Abstract
Ketohexokinase (KHK, also known as fructokinase) initiates the pathway through which most dietary fructose is metabolized. Very little is known about the cellular localization of this enzyme. Alternatively spliced KHK-C and KHK-A mRNAs are known, but the existence of the KHK-A protein isoform has not been demonstrated in vivo. Using antibodies to KHK for immunohistochemistry and Western blotting of rodent tissues, including those from mouse knockouts, coupled with RT-PCR assays, we determined the distribution of the splice variants. The highly expressed KHK-C isoform localized to hepatocytes in the liver and to the straight segment of the proximal renal tubule. In both tissues, cytoplasmic and nuclear staining was observed. The KHK-A mRNA isoform was observed exclusively in a range of other tissues, and by Western blotting, the presence of endogenous immunoreactive KHK-A protein was shown for the first time, proving that the KHK-A mRNA is translated into KHK-A protein in vivo, and supporting the suggestion that this evolutionarily conserved isoform is physiologically functional. However, the low levels of KHK-A expression prevented its immunohistochemical localization within these tissues. Our results highlight that the use of in vivo biological controls (tissues from knockout animals) is required to distinguish genuine KHK immunoreactivity from experimental artifact. (J Histochem Cytochem 57:763–774, 2009)
Keywords: ketohexokinase, fructokinase, fructose, alternative splicing, fructosuria, immunohistochemistry
Ketohexokinase (KHK) initiates dietary fructose catabolism through the specialized fructose pathway by phosphorylating it to fructose-1-phosphate. This pathway bypasses the major glycolytic checkpoint (at phosphofructokinase), and has attracted recent attention because of epidemiological evidence that high-fructose Western diets are a risk factor for development of the “diseases of affluence”—diabetes, hypertension, and gout (Basciano et al. 2005; Johnson et al. 2007; Miller and Adeli 2008).
Most enzymatic studies of KHK have focused on the liver, the tissue in which its major isoform (also known as fructokinase) is most abundant. However, KHK enzyme activity is also present in other tissues, including kidney, intestine, brain, pancreas, lung, muscle, and optic nerve (Ballard and Oliver 1964; Adelman et al. 1967; Weiser and Quill 1975; Raushel and Cleland 1977a; Das et al. 1984; Bais et al. 1985; Malaisse et al. 1989; Meakin et al. 2007). Despite this, very little is known about the distribution of KHK-producing cells within any of these tissues. Also, some biochemical studies must be interpreted with caution. Often, crude protein extracts have been assayed, in conjunction with acid or heat treatment to eliminate fructose metabolism through other pathways. These include hexokinases I, II, and III, which can convert fructose to fructose-6-phosphate with Km values of ∼1–10 mM. It is believed that fructose metabolism by these enzymes is suppressed if glucose is available, due to much lower Km values for glucose of around 0.05 mM. In the liver, other pathways may be less of a confounding factor, because hexokinase IV/glucokinase (GK) predominates over the other hexokinases, and has a fructose Km of >100 mM. Other possible routes for fructose metabolism also involve high Km values for fructose, e.g., fructose dehydrogenase (5 mM), fructose 3-phosphokinase (30 mM), and sorbitol dehydrogenase (100 mM). These pathways may, however, contribute to a minor degree to perceived KHK activity.
A further complication, unrecognized in early studies of KHK activity, is that the KHK gene encodes two forms of the enzyme as a result of alternative splicing, KHK-A and KHK-C (Hayward and Bonthron 1998). These two isoforms differ by mutually exclusive inclusion of either of two adjacent duplicated 135-nt exons (exons 3a and 3c).
Only one of these mRNA variants was detected in each of a range of human and rat tissues, indicating “all or none” splicing in favor of one variant (Hayward and Bonthron 1998). Furthermore, the KHK-C mRNA was expressed at high levels, and KHK-A at much lower levels. The exception to this general pattern was pancreas, in which, although KHK-C predominated, some KHK-A was coexpressed.
The enzymatic properties of purified recombinant KHK-A and KHK-C differ markedly. In particular, KHK-A has a 10-fold higher Km for fructose (8 mM), suggesting that it phosphorylates fructose poorly at physiological concentrations (Asipu et al. 2003).
The major enzymatic studies of KHK were carried out before the KHK-A splice variant was identified, and focused primarily on tissues such as liver, kidney, and duodenum, which contain high levels of the KHK-C splice variant only. In other tissues, which express a low level of the KHK-A variant, these earlier studies did not take into account the different kinetic properties of KHK-A, and thus may not accurately reflect the amount of KHK protein.
In the present study, we have examined KHK expression levels, splice variant usage, and cellular localization. Given the drawbacks outlined above, rather than measuring enzyme activity, we have used RT-PCR and immunohistochemistry. Interpretation was aided by comparison with tissues from animals with defined lesions of the Khk gene.
Materials and Methods
Normal and Genetically Modified Mice
All procedures involving mice were carried out in accordance with the Home Office Animal Scientific Procedures Act 1986, United Kingdom. Approval for the work was granted by the University of Leeds Ethical Review Committee. Tissues from two strains of genetically modified mice were used. The generation and characterization of these animals will be described in detail elsewhere. In brief, the Khk3a/3a mouse lacks the KHK-A protein, as a result of a stop codon introduced into exon 3a, whereas the KhkΔ/Δ mouse lacks both KHK-A and KHK-C, as a result of a deletion of exons 4–7. These lines were established following injection of correctly targeted embryonic stem cells of strain 129 into C57BL/6 blastocysts. The tissues used in this study were obtained from wild-type and homozygous mutant littermates derived from matings of heterozygous animals. All animals were maintained on a standard chow diet, without fructose supplementation, and were externally indistinguishable in regard to breeding and maintenance, regardless of genotype. Tissues were taken from both male and female animals, aged 8–12 weeks. Some additional tissues of wild-type Khk genotype were obtained from various other strains (Balb/c, CD-1, C3H, DBA, FVB) at 3–7 weeks old. No strain-specific differences in Khk immunostaining were observed.
