Abstract
We have devised a simple high throughput screening compatible fluorescence polarization based assay that can be used to detect the elongation activity of nucleic acid polymerase enzymes. The assay uses a 5’ end labeled template strand and relies on an increase in the polarization signal from the fluorescent label as it is drawn into the active site by the action of the enzyme. If the oligonucleotide is sufficiently short, then fluorescence polarization signal can also be used to detect binding prior to elongation activity. We refer to the nucleic acid substrate as a polymerase elongation template element (PETE) and demonstrated the utility of this “PETE assay” in a microtiter plate format using the RNA-dependent RNA polymerase from poliovirus to extend a self-priming hairpin RNA. The PETE assay provides an efficient method for screening compounds that may inhibit the nucleic acid binding or elongation activities of polymerases.
Keywords: High-throughput screening, polymerase, activity assay, fluorescence polarization, poliovirus
Nucleic acid polymerases present an attractive target for the development of pharmacological agents because they can be exploited to specifically inhibit growth of infectious cells or pathogens [1]. RNA viruses, for example, often package or encode for their own reverse transcriptase or RNA-dependent RNA polymerase enzymes that enable the virus to replicate in their host cells. Likewise, many bacterial or fungal pathogens have polymerase molecules whose structure is sufficiently different from those of their eukaryotic counterparts that it may be possible to design drugs to selectively inhibit these enzymes. However, the identification of lead compounds for such drugs and the utility of large-scale structure activity relationship (SAR) studies are hindered by the lack of a fast, inexpensive, and non-radioactive assay for measuring in vitro enzyme activity in a high throughput screening (HTS) format.
Here we present a very simple fluorescence polarization (FP) based assay that can be used to detect both the binding and the elongation activities of nucleic acid polymerases in a flexible solution based assay. The assay relies on presenting the polymerase with a primer-template oligonucleotide substrate that is labeled at its 5’ end with an FP probe such as fluorescein. When this substrate is initially bound by the polymerase, the FP signal will increase because the probe is no longer free in solution, as per a classical FP based binding assay. However, the very 5’ end of the oligonucleotide is far away from the enzyme and it therefore retains significant local mobility at the end of its nucleic acid tether. In a second step, the polymerase-substrate complex is presented with nucleotide triphosphates, leading to elongation of the substrate. This will result in the 5’ end of the template being drawn into the enzyme active site, reducing the mobility of the fluorophore and further increasing the FP signal. Thus, the FP signal from this straightforward assay can monitor both the binding of nucleic acid substrate and the elongation activity of the enzyme. Importantly, the enzymatic activity occurs in a native active site and the only modification is on the substrate at a site far removed from the catalytic center where it is unlikely to perturb the catalytic mechanism or interfere with binding of other ligands.
We demonstrate this assay in microplate format using a short self-priming hairpin RNA that is elongated by the RNA-dependent RNA polymerase from poliovirus, where we observe significant increases in the FP signal from both the RNA binding and the RNA elongation steps. We also show that the assay can be used to detect the incorporation of nucleoside analogs or the mis-incorporation of non-cognate bases, and as such can be used to assess polymerase fidelity.
Materials and methods
RNA binding assay
The RNA substrates with fluorescein labels attached via a 6-carbon 5’ linker (Fig. 1, Table 1) were synthesized by IDT (www.idtdna.com), solubilized in 10 mM Tris, 1 mM EDTA, pH 8 made with DEPC treated water, and annealed as 10 uM stock solutions by slow cooling from 90°C. The poliovirus RNA-dependent RNA polymerase was expressed and purified as previously described [2]. The RNA binding assays were carried out using 10 nM RNA in a standard assay buffer of 50 mM NaCl, 50 mM HEPES (pH 8.0), 1.5 mM magnesium acetate, 60 uM ZnCl2, 0.1% NP40. The polymerase-RNA complexes were assembled and pre-incubated for 30 minutes at 4°C in the presence of ATP before being warmed to room temperature for 20 minutes prior to staring the FP data collection. These incubation steps allow the polymerase to bind the RNA and then incorporate the first nucleotide as soon as the sample is warm enough to allow catalysis to take place. In the case of poliovirus polymerase, this traps the protein-RNA complex in a long-lived elongation conformation whose dissociation rate is on the order of hours [3]. All data were collected using black 384 well polystyrene plates (Corning 3710) in a Perkin-Elmer Victor V multi-mode microplate reader operating at room temperature.
