Abstract
Williams syndrome (WS) is a neurodevelopmental disorder associated with deletion of ~20 contiguous genes in chromosome band 7q11.23. Individuals with WS exhibit mild to moderate mental retardation, but are relatively more proficient in specific language and musical abilities. We used tensor-based morphometry (TBM) to visualize the complex pattern of gray/white matter reductions in WS, based on fluid registration of structural brain images.
Methods
3D T1-weighted brain MRIs of 41 WS subjects (age: 29.2±9.2SD years; 23F/18M) and 39 age-matched healthy controls (age: 27.5±7.4 years; 23F/16M) were fluidly registered to a minimum deformation target. Fine-scale volumetric differences were mapped between diagnostic groups. Local regions were identified where regional structure volumes were associated with diagnosis, and with intelligence quotient (IQ) scores. Brain asymmetry was also mapped and compared between diagnostic groups.
Results
WS subjects exhibited widely distributed brain volume reductions (~10–15% reduction; P < 0.0002, permutation test). After adjusting for total brain volume, the frontal lobes, anterior cingulate, superior temporal gyrus, amygdala, fusiform gyrus and cerebellum were found to be relatively preserved in WS, but parietal and occipital lobes, thalamus and basal ganglia, and midbrain were disproportionally decreased in volume (P < 0.0002). These regional volumes also correlated positively with performance IQ in adult WS subjects (age ≥ 30 years, P = 0.038).
Conclusion
TBM facilitates 3D visualization of brain volume reductions in WS. Reduced parietal/occipital volumes may be associated with visuospatial deficits in WS. By contrast, frontal lobes, amygdala, and cingulate gyrus are relatively preserved or even enlarged, consistent with unusual affect regulation and language production in WS.
1. INTRODUCTION
Williams Syndrome (WS) is a neurodevelopmental microdeletion disorder, resulting in characteristic cardiovascular abnormalities and typical facial features. WS is also an informative model for studying the linkage between genetic deficits and their neurobiological effects on the brain and behavior. The syndrome is associated with deletion of a 1–2 Mb contiguous genomic region containing about 25 genes in chromosome band 7q11.23, including the genes for elastin which has been implicated in congenital heart deficits in persons with WS (Korenberg et al., 2000), such as septal defects or pulmonary/aortic stenosis. Neurobiologically, individuals with WS exhibit mild to moderate mental retardation and learning difficulties, yet there are nonetheless distinctive patterns of relative strengths and weaknesses. Individuals with WS are profoundly impaired in visuoconstructive spatial abilities, impaired in problem solving, but show relative strengths in language, particularly expressive language and face processing. In addition, individuals with WS tend to exhibit a characteristic personality that includes overfriendliness as well as the use of language for social purposes (Bellugi et al., 1999; Bellugi et al., 2001).
Such an imbalance in intellectual ability has been associated with an uneven pattern of relative deficits and excesses in the volume of specific brain regions. Volumetric studies have shown that the volume of the frontal lobes in WS is relatively preserved compared with individuals with Down syndrome or healthy controls (Jernigan et al., 1993; Reiss et al., 2000), but occipital and superior parietal cortices are disproportionally reduced in volume (Reiss et al., 2004; Eckert et al., 2005). Volumetric studies can be labor-intensive, and the spatial detail obtained is limited by the manual delineation of pre-selected regions of interest so it cannot visualize the profile of volume differences at the voxel-by-voxel level. Voxel-based morphometry (VBM) provides more spatial detail regarding the profile of tissue excess and deficits throughout the brain region, by comparing the gray matter density (smoothed gray matter maps) in spatially normalized MR images at each voxel. VBM studies have detected systematic differences in brain morphology between WS and normal subjects, such as reduced volumes in parieto-occipital cortex (Meyer-Lindenberg et al., 2004; Reiss et al., 2004; Boddaert et al., 2006; Eckert et al., 2006a). Nevertheless, these studies have often been based on small samples. Even so, effect sizes in VBM studies are typically small, and may be insufficient to describe morphological variations that may be broadly distributed throughout the entire brain in this neurodevelopmental disorder. More recent work has examined cortical gray matter thickness, using surface based modeling and elastic warping of sulcal patterns to integrate information across subjects (Thompson et al., 2005). This work identified a circumscribed area of the right perisylvian and inferior temporal regions where the cortex was on average 5–10% thicker in Williams syndrome, suggesting a localized failure of cortical maturation. Other studies have detected temporo-parietal gyrification differences (Eckert et al., 2006b), and increased cortical complexity quantified using fractal dimension measures (Thompson et al., 2005), mean curvature maps (Gaser et al., 2006; Tosun et al., 2006; Tosun et al., 2007), or by counting sulcal branches identified automatically as a graph of connected curves on the cortex (Shi et al., 2007).
In this paper, we apply tensor-based morphometry (TBM), see (Davatzikos et al., 1996; Thompson et al., 2000; Davatzikos et al., 2001; Fox et al., 2001; Shen and Davatzikos, 2003; Chung et al., 2004; Studholme et al., 2004; Chiang et al., 2007) for related work, to detect and automatically quantify subtle and distributed patterns of brain volume differences between 41 WS and 39 normal subjects. In the TBM approach, all images are nonlinearly deformed to match a preselected brain image, which acts as a template. Then, the Jacobian determinant (i.e., the local expansion factor) of the deformation fields is used to gauge the local volume differences between the individual images and the template, and these can be analyzed statistically to identify group differences or localized volume increases/reductions at the voxel level. In our implementation of TBM, which we have previously applied to examine brain atrophy in HIV/AIDS (Chiang et al., 2007), nonlinear image deformation is based on a fluid-deformation algorithm by maximizing the Jensen-Rényi divergence (JRD) of the joint intensity histogram (Chiang et al., 2007). Our results demonstrate that TBM may more clearly visualize the unique morphological profile in WS brains, identifying a more extensive but complementary pattern of differences relative to prior work using voxel-based morphometry or volumetric parcellation. Among adult WS subjects, we also examined links between volumes of specific brain regions and intellectual performance. Finally, we examine the distribution of brain volume asymmetry in both WS and control subjects, mapping the profile of group differences.
2. METHODS
2.1. WS and Control Subjects
The study analyzed the data of 41 subjects with genetically confirmed Williams syndrome (mean±SD, 29.2±9.2 years of age; range, 12–50 years of age; 18 males, 23 females; diagnosis confirmed by fluorescent in situ hybridization tested for deletion of one copy of the elastin gene on chromosome 7) and 39 age-matched healthy controls (27.5±7.4 years of age; range, 18–49 years of age; 16 males, 23 females). These subjects were included in Reiss et al. (2004); the same cohort was studied in our prior reports (Thompson et al., 2005; Gaser et al., 2006), but in this study we excluded one WS and one control subject whose scans had relatively poor image contrast. Given the large effect sizes and relatively large sample, we preferred to exclude these two scans, as the sample size was more than sufficient to model the effects of interest. Nevertheless, the age distribution (mean, SD, and range) of the subjects in this paper is almost identical to that in Thompson et al. (2005) and Gaser et al. (2006). (WS: mean±SD, 29.2±9.0 years of age; range, 12–50 years of age; 19M/23F; controls: 27.5±7.4 years of age; range, 18–49 years of age; 16M/24F). Exclusion criteria included a history of medical conditions not typically associated with WS, such as epilepsy or other neurological conditions. All WS participants were evaluated at the Salk Institute (La Jolla, CA) as part of a program project on genetics, neuroanatomy, neurophysiology, and cognition. Wechsler intelligence quotient (IQ) scores (full-scale, verbal, and performance) were available for 39 of the 41 WS subjects; the two untested subjects exhibited similar levels of cognitive function on other measures. As in the study by Reiss et al. (2004), healthy control subjects (with no history of major psychiatric, neurological, or cognitive impairment) were recruited at both the Salk Institute and Stanford University. Control subjects were further screened to rule out any history of learning, language, or behavioral disorder. The majority of controls in the study did not have IQ testing performed. All procedures were approved by the Institutional Review Boards of both institutions, and all participants provided informed consent (and parents or guardians provided written consent where appropriate).
2.2. Image Scanning and Registration
Image acquisition and registration
All participants received 3D spoiled gradient recovery (SPGR) anatomical brain MRI scans (256×192×124 matrix, TR = 24 ms, TE = 5ms; 24-cm field of view; 1.2-mm slices, zero gap; flip angle = 45°) using a GE-Signa 1.5 T scanner (General Electric, Milwaukee, MI). The same scanner type and exactly the same SPGR pulse sequence were used at the Salk Institute and at Stanford University. Following removal of extracerebral tissues (e.g., scalp and meninges), the MRI brain scan of each subject was co-registered to the ICBM53 average brain template (Mazziotta et al., 2001) using 9-parameter affine registration with translations, rotations, and independent scalings in the x-, y-, and z- directions. The template is one of several standardized adult brain templates and was generated by averaging high-resolution brain MRI scans (n = 53) in the ICBM standard space to improve the signal-to-noise ratio, after nonlinear image registration using the ANIMAL algorithm ((Collins et al., 1995), Montreal Neurological Institute, McGill University, Canada); the template is part of the MINC distribution of brain templates, image analysis algorithms and display software. To retain information on the overall change in brain volume incurred by scaling images from scanner space to the ICBM space (see below), a zoom factor was computed from the determinant of the global linear transformation.