RNA Isolation and Reverse Transcription
For each tissue (liver, kidney, duodenum, pancreas, spleen, lung, heart, brain, eye, skeletal muscle, adipose, uterus, and adrenal gland) analysis was performed on samples from at least two wild-type animals. RNA was extracted using Trizol (Invitrogen Life Technologies; Paisley, UK). RT-PCR employed the Thermoscript RT-PCR system (Invitrogen Life Technologies), as per the manufacturer's instructions. For mouse tissues, a species-specific forward primer (dTCCTGCACTGTCCTTTCCTTG) and a 5′ fluorescein-labeled reverse primer (dGCCCTCAATGTGGATCCACTT) were used. Other primers and methods for PCR product digestion were used as previously described (Hayward and Bonthron 1998). Undigested or digested samples were fractionated by capillary electrophoresis on a MegaBACE 500 (GE Healthcare; Little Chalfont, UK) or a 3130 Genetic Analyzer (ABI; Warrington, UK) and analyzed using the genotyping and quantification software PeakHeights (http://dna.leeds.ac.uk/peakheights/).
Immunohistochemistry
Most experiments were performed on paraffin-embedded tissue sections fixed in 4% paraformaldehyde. Tissues from both male and female wild-type animals of various strains, described above, were used for the analysis of liver, kidney, and pancreas, in addition to the same tissues from knockout animals. Following dewaxing, rehydration, and endogenous peroxidase blocking by a 3% solution of H2O2 for 20 min, nonspecific antibody binding was blocked by incubation with casein in TBS (145 mM NaCl, 0.83 mM Tris-HCl, pH 7.6) for 20 min (Vector Laboratories, Ltd.; Peterborough, UK). Optimization of staining was performed on all primary KHK antibodies at various dilutions, with and without heat-induced epitope recovery (HIER). For HIER, sections were placed in preheated 100-mM pH 6.0 citrate buffer in a microwave and heated for 12 min, so that the solution simmered rather than boiled. Sections were then left to stand for 20 min to cool in the hot buffer.
Primary antibodies diluted in TBS were applied overnight at 4C. The primary antibodies were: (1) chicken anti-KHK (human) affinity-purified polyclonal IgY used at 1:200 dilution (Genway Biotech, Inc., San Diego, CA; catalog code 15-288-22373), referred to below as the KHK “N-terminal antibody”; (2) a chicken antibody (Genway Biotech, Inc.) raised against a KHK-C–specific peptide, LVADFRRRGVDVSQ, and used at 1:50; (3) rabbit purified polyclonal KHK (C-terminal) antibody (Abgent, Abingdon, UK; catalog code AP7069b), referred to below as the “C-terminal antibody,” used at 1:20; and (4) goat anti-aldehyde reductase (Abcam Plc., Cambridge, UK; catalog code ab11802) used at 1:200. Because detection of the latter antibody used a biotin–streptavidin system, avidin and biotin blocking was performed prior to detection (Vector Laboratories, Ltd.). Following three washes in TBS for 5 min, the secondary antibodies were applied. The antibodies used were: horseradish peroxidase (HRP)-labeled rabbit anti-chicken (Abcam Plc.), a rabbit EnVision system (Dako UK, Ltd.; Ely, UK), or biotin-labeled rabbit anti-goat immunoglobulin followed by HRP–streptavidin complex (Dako UK, Ltd.). Following appropriate washing, DAB color development was used to detect immunolabeling.
Additional experiments were performed using silver-enhanced immunogold staining. After incubation with primary antibody, sections were washed three times in PBS, and then incubated in 1:100–diluted rabbit anti-chicken IgG 10-nm gold conjugate (British Biocell International; Cardiff, UK) for 1 hr at 20C. Following thorough washing in deionized water, silver enhancement was carried out for 18 min in the dark as described (Holgate et al. 1986). Sections were counterstained in hematoxylin, and following dehydration and clearing in xylene, were mounted in distyrene/plasticizer/xylene.
Chicken preimmune IgY (Genway Biotech, Inc.) and normal rabbit IgG (Dako UK, Ltd.) were used as negative controls instead of primary antibody.
For experiments in which the primary antibody was preincubated with peptide, recombinant protein lysate, or control protein prior to incubation on tissue sections, the preincubation was either overnight or for 3 hr, and the antibody-to-protein ratio was 1:5 (w/w).
An alternative fixative, Carnoy's (ethanol 60% v/v, chloroform 30% v/v, acetic acid 10% v/v), was used in place of formalin to determine whether the staining pattern seen in normal kidney and liver samples was a formalin-induced artifact.
Cell Culture
The human hepatocarcinoma cell line HepG2 (ECACC; Salisbury, UK) was cultured in DMEM with glutamine, 10% fetal calf serum, and penicillin-streptomycin. The mouse insulinoma line MIN6 was grown in DMEM with glutamine, 15% fetal calf serum, and penicillin-streptomycin (Miyazaki et al. 1990).
Escherichia coli Crude Extracts
Recombinant human and mouse KHK proteins were expressed using the T7 polymerase-based pET vector system. Bacterial cells were pelleted and resuspended in lysis buffer overnight, followed by centrifugation for 30 min at 13,000 × g as previously described (Asipu et al. 2003).
Western Blotting
Tissue extracts were prepared by homogenizing samples of liver, kidney, spleen, or brain in radioimmunoprecipitation assay buffer (500 mM Tris HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100) containing 1 mM phenylmethylsulphonylfluoride, 1 mM dithiothreitol, and a protease inhibitor cocktail [1 mM 4-(2-aminoethyl) benzenesulphonyl fluoride, 0.8 μM aprotinin, 20 μM leupeptin, 40 μM bestatin, 15 μM pepstatin A, 14 μM E-64] (Sigma-Aldrich, Ltd.; Gillingham, UK). Recombinant protein lysates and tissue extracts were diluted in loading buffer and separated by SDS-PAGE. Proteins were transferred to polyvinylidene difluoride membranes and probed with primary and secondary antibodies according to the manufacturer's instructions (GE Healthcare). Briefly, the membrane was blocked with 5% non-fat milk powder in TBS with 0.05% Tween 20 (TBS-T). Primary antibodies were incubated with the membrane in the same buffer for 1 hr, followed by washing in TBS-T. The secondary antibodies were a goat anti-rabbit HRP used at 1:3000 (Abcam Plc.), or a goat anti-chicken HRP used at 1:20,000 (Genway Biotech, Inc.). These were incubated with the membrane in TBS-T containing 0.5% non-fat milk powder for 1 hr. The membrane was again washed in TBS-T before signal development using the SuperSignal West Pico Chemiluminescent Substrate according to the manufacturer's instructions (Pierce; Cramlington, UK).
