1 showed the highest sensitivity (82.5%) and specificity (95.2%) for CCC. Based on our previous data showing that HSulf-1 expression seems to be downregulated or lost in a much higher proportion of clear cell tumors of the ovary compared to other histological subtypes (1), we surmised that expression of HSulf-1 could be regulated by vHNF1. Consistent with this hypothesis, sequence analysis showed the presence of three vHNF1 responsive elements upstream of the transcriptional start site of HSulf-1 promoter.
Immunoblot analysis of vHNF1 expression seemed to indicate that loss of HSulf-1 could be associated with expression of vHNF1. Of interest to this study is the observation that the HSulf-1 expressing cell lines (OSE, OV167 and OV202) had complete loss of vHNF1 expression, suggesting that vHNF1 could be a repressor of HSufl-1 expression. Contrary to the report by other investigators (8, 13), we found that non clear cell derived cell lines such as SKOV3 cells also expressed significant amounts of vHNF1 and we also found lower levels of vHNF1 in OVCAR5 and A2780 cells in addition to robust expression of vHNF1 in cell lines derived from clear cell tumors, OV207 and TOV-21G (Fig. 1A). However, Tomassetti et al (26) and Terasawa et al (27) reported that other non clear cell derived cell lines such as IGROV1, SKOV3 and OVCAR3 cell lines express vHNF1.
Consistent with these observations, several findings from this study indicate a regulatory role for vHNF1 in the control of HSulf-1 transcription. By shRNA-mediated downregulation of vHNF1 in TOV21-G cells and transient enhanced vHNF1 expression in OV202 cells, we showed that vHNF1 suppresses HSulf-1 expression in ovarian cancer cell lines. We have also shown a decrease in HSulf-1 transcriptional activity in luciferase reporter assays following overexpression of vHNF1 protein in OV202 cells and that it is mediated through two different vHNF1 responsive elements (vHRE2 and vHRE3) in HSulf-1 promoter, which was further confirmed by ChIP assays. However, we did not see a significantly more transcriptional activation of HSulf-1 upon simultaneous mutation of both sites, implicating that other co-factors could be involved in regulating the activity of vHNF1 as observed for HNF-1α, where a dimerization co-factor DcOH selectively stabilized HNF-1α dimers and enhanced its transcriptional activity in a tissue specific manner (28).
Other studies indicate that members of the HNF family, for example, a closely related member of the family of HNF proteins, HNF1α mediate transcriptional activation (29). HNF1-α is a homeo domain-containing protein that acts as a transcriptional activator to regulate a large group of genes and are associated with development, metabolism and cancer. The homeo domain and dimerization domain, but not the activation domain, are highly conserved between HNF1α and vHNF1 (HNF1-β). Since vHNF1 lacks transcription activation domains, it is not surprising to note that it has no or lower transactivation potency then that of HNF1α (29). In addition, vHNF1 has been associated with repression of a subset of hepatocyte-specific genes (30). It seems that HNF1α and vHNF1 may regulate a different set of genes in vivo, although they do not distinguish between the DNA-binding sites in vitro (29). We also explored the potential regulatory role of HNF1α in HSulf-1 expression and found that overexpression of HNF1α could change neither protein/mRNA level nor promoter activity of HSulf-1 in ovarian cancer cell lines. Moreover, ChIP assay showed that HNF1α does not bind to HSulf-1 promoter region in vivo (data not shown). These results indicate that the inhibitory effect of vHNF1 on HSulf-1 expression may be independent of HNF1α.
Additional studies indicate that stable downregulation of vHNF1 sensitizes OV207 and TOV-21G cells to cisplatin- or paclitaxel-induced cytotoxicity both in vitro and in vivo (Fig. 3 and Fig. 5). As shown in Fig. 5, mouse bearing stably depleted vHNF1 TOV-21G xenografts were sensitive to both cisplatin and paclitaxel treatment compared to vHNF1 proficient TOV-21G xenografts. Consistent with this, we found elevated levels of HSulf-1 in the tumor xenografts (Figs. 5C and D) lending additional support to our previous finding that HSulf-1 is a critical determinant for increased sensitivity towards both the drugs in vivo (16). More importantly, downregulation of HSulf-1 in vHNF1 downregulated clones restored cisplatin resistance to a certain extent.
