Abstract
Peripheral nerve regeneration can be enhanced by the stimulation of formation of bands of Büngner prior to implantation. Aligned electrospun poly(ε-caprolactone) (PCL) fibers were fabricated to test their potential to provide contact guidance to human Schwann cells. After 7 days of culture, cell cytoskeleton and nuclei were observed to align and elongate along the fiber axes, emulating the structure of bands of Büngner. Microarray analysis revealed a general down-regulation in expression of neurotrophin and neurotrophic receptors in aligned cells as compared to cells seeded on two-dimensional PCL film. Real-time-PCR analyses confirmed the up-regulation of early myelination marker, MAG, and the down-regulation of NCAM-1, a marker of immature Schwann cells. Similar gene expression changes were also observed on cells cultured on randomly-oriented PCL electrospun fibers. However, up-regulation of the myelin-specific gene, P0, was observed only on aligned electrospun fibers, suggesting the propensity of aligned fibers in promoting Schwann cell maturation.
Keywords: Electrospinning, Peripheral nerve regeneration, Contact guidance, Neural tissue engineering, Schwann cell
1. INTRODUCTION
Electrospinning is a versatile technique of producing micro- and nano-fibrous scaffolds. Its popularity in tissue engineering has increased dramatically over recent years [1]. With the possibility of generating fibers in the nanoscale and the resulting scaffolds with organized architecture, electrospun fibrous meshes may mimic the extracellular matrix (ECM). Such scaffolds hold the promise to provide the topographic cues to the seeded cells and may potentially enhance tissue regeneration. A few studies have shown that aligned electrospun scaffolds are able to provide contact guidance to cultured cells, resulting in an elongation and alignment of cells along the axes of the fibers [2–4]. The majority of the studies revolve around the evaluation of cell morphological changes, whilst some assess the preservation of cell phenotype via gene or protein expression analysis [5, 6]. Quantitative evaluation of changes in cellular function under the influence of topographic cues provided by electrospun fibers is still limited [3, 7, 8].
The potential of electrospun fibrous scaffolds in enhancing nerve regeneration was demonstrated previously [9]. Electrospun fibers can encapsulate bioactive drugs and proteins [10–12] and may be used as tissue scaffolds for direct in vivo applications [9]. Inclusion of aligned electrospun fibers in a nerve conduit could enhance sciatic nerve regeneration over a 15 mm critical size defect in rats after 3 months post-implantation [8]. While tissue regeneration is clearly observed in the presence of aligned electrospun fibers, the possible mechanism behind the enhanced nerve regeneration remains unclear. Since Schwann cells play a crucial role in nerve regeneration and are likely to be in close contact with the aligned electrospun fibers, it would be informative to understand the functional changes in Schwann cells with respect to the contact guidance offered by the aligned fibers. This understanding may also serve as a good starting point for future analyses involving the use of topographical cues to accelerate the formation of bands of Bünger to enhance nerve regeneration.
In this study, human Schwann cells (hSCs) were cultured on electrospun poly(ε-caprolactone) (PCL) fibers for a period of 7 days. The morphological and gene expression changes in the cells under the influence of topographic cues from the electrospun fibers were evaluated.
2. MATERIALS AND METHODS
Scaffold Fabrication
PCL films were fabricated by compression molding, subjecting 0.5 g of PCL polymer (Mw: 60,000; Sigma-Aldrich Corporation) to a uniaxial compression load of 8 × 103 kg for 2 minutes at 65°C. A mixture of organic solvents comprising dichloromethane and methanol (Sigma-Aldrich Corporation) at a volume ratio of 8:2 was used to dissolve PCL. In order to obtain aligned PCL fibers, a solution of 16 wt% of PCL was dispensed at a flow rate of 4 ml/h and electrospun under a voltage of 7 kV. The PCL fibers were collected on a PCL film mounted on a grounded target rotating at ~2200 rpm. For scaffolds comprising randomly oriented PCL fibers, 14 wt% of PCL was dispensed at 3 ml/h under a voltage of 7 kV and collected on a grounded rotating PCL film (~200rpm). The polymer supply-to-target distance was set at 5 cm in both cases.
2.2. Scaffold Structure Evaluation
All scaffolds were sputter-coated with ~2.5 – 3 nm in thickness of chromium (Denton Vacuum, DV-502A), prior to observation under the scanning electron microscope (Leo Field Emission SEM, Leo 1530) at an accelerating voltage of 1kV. The average diameters of the electrospun fibers were determined by measuring at least 130 fibers using ImageJ 1.30v (National Institutes of Health, USA).
2.3. Human Schwann Cell Isolation
Primary human Schwann cells were isolated from the sciatic nerves of human fetuses therapeutically aborted within the first trimester. The partial nervous system tissues were obtained from de-identified aborted fetuses with local IRB (Institutional Review Board) approval. The Schwann cells were then purified by a modified Brockes method [13, 14]. Briefly, the dissected nerves were incubated in Hanks Plus (HBSS, 10 mM HEPES, 33.3 mM glucose and 5 μg/ml gentamicin) (Invitrogen Corporation) and collagenase solution for 45 min at 37°C. Thereafter, the collagenase mixture was aspirated and the nerves were digested in a mixture of 0.25% Trypsin and 1 ml of DNase by incubating at 37°C for 15 min. Fetal bovine serum (heat inactivated) was then added to end the digestion process and the nerves were washed in ice-cold dissection media (Hanks Plus, 0.3% bovine serum albumin (BSA), 12 mM MgSO4) twice. Following that, the tissue was gently dissociated through a plastic pipette tip. The solution was then filtered and the cells obtained were resuspended and plated onto tissue culture dish in Schwann cell medium (ScienCell Research Laboratories) for 24 h. On the second day, half of the volume of Schwann cell medium was aspirated and replaced with an equal volume of fresh medium along with Ara-C (10 μM) (Sigma). The concentration of Ara-C was decreased by half on the third day by aspirating half the volume of medium and replacing it with an equal volume of fresh medium. By the fourth day, the hSCs were washed with phosphate buffered saline (PBS, pH 7.4) warmed to 37°C for three times prior to adding fresh Schwann cell medium. Culturing of hSCs continued until the cell number was sufficient for seeding onto the PCL scaffolds. The medium was changed every 3 days, and half the volume of medium was exchanged with fresh medium each time. According to previous experiments, following this protocol, the purity of GFAP-expressing Schwann cells harvested is more than 97% after 2–3 weeks in culture.