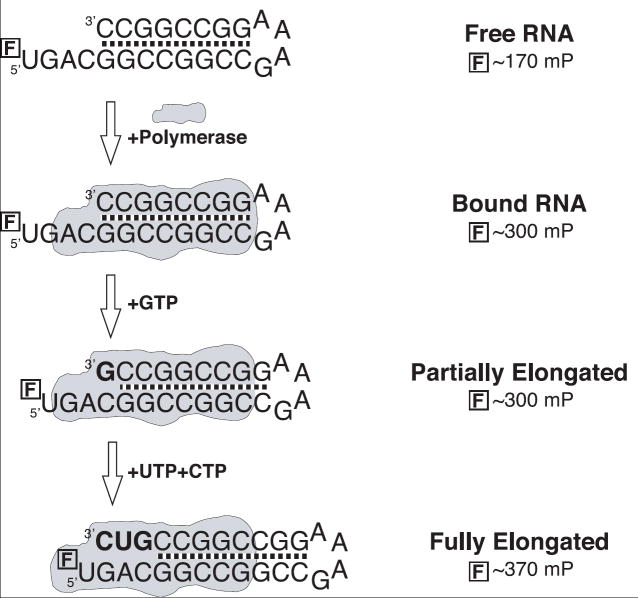
Figure 1.
Schematic illustration of the RNA oligonucleotides used to measure RNA binding and elongation by the poliovirus polymerase with fluorescence polarization. The hairpin structures are followed by a single stranded template sequence that is labeled with a fluorescein molecule at its 5’ end. The addition of NTPs leads to elongation and translocation of the template RNA and the fluorescein label eventually becomes immobilized as it contacts the protein surface, increasing its FP signal in an enzyme activity dependent manner.
Table 1.
Sequences of the self-priming RNA hairpin oligonucleotides used in the binding and elongation experiments. The RNA constructs are named with an “X-Y” convention where X is the length of the duplex region and Y is the length of the templating (boldface) single stranded 5’ extension. The fluorescein label,  , at the end of the templating sequence is attached via a 6-carbon linker.
, at the end of the templating sequence is attached via a 6-carbon linker.
| Name | Kd (μM) | Sequence | Bases to be added |
|---|---|---|---|
| 8-4 | 0.250±0.01 | ACUG | |
| 8-6 | 0.95±0.09 | AUUGCA | |
| 8-8 | 1.9±0.5 | AUUUUGCA | |
| 8-10 | 1.20±0.02 | AUUUUUUGCA | |
| 4-6 | 0.69±0.02 | AUUGCA | |
| 6-6 | 0.30±0.01 | AUUGCA | |
| 8-6 | 0.95±0.09 | AUUGCA | |
| 12-6 | 1.3±0.2 | AUUGCA | |
Polymerase elongation assay
The FP detected elongation studies were carried out using 10 nM RNA with 1 uM 3Dpol in the binding assay buffer. The 3Dpol-RNA complexes were incubated in the presence of only the first nucleotide to be incorporated (ATP), the first and second nucleotide (ATP & UTP), the first three nucleotides (ATP, UTP & GTP), or all four nucleotides. All NTPs were added during the 4°C incubation step to a final concentration of 200 nM and elongation by the requisite number of bases occurred as the microplates were warmed to room temperature prior to reading the endpoint FP signals (see Table 1). The low 200 nM NTP concentration is sufficient to prevent misincorporation of non-cognate bases (see below) and the resulting FP signals are from elongation complexes that are stalled a certain number of bases from the 5’ end of the template strand, depending on which NTPs were added.
