Abstract
Background
Cyclophilins (Cyps) are peptidyl cis/trans isomerases implicated in diverse processes such as protein folding, signal transduction, and RNA processing. They are also candidate drug targets, in particular for the immunosuppressant cyclosporine A. In addition, cyclosporine is known to exhibit anti-parasitic effects on a wide range of organisms including several apicomplexa. In order to obtain new non-immunosuppressive drugs targeting apicomplexan cyclophilins, a profound knowledge of the cyclophilin repertoire of this phylum would be necessary.
Results
BLAST and maximum likelihood analyses identified 16 different cyclophilin subfamilies within the genomes of Cryptosporidium hominis, Toxoplasma gondii, Plasmodium falciparum, Theileria annulata, Theileria parva, and Babesia bovis. In addition to good statistical support from the phylogenetic analysis, these subfamilies are also confirmed by comparison of cyclophilin domain architecture. Within an individual genome, the number of different Cyp genes that could be deduced varies between 7–9 for Cryptosporidia and 14 for T. gondii. Many of the putative apicomplexan cyclophilins are predicted to be nuclear proteins, most of them presumably involved in RNA processing.
Conclusion
The genomes of apicomplexa harbor a cyclophilin repertoire that is at least as complex as that of most fungi. The identification of Cyp subfamilies that are specific for lower eukaryotes, apicomplexa, or even the genus Plasmodium is of particular interest since these subfamilies are not present in host cells and might therefore represent attractive drug targets.
Background
Cyclophilins (Cyps) represent an ancient protein family with peptidyl-prolyl cis/trans isomerase (PPIase), also called rotamase, activity (EC 5.2.1.8) that can be found in archea, prokaryotes and eukaryotes [1,2]. PPIases catalyze the cis/trans isomerization of peptide bonds preceding a prolyl residue in polypeptides. Although ribosomes synthesize proteins with peptidyl-prolyl bonds in the lower energy trans state, about 5–7% of these bonds are estimated to occur in the unfavorable cis conformation [3]. PPIases are thought to be important for establishing this conformation during protein folding or refolding after transport of proteins into organelles [1] by stabilizing the cis/trans transition state [4]. Moreover, some Cyps possess chaperone activity that is independent from their PPIase activity [5]. Many Cyps are able to bind the widely used immunosuppressant cyclosporin A (CsA) that on one hand inhibits their PPIase activity but on the other hand results in a gain of function phenotype due to binding of Cyp/CsA complexes to calcineurin-like phosphatases resulting in inhibition of phosphatase activity. In mammalian T cells, inhibition of calcineurin by Cyp/CsA complexes after T cell receptor stimulation prevents transcription of the autocrine growth factor IL-2 resulting in immunosuppression.
In addition to cyclophilins, two also widely spread but structurally unrelated protein families, FK506-binding proteins (FKBP) and parvulins, also exhibit PPIase activity [3].
Eukaryotic genomes usually encode several Cyps. Small Cyps containing only a single Cyp domain are present along with larger multi-domain proteins containing a Cyp domain in addition to one or several unrelated domains. For instance, the genome of the fission yeast Schizosaccharomyces pombe contains four single domain Cyps – including SpCyp4 which has a signal peptide and can be found in the ER – and five multi domain Cyps [6]. Two (Encephalitozoon cuniculi), eight (Saccharomyces cerevisiae) and 17 (Rhizopus oryzae) Cyps could be identified [7-9]e.g. in the genomes of representative microsporidia and fungi. Despite their ubiquitous expression and high evolutionary conservation, convincing evidence for the importance of Cyps for cellular homeostasis is largely missing. In S. cerevisae, for instance, none of the eight Cyps is essential, and even a mutant lacking all eight Cyps and four FKBPs simultaneously has only a subtle phenotype [10].
Parasite Cyps have received increasing attention in recent years (see [11] for review) in particular because CsA has not only immunosuppressive but also anti-parasitic activity as already demonstrated in 1981 for schistosoma and murine malaria infections [12,13]. Since then, anti-parasitic activity of CsA has been demonstrated for numerous protozoan and helminth parasites [11,14]. Because the anti-parasitic effects of CsA can be superimposed in vivo by its immunosuppressive action, treatment of infected animals with CsA may either result in resolution/amelioration or aggravation of the clinical course [11]. However, the development of non-immonosuppressive CsA analogs that retain anti-parasitic activity shows that parasite Cyps may well be attractive drug targets [15].
Since the discovery of CsA sensitivity of Plasmodium chabaudi and Plasmodium berghei [13], development of several other apicomplexa has been described to be inhibitable by CsA including Plasmodium falciparum [16], Toxoplasma gondii [17], Eimeria tenella [18], Eimeria vermiformis, Eimeria mitis [19], and Cryptosporidium parvum [20]. In contrast, Theileria annulata schizonts appear to be unaffected by CsA though the drug inhibits proliferation of Theileria-transformed lymphocytes – presumably by acting on host cell Cyps [21].
Despite the long time since discovery of CsA effects on these important parasites, current knowledge about the anti-parasitic mechanisms of CsA is rather limited. For P. falciparum, two major small cytosolic Cyps and their inhibition by CsA and CsA derivates have been described [22-24]. Inhibition of P. falciparum calcineurin by a complex of CsA and PfCyp19 (= PfCyp19A in reference [25]) has also been demonstrated biochemically [26]. Using sequence analysis of highly CsA-resistant mutant lines of P. falciparum, Kumar et al. [25] could show that point mutations in the regulatory or the catalytic subunit of calcineurin or in PfCyp19 or PfCyp21.7 (= PfCyp19B) are sufficient to induce CsA resistance. In contrast, no mutations in the PfCyp24.6 (= PfCyp24) gene were identified. However, since CsA resistance in five out of nine mutant lines was not associated with changes in the sequence of any of these four genes, additional gene products can be expected to be involved in CsA action in P. falciparum. The situation is even more complicated by the fact that at least certain non-immunosuppressive CsA derivates have been shown to have profound anti-parasitic effects possibly by acting on ABC transporters of the multi-drug-resistance protein family in T. gondii and P. faciparum [15,27].
In addition to their role as putative drug targets, cyclophilins of apicomplexan parasites are also interesting from an evolutionary point of view, since a novel group of dual family PPIases has been recently described for T. gondii, which contain both a Cyp and an FKBP domain in the same protein [28]. Such FCBPs (FK506- and cyclosporin-binding proteins) appear to be present in the genomes of archae- and eubacteria as well [5], and the phylogenetic relationship of apicomplexan FCBP with such non-eukaryotic enzymes remains to be addressed.