All subjects’ images were then registered to an optimized target (see below for target selection) using a nonlinear algorithm based on maximizing the Jensen-Rényi divergence (JRD) of the joint intensity histogram, with the deformation field regularized in a fluid scheme (Chiang et al., 2007).
Target selection
To select an optimized target, we adopted the “minimum-deformation target” (MDT) proposed by Kochunov et al. (2001). For subject image i (1 ≤ i ≤ n) initially serving as a registration target, a target quality score (TQS), summarizing its deviation from all the other images, is given by:
| (1) |
At each voxel, S and D were computed from the deformation fields mapping all other subjects to the target (with uj denoting the deformation vector for subject j) fluidly mapped to the target by maximizing the Jensen-Rényi divergence (Chiang et al., 2007). The subject with the lowest TQS was then defined as the best individual target (BIT), denoted by IBIT, then the MDT brain (IMDT) is defined by:
| (2) |
where is the average displacement field for all the other subjects mapped to the BIT, and h̄−1(x) was obtained from h̄(x) following the iterative method in (Christensen and Johnson, 2001). We preferred the optimized target computed from the average transformation for a typical image (e.g., the BIT) rather than building a multi-subject average intensity atlas, as the former has sharper features and, in general, leads to larger effect sizes when used in a morphometric study (Christensen et al., 2006; Chiang et al., 2007). The MDT method is closest to what proposed in (Christensen et al., 2006) and (Park et al., 2003). Other template optimization methods (Avants and Gee, 2004; Joshi et al., 2004; Studholme and Cardenas, 2004; Twining et al., 2005) are also possible.
2.3. Tensor-based Morphometry and Statistical Comparisons
Comparison of regional brain volumes between WS subjects and controls
The Jacobian tensor in the ICBM space was obtained by taking the spatial gradient of the deformation field from the fluid registration. Then the determinant of the Jacobian tensor in the native space (i.e. before the brain was affinely registered to the ICBM53 template), denoted by Jnative, was computed from the Jacobian determinant in the ICBM space (JICBM) using Jnative = JICBM/zoom factor, where the zoom factor was defined in the previous section. The determinant of the Jacobian tensor was used as a local index of tissue expansion (Jacobian > 1) or shrinkage (Jacobian < 1) relative to the target (Davatzikos et al., 1996; Chung et al., 2001). The percentage difference in mean volume at each voxel was estimated from the ratio of the mean Jnative in WS versus control subjects. This indicates the degree to which the volume of a specific region is higher or lower in WS than in matched controls, and the ratio can be turned into a percentage deficit. The Jacobian determinant values were first subjected to a log transformation because the null distribution of the log(Jacobian) is closer to Normal than that of the Jacobian determinant, which is skewed and bounded below by zero (Ashburner and Friston, 2000; Woods, 2003; Arsigny et al., 2005; Avants et al., 2006); see (Leow et al., 2005; Leow et al., 2006) for a detailed analysis of the effects of log-transformation on the Jacobian distribution. We tested the significance of difference between the mean log(Jnative) of the WS and control groups using the Mann-Whitney U test voxelwise. Since the total brain volume in WS subjects is less than that in control subjects (see Results), the difference of the mean log(JICBM) between the two groups was also compared by the Mann-Whitney U test, after the global brain volume differences had been adjusted out by transformation to the ICBM space. Furthermore, to detrend any possible confounding effect of subjects’ age, analysis of covariance (ANCOVA) was performed at each voxel, with each participant’s age treated as a covariate, from which we derived a percentage volume difference map adjusted for subjects’ age and the total brain volume (see APPENDIX A for details).
Correlations of regional brain volume with IQ scores
Spearman’s rank test was applied at each voxel to identify regions where regional brain volume (Jnative) correlated with verbal or performance IQ in WS (n = 39) and control (n = 16) subjects separately. As the regions showing volumetric correlations with IQ may be different in different age groups (Shaw et al., 2006), we further divided the WS subjects into adolescents (range 12–16.9 years, n = 2), young adults (range 17–29.9 years, n = 19), and those in middle-adulthood (age ≥ 30 years, n = 18), such that the number of subjects in the two adult groups was roughly equal, and tested the IQ correlations in the latter two groups. (There were too few control subjects with IQ scores available to allow comparable subdivisions for the controls.)
Brain asymmetry
We followed the method proposed by Park et al. (2004), where brain asymmetry was detected by mapping all the subjects to a symmetric brain template, and then regional volume differences between left and right hemispheres can be identified by comparing the Jacobian determinant at corresponding voxels in each hemisphere. Briefly, if h(x) = x − u(x) is the displacement field for the MDT IMDT(x) mapped to its flipped image (across the midsagittal plane) IfMDT(x), and R(x) is the deformed image, i.e. R(x) = IMDT (h(x)), then with , the symmetric template S(x) is given by S(x) = S̄(h̄−1(x)), where h̄−1(x) is the inverse of h̄(x), and . We then registered all the brains to the symmetric template S(x) using the fluid JRD algorithm, and the difference between log(JICBM) between corresponding voxels (which are mirror images of each other with respect to the midsagittal plane) in the left and right hemispheres of the symmetric template brain was compared using the Wilcoxon signed rank test, using two separate one-tailed tests for right hemisphere volume greater than left one and vice versa (this is appropriate, as the literature on hemispheric petalia - reviewed in (Toga and Thompson, 2003) -leads to pre-existing one-tailed hypotheses regarding normal frontal lobe volumetric excess on the right and occipital lobe excess in the left hemisphere). We also evaluated if there was any difference in degree of asymmetry between WS and control groups, using repeated measures analysis of variance (ANOVA) to test the group × side interaction, with diagnosis (WS vs. control) as the between-subject factor, and side (left vs. right) as the within-subject factor (Carrion et al., 2001) (see APPENDIX B for details).
Statistical procedures for correction of multiple comparisons
Overall significance was assessed by permutation methods (Nichols and Holmes, 2001) to correct for multiple comparisons. Statistical testing was performed on each random permutation of subjects’ labels (e.g. “disease” or “control” in Mann-Whitney U tests, IQ scores in the correlation tests, or the ordering of “left” and “right” in the Wilcoxon signed rank test) to construct a null distribution for the number of voxels more significant than a fixed primary threshold applied at the voxel level (here set to P = 0.01). The omnibus probability (i.e., corrected for multiple comparisons) was determined by comparing the number of suprathreshold voxels in the true labeling to the permutation distribution. The number of permutations N was chosen to control the standard error SEp of omnibus probability P, which follows a binomial distribution B(N, P) with (Edgington, 1995). We selected N = 8,000 tests out of the total number of possible permutations (≈1023) such that the approximate margin of error (95% confidence interval) for P was around 5% of P, and 0.05 was chosen as the significance level.
For stronger control over the likelihood of false rejections of the null hypotheses with multiple comparisons, we adopted the method by Storey (2002), which directly measures the positive false discovery rate (pFDR) under a given primary threshold. The pFDR method is more powerful than the popular sequential p-value FDR method (Benjamini and Hochberg, 1995; Genovese et al., 2002), as it estimates the probability that the null hypothesis is true, from the empirical distribution of observed p-values (Manly et al., 2004). The pFDR measure is theoretically more suitable for representing the “rate at which discoveries are false” than the FDR measure when the primary rejection region is relatively small (Storey, 2002). Briefly, pFDR is the false discovery rate conditioned on the event that positive findings, rejecting the null hypothesis, have occurred, and is given by
| (3) |
where π0 = Pr(H = 0) is the probability that the null hypothesis is true, and γ is the rejection threshold for the individual hypothesis, which was set to 0.01 in our experiments. We refer readers to (Storey and Tibshirani, 2001; Storey, 2002) for details of the estimation procedures to obtain pFDR for statistical maps. By convention, a statistical map with pFDR < 0.05, i.e. a false discovery rate less than 5%, was considered to be significant.
Permutation tests and pFDR measures were performed on combined gray and white matter masks to generate P values that were corrected for multiple comparisons within regions containing brain tissue. White matter and gray matter masks were automatically segmented using an unsupervised Gaussian mixture classifier, after adjusting for spatial intensity inhomogeneity, using the software package “BrainSuite” (Shattuck and Leahy, 2002).
3. RESULTS
3.1. Target Selection and Symmetric Brain Template
Fig. 1 shows brain MR images with lowest and highest TQS, the MDT, and the symmetric template. TQS is a measure of how much each brain deviates from all the others in the sample (including WS and controls), and there was no difference in TQS between WS and control subjects (WS: 2.48±0.19 voxels, ranging from 2.23 to 2.88 voxels; controls: 2.45±0.20 voxels, ranging from 2.12 to 2.97 voxels; P = 0.3 for a group difference). The BIT happened to be one of the control subjects (TQS = 2.12 voxels). The TQS of the MDT was 1.48 voxels.
Fig. 1.
This figure shows, from left to right, the brain with highest TQS (a WS subject), the lowest TQS (a control subject), the MDT, and the symmetric template derived from the MDT. The mean deformation distance between the registration target and the population of WS and control brains is greatly reduced by averaging the deformation fields, and the MDT, compared with the lowest TQS brain from which it is derived, has a smoother gyral morphology, and is close to symmetric.