Nucleotide mis-incorporation assay
For the misincorporation experiments, the 3Dpol-RNA complex was preincubated in binding buffer with 200 nM each of the first, third, and fourth nucleotides to be incorporated, creating a complex that is poised for the incorporation of the omitted second nucleotide. The successful incorporation of the second nucleotide is then rapidly followed by the incorporation of the third and fourth bases to give a maximal FP signal from the now fully elongated RNA. The rate of the FP signal change is thus limited by the rate of the missing nucleotide incorporation and mis-incorporation of non-cognate bases at this position is slow enough to be followed by the kinetic data collection mode of the Victor microplate reader. Addition of the correct second nucleotide, on the other hand, results in very rapid elongation in a reaction whose kinetics are too fast to be measured by this equipment.
Results
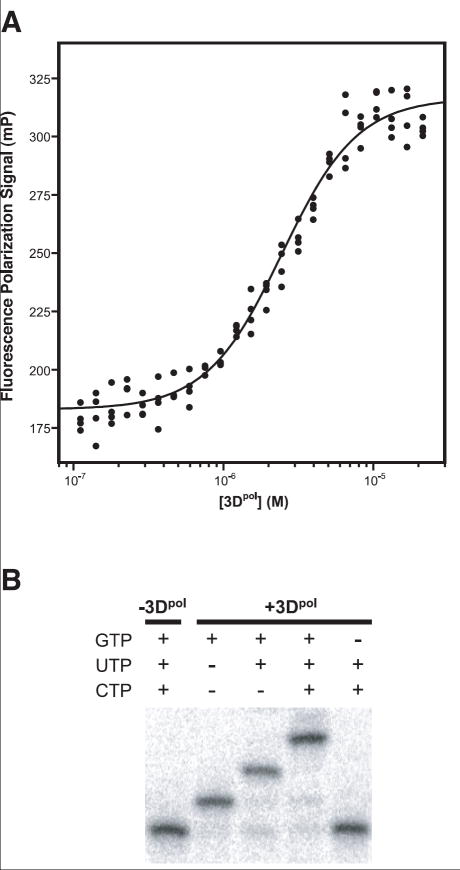
Using a fluorescence polarization based binding assay, a series of different RNA hairpins were tested to determine the minimal RNA length needed to form a stable complex with 3Dpol (Fig. 2A, Table 1). The results show that RNAs with a minimal stem length of 8 basepairs and a minimal single stranded extension of 6 nucleotides bind the polymerase with well behaved binding isotherms and essentially equivalent ~1 μM affinities. Substrates with shorter 4 and 6 basepair stems (4-6 & 6-6) resulted in binding curves with higher affinities, slight positive cooperativity in the binding reaction, and lower plateau FP values suggesting increased local motion in the 3Dpol complex. This suggests to us that these RNA oligonucleotides may be binding the polymerase in a different conformation, perhaps as a result of unfolding of their short helices to form more stable homodimers. Such dimers would have longer 12-16 basepair helices with a pair of G-A bulges at the tetraloop sequences, and the fluorescein probe at the end that was not binding 3Dpol would be far from the polymerase, resulting in the lower average FP signal. Nonetheless, these RNAs could be elongated with the expected sequence specificity for the 5’ templating sequence, indicating their primer/template junction remained intact and properly bound in the active site.
Figure 2.
A) Binding of an “8-6” RNA hairpin to poliovirus polymerase as detected by fluorescence polarization of a fluorescein label on the 5′ end of the RNA. Experiments were carried out using 10 nM RNA and the dissociation constants resulting from fitting the data to a binding isotherm are listed in Table 1. B) Gel electrophoresis analysis showing 3Dpol dependent single nucleotide elongation steps of a 32P 5′ end labeled RNA hairpin with a 3′CAG5′ templating sequence. In the presence of enzyme the RNA is elongated stepwise from a 26mer to a 29mer by the incorporation of G, U, and C, and the last lane shows that the 2nd and 3rd nucleotides are not added if the first nucleotide triphosphate (GTP) is omitted from the reaction.