Results
Tissue Distribution of KHK Splice Variants
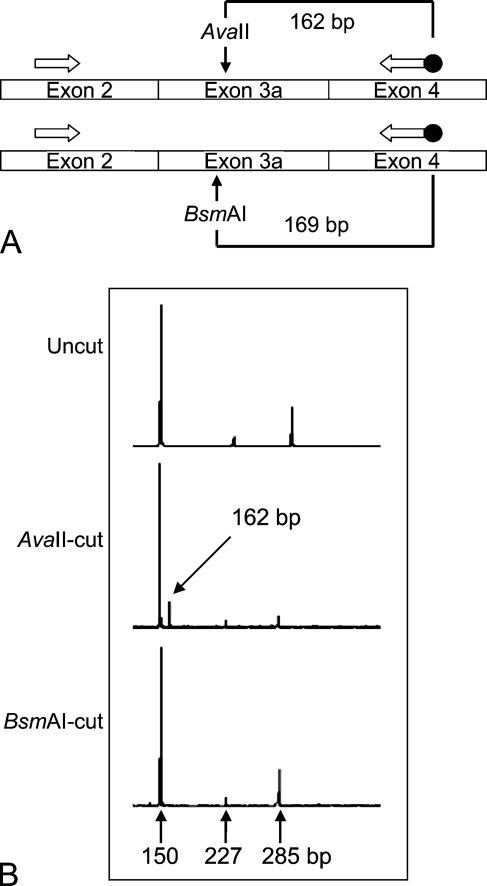
We first investigated the KHK splice variant usage in different mouse tissues. The method relies on RT-PCR with primers in exons 2 and 4, allowing coamplification of both exon 3a- and 3c-containing cDNAs (Figure 1A). The presence of exon 3a or exon 3c was determined using restriction enzymes AvaII (which cuts exon 3a) and BsmAI (which cuts exon 3c). The splicing patterns in the mouse tissues and human hepatocarcinoma line HepG2 (Table 1) were similar to those previously observed in rat and human tissues (Hayward and Bonthron 1998). Exon 3c–containing transcripts were identified in four tissues (liver, kidney, duodenum, and pancreas), and exon 3a in the remainder. As in rat and human, KHK-A expression was also accompanied by the expression of the minor aberrant splice variant KHK-3− (in which exon 2 is spliced in-frame to exon 4; Figure 1B). In mouse pancreas alone, all three splice variants were present, with KHK-C predominating. This unique expression pattern was also seen in the pancreatic insulinoma cell line MIN6. Eye and adipose tissue had not previously been studied in any species; both express KHK-A and KHK-3−. Finally, in some samples, traces of a novel aberrant splice variant were identified (Table 1 and Figure 1B). Sequencing (not shown) demonstrated that this results from use of an aberrant splice acceptor site within exon 3a, omitting the first 58 nt of exon 3a, shifting the reading frame of the resulting mRNA, and introducing an early stop codon.
Figure 1.
PCR analysis of splice variant usage. (A) Schematic of the assay. (B) Typical profile obtained. Using heart cDNA, we identified the uncut PCR product (285 bp), the 3− splice variant (150 bp), and an additional aberrantly spliced product (227 bp).
Table 1.
Splice variants identified in various cell lines and mouse tissues using RT-PCR
| Tissue/cell type | KHK-C | KHK-A | 3− | Additional |
|---|---|---|---|---|
| Liver | + | − | − | − |
| Kidney | + | − | − | − |
| Pancreas | + | + | + | − |
| Duodenum | + | − | − | − |
| Brain | − | + | + | + |
| Adrenal gland | − | + | + | − |
| Uterus | − | + | + | + |
| Lung | − | + | + | + |
| Heart | − | + | + | + |
| Skeletal muscle | − | + | + | + |
| Spleen | − | + | + | − |
| Adipose tissue | − | + | + | − |
| Eye | − | + | + | + |
| MIN6 cells | + | + | + | − |
| HepG2 cells | + | + | + | − |
KHK, ketohexokinase. KHK-C and KHK-A are the two protein-coding mRNA splice variants. 3− refers to an mRNA splice variant in which exon 2 is spliced to the acceptor site of exon 4 and “additional” describes a further splice variant using an aberrant splice acceptor site, through which the first 58 nt of exon 3a are omitted. In the Table body, + and − indicate respectively that the splice variant was detectable or undetectable, using the assay shown in Figure 1.
Expression of KHK in Tissue and Cell Lysates
To determine the histological localization of KHK, we assessed three antibodies. Two were commercially obtained, raised against either N-terminal (a recombinant protein containing aa 21–178) or C-terminal (a peptide containing aa 266–281) regions of human KHK. The third was a custom-made antibody to an epitope encoded by human exon 3c (a peptide containing aa 72–85), which should therefore only detect the KHK-C isoform. (An antibody specific for KHK-A was not raised, because the unique exon 3a sequence was predicted not to be very immunogenic.)
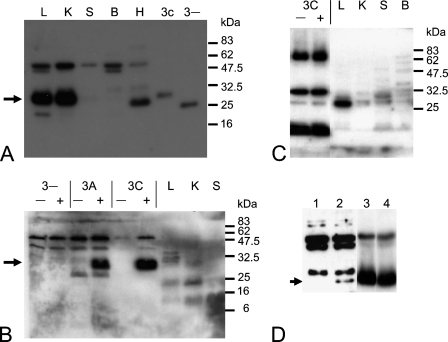
The specificity of the antibodies for KHK was first characterized by Western blotting (Figure 2). Recombinant mouse proteins (KHK-A, KHK-C, and KHK-3−) were expressed in E. coli, and crude extracts of soluble protein were prepared. Expression levels varied considerably, with KHK-A expressed at the highest level, whereas very little KHK-3− protein was recovered, mainly owing to its insolubility (not shown). This suggests that the KHK-3− protein (if translated in vivo) is not likely to be functional. The N-terminal antibody detected all three recombinant proteins. (Loading of the crude E. coli protein extracts was adjusted to obtain similar levels of each recombinant protein.) The estimated size of the KHK-C and KHK-A proteins was ∼30 kDa (KHK-A not shown), and that of KHK-3− was ∼28 kDa (Figure 2A). The C-terminal antibody, too, recognized the KHK-C and KHK-A recombinant proteins; however, larger quantities of recombinant protein were required, and additional bands were also detected in the E. coli extracts (Figure 2B). This antibody failed to detect the KHK-3− protein (which, as mentioned above, was present at much lower levels in the E. coli extract). Finally, Western blotting of induced and uninduced KHK-3C E. coli extracts indicated that the custom-made KHK-C antipeptide antibody was unable to detect the recombinant KHK protein, although it did react against a number of E. coli proteins (Figure 2C, lanes 1–2).