Up to now, research on apicomplexan Cyps has focused on small, abundant single-domain Cyps. Only recently, a multi-domain WD40 repeat containing Cyp has been described for E. tenella [29]. The progress in genome sequencing projects for several apicomplexan parasites allows now for systematic searches for cyclophilins and will presumably bring the multi-domain Cyps more into the focus of research. This work is aimed to provide a framework for such analysis by identifying and comparing the cyclophilin repertoire of the important apicomplexan pathogens T. gondii, P. falciparum, Theileria parva, T. annulata, Babesia bovis, and Cryptosporidium hominis.
Results and discussion
Identification of open reading frames for Cyps
In order to identify open reading frames (ORFs) encoding putative Cyps, BLAST and TBLASTn analyses against GenBank®, genomic sequence data and deduced coding sequences were performed. The Cyp proteins deduced from T. gondii, P. falciparum, T. annulata, T. parvum, B. bovis, and C. hominis are listed in Tables 1, 2, 3, 4, 5, 6, respectively. Two putative Cyps from Cryptosporidium muris were included in the analysis, because the orthologous Cyps could not be identified in the genome of C. hominis. Moreover, two Cyps deduced from the Plasmodium yoelii genome were included as the corresponding P. falciparum are quite unusual. Table S1 – in Additional file 1 in the supplemental online material – lists all Cyp proteins encoded in the genomes of S. pombe and Homo sapiens that were used for comparison with the apicomplexan Cyp repertoire.
Table 1.
Cryptosporidium cyclophilin proteins.
| Namea | Accession-no.b | Amino acids | MWc (kDa) | Motifs/Domainsd |
| ChCyp17.9 | 3413232 | 167 | 17.9 | Cyp |
| ChCyp18.4 | 3415531 | 172 | 18.4 | Cyp |
| ChCyp18.9 | 3415323 | 169 | 18.9 | Cyp |
| ChCyp21.2 | 3412170 | 189 | 21.2 | Cyp |
| ChCyp22.9 | 3413640 | 210 | 22.9 | SP, Cyp |
| ChCyp34.5 | 3411992 | 302 | 34.5 | Cyp, RRM |
| CmCyp44.6 | 6997393 | 391 | 44.6 | Cyp |
| CmCyp48.8 | 6997190 | 417 | 48.8 | SYF2, Cyp |
| ChCyp88.9 | 3413617 | 77.4 | 88.9 | WD40, Cyp |
a Deduced from the genome of Cryptosporidium hominis (ChCyp) or Cryptosporidium muris (CmCyp).
bAccession number in Entrez Gene.
cMW, molecular weight.
dCyp, cyclophiln domain; SP, signal peptide; RRM, RNA recognition motif; SYF2, SYF2 splicing factor; WD40, WD40 repeat; FKBP, FK506-binding domain; TRP, Tetratricopeptide repeat.
Table 2.
Toxoplasma gondii cyclophilin proteins.
| Name | Accession-no. | Amino acids | MWa (kDa) | Motifs/Domainsb |
| TgCyp18.7e | EEA99351c | 163 | 18.7 | Cyp |
| TgCyp18.8 | EEB00823c | 172 | 18.8 | Cyp |
| TgCyp19.6 | AAA17997c | 179 | 19.6 | SP, Cyp |
| TgCyp21 | EEA98581c | 195 | 21.0 | Cyp |
| TgCyp21.7 | EEB01220c | 197 | 21.7 | Cyp |
| TgCyp23 | EEB02074c | 211 | 23.0 | Cyp |
| TgCyp31.8e | TGGT1_052840d | 283 | 31.8 | Mito, Cyp |
| TgCyp36.7f | ORFf | 335 | 36.7 | Cyp |
| TgCyp38.2 | EEB00661c | 348 | 38.2 | Cyp |
| TgCyp64.5 | EEA99592c | 575 | 64.5 | Cyp |
| TgCyp66.2 | EEB00778c | 587 | 66.2 | Cyp, SYF2 |
| TgCyp66.3g | EEA99267c | 592 | 66.3 | Cype, RRM |
| TgCyp72.9 | EEB03226c | 612 | 72.9 | RING, Cyp |
| TgCyp86 | EEB02781c | 764 | 86 | WD40, Cyp |
| TgFCBP57.3 | AAX51680c | 521 | 57.3 | FKBP, TRP, Cyp |
aMW, molecular weight.
bCyp, cyclophiln domain; SP, signal peptide; Mito, mitochondrial targeting sequence; SYF2, SYF2 splicing factor; RRM, RNA recognition motif; RING, RING finger domain; WD40, WD40 repeat; FKBP, FK506-binding domain; TRP, Tetratricopeptide repeat.
c, dAccession number in Entrez Protein and ToxoDB, respectively.
eTgCyp18.7 in Entrez Protein contains only an NH2-terminally truncated Cyp domain. In all further analyses, TgCyp31.8 was used instead. This deduced protein contains the same COOH-terminus but additional NH2-terminal sequences.
fTgCyp36.7 was deduced from the T. gondii VEG genome sequence, chromosome VIIa, between position 1860418 and 1864189.
gTgCyp66.3 contains a protein containing a RNA recognition motif. However, the Cyp domain appears to be disrupted, maybe due to misprediction of the ORF.
Table 3.
Plasmodium cyclophilin proteins.
| Name | Accession-no.a | Amino acids | MWb (kDa) | Motifs/Domainsc |
| PfCyp18.6 | 810717 | 167 | 18.6 | Cyp |
| PfCyp19 | 814534 | 171 | 19 | Cyp |
| PfCyp21.7 | 810711 | 195 | 21.7 | SP, Cyp |
| PfCyp23.2 | 813100 | 204 | 23.2 | Cyp |
| PfCyp24.9 | 2655320 | 217 | 24.9 | Cyp |
| PfCyp26.4 | 811077 | 226 | 24.6 | Cyp |
| PfCyp32.3 | 811200 | 280 | 32.3 | Mito, Cyp |
| PfCyp51.8 | 811805 | 440 | 51.8 | Cyp |
| PfCyp72.5 | 813578 | 609 | 72.5 | Cyp |
| PfCyp80.9 | 2655327 | 677 | 80.9 | Cyp, SYF2 |
| PfCyp87 | 812836 | 747 | 87 | WD40, Cyp |
| PyCyp69.8 | 3830381 | 587 | 69.8 | Cyp |
| PyCyp74 | 3791457 | 621 | 74 | Cyp, SYF2 |
a Accession number in Entrez Gene.
b MW, molecular weight.
c Cyp, cyclophilin domain; SP, signal peptide; Mito, mitochondrial targeting sequence; SYF2, SYF2 splicing factor; WD40, WD40 repeat; FKBP, FK506-binding domain; TRP, Tetratricopeptide repeat.