The use of the hairpin RNA as a substrate for polymerase elongation was validated using a 32P end labeled RNA equivalent to the “8-6” construct that required the incorporation of G, U, and C as the first three nucleotides. As shown by the data in Fig. 2B, this substrate is efficiently elongated from the initial 26mer length to 27, 28, and 29 nucleotides in length by the addition of GTP, CTP, and UTP. Furthermore, the omission of GTP from the reaction prevents the addition of cytosine and uracil, showing that the elongation of this substrate is sequence specific.
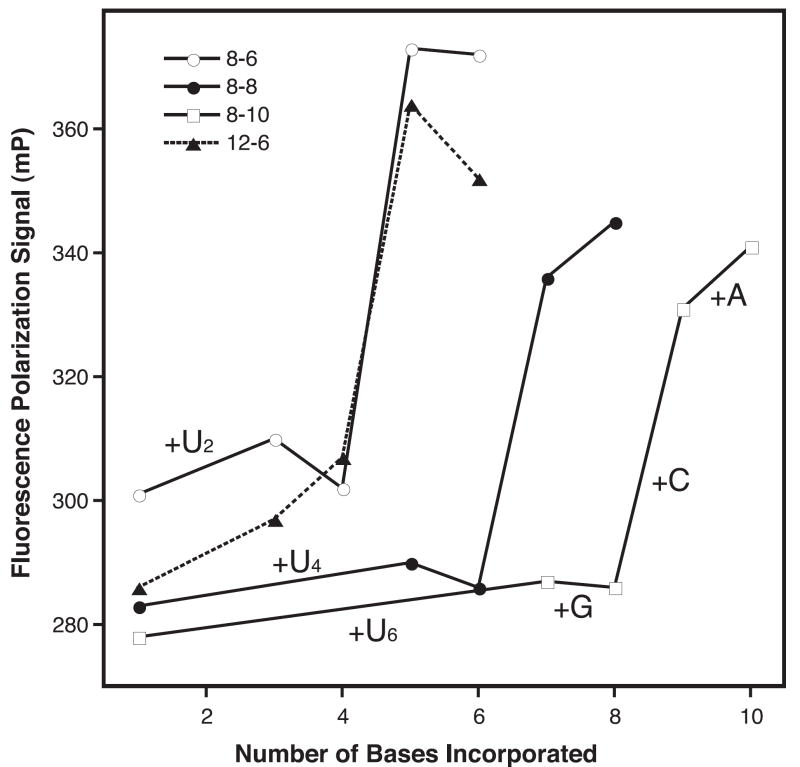
The results from the FP elongation experiments show a clear increase in the FP signal from the fluorescein at the 5’ end of the templating strand when cognate NTPs are added to the 3Dpol-RNA complex (Fig. 3). Using a series of hairpins containing the same 8 base pair stem and different single stranded UAnCGU extensions with a variable length run of n adenosines, we observe the greatest change in the FP signal from incorporation opposite the penultimate 5’ base of the templating strand, regardless of template length (Fig. 3). At a molecular level, this would correspond to a significant immobilization of the fluorescein probe as the priming strand is translocated from having the -3 to having the -2 position in the active site of the enzyme. Significantly, the incorporation of a variable number of uracils (2, 4, or 6) prior to the guanosine at the -3 position does not change the FP signal significantly, indicating that the FP assay is not very sensitive to elongation of sequence that is not proximal to the 5’ end of the template strand.
Figure 3.