In the present study we also determined the expression levels of vHNF1 and HSulf-1 on 501 ovarian carcinomas on a tissue microarray. 20.6% of ovarian tumors on this tissue microarray showed high levels of vHNF-1 staining and this was associated with none to lower levels of HSulf-1 expression. Conversely, higher level of HSulf-1 was detected in 61.8% of samples which had negative vHNF1 staining. Moreover, 68.3% of patients with serous ovarian cancer expressed HSulf-1 protein and showed a trend toward improved survival.
vHNF1 might directly or indirectly regulate unknown target genes involved in survival and apoptosis. Our studies indicate that one of these genes is HSulf-1, a putative tumor suppressor that is lost in a substantial portion of ovarian tumors of both clear cell and non-clear cell origin. Given the role that HSulf-1 plays in conferring sensitivity to commonly used chemotherapeutic agents, and the fact that it is lost in a higher proportion of ovarian tumors of clear cell origin, our data suggest that the chemoresistance of CCC could be associated with the overexpression of vHNF1 acting as a repressor of HSulf-1. Therefore, Targeting vHNF1 to upregulate HSulf-1 could have therapeutic implications in CCC of ovary.
Supplementary Material
Acknowledgments
We would like to acknowledge Dr. Lynn Hartmann and Dr. Kimberly Kalli for providing us with the ovarian tissue microarray.
Financial Support: All to V.S., US National Cancer Institute grant CA106954-04, Minnesota Ovarian Cancer Alliance (MOCA) grant Edith and Bernard Waterman Foundation
References
- 1.Lai J, Chien J, Staub J, et al. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem. 2003;278:23107–23117. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- 2.Narita K, Chien J, Mullany SA, et al. Loss of HSulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J Biol Chem. 2007;282:14413–14420. doi: 10.1074/jbc.M611395200. [DOI] [PubMed] [Google Scholar]
- 3.Narita K, Staub J, Chien J, et al. HSulf-1 inhibits angiogenesis and tumorigenesis in vivo. Cancer Res. 2006;66:6025–6032. doi: 10.1158/0008-5472.CAN-05-3582. [DOI] [PubMed] [Google Scholar]
- 4.Bach I, Mattei MG, Cereghini S, Yaniv M. Two members of an HNF1 homeoprotein family are expressed in human liver. Nucleic Acids Res. 1991;19:3553–3559. doi: 10.1093/nar/19.13.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenfeld M, Maury M, Chouard T, Yaniv M, Condamine H. Hepatic nuclear factor 1 (HNF1) shows a wider distribution than products of its known target genes in developing mouse. Development. 1991;113:589–599. doi: 10.1242/dev.113.2.589. [DOI] [PubMed] [Google Scholar]
- 6.Weng JP, Lehto M, Forsblom C, Huang X, Li H, Groop LC. Hepatocyte nuclear factor-1 beta (MODY5) gene mutations in Scandinavian families with early-onset diabetes or kidney disease or both. Diabetologia. 2000;43:131–132. doi: 10.1007/s001250050018. [DOI] [PubMed] [Google Scholar]
- 7.Gagnon A, Ye B. Discovery and application of protein biomarkers for ovarian cancer. Curr Opin Obstet Gynecol. 2008;20:9–13. doi: 10.1097/GCO.0b013e3282f226a5. [DOI] [PubMed] [Google Scholar]
- 8.Tsuchiya A, Sakamoto M, Yasuda J, et al. Expression profiling in ovarian clear cell carcinoma: identification of hepatocyte nuclear factor-1 beta as a molecular marker and a possible molecular target for therapy of ovarian clear cell carcinoma. Am J Pathol. 2003;163:2503–2512. doi: 10.1016/s0002-9440(10)63605-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goff BA, Sainz de la Cuesta R, Muntz HG, et al. Clear cell carcinoma of the ovary: a distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy in stage III disease. Gynecol Oncol. 1996;60:412–417. doi: 10.1006/gyno.1996.0065. [DOI] [PubMed] [Google Scholar]
- 10.Tammela J, Geisler JP, Eskew PN, Jr, Geisler HE. Clear cell carcinoma of the ovary: poor prognosis compared to serous carcinoma. Eur J Gynaecol Oncol. 1998;19:438–440. [PubMed] [Google Scholar]
- 11.Behbakht K, Randall TC, Benjamin I, Morgan MA, King S, Rubin SC. Clinical characteristics of clear cell carcinoma of the ovary. Gynecol Oncol. 1998;70:255–258. doi: 10.1006/gyno.1998.5071. [DOI] [PubMed] [Google Scholar]