2.4. Human Schwann Cell Culture
Human Schwann cells (P3) were cultured in three experimental groups comprising of PCL film (F), PCL film with aligned PCL fibers (A) and PCL film with randomly-oriented PCL fibers (R). Cells seeded on the PCL film were treated as the control group. The PCL scaffolds were cut to fit into the wells of a 12-well plate. The scaffolds were then sterilized by soaking in 70% ethanol for 30 min followed by exposure to ultra-violet radiation for another 30 min. The scaffolds were then rinsed with PBS three times prior to seeding the hSCs. The cell seeding density was 2 × 105 cells per scaffold. Each well was filled with 3 ml of Schwann cell medium. The medium was changed every 2 days by replacing half the original volume with an equal volume of fresh medium.
2.5. Morphological Evaluation
PCL samples were retrieved at days 3 and 7 and fixed in 4% paraformaldehyde. In staining for actin cytoskeleton, the cells were permeabilized in 0.05% Triton-X and 50 mM glycine in PBS for 20 min. The samples were then incubated in Oregon Green phalloidin (Molecular Probes) (1:500 dilution) and DAPI (Molecular Probes) (1:5000 dilution) for 30 min. The samples were washed three times in PBS between each step and incubation was carried out at room temperature.
For glial fibrillary acidic protein (GFAP) immunostaining, the samples were permeabilized in 0.2% Triton-X in PBS for 30 min and then blocked overnight at 4°C with 5% horse serum. Following that, the samples were incubated in a 1:200 dilution of GFAP mouse anti-human primary antibody (Dako) for 1 h, and thereafter, a mixture of fluorescein horse anti-mouse secondary antibody (Molecular Probes) (1:500 dilution) and DAPI (1:3000) for 2 h. All incubation steps, except overnight incubation, were carried out at room temperature. The samples were rinsed three times in PBS between each step. All samples were imaged on a Perkin Elmer UltraVIEW spinning disk confocal microscope.
2.6. Microarray Analysis
On Day 7, the hSCs on PCL scaffolds were lysed and the total RNA for each experimental group was extracted using Trizol reagent (Invitrogen Corporation) according to manufacturer’s protocol. The extracted total RNA was reconstituted in a mixture of DEPC water and RNase inhibitor (Roche Molecular Biochemical) (1:20 dilution). For each experimental group, the extracted RNA from 10 scaffolds was pooled to enhance the quantity of RNA available for microarray and subsequent real-time PCR analyses. The quality and quantity of the extracted RNA were evaluated using UV-spectrophotometry (SmartSpec Plus Spectrophotometer, Biorad Laboratories) and gel electrophoresis. For each experimental group, the concentration of total RNA was greater than 60ng/μl, the A260: A280 ratio was above 1.8 and distinct bands for 18S and 28S ribosomal RNA were observed. The RNA was labeled using the AmpoLabeling-LPR kit (SuperArray Bioscience Corporation) and hybridized onto the GEArray Q series human neurotrophin and receptors gene array (HS-018, SuperArray). For each experimental group, two microarrays were used. The arrays were detected using the Chemiluminescent Detection kit (SuperArray). The labeling of RNA, RNA hybridization and detection of the microarrays were carried out according to manufacturer’s protocols.
Fluorescence intensities of the microarrays were captured using x-ray films (Kodak) and a desktop scanner. Expression data were analyzed using GEArray Expression Analysis Suite (SuperArray) with β-actin as the housekeeping gene, and the gene expression of cells cultured on PCL film as the control for normalization. The analysis software computed the relative intensity ratios between samples using SuperArray’s proprietary calculation algorithm. The program also analyzed the signal intensities of each gene spot on the microarrays. Only signals with intensity within the detection range were analyzed; and among which only results corresponding to gene expression changes that were either two times greater or two times lower than the control data were accepted.
2.7. Real-time PCR Analyses
To validate the microarray results, real-time PCR analyses were conducted for a set of 6 selected genes as shown in Table 1. The exact sequences of the primers (Integrated DNA Technologies and SuperArray Bioscience Corporation) are also listed in the table. Table 2 shows the list of myelin-specific genes and immature Schwann cell marker, NCAM-1. This list of genes was not found on the microarray, but was a separate group of genes chosen to evaluate the maturation of Schwann cells. The same total RNA used for microarray analysis was reverse-transcripted into single-stranded cDNA using the Sensiscript® RT kit (Qiagen) following manufacturer’s protocol. A total sample volume of 50μl, which comprised of 25μl of SYBR Green mastermix (Applied Biosystems); 1.5μl of forward primer (10μM); 1.5μl of reverse primer (10μM); 1μl of cDNA; and 21μl of DEPC water, was used for the real-time PCR experiments. Each unknown sample was tested in triplicates and non-template controls were analyzed in duplicates. Real-time PCR analyses were carried out using Applied Biosystems 7300 Real-Time PCR System, with the following thermal cycle conditions: 50°C for 2min; 95°C for 10min; and 40 repeated cycles of 95°C for 15sec followed by 60°C for 1min. At the end of the program, a melting curve analysis was done and the PCR products were also analyzed using gel electrophoresis to ensure the absence of primer dimmers and non-specific PCR product amplification. β-actin was used as the housekeeping gene for the normalization of the gene expression data. Gene expressions of cells cultured on PCL film were used as the control. The replication efficiencies of the genes of interest and the housekeeping gene were verified as similar. Therefore, the comparison Ct (ΔΔCt) method was adopted for gene expression analyses.
Table 1.