3.2. Visualization of the Profile of Volume Alterations in WS
The total brain volume was 13% smaller in WS than control subjects (WS: 925.65±76.26 cc; controls: 1062.54±115.95 cc; P < 0.001). This is apparent in the volume (percentage reduction) map comparing the group differences of Jnative in Fig. 2, where WS subjects exhibited widespread volume reduction over the whole brain (permutation test P < 0.0002, pFDR = 0.0002). To control for this overall difference in total brain volume between groups, JICBM was compared voxelwise. Fig. 3 shows that, in WS subjects, the volume of prefrontal and orbitofrontal areas, the anterior cingulate gyrus, inferior parietal regions at the parietooccipital junction, the superior temporal gyrus, amygdala and part of the hippocampus (most prominently on the right side), fusiform gyrus, and cerebellum were relatively preserved (which is represented as an ‘increase’ or ‘relative excess’ in the volume percentage maps; permutation test P < 0.0002, pFDR = 0.01). However, occipital areas, parietal lobe regions close to the temporoparietal junction, the splenium and posterior body of the corpus callosum, thalamus and basal ganglia (including globus pallidus, putamen, and caudate nucleus), and midbrain were disproportionally reduced in WS, i.e. reduced to an even greater extent than the level of overall brain volume reduction (permutation test P < 0.0002, pFDR = 0.01). The volume increase/reduction in the native and the ICBM spaces was also found to be significant after adjusting for effects of age using ANCOVA (native space: permutation test P < 0.0002, pFDR = 0.03; ICBM space: permutation test P < 0.0002, pFDR = 0.02).
Fig. 2.
Voxelwise comparisons of WS and control brain volumes in the native space show widespread volume reductions throughout the entire brain, which reflects the fact that total brain volume in WS subjects is less than controls. Nevertheless, the volumes of the frontal lobes and cerebellum are relatively preserved.
Fig. 3.
Volumetric maps show systematic differences in brain morphology in WS. WS and control brain volumes were compared in the ICBM space to adjust for individual differences in total brain volumes. The ratio of the mean JICBM in WS to the mean JICBM in control subjects was computed voxelwise to map the 3D profile of brain volume increases (color-coded as red) or reductions (color-coded as blue) (first row). Panels (a) to (d) show relative preservation of the volume (which is shown as increase in the volume percentage maps) in prefrontal and orbitofrontal areas, anterior cingulate gyrus, inferior parietal regions at the parietooccipital junction, superior temporal gyrus, amygdala and part of hippocampus (especially on the right side), fusiform gyrus, and cerebellum. Occipital areas, parietal lobes close to temporoparietal junction, splenium and posterior body of the corpus callosum, thalamus and the basal ganglia (including globus pallidus, putamen, and caudate nucleus), and midbrain are disproportionally reduced. The Mann-Whitney U test was used to obtain the significance maps for the volume increase (second row) or reduction (third row). Abbreviations: aCG: anterior cingulate gyrus, Amy: amygdala, BG: basal ganglia, FG: fusiform gyrus, HP: hippocampus, IP: inferior parietal region, MiB: midbrain, OF: orbitofrontal area, OL: occipital lobe, PF: prefrontal area, PL: parietal lobe, SCC: splenium of the corpus callosum, STG: superior temporal gyrus, TL: thalamus.
3.3. Correlations of Regional Brain Volume with IQ Scores
39 WS and 16 control subjects underwent IQ testing. WS subjects had lower full-scale, verbal and performance IQ scores than control subjects (full-scale IQ: 68.7±8.4 vs. 104.3±12.4, mean±SD, P < 0.001; verbal IQ: 72.1±7.4 vs. 103.3±11.0, P < 0.001; performance IQ: 67.7±8.6 vs. 104.6±12.7, P < 0.001; Mann-Whitney U tests). However, WS subjects were relatively proficient in verbal tests as the difference between verbal and performance IQ scores was larger in WS (verbal − performance IQ: 4.4±7.1 in WS vs. −1.25±9.2 in controls, P = 0.02; Mann-Whitney U tests). This trend was made clearer by comparing the index (verbal − performance IQ)/0.5(verbal + performance IQ) to adjust for the group differences in IQ scores (WS: 0.066±0.099, controls: −0.011±0.084, P = 0.005; Mann-Whitney U tests). In either group, there were no positive or negative correlations between either verbal or performance IQ scores and the overall brain volume (in WS: P = 0.90 for verbal IQ, and P = 0.56 for performance IQ; in controls: P = 0.09 for verbal IQ, and P = 0.11 for performance IQ) or the regional brain volume (in the native space) (in WS: permutation test P = 0.5, pFDR = 1.0 for verbal IQ, permutation test P = 0.2, pFDR = 0.28 for performance IQ; in controls: permutation test P = 0.07, pFDR = 0.06 for verbal IQ, permutation test P = 0.09, pFDR = 0.10 for performance IQ; figures not shown).
We further examined IQ correlations with brain morphology in the adult WS subjects, classifying them into individuals in young adulthood (range 17–29.9 years, n = 19) or middle adulthood (age ≥ 30 years, n = 18). There was no difference in verbal IQ between these two groupings (early-adult: 72.6±5.7, middle-adult: 73.1±7.9, P = 0.96; Mann-Whitney U tests), but the middle-adult individuals had higher performance IQ scores than the young-adult ones (early-adult: 71.3±8.0, controls: 66.2±6.5, P = 0.026; Mann-Whitney U tests). Furthermore, in WS subjects aged 30 or over, regional brain volumes were positively correlated with performance IQ scores in bilateral anterior cingulate gyrus, in the genu and splenium of the corpus callosum, in parietal, occipital and prefrontal areas, right amygdala, hippocampus, fusiform gyrus and orbitofrontal areas, left superior temporal gyrus, and in the cerebellum and pons (permutation test: P = 0.038, pFDR = 0.02, Fig. 4). Negative correlations were not significant. No positive or negative correlations were detected between regional brain volume and verbal IQ scores in healthy control adults aged 30 or over, and verbal and performance IQ scores in WS subjects younger than 30 (figures not shown).
Fig. 4.
In WS subjects aged 30 or over, regionally greater brain volume was associated with higher performance IQ scores in (a) bilateral anterior cingulate gyrus, in the genu and splenium of the corpus callosum, parietal, occipital and prefrontal areas, and in the cerebellum and pons (b) left superior temporal gyrus and (c) right amygdala, hippocampus and orbitofrontal areas. These correlations were assessed using Spearman’s (non-parametric) rank correlation. Abbreviations: aCG: anterior cingulate gyrus, Amy: amygdala, FG: fusiform gyrus, GCC: genu of the corpus callosum, HP: hippocampus, OF: orbitofrontal area, OL: occipital lobe, PL: parietal lobe, SCC: splenium of the corpus callosum, STG: superior temporal gyrus.
3.4. Brain Asymmetry in WS and Control Subjects
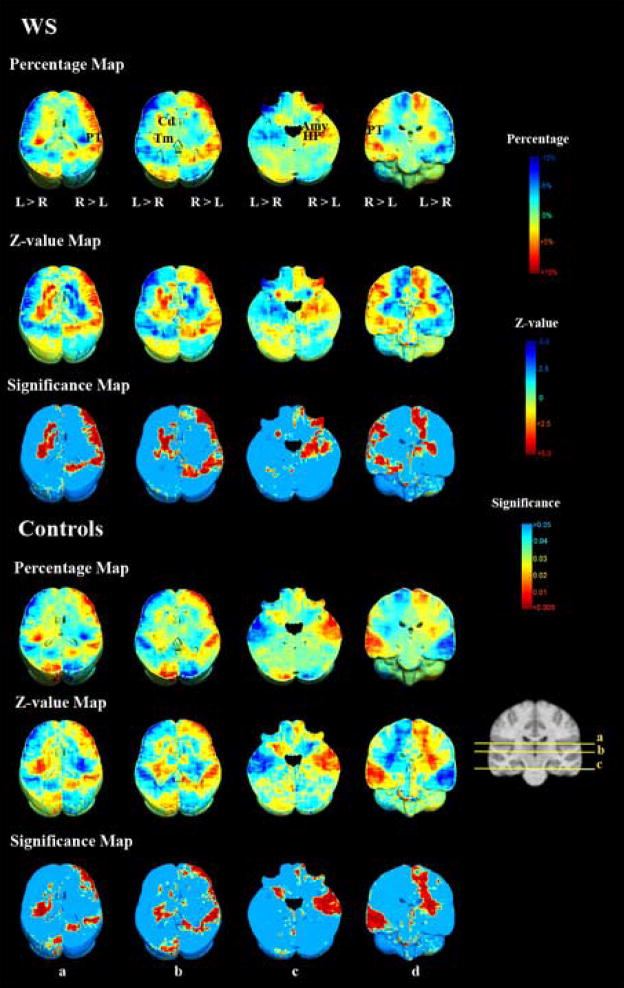
Fig. 5 shows that the patterns of brain volume asymmetry are similar for both WS and control groups (brain volume asymmetry was significant in both WS and control groups, corrected for multiple comparisons, with permutation test P < 0.0002, pFDR = 0.013 for regions with right > left volumes and permutation test P = 0.0008, pFDR = 0.017 for regions with left > right volumes in WS, and permutation test P = 0.005, pFDR = 0.013 for right > left and permutation test P = 0.001, pFDR = 0.015 for left > right in controls). As anticipated from the literature on brain torque and hemispheric petalias (Toga and Thompson, 2003), there was a rightward asymmetry (i.e., greater volume on the right) in the frontal and temporal lobes, and leftward asymmetry in occipital lobes. The thalamus, putamen, globus pallidus, and caudate nucleus were found to be larger on the left side in both groups. WS subjects had a right larger than left planum temporale. In control subjects, the planum temporale is larger in the left hemisphere, but the difference was either at trend-level or only significant for the medial part. WS subjects also differed from controls in that they displayed a rightward asymmetry in amygdala and hippocampus, and exhibited a less prominent occipital petalia. However, this group-by-side interaction does not reach statistical significance, when corrected for multiple comparisons (pFDR = 0.9).