Figure 2.
Western blot of recombinant mouse proteins and tissue lysates. (A) N-terminal antibody. Tissue and cell extracts as follows: L (liver), K (kidney), S (spleen), B (brain), H (HepG2). Induced Escherichia coli extracts expressing the KHK-3C or KHK-3− protein. The 30-kDa band is indicated with an arrow. (B) C-terminal antibody. E. coli extracts were either uninduced (−) or induced (+) with isopropyl β-d-1-thiogalactopyranoside to express the KHK-3C, -3A or -3− proteins. The 30-kDa band is indicated with an arrow. (C) KHK-C–specific antibody. (D) N-terminal antibody on Khk3a/3a mouse brain tissue extracts (1), wild-type brain tissue extracts (2), Khk3a/3a mouse liver tissue extracts (3), and wild-type liver tissue extracts (4). The 30-kDa band is indicated with an arrow.
The two commercial antibodies (neither of which appeared isoform specific from the above analysis) were next used to examine KHK expression in lysates of mouse liver and kidney (high level of KHK-C mRNA) and brain and spleen (low level of KHK-A mRNA). The N-terminal antibody detected a major band in kidney and liver, matching the ∼30-kDa size of the recombinant proteins (Figure 2A). Brain and spleen exhibited a faint band of the same size, but also additional cross-reactive bands of various sizes. The dominant band in these tissues was ∼50 kDa in size. In fact, this 50-kDa band was detected in all tissue extracts. In the HepG2 cell line, the N-terminal antibody detected a major band slightly smaller than 30 kDa and, again, a 50-kDa band. Preincubation of the N-terminal antibody with recombinant human KHK-A lysate dramatically reduced the signal intensity of the 30-kDa band in mouse tissues (data not shown), suggesting that this antibody can indeed specifically detect the 30-kDa mouse KHK protein on Western blots.
The C-terminal antibody detected multiple bands in liver and kidney extracts, one of which was ∼30 kDa in size, although this was not the dominant band (Figure 2B). The signal intensity in the liver and kidney was greater compared with that in the spleen, but this antibody produced a consistently weaker signal than did the N-terminal antibody. Thus, although the C-terminal antibody could recognize denatured recombinant mouse KHK, it appeared less specific and less sensitive at detecting KHK in tissue extracts.
Finally, the KHK-C–specific antipeptide antibody detected a single strong band in mouse liver extract only (Figure 2C), but of a different size from that detected by the N-terminal antibody (not shown). Together with its inability to detect recombinant KHK-C, this indicated that this antipeptide antibody was not useful for further tissue analysis.
From the above, only the N-terminal antibody appeared suitable for specific detection of KHK in mouse tissues; even then, in low-expressing tissues, the identities of the additional non–30-kDa bands remained unclear. To address the latter question, we examined brain extracts from mice genetically lacking KHK-A (Khk3a/3a). Only the predicted 30-kDa band was absent from these extracts, confirming its specific identity as KHK. All other bands, including the predominant one at 50 kDa, were still present in the knockout mice, and must therefore reflect nonspecific cross-reactivity (Figure 2D). Although the expression level is low, this result also provides the first direct demonstration, at the protein level, that KHK-A is the only isoform of KHK protein present in brain. In the liver, in contrast, Khk3a/3a mice exhibit normal KHK-C expression (Figure 2D).
The superior qualities of the N-antibody were confirmed in immunohistochemistry (see below). It was also the only antibody to be competed out by prior incubation with crude recombinant KHK protein extracts.
Localization of KHK in Mouse Tissues
We examined the histological distribution of KHK within highly expressing tissues (liver and kidney), in addition to one lower expressing tissue (pancreas).
Kidney
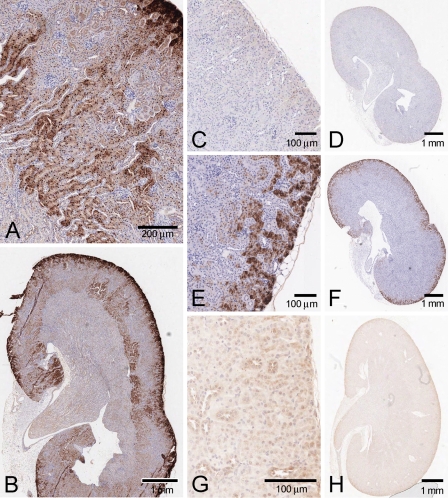
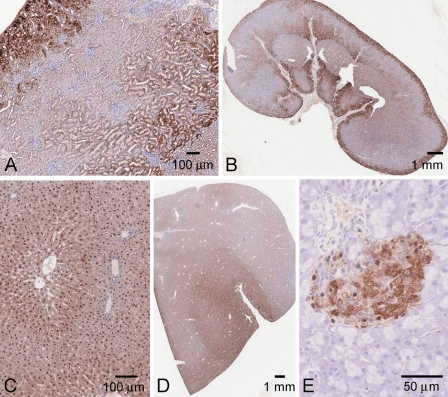
In mouse kidney, KHK detected by the N-terminal antibody was localized mostly to the proximal tubules (Figures 3A and 3B). Staining in the straight segment of the proximal tubules was found in the outer cortex and extended into the outer medulla. There was minimal staining in the distal tubules, and virtual absence from the glomerulus, although there were occasional immunolabeled patches in the surrounding Bowman's capsule epithelium. Staining was virtually absent from collecting ducts and thick ascending limb.
Figure 3.
KHK localization in mouse kidney. (A,B) N-terminal antibody, precompeted with control E. coli extract. (C,D) Same antibody, precompeted with E. coli extract containing mouse KHK-A. (E,F) Aldehyde reductase (proximal tubule marker). (G,H) N-terminal antibody, tissue from KhkΔ/Δ mice.
Both cytoplasmic staining and intense nuclear staining were seen in proximal tubule epithelial cells. In the outer cortex, there tended to be high levels of cytoplasmic staining, whereas intense nuclear staining was more apparent in the tubules deeper into the cortex and outer medulla.