Stepwise elongation of the hairpin primer-template RNAs monitored by fluorescence polarization. The RNAs were pre-incubated with 3Dpol and ATP at 4°C for 30 minutes prior to being warmed up to room temperature for the FP assays. UTP was then added, triggering the addition of 2, 4, or 6 uracils depending on the length of the template sequence (Table 1) and stalling the 3Dpol-RNA complexes three nucleotides from the 5′ end of the template strand. The RNA was then fully elongated by the stepwise addition of the remaining NTPs. Note that regardless of the RNA length, only the incorporation of the penultimate nucleotide leads to a significant increase in the FP signal. We attribute this to an immobilization of the fluorescein probe on the protein surface as the 5′ end of the templating strand is translocated into the active site.
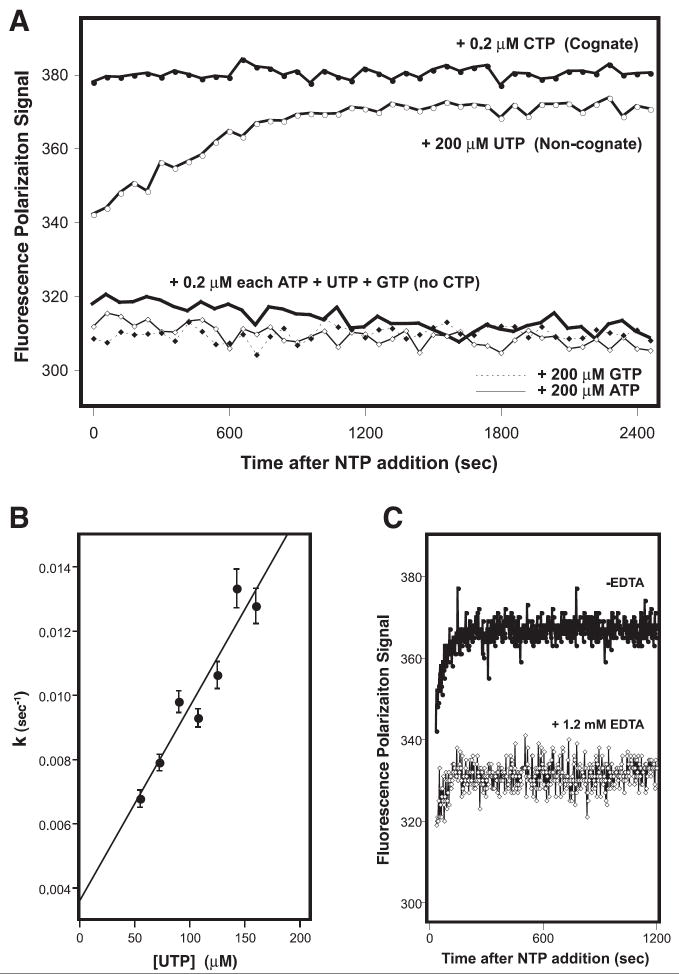
The microplate based FP elongations assay is also capable for measuring the relatively slow mass-action driven mis-incorporation of non-cognate bases. The data in Fig. 4A show kinetic traces for the elongation of the 8-4 RNA where proper elongation involves the sequential addition of an ACUG sequence. When the RNA was incubated with 200 nM each of each NTP there is an immediate elongation of the RNA that manifests itself as an increase of the FP signal from ~300 to ~380 mP in a reaction whose kinetics are too fast to observe. In contrast, when CTP is omitted from the reactions then the FP signal does not increase, showing that there is no detectable elongation due to mis-incorporation of UTP or GTP at the 200 nM concentrations used in the experiment. The addition of a one-thousand fold higher UTP concentration (200 μM) results in an exponential increase in the FP signal over time that can easily be followed with the kinetic data collection capabilities of the plate reader (Fig. 4A). In contrast, no increase in the signal is observed when 200 μM ATP or GTP are added. These data demonstrate that poliovirus 3Dpol will incorporate a uracil opposite the guanosine at the second position in the templating sequence, but it does not efficiently incorporate the larger adenosine or guanosine bases. The uracil mis-incorporation reaction also shows apparent first order kinetics up to at least 200 μM UTP, the highest concentration for which we could obtain reliable kinetic data in the plate reader (Fig. 4B). Last, the NTP dependent signal change is abolished by adding EDTA to inhibit elongation by chelating the Mg2+ ions required for polymerase activity (Fig. 4C).