Fig. 5.
Visualization of the brain asymmetry pattern in WS subjects and controls. In each group, the percentage maps (showing relative volume) were computed based on the difference in the log(JICBM) between the ipsilateral voxel and its mirror image (with respect to the midsagittal plane) in the contralateral hemisphere, with an asymmetry percentage value defined as 100% × (JL−JR)/[(JL+JR)/2] for voxels in the left hemisphere, and 100% × (JR−JL)/[(JL+JR)/2] for voxels in the right hemisphere, where JR and JL are the corresponding Jacobian determinants in left and right hemispheres (Steinmetz, 1996). The difference in the log(JICBM) between left and right sides was tested nonparametrically to obtain z-value maps using the normal approximation to the Mann-Whitney distribution, with z > 0 indicating ipsilateral > contralateral, colored-coded in red, and z < 0 for ipsilateral < contralateral, colored-coded in blue. The z-value maps are associated with probability maps that indicate the significance of z-values greater than zero. Both WS and control subjects exhibited frontal and occipital petalias (rightward asymmetry in frontal and temporal lobes, and leftward asymmetry in occipital lobes), though the occipital petalia was less apparent in WS. The thalamus, putamen, globus pallidus, and caudate nucleus were larger in the left hemisphere in both groups. The planum temporale was larger in the right hemisphere in WS subjects, but larger in the left hemisphere in control subjects. Even so, this difference was either at trend-level or only significant in the medial part. Compared with control subjects, WS participants showed a trend for a larger right amygdala and hippocampus, but this group-by-side interaction did not reach statistical significance when formally tested by ANOVA. Abbreviations: Amy: amygdala, Cd: caudate nucleus, HP: hippocampus, PT: planum temporale, Tm: thalamus.
3.5. Scanner Effect
78 subjects were scanned at the Salk Institute (with 24 WS and 16 controls at one scanner, and 17 WS and 21 controls at the other scanner), and 2 subjects were scanned at Stanford University (both were controls), using the same GE-Signa 1.5 T scanner type and the same SPGR pulse sequence. For the subjects imaged at the Salk Institute, there was no difference in the distributions of diagnosis (chi-square test: P = 0.257), age (28.4±8.6 vs. 28.4±8.2, mean±SD, P = 0.987), or gender (14M/26F vs. 18M/20F, P = 0.358) between those who were assigned to each of the two scanners. We further covaried out the scanner effect from the volumetric results by comparing JICBM and Jnative (the ICBM space and native space Jacobian maps) between WS and control subjects using ANCOVA, with the scanner treated as a confounding variable (the two scanners at the Salk Institute were coded as 1 and 2 respectively, and the scanner at Stanford University as 3). We found that volume increase/reduction in the native and the ICBM spaces was still highly significant in the presence of the scanner covariate (native space: permutation test P < 0.0002, pFDR = 0.0008; ICBM space: permutation test P < 0.0002, pFDR = 0.018).
4. DISCUSSION
Our study demonstrated that TBM based on fluid registration is helpful to visualize the profile of 3D volumetric differences in WS. Our findings corroborate prior work that used VBM and volumetric assessments (Reiss et al., 2000; Schmitt et al., 2001b, a; Tomaiuolo et al., 2002; Meyer-Lindenberg et al., 2004; Reiss et al., 2004; Meyer-Lindenberg et al., 2005b; Boddaert et al., 2006; Eckert et al., 2006a), showing disproportionate reduction in parieto-occipital lobes, posterior corpus callosum, thalamus and midbrain, but preservation in prefrontal and orbitofrontal areas, anterior cingulate, superior temporal gyrus, amygdala and hippocampus, fusiform gyrus and cerebellum. Reduction in the volume of the parieto-occipital regions, as well as the posterior corpus callosum where white matter fibers interconnect opposite visual and visual association cortices (de Lacoste et al., 1985), may be causally implicated in the visuospatial dysfunction observed in WS subjects (Bellugi et al., 1999; Bellugi et al., 2000; Brown et al., 2003). Relative preservation of the right fusiform gyrus and amygdala, which are involved in visual analysis of faces among other tasks (Haxby et al., 1999; Kawashima et al., 1999; Hoffman and Haxby, 2000; Mobbs et al., 2004; Meyer-Lindenberg et al., 2005a; Haxby, 2006), may be linked to the near-normal level of face perception and recognition in WS subjects (Bellugi et al., 2000). The right amygdala was also found to be more active in WS subjects than controls in response to musical stimuli, which may be relevant to their exceptional sensitivity to music (Levitin et al., 2003). Reductions in the basal ganglia in WS have not been emphasized in prior studies. Jernigan et al. (1993) found that, based on MR volumetry, the lenticular nucleus to cerebral gray matter proportion is smaller in WS compared with Down syndrome, but is not different than in controls. Reiss et al. (2004) detected a single cluster with reduced gray matter density in the left caudate nucleus by applying VBM. Using TBM here, we showed that the basal ganglia, including the caudate nucleus, putamen and globus pallidus, is significantly reduced in WS. This is congruent with a recent functional MRI study (Mobbs et al., 2006), which showed that neural activity in bilateral striatum, especially in the caudate nucleus and putamen, is significantly lower in WS than in typically developing subjects during a response inhibition task, suggesting striatal dysfunction. Although TBM is based on using a deformation algorithm to infer local volume increase or decrease based on changes in tissue boundaries, rather than true changes in cell/fiber density or physiological integrity of neural tissues, it is sensitive enough to visualize structural abnormalities that may be linked with these functional alterations.
The WS and control subjects were examined in Reiss et al. (2004), Thompson et al. (2005), and Gaser et al. (2006); however, this paper is partially independent of the previous ones in both the methodologies and in regions and signals assessed. Reiss et al. (2004) compared total and regional gray and white matter volume and density using volumetric measurements and the VBM method. Thompson et al. (2005) identified regionally specific alterations in cortical thickness, using surface-based cortical models and sulcal landmark-guided registration. This paper uses the TBM method, which focuses mainly on mapping the 3D profile of volume reductions in subcortical gray and white matter in WS. As such, while our results are largely congruent with earlier findings of subcortical volume reductions, they should be viewed as presenting complementary measures; the study cannot strictly speaking be viewed as an independent replication of earlier reports as the sample of subjects assessed is the same.
If cortical gray matter distribution is compared across subjects, clearly the geometry of the gyri and sulcal features is highly convoluted and individual gyri or sulci are not readily distinguished based on image intensity alone. As a result, most intensity-based automated registration algorithms (for both TBM and VBM) are unable to match them accurately for purposes of morphometric comparisons, unless specific cortical features are added to constrain the correspondence (e.g., (Fischl et al., 1999; Thompson et al., 2000)). In particular, a surface-based warping method, using explicitly defined cortical landmarks, may be more appropriate for detecting cortical thickening or thinning in WS, as landmark points defined on the cortical surfaces can be explicitly matched when compared data across subjects and groups (Thompson et al., 1996; Thompson et al., 2005). Nevertheless, surface-based warping methods provide no information below the cortex; manual labeling of gyral and sulcal landmarks can also be labor-intensive. If anatomical correspondences are defined automatically to increase efficiency, algorithms typically assume that aligning 3D features, such as mean curvature or conformal factor measures, increases anatomical homology across subjects (Fischl et al., 1999; Wang et al., 2005; Lui et al., 2007). Therefore, cortical differences in WS may be better detected using surface-based modeling methods, but TBM complements traditional volumetric studies in WS with greater automation, additional detail in the form of 3D maps, and improved 3D visualization of deficits in subcortical gray and white matter regions.
Compared with the unmodulated (Reiss et al., 2004; Boddaert et al., 2006) or modulated (Meyer-Lindenberg et al., 2004; Eckert et al., 2006a) VBM methods, in which the “gray matter density” or “gray matter volume” is compared across groups of subjects, the TBM method directly detects volume reductions in gray and white matter. Because VBM and TBM differ in the signals they examine, and in their power to detect differences in different regions of the brain, there are some differences in volumetric findings using these methods. Specifically, our TBM results here are consistent with those found by Reiss et al. (2004) using unmodulated VBM, in that gray matter volume (TBM) or density (unmodulated VBM) was reduced in parietal-occipital gyri, but preserved in orbitofrontal gyri, cerebellar vermis and hemispheres, and cingulate and fusiform gyri. Nevertheless, TBM found additional volume reductions in the basal ganglia and the thalamus, while unmodulated VBM detected reduced gray matter density in the parahippocampal gyri bilaterally. The unmodulated, or standard VBM method measures the voxel intensity of smoothed, spatially normalized gray matter maps, which is defined as the locally weighted average of gray matter density in a region defined by a smoothing filter (Ashburner and Friston, 2000; Good et al., 2001b). If no modulation step is applied, comparisons of gray matter density alone do not fully reflect volume differences between subjects, as they do not contain information on the expansions and contractions needed to match the subject to the template (the Jacobian maps). The modulated, or optimized VBM method (Good et al., 2001b) addresses this issue by multiplying (modulating) the voxel intensity value of the spatially normalized gray matter maps by the corresponding Jacobian determinant of the deformation fields, so that the measured amount of gray matter at each voxel, which is the product of the gray matter density and the volume of the deformed voxel, remains unchanged during warping. Davatzikos et al. (2001) have proposed a conceptually related approach. Nevertheless, if gray matter density and volumetric expansion are multiplied, the resulting compound measure at each voxel represents the mixed influence of regional gray matter volume, thickness and volumetric averaging by the smoothing filter (Eckert et al., 2006a), and thus may be somewhat less explicit in its physical meaning than using the Jacobian maps directly, which measure volume differences.