No staining was seen with control preimmune purified total IgY (not shown), and preincubation of the N-terminal antibody with either mouse or human KHK-A recombinant protein extract reduced the immunostaining considerably (Figures 3C and 3D).
In the outer cortex, we confirmed that the expression was predominantly in the proximal tubules by comparison to the localization of aldehyde reductase, which has been shown to be proximal tubule–specific in this region (Barski et al. 2005) (Figures 3E and 3F).
Using the C-terminal antibody, even at high concentration, immunohistochemical staining was considerably weaker and more variable. Preincubation of the C-terminal antibody with either mouse or human recombinant KHK-A proteins also failed to quench the immunostaining (not shown). This antibody was therefore not considered useful for further immunohistochemistry.
The renal localization defined by the N-terminal antibody was further validated using tissue from the KhkΔ/Δ knockout mice. There was dramatically reduced proximal tubule staining (both cytoplasmic and nuclear) in the cortex and outer medulla. In the distal tubule, knockout animals showed a somewhat enhanced background cytoplasmic immunolabeling, although the nuclei (as in wild-type animals) remained unstained (Figures 3G and 3H).
Overall, these experiments imply that endogenous renal KHK protein is most likely confined to the proximal tubule; this pattern was consistently observed for tissue samples obtained from both sexes and a range of ages.
Liver
In mouse liver, staining by the N-terminal antibody was variable (Figure 4); in some areas, the hepatocytes were more intensely immunolabeled than in others, with staining of both the nucleus and the cytoplasm, although not all nuclei in an intensely immunolabeled region were positive. The strongest expression was generally seen in perivascular hepatocytes, although this too was variable, with highest expression in some cases around the central venule and in others around portal tracts. Preincubation of the antibody with either mouse or human KHK-A recombinant protein again reduced the level of immunolabeling considerably (not shown), supporting its specificity. Also, in KHK-null mice, the N-terminal antibody staining in hepatocytes, both cytoplasmic and nuclear, was again dramatically reduced (data not shown).
Figure 4.
KHK localization in mouse liver (N-terminal antibody). (A) liver lobe, low power, (B) high-power view.
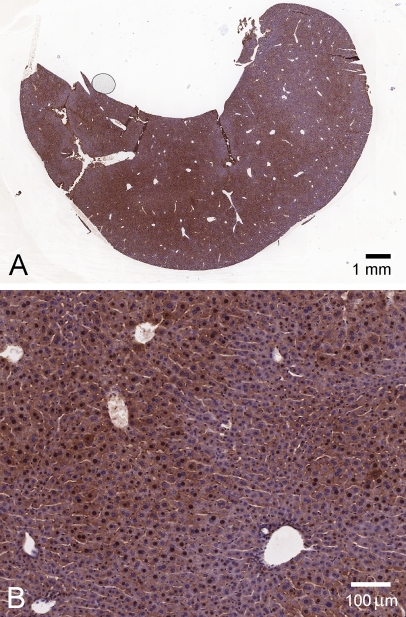
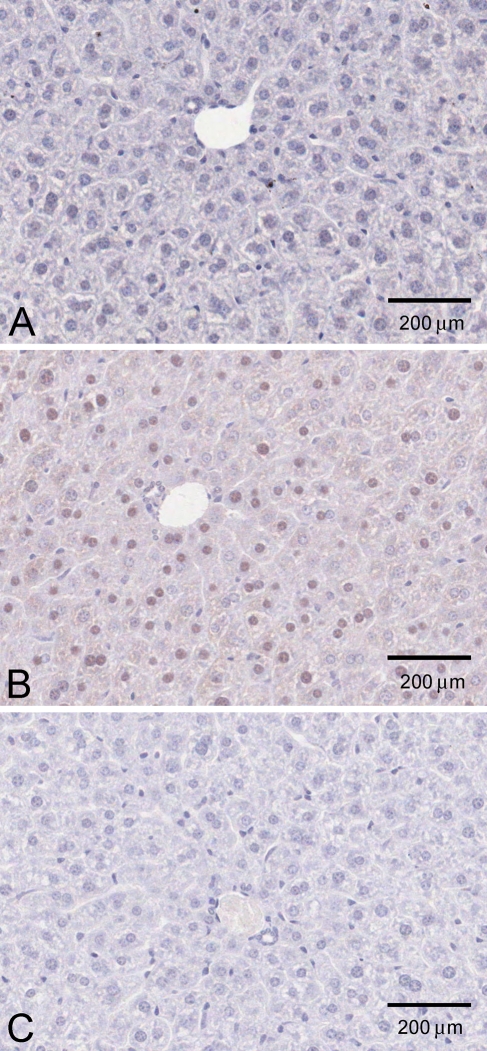
We performed additional experiments to address the possibility that the nuclear localization of KHK immunoreactivity was an artifact, either of formalin fixation or of preferential enzyme-mediated DAB production in the nucleus. Using Carnoy's fixative, both cytoplasmic staining and nuclear staining were again observed, in both kidney and liver (Figure 5). We also used a gold-conjugated secondary antibody to examine liver. Although much fainter than the HRP-mediated DAB staining, brown staining of nuclei and cytoplasm was again clearly detectable in normal liver (Figure 6B) but not liver of KhkΔ/Δ animals (Figure 6A).
Figure 5.
KHK localization in mouse kidney (A,B) and liver (C,D), using Carnoy's fixative (N-terminal antibody).
Figure 6.
Silver-enhanced immunogold detection of KHK in liver (N-terminal antibody). (A) KhkΔ/Δ knockout animal. (B) Wild-type animal. (C) Wild-type animal, primary antibody omitted.
Pancreas
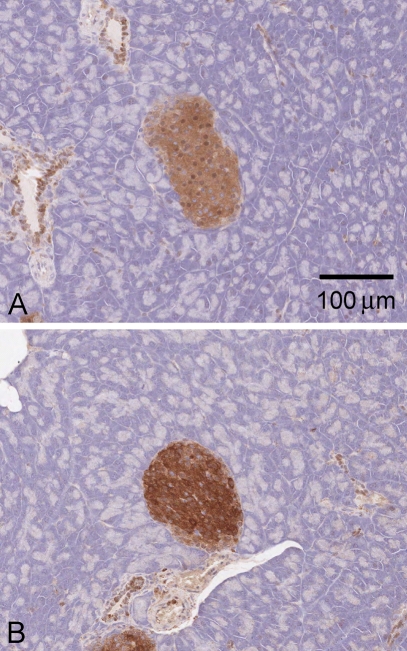
In mouse pancreas, the N-terminal antibody predominantly stained islets, with some labeling also in connective tissue; staining was generally absent from the surrounding acinar cells (Figure 7A). Both cytoplasm and nucleus were labeled, and staining was blocked by prior incubation with recombinant protein (not shown). Surprisingly, however, the islet staining was retained in KhkΔ/Δ knockout animals, suggesting that it does not represent true KHK immunoreactivity (Figure 7B). Immunolabeling in the acinar and connective tissue was also still present, suggesting that these were similarly nonspecific.