Figure 4.
A) Kinetic analysis of cognate and non-cognate base incorporation by 3Dpol. Polymerase was pre-incubated with the “8-4” RNA hairpin and the 1st, 3rd, and 4th required NTPs at concentrations of 0.2 μM each. This results in a stable complex that has incorporated the first base (A) and is poised for the addition of the missing second nucleotide (C) before fully elongating by incorporating U and G. Addition of the cognate CTP at 0.2 μM concentration results in complete elongation at a rate too rapid to detect by the microplate reader. The incorporation of non-cognate uracil can be driven by increasing the UTP concentration one thousand-fold to 200 μM, resulting in a time dependent exponential increase in the FP signal, while addition of the bulkier ATP or GTP does not drive mis-incorporation. B) Plot showing that the uracil incorporation rate constant is linear with respect to UTP concentration. The rates were obtained by fitting the temporal uracil incorporation curves (e.g. panel A) to a single exponential function. C) Data showing an absence of an increase in the FP signal when the polymerase is inhibited by adding EDTA to chelate the Mg2+ ions required for elongation activity.
Discussion
In these experiments we have outlined a high throughput screening compatible assay that can be used to examine both the nucleic acid binding and elongation activities of polymerases. The assay relies on using an RNA substrate will call a Polymerase Elongation Template Element (PETE) and we demonstrate the utility of this “PETE assay” using a self-priming RNA hairpin and the 3Dpol RNA-dependent RNA polymerase from poliovirus.
We first examined the RNA length requirements needed for binding to 3Dpol using a series of RNAs that differ in the lengths of their duplex and 5’ extension sequences. The binding data indicate that an eight base pair duplex length and six base single stranded templating sequence are needed to get stable binding of the RNA to poliovirus 3Dpol. Further increases in the length of the duplex or templating regions do not significantly affect the binding equilibrium (Table 1). These RNA lengths are consistent with those obtained by modeling a RNA stem-loop structure onto the 3Dpol structure using the coordinates of the closely related foot-and-mouth disease virus polymerase-RNA complex [4] and bacteriophage T7 RNA polymerase elongation complex [5] as guides for the path or the RNA on the 3Dpol molecule. We expect that the specific lengths of the duplex region and single stranded template regions need to be optimized for other polymerases, but this should be straightforward because the minimal “8-6” construct we find is very similar to hairpin RNAs that have been used to examine hepatitis C virus polymerase activity [6; 7].
The novel aspect of this polymerase assay relies on detecting polymerase elongation activity via a further increase in FP signal that results from translocation of the template after addition of NTPs. The data show that this FP increase occurs as the fluorophore on the 5’ end of the templating strand becomes immobilized on the surface of the polymerase as a result of translocation of the substrate. The FP data from a series of RNAs with single stranded templating regions of variable length show an initial FP value of 280-300 mP units that remains fairly constant as 2, 4, or 6 uracils are added to the RNA (Fig. 3). The end point for all these elongation reactions is such that there are always three unpaired bases remaining on the template strand after addition of the variable number of uracils. Addition of the next nucleotide triphosphate (GTP) in the assay causes the template to be translocated one more base and this also has little effect on the FP signal. However, the further inclusion of the one more nucleotide, CTP, leads to a significant ~60 mP increase in the FP signal. This increase results from the addition of a base opposite the penultimate nucleotide of the templating strand and we observe a similar increase regardless of the starting length of the templating single stranded sequence. Further addition of ATP, the last nucleotide to be incorporated, has only a small additional effect on the FP signal.