Based on comparing the ratio of verbal-peformance IQ difference to the average of both test scores, the intellectual ability in WS subjects is more skewed toward language than visuospatial processing, compared with normal individuals. This is also evident in their relative facility in grammar, proclivity for unusual vocabulary, and enrichment in language affect, which is remarkable given their impairments in general cognition (Bellugi et al., 1999; Bellugi et al., 2001). Performance IQ in WS subjects aged 30 or over was found to be related to the volume of brain structures that have been previously reported to correlate with full-scale or performance IQ, such as prefrontal, parietal and occipital regions (Reiss et al., 1996; Flashman et al., 1997; Nguyen and McDaniel, 2000; Thompson et al., 2001; Posthuma et al., 2002; Gray and Thompson, 2004; Haier et al., 2004; McDaniel, 2005; Shaw et al., 2006). This may reflect a possible benefit of greater gray and/or white volume for visuospatial processing (perhaps via increased myelination or conduction velocity). The linkage of performance IQ with volumes of limbic structures, such as anterior cingulate gyrus, amygdala, hippocampus, and orbitofrontal areas may be of special interest. The anterior cingulate gyrus, particularly the dorsal part, is an important component of parallel distributed attentional and emotional networks that are involved in cognitive processing by handling conflicting or competitive stimuli coming from different brain areas (Bush et al., 2000). Its gray matter volume is also correlated with performance IQ (Wilke et al., 2003), although it is not clear whether this is an anatomically specific correlation or mediated by the overall relationship between brain volume and IQ. The right hippocampus is activated in topographical learning and memory retrieval tasks, showing increased regional blood flow on positron emission tomography (Maguire et al., 1996; Maguire, 1997; Meyer-Lindenberg et al., 2005b). Nevertheless, the mechanisms that associate brain structure volumes and intellectual ability, especially in the presence of disease interactions such as Williams syndrome, remain largely unknown. Alternatively, regions with positive correlations between performance IQ and brain volume may coincide with those that show significant volume differences between WS and controls. In that case, a greater relative preservation in brain volume may indicate less disease burden, and thus better intellectual performance. It is not surprising that we found no correlation between IQ and overall or regional brain volume for the whole WS or control group, as a large sample size is commonly required to detect such correlation in normal subjects (Nguyen and McDaniel, 2000; Thompson et al., 2001; Gray and Thompson, 2004), and the pattern of correlation may be different in different age groups (Wilke et al., 2003; Shaw et al., 2006). We also cannot rule out that there is an IQ × disease interaction, but there were not enough control subjects with IQ scores available to determine this.
The results from our analysis of brain asymmetry showed the well-known frontal and occipital petalia in control subjects, consistent with other VBM studies (Good et al., 2001a; Watkins et al., 2001). To our knowledge, this asymmetry pattern has not been mapped before with TBM. WS subjects exhibited frontal petalia, but the occipital petalia was somewhat less prominent (albeit not significantly so). Prior volumetric analyses on this subject population showed a significant group-by-side interaction with leftward asymmetry for occipital gray matter volume in WS subjects (Reiss et al., 2004); however, our TBM study did not detect this. Possible reasons include (i) TBM is less sensitive in detecting the relative extent of gray/white matter volume contributions; (ii) intensity-based registration may not match cortical gray matter as well as subcortical gray or white matter. In the normal population, the planum temporale is greater in volume in the left hemisphere (Geschwind and Levitsky, 1968; Galaburda et al., 1978; Good et al., 2001a; Watkins et al., 2001), although some reports found that such leftward asymmetry was not significant, which may be due to different anatomical criteria and parcellation methods, measurement of surface area rather than gray matter volume, power limitations in small samples, and the effects of brain size correction (Beaton, 1997; Westbury et al., 1999). Moreover, planum temporale is somewhat difficult to localize consistently on brain MRI and there is substantial volume variation and some cytoarchitectural variation of the histologically defined planum temporale with respect to grossly observable anatomical landmarks (Steinmetz et al., 1989). In WS subjects, we found that the right planum temporale was larger in volume, which is at variance with the leftward asymmetry in surface areas detected in a largely overlapping sample of subjects (Eckert et al., 2006b), and in a previous small-sample study (Hickok et al., 1995). This discrepancy may result from: (i) the volume comparison computed from the Jacobian determinant of registration deformation fields differing in definition from the volume or surface area measured in volumetric analysis; (ii) adjustment for overall brain volume by mapping data to the ICBM stereotaxic space may not have the same effect as linear detrending of the overall volume using statistical procedures such as ANCOVA, and (iii) intensity-based registration accuracy may be not optimal for small substructures such as the planum temporale, which lack sharply contrasting boundaries on MRI. Nevertheless, the leftward asymmetry in WS subjects was greatly reduced, and WS subjects had a significantly larger right planum temporale, but not left planum temporale, than the control subjects, whether the planum areas were adjusted for cerebral tissue volume or not (Eckert et al., 2006b). This may be linked with the thickening in the language cortex of the right hemisphere found in our previous study (Thompson et al., 2005).
In this paper we used the “minimum-deformation target” (MDT) as the common template for image registration and statistical comparisons, which is helpful to reduce any bias in the registration process towards the particular geometry of a single subject. We built the MDT by averaging the deformation fields rather than averaging the voxel intensities of the registered images (Evans et al., 1992; Good et al., 2001a; Watkins et al., 2001), as the effect sizes for group comparisons based on using an MDT with intensity distributions similar to a single brain have been found to be greater than using an average intensity atlas (Chiang et al., 2007). This is apparent in the subcortical gray matter where the intensity contrast with the surrounding white matter is lower when using an average intensity template. The MDT was derived from the best individual target (BIT) with lowest TQS among all study participants, which we considered as a better target than any individual subject because the geometry of the BIT is the most representative of the whole group and deviates the least from all other brains; i.e., the warping required for other subjects to be mapped to the BIT is minimal on average. Furthermore, using the BIT to make the MDT may be theoretically better than using a single-subject standard template, e.g., the Colin27 brain template (Holmes et al., 1998), as in our previous approach (Lee et al., 2007), as the BIT was scanned with the same imaging parameters and has a similar morphology to the study subjects. Nevertheless, the BIT-MDT method suffers from a relatively high computational load (it requires N(N−1) or ~N2 nonlinear registrations for N subjects to determine the BIT), and further comparisons of these two approaches on the effect sizes for disease detection are required (see (Chou et al., 2007) for related work).
Prior to nonlinear deformation, all images were first aligned to the ICBM template using affine registration with scaling in x-, y-, and z- directions, such that the linear term was fitted before estimating the nonlinear parameters. Although the warping can be performed on the native space data, an initial 9-parameter transformation helps to avoid misregistrations due to the deformation field settling in local minima of the cost function, and to accelerate convergence. JICBM obtained from fluid mapping in the ICBM space for an individual subject to the MDT is then convertible to Jnative in the native space by dividing by the zoom factor resulting from the affine transformation. Comparing JICBM can be considered as comparing the relative (proportional) volumes of regional brain structures, as the difference in global brain volume has been scaled out, while Jnative refers to the regional volume of structures at their original scale (native space or ‘scanner space’). We studied brain asymmetry using JICBM in order to avoid giving greater weights on more-symmetric brains with greater global volume in statistical tests than more-asymmetric ones with smaller global volume. This also ensured that regional differences were not confounded by global brain differences when group-by-side interactions are tested. On the other hand, we linked Jnative with IQ scores, as we were interested in detecting the specific regions where their native volume is positively correlated to subjects’ IQ, on the basis that greater total brain volume is correlated with higher IQ and may be functionally beneficial for physiological reasons (McDaniel, 2005).
Although this study, as well as others (Schmitt et al., 2001b; Meyer-Lindenberg et al., 2004; Reiss et al., 2004), suggests a seemingly natural rule that reduction and preservation of volume may be linked with the functional integrity or capacity of brain structures in WS and normal subjects, this may imply an overly simplistic relationship between structure and function. For instance, increased thickness in the perisylvian region of the parieto-occipital cortex in WS was not clearly associated with any apparent functional gain but mostly likely with aberrant packing of cortical neurons (Thompson et al., 2005). A combined VBM and functional MRI study on WS subjects revealed that even though the volume of the hippocampal formation was preserved, its function was impaired (Meyer-Lindenberg et al., 2005b). To understand relations between white matter volume and its functional integrity, different types of investigations are required. A preliminary diffusion tensor imaging study showed reduced diffusivity indices (fractional anisotropy and trace) in the right parieto-occipital regions, indicating altered white matter integrity (Marenco et al., 2004). We expect that a future combination of TBM and diffusion tensor imaging will be advantageous in unraveling the underlying white-matter structural-functional connection in WS.