Figure 7.
KHK immunostaining of mouse pancreas (N-terminal antibody). (A) Normal mouse pancreas. (B) Pancreas of homozygous KhkΔ/Δ knockout mouse.
Localization of KHK in Rat Tissues
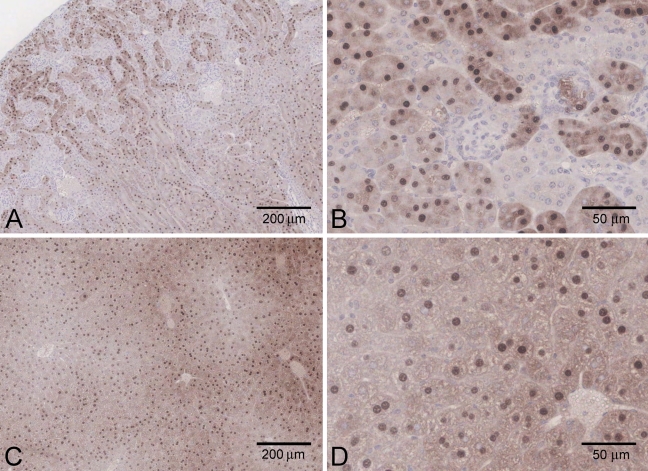
Using the N-terminal antibody, localization patterns in rat kidney, liver, and pancreas appeared very similar to those seen in mouse tissues (Figure 8).
Figure 8.
Immunohistochemical staining of rat tissues, using the N-terminal antibody. (A,B) Kidney. (C,D) Liver. (E) Pancreas.
In the kidney, strongest expression was again seen in proximal tubules in the outer cortex, with intense nuclear reactivity. As in the mouse, there was a further zone of intense tubular staining at the cortico–medullary border (Figures 8A and 8B).
In hepatocytes, similarly to the mouse, there was cytoplasmic and nuclear immunolabeling, with the cytoplasmic staining somewhat less intense in the periportal region (Figures 8C and 8D).
In the pancreas, staining was again restricted to the islets (Figure 8E), and was blocked by preincubation of the N-terminal antibody with the recombinant proteins. However, in light of the result obtained with the knockout mouse pancreas, the reliability of this result is unclear.
Discussion
Our results indicate that considerable care is needed in the interpretation of KHK immunolocalization data. Of the three antibodies assessed, only the N-terminal antibody proved specific for detection of KHK in tissues. This antibody detects all three recombinant KHK proteins in crude E. coli extracts and an endogenous protein of the predicted size, and of an intensity correlating well with previous measurements of transcript levels in different tissues (Hayward and Bonthron 1998). The remaining two antibodies, both raised to peptide epitopes, were disappointing. This was especially true of the KHK-C–specific antibody, which was unable to detect the recombinant protein at all. Our inability to obtain an isoform-specific reagent for use in immunohistochemistry means that we have had to infer the identity of KHK-immunoreactive material in brain tissue genetically, by observing the loss of immunoreactivity in mice with a selective deficiency of KHK-A (Figure 2D). The C-terminal antibody did appear to have a low affinity for KHK by Western blotting and immunohistochemistry, but was unable to interact with the recombinant proteins in competition experiments. The reason for this is unknown, inasmuch as the epitope used to produce this antibody is not buried, but accessible on the surface of the KHK dimer (PDB accession 2HW1, 2HQQ; Trinh et al. 2009). However, the peptide immunogen was derived from the human sequence, which differs from the mouse sequence at four amino acids, perhaps explaining the apparent lack of specificity.
The strong KHK immunostaining in hepatocytes is consistent with the abundance of fructokinase activity in the liver (Weiser and Quill 1975; Raushel and Cleland 1977a,b; James et al. 1982; Bais et al. 1985; Werman and Bhathena 1995; Giroix et al. 2006; Miller and Adeli 2008), which is the primary site of dietary fructose metabolism following intestinal absorption and transport via the portal vein. Some liver samples displayed strongest immunolabeling around the portal tract, whereas others had more immunolabeling surrounding the central venule. Expression patterns of other metabolic enzymes are known to vary within the hepatic lobule, with some gluconeogenic enzymes more highly expressed around the portal tract, and some glycolytic enzymes around the central venule (Braeuning et al. 2006).
In mouse and rat kidney, KHK was predominantly localized to the straight segments of the proximal tubules, which lie in medullary rays, extending through the full cortical thickness. These findings agree with data obtained from rats fed a high-fructose diet; in these animals, it was the proximal straight segment of the tubule that contained the highest levels of fructose-1-phosphate and fructokinase activity (Burch et al. 1980). The immunohistochemical data are also consistent with transcript localization by RNA in situ hybridization (Hwa et al. 2006). The function of KHK in the renal tubule is currently unknown. Although glucose is known to be reabsorbed in the proximal tubule, the level of fructose in non-portal blood is very low, suggesting that tubular fructose reabsorption is unlikely to be significant. An alternative possibility is that KHK may metabolize fructose generated in the proximal tubule itself through the polyol pathway. In this pathway, glucose is converted to sorbitol and then fructose by the action of aldose reductase and sorbitol dehydrogenase, both present in the proximal tubule (Corder et al. 1977; Sands et al. 1989; Lee et al. 1995; Burger-Kentischer et al. 1999).
The attempt to immunolocalize KHK in mouse pancreas clearly illustrates the potential pitfalls in such studies. Even though the apparently islet-specific staining was competed by recombinant KHK, its retention in the the KhkΔ/Δ knockout animals indicated its artifactual nature. Consistent with this finding, Western blot analysis of wild-type and knockout pancreas tissue using the same antibody failed to identify a KHK-specific band, owing to the many artifactual bands detected and their position (data not shown).