- 12.Crotzer DR, Sun CC, Coleman RL, Wolf JK, Levenback CF, Gershenson DM. Lack of effective systemic therapy for recurrent clear cell carcinoma of the ovary. Gynecol Onol. 2007;105:404–408. doi: 10.1016/j.ygyno.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 13.Kato N, Toukairin M, Asanuma I, Motoyama T. Immunocytochemistry for hepatocyte nuclear factor-1beta (HNF-1beta): a marker for ovarian clear cell carcinoma. Diagn Cytopathol. 2007;35:193–197. doi: 10.1002/dc.20623. [DOI] [PubMed] [Google Scholar]
- 14.Köbel M, Kalloger SE, Carrick J, et al. A limited panel of immunomarkers can reliably distinguish between clear cell and high-grade serous carcinoma of the ovary. Am J Surg Pathol. 2009;33:14–21. doi: 10.1097/PAS.0b013e3181788546. [DOI] [PubMed] [Google Scholar]
- 15.Kato N, Tamura G, Motoyama T. Hypomethylation of hepatocyte nuclear factor-1beta (HNF-1beta) CpG island in clear cell carcinoma of the ovary. Virchows Arch. 2008;452:175–180. doi: 10.1007/s00428-007-0543-z. [DOI] [PubMed] [Google Scholar]
- 16.Staub J, Chien J, Pan Y, et al. Epigenetic silencing of HSulf-1 in ovarian cancer:implications in chemoresistance. Oncogene. 2007;26:4969–4978. doi: 10.1038/sj.onc.1210300. [DOI] [PubMed] [Google Scholar]
- 17.Chien J, Aletti G, Baldi A, et al. Serine protease HtrA1 modulates chemotherapy-induced cytotoxicity. J Clin Invest. 2006;116:1994–2004. doi: 10.1172/JCI27698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chien J, Staub J, Avula R, et al. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24:5089–5100. doi: 10.1038/sj.onc.1208700. [DOI] [PubMed] [Google Scholar]
- 19.Senkel S, Lucas B, Klein-Hitpass L, Ryffel GU. Identification of target genes of the transcription factor HNF1beta and HNF1alpha in a human embryonic kidney cell line. Biochim Biophys Acta. 2005;1731:179–190. doi: 10.1016/j.bbaexp.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Lai JP, Chien J, Strome SE, et al. HSulf-1 modulates HGF-mediated tumor cell invasion and signaling in head and neck squamous carcinoma. Oncogene. 2004;23:1439–1447. doi: 10.1038/sj.onc.1207258. [DOI] [PubMed] [Google Scholar]
- 21.Lai JP, Chien JR, Moser DR, et al. hSulf1 Sulfatase promotes apoptosis of hepatocellular cancer cells by decreasing heparin-binding growth factor signaling. Gastroenterology. 2004;126:231–248. doi: 10.1053/j.gastro.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy AW, Biscotti CV, Hart WR, Webster KD. Ovarian clear cell adenocarcinoma. Gynecol Oncol. 1989;32:342–349. doi: 10.1016/0090-8258(89)90637-9. [DOI] [PubMed] [Google Scholar]
- 23.Omura GA, Brady MF, Homesley HD, et al. Long-term follow-up and prognostic factor analysis in advanced ovarian carcinoma: the Gynecologic Oncology Group experience. J Clin Oncol. 1991;9:1138–1150. doi: 10.1200/JCO.1991.9.7.1138. [DOI] [PubMed] [Google Scholar]
- 24.Yamada K, Tachibana T, Hashimoto H, et al. Establishment and characterization of cell lines derived from serous adenocarcinoma (JHOS-2) and clear cell adenocarcinoma (JHOC-5, JHOC-6) of human ovary. Hum Cell. 1999;12:131–138. [PubMed] [Google Scholar]
- 25.Kato N, Sasou S, Motoyama T. Expression of hepatocyte nuclear factor-1beta (HNF-1beta) in clear cell tumors and endometriosis of the ovary. Mod Pathol. 2006;19:83–89. doi: 10.1038/modpathol.3800492. [DOI] [PubMed] [Google Scholar]
- 26.Tomassetti A, Mangiarotti F, Mazzi M, et al. The variant hepatocyte nuclear factor 1 activates the P1 promoter of the human alpha-folate receptor gene in ovarian carcinoma. Cancer Res. 2003;63:696–704. [PubMed] [Google Scholar]
- 27.Terasawa K, Toyota M, Sagae S, et al. Epigenetic inactivation of TCF2 in ovarian cancer and various cancer cell lines. Br J Cancer. 2006;94:914–921. doi: 10.1038/sj.bjc.6602984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendel DB, Khavari PA, Conley PB, et al. Characterization of a cofactor that regulates dimerization of a mammalian homeodomain protein. Science. 1991;254:1762–1767. doi: 10.1126/science.1763325. [DOI] [PubMed] [Google Scholar]
- 29.Mendel DB, Hansen LP, Graves MK, Crabtree GR. HNF-1 alpha and HNF-1 beta (vHNF-1) share dimerization and homeo domains, but not activation domains, and form heterodimers in vitro. Genes Devq. 1991;5:1042–1056. doi: 10.1101/gad.5.6.1042. [DOI] [PubMed] [Google Scholar]
- 30.Baumhueter S, Courtois G, Crabtree GR. A variant nuclear protein in dedifferentiated hepatoma cells binds to the same functional sequences in the beta fibrinogen gene promoter as HNF-1. EMBO J. 1988;7:2485–2493. doi: 10.1002/j.1460-2075.1988.tb03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.