Acknowledgments
This work was funded by grants from the National Institute for Biomedical Imaging and Bioengineering, the National Center for Research Resources, and the National Institute on Aging, (EB01651, RR019771, AG016570 to PT), from the National Institute of Mental Health (K02 MH01142; to ALR), and the National Institute of Child Health and Human Development (R01 HD31715 to ALR and P01 HD33113 to ALR, JRK, DM, AG and UB). Additional support was provided by NCRR Resource grant P41 RR13642 to AWT, and a fellowship from the Government of Taiwan (to M.C.C.).
APPENDIX
A. Analysis of covariance for the group difference of regional brain volume
To detrend any possible confounding effect of subjects’ age on the regional volumetric differences observed between groups, analysis of covariance was performed at each voxel by least-squares fitting of coefficients βi in the following general linear model:
| (A1) |
Here diagnosis was coded as a binary variable (0 for control and 1 for WS), and age was detrended as a covariate. ε is an error term. We then tested H0: β1 = 0 by defining an F statistic (Rencher, 2002), distributed approximately as Fh,n−q−1:
| (A2) |
where is the regression sum of squares, and is the sum of squares of error for the full model (Eq. A1). is the regression sum of squares for the reduced model:
| (A3) |
Here n is the total number of subjects, q is the number of variables in the full model (= 2 for diagnosis and age), and h is the number of variable to be tested (=1 for diagnosis). represents log(JICBM), β̂j β̂rj the estimated regression coefficient, and xij or xrij the independent variables of subject i, with the subscript r for the reduced model; for instance, xi1 indicates the value coding for diagnosis (0 or 1), and xi2 the age of subject i, and xi0 = 1 is the regression constant. We rejected H0 if F > Fα, where α = 0.01.
B. Repeated measures analysis of variance for the group by side interaction effect
We compared the difference in asymmetry between WS and control groups by voxelwise testing the following null hypothesis H0:
| (B1) |
where μij is the mean of log(JICBM) at the voxel for diagnosis group i and side j, with i ∈ {0, 1} = {control, WS} and j ∈ {0, 1} = {left, right}. Writing Eq. B1 in matrix-vector form, we defined a contrast vector c = [1 − 1]T, then the above H0 becomes:
| (B2) |
where μi = [μi0 μi1]T. The group by side interaction effect can be tested by the Wilks’ Λ (Rencher, 2002):
| (B3) |
Here and is the voxel value vector for log(JICBM) at the left (subscript = 0) and right (subscript = 1) sides of subject l in diagnosis group k, , and , while is the total number of subjects, which were assigned to K diagnostic groups with each group composed of nk subjects. Λ is distributed as Λp,νH, νE, with p = number of sides (left and right) − 1 = 1, νH = K − 1, and νE = N − K. We rejected H0 if Λ ≤ Λα,p, νH, νE, where α was set to 0.01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arsigny V, Fillard P, Pennec X, Ayache N. Fast and simple calculus on tensors in the log-Euclidean framework. Int Conf Med Image Comput Comput Assist Interv (MICCAI); 2005. pp. 115–122. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Avants B, Gee JC. Geodesic estimation for large deformation anatomical shape averaging and interpolation. Neuroimage. 2004;23(Suppl 1):S139–150. doi: 10.1016/j.neuroimage.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Avants BB, Schoenemann PT, Gee JC. Lagrangian frame diffeomorphic image registration: Morphometric comparison of human and chimpanzee cortex. Med Image Anal. 2006;10:397–412. doi: 10.1016/j.media.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Beaton AA. The relation of planum temporale asymmetry and morphology of the corpus callosum to handedness, gender, and dyslexia: a review of the evidence. Brain Lang. 1997;60:255–322. doi: 10.1006/brln.1997.1825. [DOI] [PubMed] [Google Scholar]
- Bellugi U, Korenberg JR, Klima ES. Williams syndrome: an exploration of neurocognitive and genetic features. Clinical Neuroscience Research. 2001;1:217–229. [Google Scholar]
- Bellugi U, Lichtenberger L, Mills D, Galaburda A, Korenberg JR. Bridging cognition, the brain and molecular genetics: evidence from Williams syndrome. Trends Neurosci. 1999;22:197–207. doi: 10.1016/s0166-2236(99)01397-1. [DOI] [PubMed] [Google Scholar]
- Bellugi U, Lichtenberger L, Jones W, Lai Z, St George M. I. The neurocognitive profile of Williams Syndrome: a complex pattern of strengths and weaknesses. J Cogn Neurosci. 2000;12(Suppl 1):7–29. doi: 10.1162/089892900561959. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Boddaert N, Mochel F, Meresse I, Seidenwurm D, Cachia A, Brunelle F, Lyonnet S, Zilbovicius M. Parieto-occipital grey matter abnormalities in children with Williams syndrome. Neuroimage. 2006;30:721–725. doi: 10.1016/j.neuroimage.2005.10.051. [DOI] [PubMed] [Google Scholar]
- Brown JH, Johnson MH, Paterson SJ, Gilmore R, Longhi E, Karmiloff-Smith A. Spatial representation and attention in toddlers with Williams syndrome and Down syndrome. Neuropsychologia. 2003;41:1037–1046. doi: 10.1016/s0028-3932(02)00299-3. [DOI] [PubMed] [Google Scholar]
- Bush G, Luu P, Posner MI. Cognitive and emotional influences in anterior cingulate cortex. Trends Cogn Sci. 2000;4:215–222. doi: 10.1016/s1364-6613(00)01483-2. [DOI] [PubMed] [Google Scholar]
- Carrion VG, Weems CF, Eliez S, Patwardhan A, Brown W, Ray RD, Reiss AL. Attenuation of frontal asymmetry in pediatric posttraumatic stress disorder. Biol Psychiatry. 2001;50:943–951. doi: 10.1016/s0006-3223(01)01218-5. [DOI] [PubMed] [Google Scholar]
- Chiang M-C, Dutton RA, Hayashi KM, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. 3D pattern of brain atrophy in HIV/AIDS visualized using tensor-based morphometry. Neuroimage. 2007;34:44–60. doi: 10.1016/j.neuroimage.2006.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y, Leporé N, de Zubicaray G, Rose S, Carmichael O, Becker J, Toga A, Thompson P. Automatic 3D mapping and analysis of the lateral ventricles using fluid registration of multiple labeled atlases. IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI).2007. [Google Scholar]
- Christensen GE, Johnson HJ. Consistent image registration. Medical Imaging, IEEE Transactions on. 2001;20:568–582. doi: 10.1109/42.932742. [DOI] [PubMed] [Google Scholar]
- Christensen GE, Johnson HJ, Vannier MW. Synthesizing average 3D anatomical shapes. Neuroimage. 2006;32:146–158. doi: 10.1016/j.neuroimage.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Chung MK, Dalton KM, Alexander AL, Davidson RJ. Less white matter concentration in autism: 2D voxel-based morphometry. Neuroimage. 2004;23:242–251. doi: 10.1016/j.neuroimage.2004.04.037. [DOI] [PubMed] [Google Scholar]
- Chung MK, Worsley KJ, Paus T, Cherif C, Collins DL, Giedd JN, Rapoport JL, Evans AC. A unified statistical approach to deformation-based morphometry. Neuroimage. 2001;14:595–606. doi: 10.1006/nimg.2001.0862. [DOI] [PubMed] [Google Scholar]
- Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3-D model-based neuroanatomical segmentation. Hum Brain Mapp. 1995;3:190–208. [Google Scholar]
- Davatzikos C, Genc A, Xu D, Resnick SM. Voxel-based morphometry using the RAVENS maps: methods and validation using simulated longitudinal atrophy. Neuroimage. 2001;14:1361–1369. doi: 10.1006/nimg.2001.0937. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Vaillant M, Resnick SM, Prince JL, Letovsky S, Bryan RN. A computerized approach for morphological analysis of the corpus callosum. J Comput Assist Tomogr. 1996;20:88–97. doi: 10.1097/00004728-199601000-00017. [DOI] [PubMed] [Google Scholar]
- de Lacoste MC, Kirkpatrick JB, Ross ED. Topography of the human corpus callosum. J Neuropathol Exp Neurol. 1985;44:578–591. doi: 10.1097/00005072-198511000-00004. [DOI] [PubMed] [Google Scholar]
- Eckert MA, Tenforde A, Galaburda AM, Bellugi U, Korenberg JR, Mills D, Reiss AL. To modulate or not to modulate: Differing results in uniquely shaped Williams syndrome brains. Neuroimage. 2006a;32:1001–1007. doi: 10.1016/j.neuroimage.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Eckert MA, Hu D, Eliez S, Bellugi U, Galaburda A, Korenberg J, Mills D, Reiss AL. Evidence for superior parietal impairment in Williams syndrome. Neurology. 2005;64:152–153. doi: 10.1212/01.WNL.0000148598.63153.8A. [DOI] [PubMed] [Google Scholar]