The limited antibody specificity and sensitivity have proven even more problematic in assessing tissues with lower levels of KHK expression (those that express KHK-A rather than KHK-C). This is compounded in the pancreas, where both splice variants are expressed, owing to the lack of isoform-specific antibodies.
Fructokinase activity has been found previously in rat pancreas (Malaisse et al. 1989), although, consistent with our findings, at considerably lower levels than in liver (Giroix et al. 2006). It was further suggested that KHK levels were similar in partially purified β cells and non-β cells (Giroix et al. 2006). Because the KHK-A and KHK-C proteins have very different biochemical properties (Asipu et al. 2003) and both splice variants are present in the pancreas, the physiological function(s) of KHK in this organ remain unclear. One suggestion has been that the presence of KHK in the islets could indicate a role in modulating glucose-regulated (GK-dependent) insulin and glucagon secretion. Fructose has indeed been described as influencing islet GK activity (Malaisse et al. 1990; Sener and Malaisse 1996; Sener et al. 2004); however, the mechanism of this effect may not involve the GK regulatory protein (GKRP) sequestration of GK in the nucleus, inasmuch as GKRP is thought to be absent from islets (Brown et al. 1997; Tiedge et al. 1999; Zelent et al. 2006).
We observed both nuclear and cytoplasmic KHK staining in formalin- and Carnoy's-fixed liver and kidney. The intensity of nuclear staining was quite variable even in adjacent cells. This brings to mind the nuclear–cytoplasmic shuttling behavior exhibited by GK (Toyoda et al. 1994; Brown et al. 1997). We therefore attempted to determine whether KHK localization in HepG2 cells changes upon manipulating fructose or glucose levels in culture media, but were not able to obtain reliable immunofluorescence results using the N-terminal antibody. Indeed, this antibody generates high levels of background fluorescence in frozen tissue, with no differences discernable between normal and knockout tissues (not shown). This finding may be relevant to a recent study (Meakin et al. 2007) in which cytoplasmic and cell membrane localization but no nuclear labeling was observed in frozen liver sections using the same N-terminal antibody. Indeed, we emphasize from our own experience that without the use of biological controls (knockout tissues), such findings are very hard to interpret. It is only by comparing normal and knockout tissues that we have been able to conclude with certainty that the nuclear staining we observe in liver and kidney is indeed genuine KHK protein. We obtained similar appearances, whether fixation was performed with paraformaldehyde or Carnoy's fixative (Figure 5). Despite this, we cannot rule out absolutely the possibility that even though at over 60 kDa the KHK dimer is not a small molecule, it may have diffused into the nucleus during tissue fixation.
KHK occupies a crucial metabolic position because of its rapid handling of large dietary fructose loads, which have been shown in both epidemiological and animal studies to predispose to type 2 diabetes and metabolic syndrome (Basciano et al. 2005; Johnson et al. 2007; Miller and Adeli 2008). These effects, including the accompanying renal dysfunction, may be mediated by local or systemic fructose-induced hyperuricaemia (Cirillo et al. 2006). The poorly understood cellular physiology of KHK within the kidney is therefore of particular interest, and could have an important influence on the development of renal damage induced by high-fructose diets. By appropriate dietary manipulation of our new KHK knockout mouse models, these hypotheses should now be amenable to direct experimental testing.
Acknowledgments
This work was supported by a project grant from Diabetes UK (RD04/0002833).
MIN6 cells were kindly provided by Dr. Jun-ichi Miyazaki. We thank Dr. Nic Orsi for help with sample collection, Doreen Crellin for immunohistochemistry sample processing, Dr. Jack Leek for MIN6 cell culture, and Lydia Teboul (Medical Research Council Mammalian Genetics Unit) for mouse tissue samples.
References
- Adelman RC, Ballard FJ, Weinhouse S (1967) Purification and properties of rat liver fructokinase. J Biol Chem 242:3360–3365 [PubMed] [Google Scholar]
- Asipu A, Hayward BE, O’Reilly J, Bonthron DT (2003) Properties of normal and mutant recombinant human ketohexokinases and implications for the pathogenesis of essential fructosuria. Diabetes 52:2426–2432 [DOI] [PubMed] [Google Scholar]
- Bais R, James HM, Rofe AM, Conyers RA (1985) The purification and properties of human liver ketohexokinase. A role for ketohexokinase and fructose-bisphosphate aldolase in the metabolic production of oxalate from xylitol. Biochem J 230:53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard FJ, Oliver IT (1964) Ketohexokinase, isoenzymes of glucokinase and glycogen synthesis from hexoses in neonatal rat liver. Biochem J 90:261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski OA, Papusha VZ, Ivanova MM, Rudman DM, Finegold MJ (2005) Developmental expression and function of aldehyde reductase in proximal tubules of the kidney. Am J Physiol Renal Physiol 289:F200–207 [DOI] [PubMed] [Google Scholar]
- Basciano H, Federico L, Adeli K (2005) Fructose, insulin resistance, and metabolic dyslipidemia. Nutr Metab 2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeuning A, Ittrich C, Kohle C, Hailfinger S, Bonin M, Buchmann A, Schwarz M (2006) Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J 273:5051–5061 [DOI] [PubMed] [Google Scholar]
- Brown KS, Kalinowski SS, Megill JR, Durham SK, Mookhtiar KA (1997) Glucokinase regulatory protein may interact with glucokinase in the hepatocyte nucleus. Diabetes 46:179–186 [DOI] [PubMed] [Google Scholar]