Table 4.
Theileria parva cyclophilin proteins.
| Name | Accession-no.a | Amino acids | MWb (kDa) | Motifs/Domainsc |
| TpCyp18.4 | 3500351 | 164 | 18.4 | Cyp |
| TpCyp20.3 | 3502670 | 175 | 20.3 | Cyp |
| TpCyp21.4 | 3501804 | 196 | 21.4 | SP, Cyp |
| TpCyp24.4 | 3502715 | 217 | 24.4 | Cyp |
| TpCyp24.5 | 3503253 | 216 | 24.5 | Mito, Cyp |
| TpCyp25.5 | 3502505 | 227 | 25.5 | AP, Cyp |
| TpCyp51.4 | 3501424 | 445 | 51.4 | Cyp |
| TpCyp58.8 | 3501575 | 517 | 58.8 | RING, Cyp |
| TpCyp59.8d | 3500432 | 527 | 59.8 | Cyp |
| TpCyp61.3 | 3503208 | 539 | 61.3 | WD40, Cyp |
| TpFCBP51.4 | 3861988 | 460 | 51.4 | FKBP, TRP, Cyp |
a Accession number in Entrez Gene.
b MW, molecular weight.
c Cyp, cyclophiln domain; SP, signal peptide; Mito, mitochondrial targeting sequence; AP, apicoplast targeting sequence; RING, RING finger domain; WD40, WD40 repeat; FKBP, FK506-binding domain; TRP, Tetratricopeptide repeat.
d NH2-terminus extended according to similarity with TaCyp63.
Table 5.
Theileria annulata cyclophilin proteins.
| Name | Accession-no.a | Amino acids | MWb (kDa) | Motifs/Domainsc |
| TaCyp18.3d | 3865382 | 164 | 18.3 | Cyp |
| TaCyp21.6 | 3862085 | 196 | 21.6 | SP, Cyp |
| TaCyp22.8 | 3863397 | 205 | 22.8 | Cyp |
| TaCyp24.8 | 3864489 | 220 | 24.8 | Mito, Cyp |
| TaCyp25.7 | 3864051 | 228 | 25.7 | AP, Cyp |
| TaCyp48.8 | 3861733 | 426 | 48.8 | Cyp |
| TaCyp58.8 | 3861717 | 515 | 515 | RING, Cyp |
| TaCyp63 | 3864616 | 553 | 63.0 | Cyp |
| TaCyp70 | 3863563 | 613 | 70 | WD40, Cyp |
| TaFCBP51.3e | 3861988 | 459 | 51.3 | FKBP, TRP, Cyp |
a Accession number in Entrez Gene.
b MW, molecular weight.
c Cyp, cyclophiln domain; SP, signal peptide; Mito, mitochondrial targeting sequence; AP, apicoplast targeting sequence; RING, RING finger domain; WD40, WD40 repeat; FKBP, FK506-binding domain; TRP, Tetratricopeptide repeat.
dResidues 44–54 deleted to restore similarity to all other cyclophilins. These eleven amino acids presumably represent an intron that was not recognized by the prediction algorithm.
e NH2-terminus extended according to similarity with TpCyp51.4.
Table 6.
Babesia bovis cyclophilin proteins.
| Name | Accession-no.a | Amino acids | MWb (kDa) | Motifs/Domainsc |
| BbCyp19.2 | 5477772 | 175 | 19.2 | Cyp |
| BbCyp21.1 | 5480474 | 195 | 21.1 | SP, Cyp |
| BbCyp21.9 | 5479361 | 200 | 21.9 | Cyp |
| BbCyp23.7 | 5478663 | 217 | 23.7 | SP, Cyp |
| BbCyp26.9 | 5477314 | 242 | 26.9 | Mito, Cyp |
| BbCyp28.6 | 5477723 | 248 | 28.6 | Cyp |
| BbCyp39.4 | 5478386 | 354 | 39.4 | Cyp |
| BbCyp57.3 | 5478847 | 508 | 57.3 | Cyp, SYF2 |
| BbCyp59.4 | 5478371 | 524 | 59.4 | U box, Cyp |
| BbCyp65.8 | 5479111 | 589 | 65.8 | WD40, Cyp |
| BbFCBP51.1 | 5476893 | 460 | 51.1 | FKBP, TRP, Cyp |
a Accession number in Entrez Gene.
b MW, molecular weight.
c Cyp, cyclophiln domain; SP, signal peptide; Mito, mitochondrial targeting sequence; SYF2, SYF2 splicing factor; RING, RING finger domain; WD40, WD40 repeat; FKBP, FK506-binding domain; TRP, Tetratricopeptide repeat.
The number of putative Cyp genes identified per genome ranges from 7 to 9 for C. hominis (whether or not orthologs for CmCyp44.6 and CmCyp48.8 are assumed to be present in C. hominis) to 14 for T. gondii, while the genomes of all four haemosporidia exhibit an intermediate number of 11 putative Cyps per genome. For T. annulata, an ortholog to TpCyp20.3 appears to be present on chromosome 1, however, its complete sequence could not be deduced from the genome data. Therefore, Table 2 lists only 10 Cyps for this organism although 11 Cyps are expected to be present. The number of Cyps in apicomplexan genomes is very similar to the 6 to 11 Cyp genes in the genomes of most fungi although it should be mentioned that there are fungi with extreme low (2 Cyps in the microsporidium Encephalitozoon cuniculi) and extreme high (16 Cyps in Rhizopus oryzae) numbers of Cyp genes [9]. An extremely high number of 19 Cyp genes per genome can also be found in the kinetoplastid protozoan parasite Trypanosoma cruzi [30]. Similar extremes cannot be found in the genomes of the currently sequenced apicomplexa.