- Eckert MA, Galaburda AM, Karchemskiy A, Liang A, Thompson P, Dutton RA, Lee AD, Bellugi U, Korenberg JR, Mills D, Rose FE, Reiss AL. Anomalous sylvian fissure morphology in Williams syndrome. Neuroimage. 2006b;33:39–45. doi: 10.1016/j.neuroimage.2006.05.062. [DOI] [PubMed] [Google Scholar]
- Edgington ES. Randomization Tests. New York, NY: Marcel Dekker; 1995. [Google Scholar]
- Evans AC, Collins DL, Milner B. An MRI-based stereotactic brain atlas from 300 young normal subjects. Proc. 22nd Annu. Symp. Soc. Neurosci; Anaheim.. 1992. p. 408. [Google Scholar]
- Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashman LA, Andreasen NC, Flaum M, Swayze IIVW. Intelligence and regional brain volumes in normal controls. Intelligence. 1997;25:149–160. [Google Scholar]
- Fox NC, Crum WR, Scahill RI, Stevens JM, Janssen JC, Rossor MN. Imaging of onset and progression of Alzheimer’s disease with voxel-compression mapping of serial magnetic resonance images. Lancet. 2001;358:201–205. doi: 10.1016/S0140-6736(01)05408-3. [DOI] [PubMed] [Google Scholar]
- Galaburda AM, LeMay M, Kemper TL, Geschwind N. Right-left asymmetrics in the brain. Science. 1978;199:852–856. doi: 10.1126/science.341314. [DOI] [PubMed] [Google Scholar]
- Gaser C, Luders E, Thompson PM, Lee AD, Dutton RA, Geaga JA, Hayashi KM, Bellugi U, Galaburda AM, Korenberg JR, Mills DL, Toga AW, Reiss AL. Increased local gyrification mapped in Williams syndrome. Neuroimage. 2006;33:46–54. doi: 10.1016/j.neuroimage.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Geschwind N, Levitsky W. Human brain: left-right asymmetries in temporal speech region. Science. 1968;161:186–187. doi: 10.1126/science.161.3837.186. [DOI] [PubMed] [Google Scholar]
- Good CD, Johnsrude I, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. Cerebral asymmetry and the effects of sex and handedness on brain structure: a voxel-based morphometric analysis of 465 normal adult human brains. Neuroimage. 2001a;14:685–700. doi: 10.1006/nimg.2001.0857. [DOI] [PubMed] [Google Scholar]
- Good CD, Johnsrude IS, Ashburner J, Henson RN, Friston KJ, Frackowiak RS. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001b;14:21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Gray JR, Thompson PM. Neurobiology of intelligence: science and ethics. Nat Rev Neurosci. 2004;5:471–482. doi: 10.1038/nrn1405. [DOI] [PubMed] [Google Scholar]
- Haier RJ, Jung RE, Yeo RA, Head K, Alkire MT. Structural brain variation and general intelligence. Neuroimage. 2004;23:425–433. doi: 10.1016/j.neuroimage.2004.04.025. [DOI] [PubMed] [Google Scholar]
- Haxby JV. Fine structure in representations of faces and objects. Nat Neurosci. 2006;9:1084–1086. doi: 10.1038/nn0906-1084. [DOI] [PubMed] [Google Scholar]
- Haxby JV, Ungerleider LG, Clark VP, Schouten JL, Hoffman EA, Martin A. The effect of face inversion on activity in human neural systems for face and object perception. Neuron. 1999;22:189–199. doi: 10.1016/s0896-6273(00)80690-x. [DOI] [PubMed] [Google Scholar]
- Hickok G, Bellugi U, Jones W. Asymmetrical ability. Science. 1995;270:219–220. [PubMed] [Google Scholar]
- Hoffman EA, Haxby JV. Distinct representations of eye gaze and identity in the distributed human neural system for face perception. Nat Neurosci. 2000;3:80–84. doi: 10.1038/71152. [DOI] [PubMed] [Google Scholar]
- Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Evans AC. Enhancement of MR images using registration for signal averaging. J Comput Assist Tomogr. 1998;22:324–333. doi: 10.1097/00004728-199803000-00032. [DOI] [PubMed] [Google Scholar]
- Jernigan TL, Bellugi U, Sowell E, Doherty S, Hesselink JR. Cerebral morphologic distinctions between Williams and Down syndromes. Arch Neurol. 1993;50:186–191. doi: 10.1001/archneur.1993.00540020062019. [DOI] [PubMed] [Google Scholar]
- Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage. 2004;23(Suppl 1):S151–160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- Kawashima R, Sugiura M, Kato T, Nakamura A, Hatano K, Ito K, Fukuda H, Kojima S, Nakamura K. The human amygdala plays an important role in gaze monitoring. A PET study. Brain. 1999;122 (Pt 4):779–783. doi: 10.1093/brain/122.4.779. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Lancaster JL, Thompson P, Woods R, Mazziotta J, Hardies J, Fox P. Regional spatial normalization: toward an optimal target. J Comput Assist Tomogr. 2001;25:805–816. doi: 10.1097/00004728-200109000-00023. [DOI] [PubMed] [Google Scholar]
- Korenberg JR, Chen XN, Hirota H, Lai Z, Bellugi U, Burian D, Roe B, Matsuoka R. VI. Genome structure and cognitive map of Williams syndrome. J Cogn Neurosci. 2000;12(Suppl 1):89–107. doi: 10.1162/089892900562002. [DOI] [PubMed] [Google Scholar]
- Lee AD, Leow AD, Lu A, Reiss AL, Hall S, Chiang MC, Toga AW, Thompson PM. 3D pattern of brain abnormalities in Fragile X syndrome visualized using tensor-based morphometry. Neuroimage. 2007;34:924–938. doi: 10.1016/j.neuroimage.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow A, Huang S, Geng A, Becker J, Davis S, Toga A, Thompson P. Inverse Consistent Mapping in 3D Deformable Image Registration: Its Construction and Statistical Properties. Information Processing in Medical Imaging (IPMI); Colorado: Glenwood Springs; 2005. [DOI] [PubMed] [Google Scholar]
- Leow AD, Klunder AD, Jack CR, Jr, Toga AW, Dale AM, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Whitwell JL, Borowski BJ, Fleisher AS, Fox NC, Harvey D, Kornak J, Schuff N, Studholme C, Alexander GE, Weiner MW, Thompson PM. Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage. 2006;31:627–640. doi: 10.1016/j.neuroimage.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin DJ, Menon V, Schmitt JE, Eliez S, White CD, Glover GH, Kadis J, Korenberg JR, Bellugi U, Reiss AL. Neural correlates of auditory perception in Williams syndrome: an fMRI study. Neuroimage. 2003;18:74–82. doi: 10.1006/nimg.2002.1297. [DOI] [PubMed] [Google Scholar]
- Lui LM, Wang Y, Chan TF, Thompson PM. Brain anatomical feature detection by solving partial differential equations on a general manifold. Journal of Discrete and Continuous Dynamical Systems-Series B. 2007;7:605 – 618. [Google Scholar]
- Maguire EA. Hippocampal involvement in human topographical memory: evidence from functional imaging. Philosophical Transactions of the Royal Society B: Biological Sciences. 1997;352:1475–1480. doi: 10.1098/rstb.1997.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire EA, Frackowiak RS, Frith CD. Learning to find your way: a role for the human hippocampal formation. Proc Biol Sci. 1996;263:1745–1750. doi: 10.1098/rspb.1996.0255. [DOI] [PubMed] [Google Scholar]
- Manly KF, Nettleton D, Hwang JT. Genomics, prior probability, and statistical tests of multiple hypotheses. Genome Res. 2004;14:997–1001. doi: 10.1101/gr.2156804. [DOI] [PubMed] [Google Scholar]
- Marenco S, Meyer-Lindberg A, Kippenhan S, Olsen RK, Mervis CB, Morris CA, Pierpaoli C, Berman KF. Human Brain Mapping. Budapest; Hungary: 2004. Preliminary diffusion tensor imaging (DTI) observations in 5 individuals with Williams syndrome (WS) [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel MA. Big-brained people are smarter: A meta-analysis of the relationship between in vivo brain volume and intelligence. Intelligence. 2005;33:337–346. [Google Scholar]
- Meyer-Lindenberg A, Kohn P, Mervis CB, Kippenhan JS, Olsen RK, Morris CA, Berman KF. Neural basis of genetically determined visuospatial construction deficit in Williams syndrome. Neuron. 2004;43:623–631. doi: 10.1016/j.neuron.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Hariri AR, Munoz KE, Mervis CB, Mattay VS, Morris CA, Berman KF. Neural correlates of genetically abnormal social cognition in Williams syndrome. Nat Neurosci. 2005a;8:991–993. doi: 10.1038/nn1494. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Mervis CB, Sarpal D, Koch P, Steele S, Kohn P, Marenco S, Morris CA, Das S, Kippenhan S, Mattay VS, Weinberger DR, Berman KF. Functional, structural, and metabolic abnormalities of the hippocampal formation in Williams syndrome. J Clin Invest. 2005b;115:1888–1895. doi: 10.1172/JCI24892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobbs D, Garrett AS, Menon V, Rose FE, Bellugi U, Reiss AL. Anomalous brain activation during face and gaze processing in Williams syndrome. Neurology. 2004;62:2070–2076. doi: 10.1212/01.wnl.0000129536.95274.dc. [DOI] [PubMed] [Google Scholar]