Real-time PCR primer sequences of selected genes from microarray.
| Gene | Refseq Accession No. | Primer Sequence | Amplicon (bp) |
|---|---|---|---|
| β-actin | NM_001101 | F:5′-GGC ACC CAG CAC AAT GAA GAT CAA -3′ R:5′-ACT CGT CAT ACT CCT GCT TGC TGA -3′ |
134 |
| VGF | NM_003378 | F: 5′-TGC TCC AGG AAC TGC GAG ATT TCA -3′ R: 5′-TCT CCA GAT TCA CTC GGG TCA GC-3′ |
111 |
| IL-6 | NM_000600 | F: 5′-AGC CAC TCA CCT CTT CAG AAC GAA -3′ R: 5′-CAG TGC CTC TTT GCT GCT TTC ACA -3′ |
122 |
| BDNF | NM_001709 | F: 5′-AAG AGC TGT TGG ATG AGG ACC ACA -3′ R: 5′-AGG CTC CAA AGG CAC TTG ACT ACT -3′ |
110 |
| STAT 1 | NM_007315 | F: 5′-CAG CAA GTT CGG CAG CAG CTT AAA -3′ R: 5′-AAA GAC TGA AGG TGC GGT CCC ATA -3′ |
115 |
| PTN | NM_002825 | F: 5′-GTG CAA GCA AAC CAT GAA GAC CCA -3′ R: AGG GCT GTG TTC AGG TCA CAT TCT -3′ |
120 |
| CD40 | NM_001250 | F: 5′-AGC TGT GAG ACC AAA GAC CTG GTT -3′ R: TGG CAA ACA GGA TCC CGA AGA TGA -3′ |
127 |
Table 2.
Real-time PCR primer sequences of myelin specific proteins and immature Schwann cell marker.
| Gene name | Refseq Accession No. | Primer Sequence | Amplicon (bp) |
|---|---|---|---|
| PMP22 | NM_153321 | F: 5′-AAC TCG GAT TAC TCC TAC GGT TTC GC -3′ R: 5′-CAT TCG CGT TTC CGC AAG ATC ACA -3′ |
104 |
| P0 | NM_000530 | F: 5′-AAT TGC ACA AGC CAG GAA AGG ACG -3′ R: 5′-TCA CTG ACA GCT TTG GTG CTT CTG -3′ |
100 |
| MAG | NM_002361 | SuperArray Cat # PPH07097A | 100 – 200 |
| MBP | NM_002385 | SuperArray Cat # PPH02627A | 100 – 200 |
| NCAM 1 | NM_000615 | SuperArray Cat # PPH00639A | 100 – 200 |
2.8. Statistical Analysis
All results shown are expressed as mean ± standard error of mean (SE). Student t-test was used for the analysis of real-time PCR gene expression data.
3. RESULTS
3.1. Scaffold and Cell Morphology Characterization
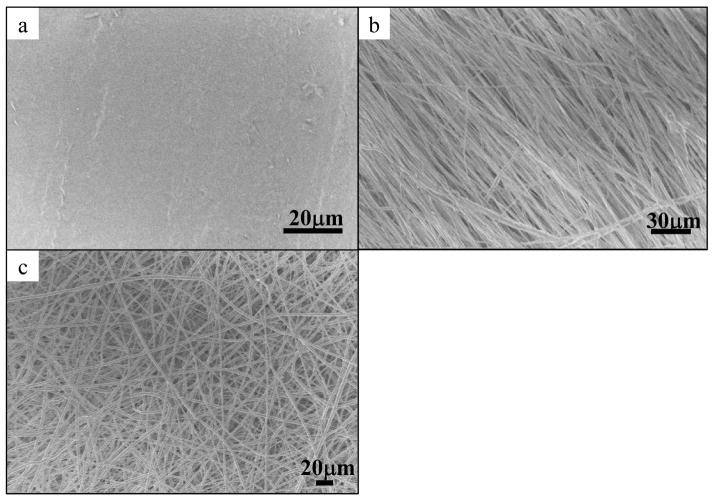
Figure 1 shows the SEM micrographs of PCL scaffolds. The surface of the PCL film sample was generally smooth as compared to the electrospun scaffolds. The average fiber diameters of aligned and randomly-oriented PCL fibers were φ = (1.03 ± 0.03) μm and φ = (2.26 ± 0.08)μm respectively.
Figure 1.
SEM micrographs of PCL scaffolds for hSC culture. a) PCL film, b) aligned PCL electrospun fibers, φ = 1.03 ± 0.03μm and c) randomly oriented PCL electrospun fibers, φ = 2.26 ± 0.08 μm.
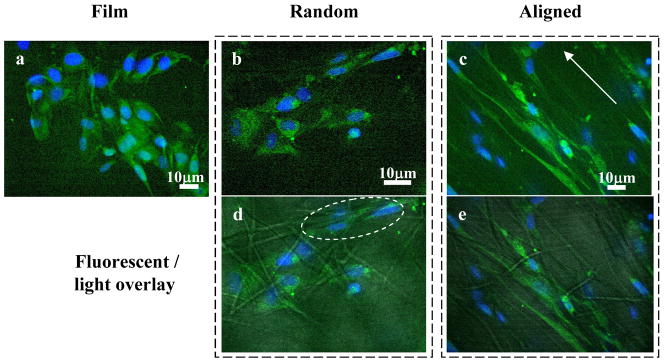
The difference in hSC morphologies when cultured on different PCL scaffolds is shown in Figure 2. On smooth PCL films, the cell actin cytoskeleton appeared more spread-out than cells cultured on electrospun fibers. Cell orientation was also haphazard on the films as well as on the randomly-oriented fibers. However, hSCs responded to the topographic cues from the randomly-oriented fibers by stretching the cell cytoskeleton across multiple fibers (Figure 2d). The effect of contact guidance provided by the aligned fibers appeared to be more dramatic than the randomly-oriented fibers. When cultured on aligned fibers, the cytoskeleton and nuclei aligned and elongated on the fiber axes. Such cellular alignment is very similar to that observed with other cell types such as human fibroblasts and Schwann cells in other studies [2, 4].
Figure 2.