Phylogenetic relationship of Cyp domains
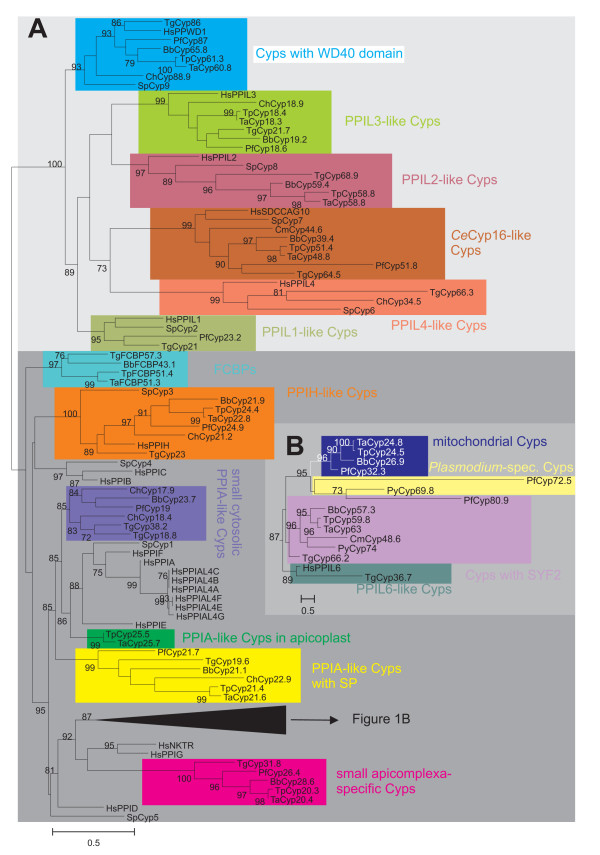
In order to identify subfamilies within the Cyp repertoire and to analyze their phylogenetic relationship, the putative Cyp domains as identified by CD-BLAST [31,32] were aligned by ClustalW2 [33]. Maximum likelihood analysis with PhyML [34] was used to calculate an unrooted tree shown in Figure 1. Statistical support values at the branches are calculated by a likelihood ratio test which produces values similar but not identical to those obtained by bootstrapping [34].
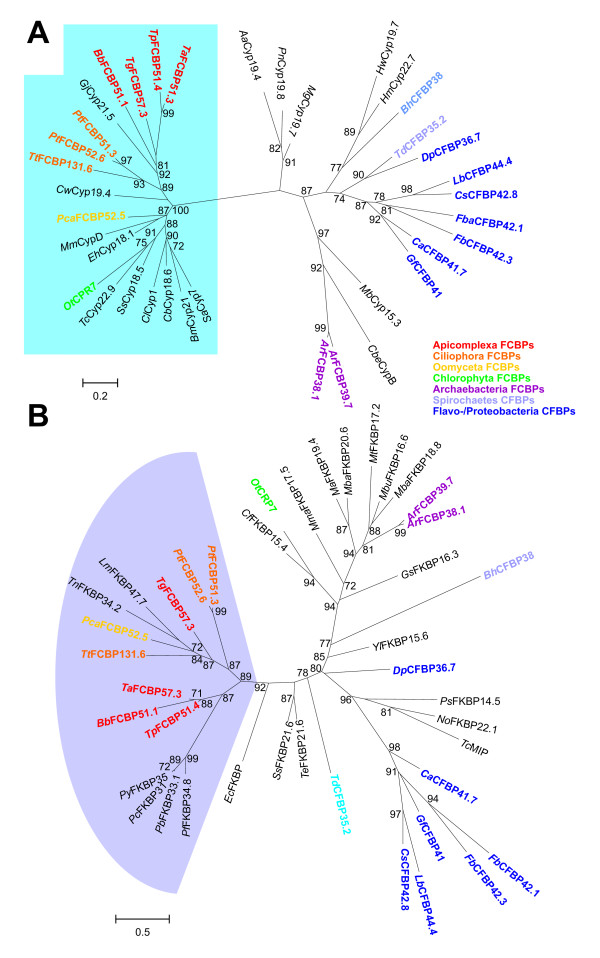
Figure 1.
Unrooted phylogram representing evolutionary relationship between apicomplexan Cyps. (A) Sequences of putative Cyp domains were aligned using ClustalW2 and an unrooted maximum likelihood tree was calculated using PhyML [34]. For comparison, the human and fission yeast Cyp repertoires were included in the analysis. Statistical support of nodes calculated as likelihood ratios is indicated for those nodes with at least 70% support. Cyp subfamilies as revealed by phylogenetic analysis and domain architecture are highlighted by different colors. The dark gray background in the lower part of the figures marks Cyp subfamilies containing Cyp_ABH type or related domains. (B) Subtree from phylogram presented only compressed in (A). The scale bars represent 0.5 substitutions per amino acid position. Ch, C. hominis; Tg, T. gondii; Pf, P. falciparum; Py, Plasmodium yoelii; Bb, B. bovis; Ta, T. annulata; Tp, T. parva; Hs, Homo sapiens; Sp, Schizosaccharomyces pombe. The identity of individual protein sequences used for analyses can be obtained from Tables 1–6 and Table S1 in Additional file 1.
Although it may be assumed that small Cyps containing only a single Cyp domain have been present early in evolution before occurrence of Cyps with one or more additional domains, it is not possible to unequivocally identify a "primitive" Cyp protein subfamily in the apicomplexa from which all other subfamilies have derived, since there are several single- and multi-domain Cyps in the genomes of all protists analyzed so far. Obviously, many subfamilies of Cyps have already evolved before spread of the major lines of eukaryotic evolution.
According to their phylogenetic relationship, 16 different Cyp protein subfamilies were defined here (Figure 1) – many of them well known from other eukaryotes. All these subfamilies exhibit a statistical support in the likelihood ratio test implemented in PhyML of at least 85% and all families containing Cyps with multiple domains are also supported by their domain architecture. The only exception is the subfamily containing putative Cyps with a so-called SYF2 domain, a domain first described in the yeast splicing factor SYF2 [35]. One of these putative SYF2-containing Cyps, i.e. PfCyp80.9, has a very divergent sequence that does not fall into the same PhyML-deduced group as the other subfamily members (Figure 1B). The corresponding protein deduced from P. yoelii (PyCyp74) was therefore also included and the latter is apparently an ortholog to the SYF2 Cyps of other apicomplexa. Since the subfamily of Cyps with SYF2 is strongly supported by domain architecture and all Plasmodium species but P. falciparum posses putative SYF2 Cyps with high similarity to PyCyp74, it appears that the putative PfCyp80.9 was either not predicted correctly or has undergone dramatic alterations after separation of P. falciparum from P. vivax and the rhodent malaria species. Instead of clustering with other SYF2 Cyps, PfCyp80.9 forms a group together with a group of large putative Cyps that can only be identified in the genus Plasmodium (Plasmodium-spec. Cyps), represented in Figure 1 by PfCyp72.9 and PyCyp69.8.
The phylogram in Figure 1 also indicates the presence of two major groups of Cyps depending on whether they contain a Cyp domain related to the Cyp_ABH subtype (CD database accession number [cd01926], drawn on dark gray background in the lower half of Figure 1A) or any of the non Cyp_ABH-like domains (on light gray background in the upper half of Figure 1A). Within the Cyp_ABH group, it is noteworthy that several important groups of well-known Cyps are absent from apicomplexan genomes whereas there are new Cyp subfamilies that appear to be specific for apicomplexa. On one hand, there are apparently no orthologs of HsPPIB or HsPPIC (both involved in protein folding in the secretory pathway [8]), PPID (function in mitochondrial permeability transition during cell death responses [36,37]), and PPIG (involved in splicing [38]). On the other hand, there are several Cyp subfamilies that are specific at least for lower eukaryotes or even for apicomplexa but do not have orthologs in their mammalian hosts and might therefore be promising drug targets in the future. This includes in particular mitochondrial Cyps, Cyps with SYF2, Cyps with signal peptide, and a group of small, presumably cytosolic Cyps specific for apicomplexa.