The assay can also be used to examine mis-incorporation of non-cognate bases. To show this we provided the 3Dpol-RNA complex with three of the four NTPs needed to incorporate an ACUG sequence opposite the template strand of the “8-4” substrate. By excluding the second nucleotide (CTP) from the reaction mixture, we enable incorporation of only the first nucleotide and stall the polymerase as it waits for the second nucleotide. Once a nucleotide is incorporated in this position, the substrate will then be rapidly fully elongated because the third and fourth nucleotides are already present in solution. The three cognate nucleotides (A, U, G) are added at a low concentration (200 nM) that will support elongation, but not promote mis-incorporation at the missing position. We can then drive mis-incorporation at the second position by simply increasing the concentration of a non-cognate base that is a candidate for incorporation at the second position. As shown by the data in Fig. 3, poliovirus polymerase readily mis-incorporates a uracil opposite the guanosine on the templating strand when the UTP concentration is increased 1000-fold to 200 uM. The rate of the mis-incorporation reaction, as determined by curve fitting the increase in FP signal to a simple single exponential curve, is linear with increasing UTP concentration (Fig. 4B), as expected for a mass action driven reaction. In contrast, similarly increasing the concentration of adenosine or guanosine trinucleotides that would require a bulky and sterically unfavorable purine-purine mismatch in the active site does not result in mis-incorporation. This format of the assay can also be used to evaluate the effectiveness of nucleoside analog compounds, such as ribavirin [8]. In this case one would simply set up the mis-incorporation type assay and drive the reaction forward with potential nucleoside analogs instead of high concentrations of the natural NTPs.
As demonstrated, the RdRP–RNA binding step results in a ~100 mP unit change in the FP signal and the subsequent elongation step results in an additional ~60 mP unit increase, placing this assay at the lower end of the HTS detection range. It may be possible to increase these FP signal changes by using different fluorophores or tethering them to the 5’ end of the template strand with linkers that are shorter than the C6 chain used here. We did not explore template strands shorter than four nucleotides because our structural modeling indicated this was the minimum number of nucleotides needed to fill the template entry channel of the polymerase. One of the advantages of the PETE assay is that the initial polymerase-RNA complex is in effect fully native and should not be affected by the fluorescein label because it is located far from the active site. Using shorter template strands or different linker lengths may be useful to increase sensitivity to a particular step in the polymerase catalytic cycle, such as a pre-translocation conformational change to position the NTP for catalysis or elongation by an initiation complex rather than an elongation complex. With shorter templates, however, it is likely that the presence of the label close to the active site will perturb the geometry and/or activity of the complex, although this many not be a major problem for a HTS format assay with a binary readout of elongation or no elongation in response to adding an inhibitor. We should also note that the poliovirus 3Dpol-RNA elongation complex is kinetically very stable, providing a long time window for readout of the data, but this many not be the case of other polymerases. As an alternative readout strategy, it may useful to design the template strand with a few cytosine residues at its 5’ end which will template guanosine residues. If the polymerase-RNA complex dissociates, then it is quite possible that the fluorescein will stack onto the end of the duplex region, and the stacking interaction with the newly incorporated guanosines is likely to result in a quenching of the steady state fluorescein emission signal that could be used as an alternative readout for successful elongation.
One potential drawback of this assay for HTS purposes is the requirement for a large amount of protein because this is a direct binding assay, rather than an enzymatic turnover based assay that can amplify small signals. However, based on our experience with viral RNA-dependent RNA polymerases this is by no means an insurmountable obstacle. Our experiments were carried out using ~1 uM protein in 50 ul reactions, but we do not foresee a problem automating the assays and scaling them down to 10 ul reaction volumes. With such volumes, a large set of 105 compounds could be tested with ~55 mg of protein that could easily be made given that ~25 mg of poliovirus polymerase can be purified from only one liter of bacterial culture. We have obtained similar yields for the coxsackie virus and bovine viral diarrhea virus polymerases, showing that viral RdRPs can often be easily overexpressed at very high levels in bacteria.