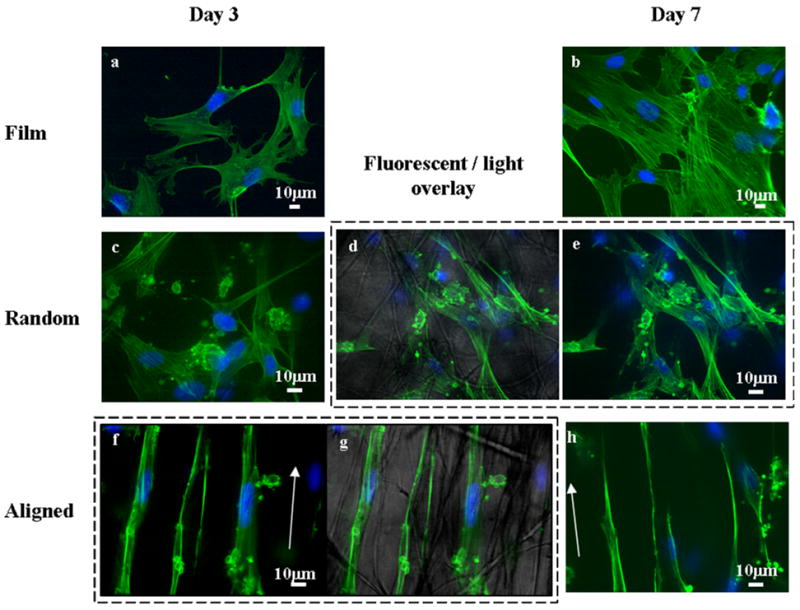
Confocal fluorescent images of human Schwann cells cultured on PCL scaffolds for 3 days (a, c and f) and 7 days (b, e and h). a) and b) PCL film; c), d) and e) randomly oriented PCL fibers; and f), g) and h) aligned PCL electrospun fibers, arrows depict directions of fiber alignment. d) and g) fluorescent-light images overlay. Green: actin cytoskeleton, blue: DAPI.
After 7 days of culture, hSCs continued to stain positively for GFAP as depicted in Figure 3. The cells appeared to respond to the contact guidance provided by the randomly-oriented and aligned electrospun fibers by aligning and elongating along the fibers as shown in Figures 3d (dashed circle) and 3e respectively.
Figure 3.
GFAP staining of human Schwann cells. hSCs still stains positively for GFAP at Day 7. Confocal immunofluorescent micrographs of hSCs cultured for 7 days on (a) PCL film, (b and d) randomly-oriented PCL electrospun fibers, and (c and e) aligned PCL fibers. (d and e) show fluorescent images (b and c respectively) superimposed on phase images of PCL electrospun fibers. Arrow in (c) indicates the general orientation of the aligned fibers. Dotted circles in (d) highlights the contact guidance provided by randomly-oriented PCL fibers. Green: GFAP, blue: DAPI
3.2. Microarray and Real-time PCR Analyses
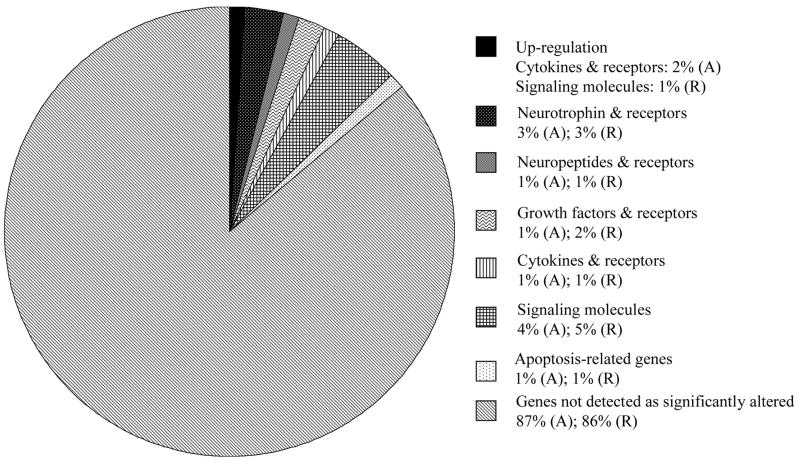
Table 3 shows the gene expression changes of hSCs cultured on PCL scaffolds, with the results being normalized with respect to the PCL film sample. (A) and (R) identifies the gene expression of cells cultured on aligned PCL fibers and randomly-oriented PCL fibers respectively. A general decrease in expression of neurotrophin and neurotrophic receptors was observed. Figure 4 shows the percentage of genes that were significantly up- or down-regulated in the microarray analyses. A total of 101 genes were analyzed using the focused microarray.
Table 3.
Gene expression changes identified by microarray analysis. Results normalized with respect to film sample. (A): gene expression of cells cultured on aligned PCL fibers; (R): gene expression of cells cultured on randomly-oriented PCL fibers.
| Genes | Fold changes with respect to FILM sample* |
|---|---|
| Neurotrophins & receptors: | |
| Brain-derived neurotrophic factor (BDNF) | 0.470 (A); 0.323 (R) |
| Nuclear receptor subfamily 1, group I, member 2(NR1I2) | 0.477 (A); 0.492 (R) |
| Fibroblast growth factor receptor substrate 2 (FRS2, SNT-1) | 0.363 (A); 0.367 (R) |
| Neuropeptides & receptors: | |
| Neuropeptide FF-amide peptide precursor (NPFF) | 0.453 (A); 0.270 (R) |
| Growth factors & receptors: | |
| Pleiotrophin (heparin binding growth factor 8, neurite growth-promoting factor 1) (PTN) | 0.206 (A); 0.107 (R) |
| Transforming growth factor, beta 1 (TGFb1) | 0.488 (R) |
| Cytokines & receptors: | |
| Interleukin 10 (IL-10) | 2.027 (A) |
| Interleukin 6 (interferon, beta 2) (IL-6) | 0.426 (A); 0.303 (R) |
| Leukemia inhibitory factor (LIF) | 2.450 (A) |
| Signaling molecules: | |
| MADS box transcription enhancer factor 2, polypeptide C (myocyte enhancer factor 2C) (MEF2C) | 0.268 (A); 0.239 (R) |
| Ribosomal protein S6 kinase 90kDa, polypeptide 6 (RSK4) | 0.251 (A); 0.207 (R) |
| Signal transducer and activator of transcription 1, 91kDa (Stat1) | 0.339 (A); 0.319 (R) |
| Signal transducer and activator of transcription 3 (acute-phase response factor) (Stat3) | 0.334 (A); 0.197 (R) |
| Signal transducer and activator of transcription 4 (STAT 4) | 2.131 (R) |
| Apoptosis related genes: | |
| Tumor necrosis factor receptor superfamily, member 6 (Fas/Apo-1/CD95) | 0.038 (A); 0.028 (R) |
Figure 4.