The following sections will describe genomic organization and protein domain architecture of these subfamilies beginning with the Cyp_ABH-containing proteins. The different subfamilies will be described in the same order in which they are presented in the phylogenetic tree in Fig. 1.
PPIA-like small cytoplasmic Cyps and apicoplast Cyps
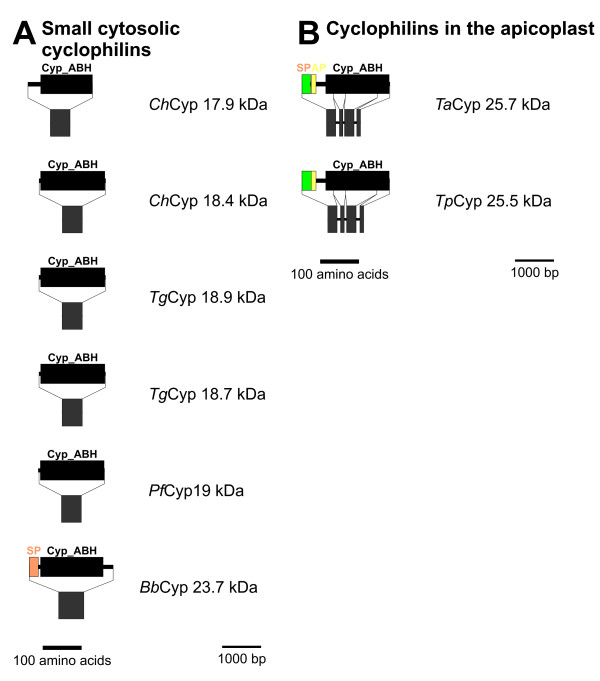
The prototypical Cyps in humans and S. pombe, HsPPIA and SpCyp1, respectively, are closely related as shown in Figure 1. They form a cluster together with additional human paralogs such as PPIE, PPIF and PPIAL4A-G. The corresponding putative Cyps in apicomplexa, ChCyp17.9, BbCyp23.7, PfCyp19, ChCyp18.4, TgCyp18.8, and TgCyp18.9 (Figure 2A), form a related but separate cluster, i.e. SpCyp1 is significantly more closely related to HsPPIA, HsPPIE, and HsPPIF than to any of the apicomplexan Cyps. C. hominis and T. gondii encode two distinct putative members of this PPIA-like subfamily in their genomes. Due to their very high expression levels and their cytoplasmic localization, cytoplasmic Cyps containing a Cyp_ABH type domain (CD database accession-no [cd01926]) are considered to be the most important receptors for CsA leading to inactivation of the cytosolic calcineurins [39]. Indeed, mutations in PfCyp19 appear to be sufficient to confer resistance to CsA to P. falciparum [25].
Figure 2.
Domain architecture and genomic organization of PPIA-like Cyps. (A) PPIA-like cytosolic Cyps. For each Cyp, proteins domains are shown in the upper and exon/intron structure in the lower panel. Proteins and genes are presented by lines, domains and exons are highlighted by boxes. Separate scale bars are given for protein and genome scemes. (B) Cyps with apicoplast localization signal. Species are abbreviated as in Fig. 1. Cyp_ABH, ABH-type Cyp domain (CD accession-no.: [cd01926]); SP, signal peptide; AP, apicolast transit signal.
All these small cytosolic Cyps have no introns interrupting their putative ORFs (Figure 2A).
Conspicuously, the putative PPIA-like Cyps of the piroplasms, BbCyp23.7 (Figure 2A), TpCyp25.5 and TaCyp25.7 (Figure 2B), are predicted to contain an NH2-terminal signal peptide (SP) for cotranslational transport into the ER and for the latter two the PATS algorithm [40] predicts an apicoplast transit signal (AP) following the SP. Such combinations of SP and AP are typical for proteins which are transported into the apicoplast [41]. TpCyp25.5 has been described to be cotranslationally transported into dog pancreas rough microsomes in a wheat germ in vitro translation system [42] demonstrating that the SP is functionally active in this heterologous system. Since no removal of the signal peptide was detectable in this system, TpCyp25.5 was proposed to be anchored to the membrane of the ER via an uncleavable signal peptide. However, since transport of proteins to the apicoplast has been shown to require passage through the ER (for review see [41]) and the properties of proteases responsible for removal of signal peptides might be quite different in apicomplexa and mammals, these results do not exclude that in vivo TpCyp25.5 is transported further from the lumen of the ER into the apicoplast. Moreover, it cannot yet be excluded that the putative BbCyp23.7 will turn out to be localized in the apicoplast as well since the sequence between the SP and the Cyp domain is long enough to function as an AP. Since the neural network analysis used in PATS has been trained only on AP of proteins from P. falciparum, it might well turn out to be less sensitive to functional AP in other apicomplexa such as B. bovis. However, TpCyp25.5 and TaCyp25.7 appear to be more closely related to HsPPIA and SpCyp1 than to the other apicomplexan Cyps of this group whereas BbCyp23.7 clusters together with the clearly cytoplasmic Cyps of other apicomplexa. Whether the position of apicoplast Cyps in the phylogram truly reflects different evolutionary origins from cytoplasmic Cyps or different selective pressures caused by localization in cytoplasm and apicoplast cannot be decided using the current dataset. In addition to the presence of a putative AP, this group also differs from the small cytosolic apicomplexan Cyps – including BbCyp23.7 – by the presence of three introns within the coding sequence. In order to evaluate whether these Cyps with AP are more closely related to Cyps from plants or algae, BLASTp analyses were performed against protein database entries from dinoflagellates, red, green, and brown algae, green plants, and Arabidopsis thaliana. However, highest similarity was always found to cytosolic PPIA-like Cyps and never to Cyps known to be localized in plastids (data not shown). Experimental evidence concerning the localization of BbCyp23.7, TaCyp25.7 and TpCyp25.5 might provide important information on the evolutionary history of these proteins as well, as they should be considered to be monophyletic if all three turn out to be localized in the apicoplast. Moreover, the fact that Theileria species do not have a prototypical cyctosolic PPIA-like Cyp might explain why T. annulata is resistant to CsA [21].