- Burch HB, Choi S, Dence CN, Alvey TR, Cole BR, Lowry OH (1980) Metabolic effects of large fructose loads in different parts of the rat nephron. J Biol Chem 255:8239–8244 [PubMed] [Google Scholar]
- Burger-Kentischer A, Muller E, Neuhofer W, Marz J, Thurau K, Beck F (1999) Expression of aldose reductase, sorbitol dehydrogenase and Na+/myo-inositol and Na+/Cl-/betaine transporter mRNAs in individual cells of the kidney during changes in the diuretic state. Pflugers Arch 437:248–254 [DOI] [PubMed] [Google Scholar]
- Cirillo P, Sato W, Reungjui S, Heinig M, Gersch M, Sautin Y, Nakagawa T, et al. (2006) Uric acid, the metabolic syndrome, and renal disease. J Am Soc Nephrol 17:S165–S168 [DOI] [PubMed] [Google Scholar]
- Corder CN, Collins JG, Brannan TS, Sharma J (1977) Aldose reductase and sorbitol dehydrogenase distribution in rat kidney. J Histochem Cytochem 25:1–8 [DOI] [PubMed] [Google Scholar]
- Das DK, Neogi A, Steinberg H (1984) Fructose utilization by lung. J Appl Physiol 56:333–337 [DOI] [PubMed] [Google Scholar]
- Giroix MH, Jijakli H, Courtois P, Zhang Y, Sener A, Malaisse WJ (2006) Fructokinase activity in rat liver, ileum, parotid gland, pancreas, pancreatic islet, B and non-B islet cell homogenates. Int J Mol Med 17:517–522 [PubMed] [Google Scholar]
- Hayward BE, Bonthron DT (1998) Structure and alternative splicing of the ketohexokinase gene. Eur J Biochem 257:85–91 [DOI] [PubMed] [Google Scholar]
- Holgate CS, Jackson P, Pollard K, Lunny D, Bird CC (1986) Effect fixation on T and B lymphocyte surface membrane antigen demonstration in paraffin processed tissue. J Pathol 149:293–300 [DOI] [PubMed] [Google Scholar]
- Hwa JS, Kim HJ, Goo BM, Park HJ, Kim CW, Chung KH, Park HC, et al. (2006) The expression of ketohexokinase is diminished in human clear cell type of renal cell carcinoma. Proteomics 6:1077–1084 [DOI] [PubMed] [Google Scholar]
- James HM, Bais R, Edwards JB, Rofe AM, Conyers AJ (1982) Models for the metabolic production of oxalate from xylitol in humans: a role for fructokinase and aldolase. Aust J Exp Biol Med Sci 60:117–122 [DOI] [PubMed] [Google Scholar]
- Johnson RJ, Segal MS, Sautin Y, Nakagawa T, Feig DI, Kang DH, Gersch MS, et al. (2007) Potential role of sugar (fructose) in the epidemic of hypertension, obesity and the metabolic syndrome, diabetes, kidney disease, and cardiovascular disease. Am J Clin Nutr 86:899–906 [DOI] [PubMed] [Google Scholar]
- Lee FK, Lee AY, Lin CX, Chung SS, Chung SK (1995) Cloning, sequencing, and determination of the sites of expression of mouse sorbitol dehydrogenase cDNA. Eur J Biochem 230:1059–1065 [DOI] [PubMed] [Google Scholar]
- Malaisse WJ, Malaisse-Lagae F, Davies DR, Vandercammen A, Van Schaftingen E (1990) Regulation of glucokinase by a fructose-1-sensitive protein in pancreatic islets. Eur J Biochem 190:539–545 [DOI] [PubMed] [Google Scholar]
- Malaisse WJ, Malaisse-Lagae F, Davies DR, Van Schaftingen E (1989) Presence of fructokinase in pancreatic islets. FEBS Lett 255:175–178 [DOI] [PubMed] [Google Scholar]
- Meakin PJ, Fowler MJ, Rathbone AJ, Allen LM, Ransom BR, Ray DE, Brown AM (2007) Fructose metabolism in the adult mouse optic nerve, a central white matter tract. J Cereb Blood Flow Metab 27:86–99 [DOI] [PubMed] [Google Scholar]
- Miller A, Adeli K (2008) Dietary fructose and the metabolic syndrome. Curr Opin Gastroenterol 24:204–209 [DOI] [PubMed] [Google Scholar]
- Miyazaki J, Araki K, Yamato E, Ikegami H, Asano T, Shibasaki Y, Oka Y, et al. (1990) Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127:126–132 [DOI] [PubMed] [Google Scholar]
- Raushel FM, Cleland WW (1977a) Bovine liver fructokinase: purification and kinetic properties. Biochemistry 16:2169–2175 [DOI] [PubMed] [Google Scholar]
- Raushel FM, Cleland WW (1977b) Determination of the rate-limiting steps and chemical mechanism of fructokinase by isotope exchange, isotope partitioning, and pH studies. Biochemistry 16:2176–2181 [DOI] [PubMed] [Google Scholar]
- Sands JM, Terada Y, Bernard LM, Knepper MA (1989) Aldose reductase activities in microdissected rat renal tubule segments. Am J Physiol 256:F563–569 [DOI] [PubMed] [Google Scholar]
- Sener A, Leclercq-Meyer V, Malaisse WJ (2004) Immediate and delayed effects of D-fructose upon insulin, somatostatin, and glucagon release by the perfused rat pancreas. Endocrine 24:73–81 [DOI] [PubMed] [Google Scholar]
- Sener A, Malaisse WJ (1996) Hexose metabolism in pancreatic islets: apparent dissociation between the secretory and metabolic effects of D-fructose. Biochem Mol Med 59:182–186 [DOI] [PubMed] [Google Scholar]
- Tiedge M, Steffeck H, Elsner M, Lenzen S (1999) Metabolic regulation, activity state, and intracellular binding of glucokinase in insulin-secreting cell. Diabetes 48:514–523 [DOI] [PubMed] [Google Scholar]
- Toyoda Y, Miwa I, Kamiya M, Ogiso S, Nonogaki T, Aoki S, Okuda J (1994) Evidence for glucokinase translocation by glucose in rat hepatocytes. Biochem Biophys Res Commun 204:252–256 [DOI] [PubMed] [Google Scholar]
- Trinh CH, Asipu A, Bonthron DT, Phillips SE (2009) Structures of alternatively spliced isoforms of human ketohexokinase. Acta Crystallogr D Biol Crystallogr 65:201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser MM, Quill H (1975) Estimation of fructokinase (ketohexokinase) in crude tissue preparations. Methods Enzymol 41:61–63 [DOI] [PubMed] [Google Scholar]
- Werman MJ, Bhathena SJ (1995) Fructose metabolising enzymes in the rat liver and metabolic parameters: interactions between dietary copper, type of carbohydrates, and gender. J Nutr Biochem 6:373–379 [DOI] [PubMed] [Google Scholar]
- Zelent D, Golson ML, Koeberlein B, Quintens R, van Lommel L, Buettger C, Weik-Collins H, et al. (2006) A glucose sensor role for glucokinase in anterior pituitary cells. Diabetes 55:1923–1929 [DOI] [PubMed] [Google Scholar]