Gene expression of cells cultured on aligned PCL fibers (A) and randomly-oriented PCL fibers (R). Results were normalized against gene expression of cells cultured on PCL film and indicated the percentage of genes on the microarray that was down-regulated on electrospun scaffolds, unless otherwise stated. The total number of genes analyzed on the microarray was 101.
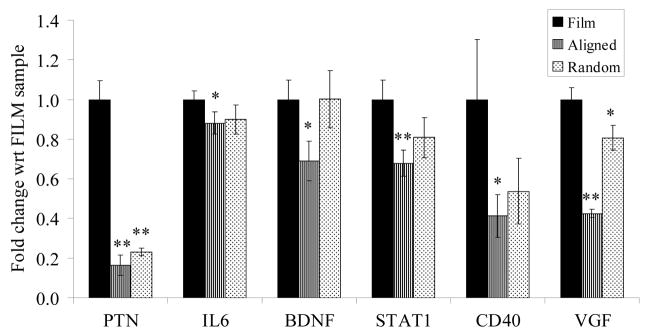
Real-time PCR shows similar trends in gene expression changes (Figure 5.) as the microarray analysis. Down-regulation of pleiotrophin (PTN), Interleukin 6 (IL-6), brain-derived neurotrophic factor (BDNF) and signal transducer and activator of transcription 1, 91kDa, (STAT1) gene expression was observed in cells cultured on electrospun fibers as compared to PCL film. CD40 antigen and VGF nerve growth factor inducible (VGF) gene expressions were also significantly decreased in hSCs cultured on electrospun fibers, although these genes were not identified as significantly expressed via microarray analysis.
Figure 5.
Real-time PCR results of selected genes from microarray. * p < 0.05; ** p < 0.01; t-test between electrospun fibers and film samples; n=3.
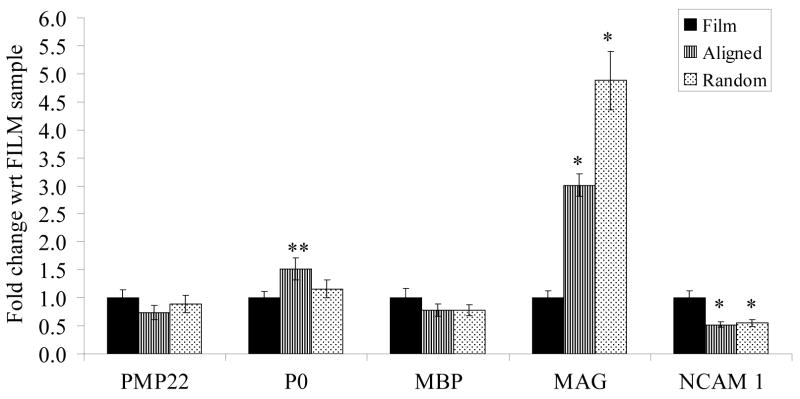
Figure 6 shows the changes in gene expression of an immature Schwann cell marker, neural cell adhesion molecule 1 (NCAM-1), and myelin-specific proteins: peripheral myelin protein (PMP22), myelin protein zero (P0), myelin basic protein (MBP) and myelin-associated glycoprotein (MAG). A significant up-regulation of MAG and a significant down-regulation of NCAM-1 were observed in hSCs cultured on electrospun fibers. P0 was significantly up-regulated in hSCs cultured on aligned PCL fibers.
Figure 6.
Real-time PCR results of myelin specific genes and immature Schwann cell marker, NCAM 1. Results are normalized with respect to film sample and suggest a possible enhancement in maturation of human Schwann cells cultured on electrospun fibers. * p < 0.05; ** p < 0.01, t-test between electrospun fibers and film samples; n=3.
4. DISCUSSION
Alignment of Schwann cells by microgrooves or micropatterned ECM molecules [15–17] are ways to imitate the formation of bands of Büngner in vitro. In vivo, nerve conduits seeded with aligned Schwann cells have been used [18, 19]. In all cases, Schwann cells are observed to elongate and align parallel to the long axes of the micropatterns or the nerve conduits. The main advantage of these aligned Schwann cells is the enhancement in the rate and extent of neurite elongation from dorsal root ganglia cultured on these aligned cells [19, 20]. Although these studies suggest the potential of contact guidance and Schwann cell alignment on enhancing nerve regeneration, the associated functional changes in Schwann cells have not been evaluated.
The effect of contact guidance on cell morphological changes is also evident in other cell types [21–23]. Functional changes corresponding to changes in cell morphology can range from apoptosis to proliferation, differentiation, contractility, rate of cell migration, and gene expression. [24–27]. The cellular response and sensitivity towards growth factors and mitogens may also be altered. In general, cells respond to a particular topography in a unique manner [21, 28, 29]. A few studies have analyzed the gene or protein expression changes of fibroblasts [30, 31], hepatocytes [32] and keratinocytes [33] in response to topographical cues, but none on human Schwann cells.
In this study, electrospun fibers were able to provide contact guidance to human Schwann cells. Although aligned fibers induced a more obvious alignment of cells, cells that were cultured on randomly-oriented fibers also appeared to respond to the topographic cues as shown in Figures 2d and 3d. As the length of a fiber between crosslinks in the random fibrous scaffold was still relatively long compared to the dimension of a Schwann cell, short-ranged topographic guidance was still possible. This partial alignment of cells may be the reason behind the similar gene expression changes observed in the two fibrous scaffolds. In contrast, Schwann cells cultured on PCL film, in the absence of topographical signals, adopted a random orientation and spread-out morphology similar to that observed on other two-dimensional smooth substrates [34–37].
During the electrospinning process, lower polymer concentration and polymer dispense rate were used for the fabrication of randomly-oriented PCL fibers. These parameters were obtained from a set of optimized conditions in order to fabricate fibers with diameters that were similar and in the same order of magnitude as the aligned fibers. The average diameters of aligned PCL fibers and randomly-oriented PCL fibers were φ = (1.03 ± 0.03) μm and φ = (2.26 ± 0.08) μm respectively. This variation is mainly attributed to the lower elongation forces experienced by the randomly-oriented fibers as they deposit onto a target that is rotating at a much slower speed. Due to the nature of the electrospinning process, it is difficult to achieve precise control over the diameter of the resulting fibers. The obtained fiber diameters were the closest that could be achieved in this study.