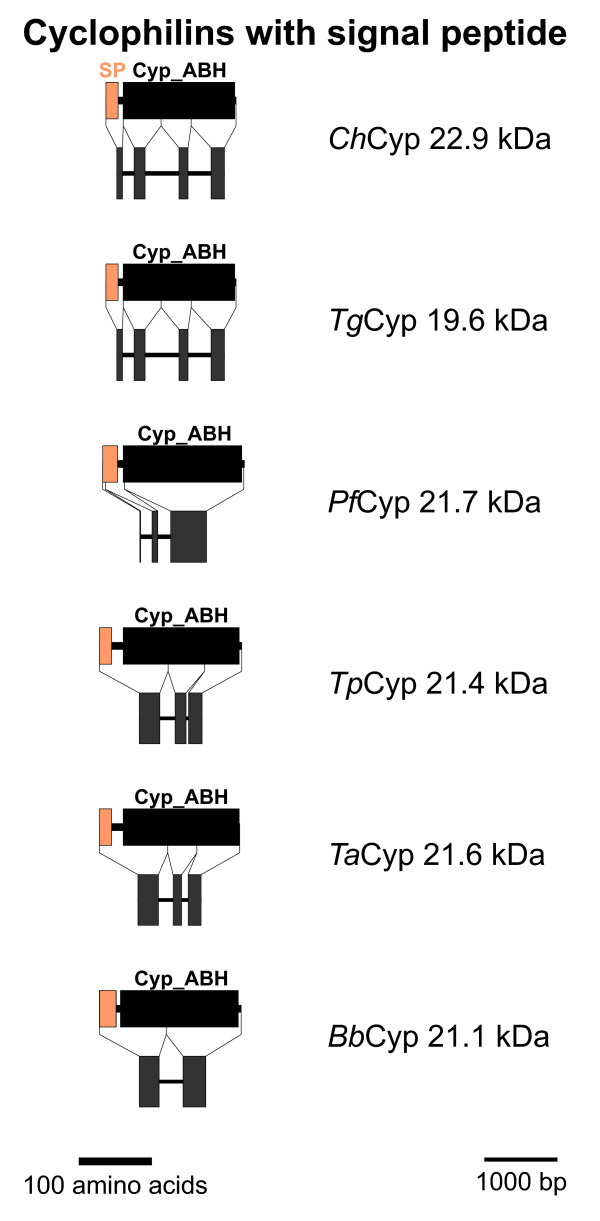
PPIA-like Cyps with signal peptide
The dendrogram in Figure 1 reveals a group of putative small Cyps with SP that also contain a Cyp_ABH type domain and are relatively closely related to the PPIA-type Cyps. The domain architecture and genomic organization of these Cyps is schematically presented in Figure 3. One putative member of this Cyp subfamily could be identified in each apicomplexan genome. In contrast to the small cytosolic PPIA-like Cyps, the coding regions of all subfamily members are interrupted by introns. Whereas C. hominis and T. gondii show a very similar exon/intron structure with 4 introns, the putative genes of both Theileria species have only 2 introns – apparently due to fusion of exons 3 and 4 – and after further fusion of exons 1 and 2 only a single intron remains in B. bovis. In PfCyp21.7, loss of introns has resulted in a Cyp domain that is encoded by a single exon. However, a new intron has also appeared within the region encoding the SP, which is encoded by a single exon in the other Cyps of this subfamily.
Figure 3.
PPIA-like Cyps with signal peptide. Domain architecture and genomic organization of Cyps with signal peptide are shown. Species are abbreviated as in Fig. 1. Cyp_ABH, ABH-type Cyp domain (CD accession-no.: [cd01926]); SP, signal peptide.
Cyps within the secretory pathway of other eukaryotes are typically of the PPIB type. The phylogram in Figure 1, however, reveals that the Cyp subfamily with putative ER-localization in apicomplexa does not form any cluster with HsPPIB and SpCyp4 and is therefore proposed not to represent orthologs of PPIB-like Cyps. Apparently, Cyps in the secretory pathway evolved independently at least twice during evolution of eukaryotic Cyps.
For some members of this Cyp subfamily experimental evidence regarding their expression and function is available. First, the PfCyp21.7 protein has been shown to be expressed at extremely high levels in blood-stage parasites, constituting up to 0.5% of total cellular protein [24]. Conspicuously, PfCyp21.7 has been been reported not to be confined to the secretory pathway but to be at least partially present in the cytosol as well [24]. This raises the possibility that this Cyp subfamily might also be able to interact with cytosolic calcineurin-like phosphatases. Indeed, genetic analysis provides evidence that a mutation in PfCyp21.7 is sufficient to confer resistance to CsA even in the presence of intact PfCyp19 [25].
Secondly, TgCyp19.6 has been shown to be secreted by the parasite and to trigger release of IL-12 from host dendritic cells. Moreover, a 19.4 kDa Cyp from Neospora caninum belongs to the same orthology group (data not shown). This protein has been described to be secreted by the parasite and to be present in large amounts in culture supernatants of cell infected with N. caninum tachyzoites [43]. NcCyp19.4 from cell culture supernatants was shown to be a very potent inducer of IFNγ production by peripheral blood mononuclear cells and CD4+ T cells. Induction of IFNγ by NcCyp19.4 could be specifically inhibited by CsA in a dose dependant manner. These results indicate that apicomplexan Cyps with signal peptide are not only involved in protein folding in the secretory pathway but can fulfill important immunomodulatory functions in infected tissues.