Last, the PETE assay can also be used to observe polymerase translocation rates by rapid stopped-flow techniques similar to the approach used in studies of the E. coli DNA polymerase Klenow fragment, where dansyl probes attached to DNA show differences in polarization and fluorescence lifetime depending on position within the protein-DNA complex [9; 10]. In such studies it is possible that the observed translocation rate may be perturbed by the presence of the fluorescent label at the end of the template strand, but this complication could be circumvented by placing the FP probe at the product end of the substrate oligonucleotide, i.e. in the loop region of the hairpin used here. The initial signal upon polymerase binding event will then results in a maximal FP value because the fluorophore is bound very close to the polymerase, and the signal will then decrease during elongation as the probe is translocated away from the polymerase.
Conclusions
Our results demonstrate the utility of a simple fluorescence polarization based high-throughput screening compatible assay for detecting the elongation activity of nucleic acid polymerases. Coupled with the classic use of polarization as a measure of ligand binding, this constitutes a rapid and convenient method for detecting both the binding to and elongation of nucleic acids by polymerases. The assay provides for the rapid functional testing of protein mutants or compounds that interfere with polymerase function. We have shown this assay with a self-priming hairpin RNA substrate and the RNA-dependent RNA polymerase from poliovirus, but it is easily adaptable to other types of polymerase systems utilizing primer-template pairs that contain DNA, RNA, or heteroduplexes. In these cases one would simply anneal two complementary oligonucleotides, only one of which is labeled with a fluorophore at an appropriate location to elicit a change in FP signal upon translocation.
Acknowledgments
This work was supported by NSF award 0213162 and NIH grant R01-AI059130 to O.B.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beaulieu PL. Finger loop inhibitors of the HCV NS5B polymerase: discovery and prospects for new HCV therapy. Curr Opin Drug Discov Devel. 2006;9:618–26. [PubMed] [Google Scholar]
- 2.Thompson AA, Peersen OB. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. Embo J. 2004;23:3462–71. doi: 10.1038/sj.emboj.7600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold JJ, Cameron CE. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub) J Biol Chem. 2000;275:5329–36. doi: 10.1074/jbc.275.8.5329. [DOI] [PubMed] [Google Scholar]
- 4.Ferrer-Orta C, Arias A, Perez-Luque R, Escarmis C, Domingo E, Verdaguer N. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J Biol Chem. 2004;279:47212–21. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- 5.Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–95. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 6.Labonte P, Axelrod V, Agarwal A, Aulabaugh A, Amin A, Mak P. Modulation of hepatitis C virus RNA-dependent RNA polymerase activity by structure-based site-directed mutagenesis. J Biol Chem. 2002;277:38838–46. doi: 10.1074/jbc.M204657200. [DOI] [PubMed] [Google Scholar]
- 7.Liu C, Chopra R, Swanberg S, Olland S, O’Connell J, Herrmann S. Elongation of synthetic RNA templates by hepatitis C virus NS5B polymerase. J Biol Chem. 2004;279:10738–46. doi: 10.1074/jbc.M310062200. [DOI] [PubMed] [Google Scholar]
- 8.Crotty S, Maag D, Arnold JJ, Zhong W, Lau JY, Hong Z, Andino R, Cameron CE. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med. 2000;6:1375–9. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- 9.Guest CR, Hochstrasser RA, Dupuy CG, Allen DJ, Benkovic SJ, Millar DP. Interaction of DNA with the Klenow fragment of DNA polymerase I studied by time-resolved fluorescence spectroscopy. Biochemistry. 1991;30:8759–70. doi: 10.1021/bi00100a007. [DOI] [PubMed] [Google Scholar]
- 10.Bailey MF, Thompson EH, Millar DP. Probing DNA polymerase fidelity mechanisms using time-resolved fluorescence anisotropy. Methods. 2001;25:62–77. doi: 10.1006/meth.2001.1216. [DOI] [PubMed] [Google Scholar]