Although a small difference in average fiber diameter existed between the aligned and randomly-oriented fibers, we believe this is not a major contributor to the differing behaviour of the Schwann cells observed in this study. Yim et al. [38] studied the effects of the size of nanogratings on the alignment of smooth muscle cells. They demonstrated that the alignment of smooth muscle cells significantly differed only when the half pitch of the nanogratings was changed from 350nm to 2μm. Wen and Tresco [39] studied the effects of polypropylene filament diameter on neurite outgrowth and Schwann cell migration distance. They demonstrated that both neurite outgrowth and Schwann cell migration distance were significantly decreased as the diameters of the filament increased by two orders of magnitude, from 5μm to 200μm. The same study also illustrated that cellular behaviour was similar on filaments with diameters between 5μm to 100μm and that these observations were consistent with raw polypropylene filaments and filaments that were precoated with laminin or fibronectin. Taken together, these studies suggest that the small difference in fiber diameter in this study would not play a prominent role.
Evaluation of the CD40 and VGF genes was an attempt to test for false negatives in the microarray analysis. This is because, results from random experimental measurements would favor the null hypothesis that gene expressions are the same between sample groups [40]. Using the same argument along with the accurate validation of the trend in gene expression by real-time PCR analyses, Table 3 accurately lists the genes that were significantly altered on electrospun fibers. This table, however, is not exhaustive and other false negatives might have been missed.
Following nerve injury, denervated Schwann cells express higher levels of neurotrophic factors such as NGF, IL-6, LIF, BDNF, PTN and NT4/5 [41] due to the need to establish a more conducive microenvironment for the regeneration, maintenance and regulation of neuronal function [42, 43]. The fact that the gene expression of neurotrophic factors is generally down-regulated on electrospun fibers in this study suggests that the seeded hSCs have adopted a more mature phenotype. The fibrous scaffolds may, therefore, be a better microenvironment for Schwann cell attachment and growth as compared to a two-dimensional film. Since it has been observed that few major quality differences exist in the molecular expression between Schwann cells in distal nerve stumps and Schwann cells cultured in vitro in the absence of neurons [44], cultured Schwann cells may serve as a model of denervated Schwann cells in the distal stump of an injured nerve. Although we do not know if the relative levels of neurotrophic expression by seeded Schwann cells versus endogenous Schwann cells are comparable; we believe that the possible enhancement in the maturation of Schwann cells in contact with electrospun fibers as compared to a flat surface may thus be one of the reasons for the enhanced sciatic nerve regeneration observed in our previous study [9].
In order to validate the enhancement of Schwann cell maturation by topographic cues from electrospun fibers, the expression of the following genes has been analyzed: MAG, P0, MBP, PMP22 and NCAM-1. The first four genes are myelin-specific genes and are significantly up-regulated during Schwann cell myelination [45, 46]. NCAM-1, on the other hand, is associated with immature Schwann cells and mature non-myelinating cells and is often down-regulated during myelination [45]. Pro-myelinating Schwann cells also show up-regulation of MAG, P0 and MBP; and a down-regulation of NCAM-1 [46, 47].
As MAG is an important promyelinogenic marker in Schwann cells [47] and an early marker for myelin-differentiation [41], the significant up-regulation of MAG expression in hSCs cultured on electrospun fibers indicates the efficacy of these fibers in driving hSCs towards the pro-myelinating state. Since P0 and MBP are later gene products of myelin-differentiation [41] and PMP22 is associated with myelin formation, the general lack of differences in gene expression between the two-dimensional film and electrospun fibers is not unexpected. The more significant up-regulation of P0 in hSCs cultured on aligned fibers, however, seems to suggest that the alignment and elongation of hSCs on aligned fibers could promote the maturation of hSCs more than the randomly oriented fibers.
Schwann cell differentiation signals do not come solely from soluble biochemical signals, but also from the extracellular matrix [46]. Progression towards myelin formation requires signals from the basement membrane. Although the exact mechanism behind the pro-myelination of Schwann cells by the ECM is unknown, it is possible that integrins may interact with the cytoskeleton or regulate myelin-related gene expressions [46]. As shown in this study, hSCs were directed towards the pro-myelinating state by topographic cues from electrospun fibers. While the exact mechanism relating the changes in cell morphology to gene expression changes remains unknown, electrospun scaffolds, particularly scaffolds with aligned fibers show potential in promoting the maturation of Schwann cells. These scaffolds may, therefore, be an attractive platform for cell transplantation application, where cells may be primed towards the pro-myelinating stage prior to transplantation. This control over the differentiation of Schwann cells may help improve the outcome of cell-based approaches to cure demyelinated lesions.
5. CONCLUSIONS
The effects of contact guidance provided by electrospun fibrous scaffolds on the functional changes in human Schwann cells were evaluated in this study. hSCs adopted a spread-out morphology on PCL films. On random PCL fibers, they were randomly oriented, but stretched across multiple electrospun fibers and elongated along the fiber axes. In contrast, when cultured on aligned fibers, all cells aligned and elongated unidirectionally along the fiber axes. Decrease in expression of neurotrophin and neurotrophic receptors from cells cultured on electrospun fibers suggested a more mature phenotype adopted by the hSCs. This conclusion was further supported by the up-regulation in MAG and down-regulation in NCAM-1 expressions. Aligned electrospun fibers appeared to enhance Schwann cell maturation more than randomly-oriented fibers, as suggested by the more significant up-regulation of P0 expression. Such electrospun scaffolds may be an interesting platform for transplantation of ‘primed’ cells for the treatment of demyelinating lesions or for enhancing peripheral nerve regeneration.
Acknowledgments
This work is partially support by NIH (EB003447), and S Y Chew would like to thank the support of the NTU Overseas Scholarship.