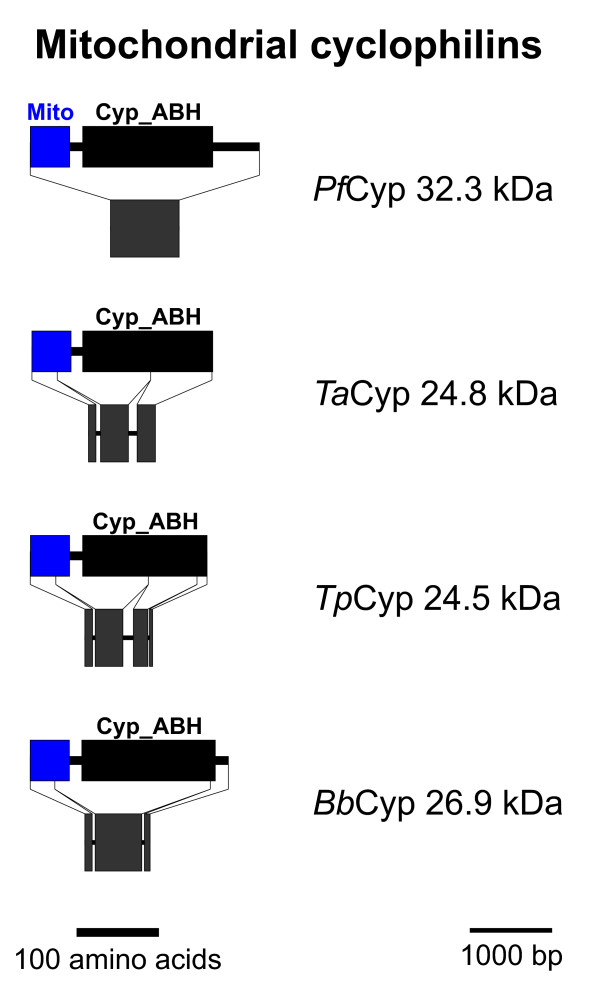
Mitochondrial Cyps
Putative Cyps with a mitochondrial localization signal at their NH2-terminus are schematically shown in Figure 4. The mitochondrial localization signal and a cleavage site were significantly predicted by MitoProt II [44] for TaCyp24.8 and TpCyp24.5. In contrast, cleavage site prediction was not possible for both PfCyp32.3 and BbCyp26.9. Nevertheless, MitoProt II predicts a high probability of mitochondrial localization and the algorithm PlasMit [45], which was specifically developed to predict mitochondrial proteins in Plasmodium, also suggest a mitochondrial localization of PfCyp32.3. Putative proteins of this Cyp subfamily could be detected only in the haemosporidia but neither in T. gondii nor in C. hominis. In T. gondii, mitochondrial PPIase activity might be achieved by the putative TgCyp31.8, a member of the subfamily of apicomplexa-specific Cyps (Figure 6). In contrast to all other members of this group, TgCyp31.8 is predicted to have an NH2-terminal mitochondrial localization signal. However, mitochondrial PPIase activity might also be dispensable in apicomplexan mitochondria as it is completely absent from the genomes of both C. hominis and C. muris.
Figure 4.
Mitochondrial Cyps. Domain architecture and genomic organization of mitochondrial Cyps. Species are abbreviated as in Fig. 1. Cyp_ABH, ABH-type Cyp domain (CD accession-no.: [cd01926]); Mito, mitochondrial localization signal.
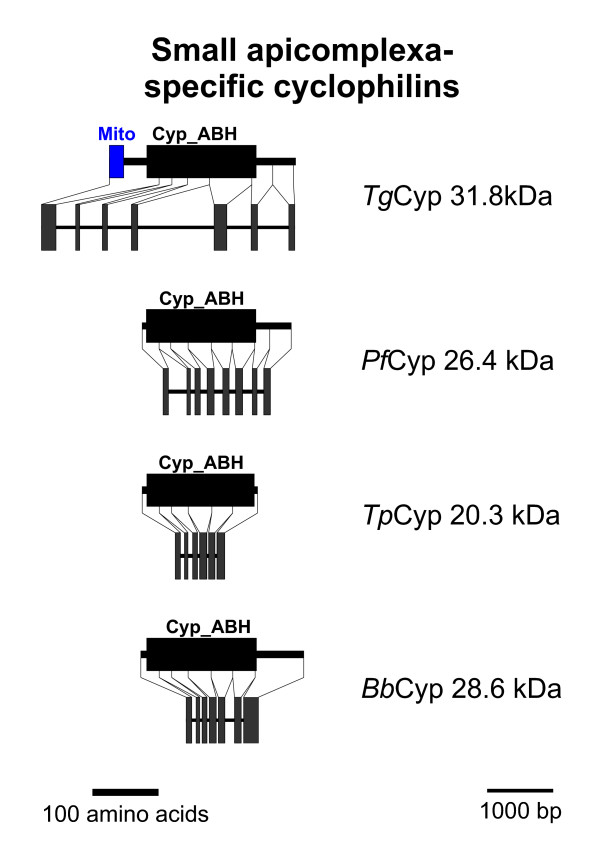
Figure 6.
Small apicomplexa-specific Cyps. Domain architecture and genomic organization of small apicomplexa-specific Cyps. Species are abbreviated as in Fig. 1. Cyp_ABH, ABH-type Cyp domain (CD accession-no.: [cd01926]); Mito, mitochondrial localization signal.
The genomic organization differs largely between the different genera with 5 exons in T. gondii, 3 or 4 in T. annulata and T. parva, respectively, and only a single large exon in P. falciparum (Figure 4).
Plasmodium-specific large Cyps
A group of putative Cyp proteins that appears to be present exclusively in Plasmodium species is shown in Figure S1 in Additional file 2 in the supplemental online material. In order to demonstrate that these proteins represent a subfamily on their own, the putative PyCyp69.8 was included in the phylogenetic analysis shown in Figure 1. Figure S1 reveals that both PfCyp72.5 and PyCyp69.8 possess several nuclear localization signals and two coiled-coil domains, which are typically involved in protein-protein interaction. Moreover, PSORT II recognizes an RNA-binding motif typical for components of ribonucleoprotein particles [46] in PfCyp72.5 further suggesting that this subfamily might somehow be involved in RNA processing.
Cyps with SYF2 domain
A multi-domain Cyp subfamily within the Cyp_ABH domain group are the predicted Cyps containing an SYF2 domain (PFAM accession-no.: [pfam08231]) (Figures 1 and 5). This subfamily does not form a monophyletic cluster in Figure 1B due to the fact that putative PfCyp80.9 is quite aberrant and therefore clusters together with the Plasmodium-specific Cyps described in the section above. However, since PfCyp80.9 contains a SYF2 domain and since its ortholog in P. yoelii, PyCyp74, is closely related to the other SYF2 domain containing Cyps, this family should nevertheless be considered to be monophyletic.
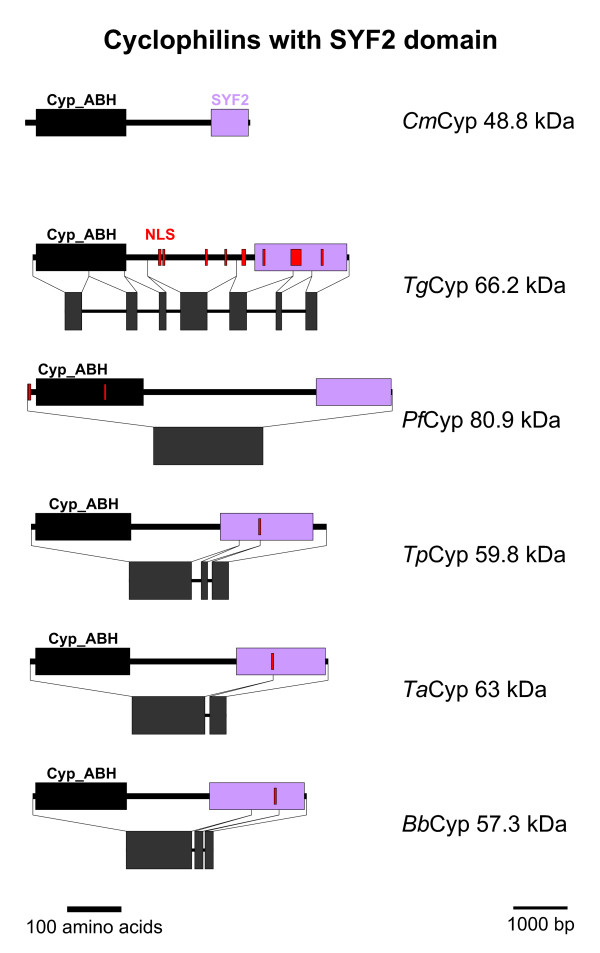
Figure 5.