- Mobbs D, Eckert MA, Mills D, Korenberg J, Bellugi U, Galaburda AM, Reiss AL. Frontostriatal Dysfunction During Response Inhibition in Williams Syndrome. Biol Psychiatry. 2006 doi: 10.1016/j.biopsych.2006.05.041. [DOI] [PubMed] [Google Scholar]
- Nguyen NT, McDaniel MA. Brain size and intelligence: A meta-analysis. First Annual Conference of the International Society of Intelligence Research; Cleveland, OH.. 2000. [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Hum Brain Mapp. 2001;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Kubicki M, Shenton ME, Guimond A, McCarley RW, Maier SE, Kikinis R, Jolesz FA, Westin CF. Spatial normalization of diffusion tensor MRI using multiple channels. Neuroimage. 2003;20:1995–2009. doi: 10.1016/j.neuroimage.2003.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Westin CF, Kubicki M, Maier SE, Niznikiewicz M, Baer A, Frumin M, Kikinis R, Jolesz FA, McCarley RW, Shenton ME. White matter hemisphere asymmetries in healthy subjects and in schizophrenia: a diffusion tensor MRI study. Neuroimage. 2004;23:213–223. doi: 10.1016/j.neuroimage.2004.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma D, De Geus EJ, Baare WF, Hulshoff Pol HE, Kahn RS, Boomsma DI. The association between brain volume and intelligence is of genetic origin. Nat Neurosci. 2002;5:83–84. doi: 10.1038/nn0202-83. [DOI] [PubMed] [Google Scholar]
- Reiss AL, Abrams MT, Singer HS, Ross JL, Denckla MB. Brain development, gender and IQ in children. A volumetric imaging study. Brain. 1996;119 (Pt 5):1763–1774. doi: 10.1093/brain/119.5.1763. [DOI] [PubMed] [Google Scholar]
- Reiss AL, Eliez S, Schmitt JE, Straus E, Lai Z, Jones W, Bellugi U. IV. Neuroanatomy of Williams syndrome: a high-resolution MRI study. J Cogn Neurosci. 2000;12(Suppl 1):65–73. doi: 10.1162/089892900561986. [DOI] [PubMed] [Google Scholar]
- Reiss AL, Eckert MA, Rose FE, Karchemskiy A, Kesler S, Chang M, Reynolds MF, Kwon H, Galaburda A. An experiment of nature: brain anatomy parallels cognition and behavior in Williams syndrome. J Neurosci. 2004;24:5009–5015. doi: 10.1523/JNEUROSCI.5272-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rencher AC. Methods of multivariate analysis. 2. New York: J. Wiley.; 2002. [Google Scholar]
- Schmitt JE, Eliez S, Warsofsky IS, Bellugi U, Reiss AL. Enlarged cerebellar vermis in Williams syndrome. J Psychiatr Res. 2001a;35:225–229. doi: 10.1016/s0022-3956(01)00024-3. [DOI] [PubMed] [Google Scholar]
- Schmitt JE, Eliez S, Warsofsky IS, Bellugi U, Reiss AL. Corpus callosum morphology of Williams syndrome: relation to genetics and behavior. Dev Med Child Neurol. 2001b;43:155–159. [PubMed] [Google Scholar]
- Shattuck DW, Leahy RM. BrainSuite: an automated cortical surface identification tool. Med Image Anal. 2002;6:129–142. doi: 10.1016/s1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- Shaw P, Greenstein D, Lerch J, Clasen L, Lenroot R, Gogtay N, Evans A, Rapoport J, Giedd J. Intellectual ability and cortical development in children and adolescents. Nature. 2006;440:676–679. doi: 10.1038/nature04513. [DOI] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. Very high-resolution morphometry using mass-preserving deformations and HAMMER elastic registration. Neuroimage. 2003;18:28–41. doi: 10.1006/nimg.2002.1301. [DOI] [PubMed] [Google Scholar]
- Shi Y, Reiss A, Lee A, Dutton R, Bellugi U, Galaburda A, Korenberg J, Mills D, Dinov I, Thompson P, Toga A. Hamilton-Jacobi skeletons on cortical surfaces with applications in characterizing the gyrification pattern in Williams syndrome. IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI).2007. [Google Scholar]
- Steinmetz H. Structure, functional and cerebral asymmetry: in vivo morphometry of the planum temporale. Neurosci Biobehav Rev. 1996;20:587–591. doi: 10.1016/0149-7634(95)00071-2. [DOI] [PubMed] [Google Scholar]
- Steinmetz H, Rademacher J, Huang YX, Hefter H, Zilles K, Thron A, Freund HJ. Cerebral asymmetry: MR planimetry of the human planum temporale. J Comput Assist Tomogr. 1989;13:996–1005. [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002;64:479–498. [Google Scholar]
- Storey JD, Tibshirani R. Technical Report 2001–28. Department of Statistics, Stanford University; 2001. Estimating false discovery rates under dependence, with applications to DNA microarrays. [Google Scholar]
- Studholme C, Cardenas V. A template free approach to volumetric spatial normalization of brain anatomy. Pattern Recognition Letters. 2004;25:1191–1202. [Google Scholar]
- Studholme C, Cardenas V, Blumenfeld R, Schuff N, Rosen HJ, Miller B, Weiner M. Deformation tensor morphometry of semantic dementia with quantitative validation. Neuroimage. 2004;21:1387–1398. doi: 10.1016/j.neuroimage.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Schwartz C, Toga AW. High-resolution random mesh algorithms for creating a probabilistic 3D surface atlas of the human brain. Neuroimage. 1996;3:19–34. doi: 10.1006/nimg.1996.0003. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Giedd JN, Woods RP, MacDonald D, Evans AC, Toga AW. Growth patterns in the developing brain detected by using continuum mechanical tensor maps. Nature. 2000;404:190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Lee AD, Dutton RA, Geaga JA, Hayashi KM, Eckert MA, Bellugi U, Galaburda AM, Korenberg JR, Mills DL, Toga AW, Reiss AL. Abnormal cortical complexity and thickness profiles mapped in Williams syndrome. J Neurosci. 2005;25:4146–4158. doi: 10.1523/JNEUROSCI.0165-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Cannon TD, Narr KL, van Erp T, Poutanen VP, Huttunen M, Lonnqvist J, Standertskjold-Nordenstam CG, Kaprio J, Khaledy M, Dail R, Zoumalan CI, Toga AW. Genetic influences on brain structure. Nat Neurosci. 2001;4:1253–1258. doi: 10.1038/nn758. [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM. Mapping brain asymmetry. Nat Rev Neurosci. 2003;4:37–48. doi: 10.1038/nrn1009. [DOI] [PubMed] [Google Scholar]
- Tomaiuolo F, Di Paola M, Caravale B, Vicari S, Petrides M, Caltagirone C. Morphology and morphometry of the corpus callosum in Williams syndrome: a T1-weighted MRI study. Neuroreport. 2002;13:2281–2284. doi: 10.1097/00001756-200212030-00022. [DOI] [PubMed] [Google Scholar]
- Tosun D, Reiss A, Lee A, Dutton R, Bellugi U, Galaburda A, Korenberg J, Mills D, Toga A, Thompson P. Use of cortical curvature features to analyze local and regional cortical folding complexity. 2007 to be submitted to NeuroImage Dec 2006. [Google Scholar]
- Tosun D, Reiss A, Lee A, Dutton R, Geaga J, Hayashi K, Eckert M, Bellugi U, Galaburda A, Korenberg J, Mills D, Toga A, Thompson P. Use of 3-D cortical morphometry for mapping increased cortical gyrification and complexity in Williams syndrome. IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI); Arlington, Virginia.. 2006. [Google Scholar]
- Twining CJ, Cootes T, Marsland S, Petrovic V, Schestowitz R, Taylor CJ. A Unified Information-Theoretic Approach to Groupwise Non-rigid Registration and Model Building. Information Processing in Medical Imaging: 19th International Conference, IPMI 2005; Glenwood Springs, CO; 2005. pp. 1–14. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chiang MC, Thompson PM. Automated surface matching using mutual information applied to Riemann surface structures. Int Conf Med Image Comput Comput Assist Interv (MICCAI); Palm Springs, CA; 2005. pp. 666–674. [DOI] [PubMed] [Google Scholar]
- Watkins KE, Paus T, Lerch JP, Zijdenbos A, Collins DL, Neelin P, Taylor J, Worsley KJ, Evans AC. Structural asymmetries in the human brain: a voxel-based statistical analysis of 142 MRI scans. Cereb Cortex. 2001;11:868–877. doi: 10.1093/cercor/11.9.868. [DOI] [PubMed] [Google Scholar]
- Westbury CF, Zatorre RJ, Evans AC. Quantifying variability in the planum temporale: a probability map. Cereb Cortex. 1999;9:392–405. doi: 10.1093/cercor/9.4.392. [DOI] [PubMed] [Google Scholar]
- Wilke M, Sohn JH, Byars AW, Holland SK. Bright spots: correlations of gray matter volume with IQ in a normal pediatric population. Neuroimage. 2003;20:202–215. doi: 10.1016/s1053-8119(03)00199-x. [DOI] [PubMed] [Google Scholar]
- Woods RP. Characterizing volume and surface deformations in an atlas framework: theory, applications, and implementation. Neuroimage. 2003;18:769–788. doi: 10.1016/s1053-8119(03)00019-3. [DOI] [PubMed] [Google Scholar]