Footnotes
This work is supported by NIH (EB003447) and the Nanyang Technological University Overseas Scholarship.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chew SY, Wen Y, Dzenis Y, Leong KW. The role of electrospinning in the emerging field of nanomedicine. Current Pharmaceutical Design. 2006;12:4751–4770. doi: 10.2174/138161206779026326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee CH, Shin HJ, Cho IH, Kang YM, Kim IA, Park KD, et al. Nanofiber alignment and direction of mechanical strain affect the ECM production of human ACL fibroblast. Biomaterials. 2005;26(11):1261–1270. doi: 10.1016/j.biomaterials.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 3.Zong X, Bien H, Chung C-Y, Yin L, Fang D, Hsiao BS, et al. Electrospun fine-textured scaffolds for heart tissue constructs. Biomaterials. 2005;26:5330–5338. doi: 10.1016/j.biomaterials.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 4.Schnell E, Klinkhammer K, Balzer S, Brook G, Klee D, Dalton P, et al. Guidance of glial cell migration and axonal growth on electrospun nanofibers of poly-epsilon-caprolactone and a collagen/poly-epsilon-caprolactone blend. Biomaterials. 2007;28(19):3012–3025. doi: 10.1016/j.biomaterials.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Xu C, Inai R, Kotaki M, Ramakrishna S. Electrospun nanofiber fabrication as synthetic extracellular matrix and its potential for vascular tissue engineering. Tissue Engineering. 2004;10(78):1160–1168. doi: 10.1089/ten.2004.10.1160. [DOI] [PubMed] [Google Scholar]
- 6.Williamson MR, Black R, Kielty C. PCL-PU composite vascular scaffold production for vascular tissue engineering: attachment, proliferation and bioactivity of human vascular endothelial cells. Biomaterials. 2006;27:3608–3616. doi: 10.1016/j.biomaterials.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 7.Li W-J, Danielson KG, Alexander PG, Tuan RS. Biological response of chondrocytes cultured in three-dimensional nanofibers poly(ε-caprolactone) scaffolds. Journal of biomedical material research. 2003;67A(4):1105–1114. doi: 10.1002/jbm.a.10101. [DOI] [PubMed] [Google Scholar]
- 8.Li W-J, Tuli R, Okafor C, Derfoul A, Danielson KG, Hall DJ, et al. A three-dimensional nanofibrous scaffold for cartilage tissue engineering using human mesenchymal stem cells. Biomaterials. 2005;26:599–609. doi: 10.1016/j.biomaterials.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Chew SY, Mi R, Hoke A, Leong KW. Aligned protein-polymer composite fibers enhance nerve regeneration: a potential tissue engineering platform. Advanced Functional Materials. 2007;17:1288–1296. doi: 10.1002/adfm.200600441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chew SY, Wen J, Yim EKF, Leong KW. Sustained release of proteins from electrospun biodegradable fibers. Biomacromolecules. 2005;6(4):2017 – 2024. doi: 10.1021/bm0501149. [DOI] [PubMed] [Google Scholar]
- 11.Chew SY, Hufnagel TC, Lim CT, Leong KW. Mechanical properties of single electrospun drug-encapsulated nanofibres. Nanotechnology. 2006;17(15):3880–3891. doi: 10.1088/0957-4484/17/15/045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao IC, Chew SY, Leong KW. Aligned core-shell nanofibers delivering bioactive proteins. Nanomedicine. 2006;1(4):465–471. doi: 10.2217/17435889.1.4.465. [DOI] [PubMed] [Google Scholar]
- 13.Brockes J, Fields K, Raff M. Studies on cultured rat Schwann cells. I. Establishment of purified populations from cultures of peripheral nerve. Brain Research. 1979;165(1):105–118. doi: 10.1016/0006-8993(79)90048-9. [DOI] [PubMed] [Google Scholar]
- 14.Hoke A, Ho T, Crawford T, LeBel C, Hilt D, Griffin J. Glial cell line-derived neurotrophic factor alters axon schwann cell units and promotes myelination in unmyelinated nerve fibers. Journal of Neuroscience. 2003;23(2):561–567. doi: 10.1523/JNEUROSCI.23-02-00561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson DM, Buettner HM. Schwann cell response to micropatterned laminin surfaces. Tissue Engineering. 2001;7(3):247–265. doi: 10.1089/10763270152044125. [DOI] [PubMed] [Google Scholar]
- 16.Miller C, Shanks H, Witt A, Rutkowski G, Mallapragada S. Oriented Schwann cell growth on micropatterned biodegradable polymer substrates. Biomaterials. 2001;22:1263–1269. doi: 10.1016/s0142-9612(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 17.Schmalenberg KE, Uhrich KE. Micropatterned polymer substrates control alignment of proliferating Schwann cells to direct neuronal regeneration. Biomaterials. 2005;26:1423–1430. doi: 10.1016/j.biomaterials.2004.04.046. [DOI] [PubMed] [Google Scholar]
- 18.Phillips JB, Bunting SCJ, Hall SM, Brown RA. Neural tissue engineering: a self-organizing collagen guidance conduit. Tissue Engineering. 2005;11(910):1611–1617. doi: 10.1089/ten.2005.11.1611. [DOI] [PubMed] [Google Scholar]
- 19.Lietz M, Dreesmann L, Hoss M, Oberhoffner S, Schlosshauer B. Neuro tissue engineering of glial nerve guides and the impact of different cell types. Biomaterials. 2006;27:1425–1436. doi: 10.1016/j.biomaterials.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Miller C, Jeftinija S, Mallapragada S. Micropatterned Schwann cell-seeded biodegradable polymer substrates significantly enhance neurite alignment and outgrowth. Tissue Engineering. 2001;7 (6):705–715. doi: 10.1089/107632701753337663. [DOI] [PubMed] [Google Scholar]
- 21.Yim EKF, Leong KW. Significance of synthetic nanostructures in dictating cellular response. Nanomedicine: Nanotechnology, Biology and Medicine. 2005;1:10–21. doi: 10.1016/j.nano.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Micropatterned surfaces for control of cell shape, position and function. Biotechnology. 1998;14:356–363. doi: 10.1021/bp980031m. [DOI] [PubMed] [Google Scholar]
- 23.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CDW, Curtis ASG. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Experimental Cell Research. 2003;284:274–282. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 24.Ingber DE, Folkman J. Mechanochemical switching between growth and differentiation during Fibroblast Growth Factor-stimulated angiogenesis in vitro: Role of extracellular matrix. The Journal of Cell Biology. 1989;109:317–330. doi: 10.1083/jcb.109.1.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong HJ, Liu J, Riddle K, Matsumoto T, Leach K, Mooney DJ. Non-viral gene delivery regulated by stiffness of cell adhesion substrates. Nature Materials. 2005;4:460–464. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- 26.Ingber DE. Mechanical control of tissue growth: function follows form. Proceedings of the National Academy of Sciences. 2005;102(33):11471–11572. doi: 10.1073/pnas.0505939102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersson A-S, Backhed F, Euler Av, Richter-Dahlfors A, Sutherland D, Kasemo B. Nanoscale features influence epithelial cell morphology and cytokine production. Biomaterials. 2003;24:3427–3436. doi: 10.1016/s0142-9612(03)00208-4. [DOI] [PubMed] [Google Scholar]
- 28.Rajnicek AM, Britland S, McCaig CD. Contact guidance of CNS neurites on grooved quartz: Influence of groove dimensions, neuronal age and cell type. Journal of Cell Science. 1997;110:2905–2913. doi: 10.1242/jcs.110.23.2905. [DOI] [PubMed] [Google Scholar]
- 29.Cutis A, Wilkinson C. Topographical control of cells. Biomaterials. 1997;18:1573–1583. doi: 10.1016/s0142-9612(97)00144-0. [DOI] [PubMed] [Google Scholar]
- 30.Dalby MJ, Riehle MO, Sutherland DS, Agheli H, Curtis ASG. Use of nanotopography to study mechanotransduction in fibroblasts - methods and perspectives. European Journal of Cell Biology. 2004;83:159–169. doi: 10.1078/0171-9335-00369. [DOI] [PubMed] [Google Scholar]
- 31.Ayutsede J, Gandhi M, Sukigara S, Micklus M, Chen HE, Ko F. Regeneration of Bombyx Mori silk by electrospinning. part 3: characterization of electrospun nonwoven mat. Polymer. 2005;46 (5):1625–1634. [Google Scholar]
- 32.Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DIC, Whiteside GM, et al. Engineering cell shape and function. Science. 1994;264:696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 33.Watt FM, Jordon PW, O’Neill CH. Cell shape controls terminal differentiation of human epidermal keratinocytes. Proceedings of the National Academy of Science, USA. 1988;85:5576–5580. doi: 10.1073/pnas.85.15.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu S-h, Chen C-Y, Lu PS, Lai C-S, Chen C-J. Oriented Schwann cell growth on microgrooved surfaces. Biotechnology and Bioengineering. 2005;92(5):579–588. doi: 10.1002/bit.20634. [DOI] [PubMed] [Google Scholar]
- 35.Eguchi Y, Ogiue-Ikeda M, Ueno S. Control of orientation of rat Schwann cells using an 8-T static magnetic field. Neuroscience Letters. 2003;351:130–132. doi: 10.1016/s0304-3940(03)00719-5. [DOI] [PubMed] [Google Scholar]
- 36.Wang D-Y, Huang Y-C, Chiang H, Wo AM, Huang Y-Y. Microcontact printing of laminin on oxygen plasma activated substrates for the alignment and growth of Schwann cells. Journal of Biomedical Materials Research Part B: Applied Biomaterials. 2007;80B:447– 453. doi: 10.1002/jbm.b.30616. [DOI] [PubMed] [Google Scholar]
- 37.Schmalenberg KE, Uhrich KE. Micropatterned polymer substrates control alignment of proliferating Schwann cells to direct neuronal regeneration. Biomaterials. 2005;26(12):1423–1430. doi: 10.1016/j.biomaterials.2004.04.046. [DOI] [PubMed] [Google Scholar]
- 38.Hu W, Yim EKF, Reano RM, Leong KW, Pang SW. Effects of nanoimprinted patterns in tissue-culture polystyrene on cell behavior. Journal of Vacuum Science and Technology B. 2005;23(6):2984–2989. doi: 10.1116/1.2121729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen X, Tresco PA. Effect of filament diameter and extracellular matrix molecule precoating on neurite outgrowth and Schwann cell behavior on multifilament entubulation bridging device in vitro. Journal of Biomedical Materials Research. 2006;76A:626–637. doi: 10.1002/jbm.a.30520. [DOI] [PubMed] [Google Scholar]
- 40.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nature Reviews Genetics. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 41.Mirsky R, Jessen PR. The neurobiology of Schwann cells. Brain Pathology. 1999;9:293–311. doi: 10.1111/j.1750-3639.1999.tb00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin Q, Kemp GJ, Frostick SP. Neurotrohpins, neurones and peripheral nerve regeneration. Journal of Hand Surgery (British and European Volume) 1998;23B(4):433–437. doi: 10.1016/s0266-7681(98)80117-4. [DOI] [PubMed] [Google Scholar]
- 43.Makwana M, Raivich G. Molecular mechanisms in successful peripheral regeneration. FEBS Journal. 2005;272:2628–2638. doi: 10.1111/j.1742-4658.2005.04699.x. [DOI] [PubMed] [Google Scholar]
- 44.Tofaris GK, Patterson PH, Jessen KR, Mirsky R. Denervated Schwann cells attract macrophages by secretion of Leukemia Inhibitory Factor (LIF) and Monocyte Chemoattractant Protein-1 in a process regulated by Interleukin-6 and LIF. The Journal of Neuroscience. 2002;22 (15):6696–6703. doi: 10.1523/JNEUROSCI.22-15-06696.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mirsky R, Jessen KR, Brennan A, Parkinson D, Dong Z, Meier C, et al. Schwann cells as regulators of nerve development. Journal of Physiology - Paris. 2002;96:17–25. doi: 10.1016/s0928-4257(01)00076-6. [DOI] [PubMed] [Google Scholar]
- 46.Ogata T, Yamamoto S-i, Nakamura K, Tanaka S. Signaling axis in Schwann cell proliferation and differentiation. Molecular Neurobiology. 2006;33:51–61. doi: 10.1385/mn:33:1:051. [DOI] [PubMed] [Google Scholar]
- 47.Gupta R, Truong L, Bear D, Chafik D, Modafferi E, Hung CT. Shear stress alters the expression of Myelin-Associated Glycoprotein (MAG) and Myelin Basic Protein (MBP) in Schwann cells. Journal of Orthopaedic Research. 2005;23:1232–1239. doi: 10.1016/j.orthres.2004.12.010. [DOI] [PubMed] [Google Scholar]