Cyps with SYF2 domain. Domain architecture and genomic organization of Cyps with SYF2 domain. Species are abbreviated as in Fig. 1. Cyp_ABH, ABH-type Cyp domain (CD accession-no.: [cd01926]); SYF2, SYF2 splicing factor domain (PFAM accession-no.: [pfam08231]); NLS, nuclear localization signal.
In the genome of C. hominis, a SYF2 Cyp could not be identified, presumably due to incomplete sequence information since a putative orthologues protein is encoded in the genome of C. muris, and this sequences was therefore included for further analyses (Figures 1 and 5). The predicted SYF2-Cyps are quite large proteins with predicted molecular weights between 48.8 kDa (C. muris) and 80.9 kDa (P. falciparum). The Cyp_ABH domain is located in the immediate NH2-terminus of the proteins while the SYF2 domain can be found close to the COOH-terminus (Figure 5). The large region between these two defined domains does not exhibit any known sequence features, and homology between individual subfamily members is very low. Only a few scattered amino acids appear to be conserved throughout the subfamily. In accordance with a suspected role in RNA processing, PSORT II predicts at least one nuclear localization signal in all putative SYF2-Cyps but CmCyp48.8. The latter is also peculiar due to its small size and the presence of only an incomplete SYF2 domain. It must at least be considered that the prediction of the protein coding region of this protein from the genomic sequence is still only partially correct.
The genomic organization of SYF2-Cyps is again characterized by progressing loss of introns. While the coding sequence of TgCyp66.2 is spread across six small exons, there is a large first exon in all other members of the subfamily encoding the Cyp domain, the intervening region and the first part of the SYF2 domain. The remaining sequence is split in two exons in TpCyp59.8 and BbCyp57.3 which have further fused in TaCyp63 to give a two exon structure. Finally, the coding sequence of PfCyp80.9 is encoded by a single very large exon. For CmCyp48.8, only parts of the coding sequence could be identified in the available genomic sequences – further challenging the reliability of the predicted protein sequence. Therefore, schematic presentation of the genomic organisation of CmCyp48.8 is not shown in Figure 6.
SYF2 is a nuclear protein described to be involved in splicing of primary transcripts in the yeast Saccharomyces cerevisiae suggesting a contribution of SYF2-Cyps in RNA processing. Cyps with an SYF2 domain have not been identified in higher eukaryotes, however, the CDART tool [47] retrieves three non-apicomplexan putative protein sequences with a similar domain architecture from the ciliates Tetrahymena thermophila (accession-no.: [XP_001019212]) and Paramecium tetraurelium [XP_001423850] as well as from the primitive green algae Ostreococcus tauri (chlorophyta) [CAL53491].
PPIL6-like Cyp TgCyp36.7
The sequence of TgCyp36.7 has been predicted by the TwinScan algorithm [48], however, this sequence (TgTwinScan_3870) is no longer available in the predicted ORFs of the T. gondii genome. Since this is the only gene prediction from this region of chromosome VII containing the complete Cyp domain, it was nevertheless included in the analyses though the correct prediction of the ORF outside the Cyp domain is quite dubious. The putative TgCyp36.7 protein (Figure S2 in Additional file 3) is very perculiar in several aspects. First, it does not have orthologues in any of the other apicomplexan genomes (Figure 1). Secondly, despite its relatively large size, the only known protein domain recognized within its sequence is a Cyp domain. Though the latter clusters together with Cyp_ABH domains in Figure 1, it is not recognized as this domain subtype by CD-BLAST but only as general Cyp domain [cd00137]. Thirdly, TgCyp36.7 and HsPPIL6 form a very significant cluster in the phylogenetic analysis (Figure 1) indicating that they might be orthologs. Indeed, size and domain architecture of both proteins are similar. However, the huge evolutionary distance between TgCyp37.7 and HsPPIL6 – indicated by the long branch leading to TgCyp36.7 in the phylogram (Figure 1) – severely questions this hypothesis. Currently, no functional data are available for either PPIL6 or TgCyp36.7.
Small apicomplexa-specific Cyps
An additional subfamily of relatively small putative Cyps containing a Cyp_ABH domain can be identified in most apicomplexan genomes with the exception of C. hominis and T. annulata (Figures 1 and 6). Since there is also no evidence for an ortholog from C. muris or Cryptosporidium parvum sequences, the conclusion that this subfamily was lost in the genus Cryptosporidium appears to be valid. In contrast, BLAST analysis indicates the presence of an orthologous gene on chromosome I of T. annulata though the coding sequence could not be completely deduced – maybe due to insufficient sequence quality of the genome sequence. Domain architecture of this Cyp subfamily (Figure 5) reveals that there is a considerably larger heterogeneity than for the two groups described above. First, the putative TgCyp31.8 sequence contains an additional NH2-terminal mitochondrial localization signal as predicted by MitoProtII [44]. Due to the fact that this signal is only observable in a single species and would indicate a significant functional difference to its orthologs in other apicomplexa, careful experimental analyses are needed to compare localization and function of this group of Cyps in different apicomplexa. Secondly, TpCyp20.3 is very small and consists of little more than a Cyp domain, while BbCyp28.6, PfCyp26.4, and TgCyp31.5 have considerable COOH-terminal extensions. Functional data on this Cyp subfamily are completely missing yet. This group of Cyps has obviously no direct orthologs in mammalian genomes and appears to be specific for apicomplexa. In BLASTp analyses, the most closely related non-apicomplexan Cyps appear to be of plant origin (data not shown). The fact that most of these proteins are predicted to be cytoplasmic and that they have no orthologs in mammalian hosts makes them an attractive target to develop drugs such as non-immunosuppressive CsA derivatives that might specifically target this Cyp subfamily.
PPIH-like Cyps
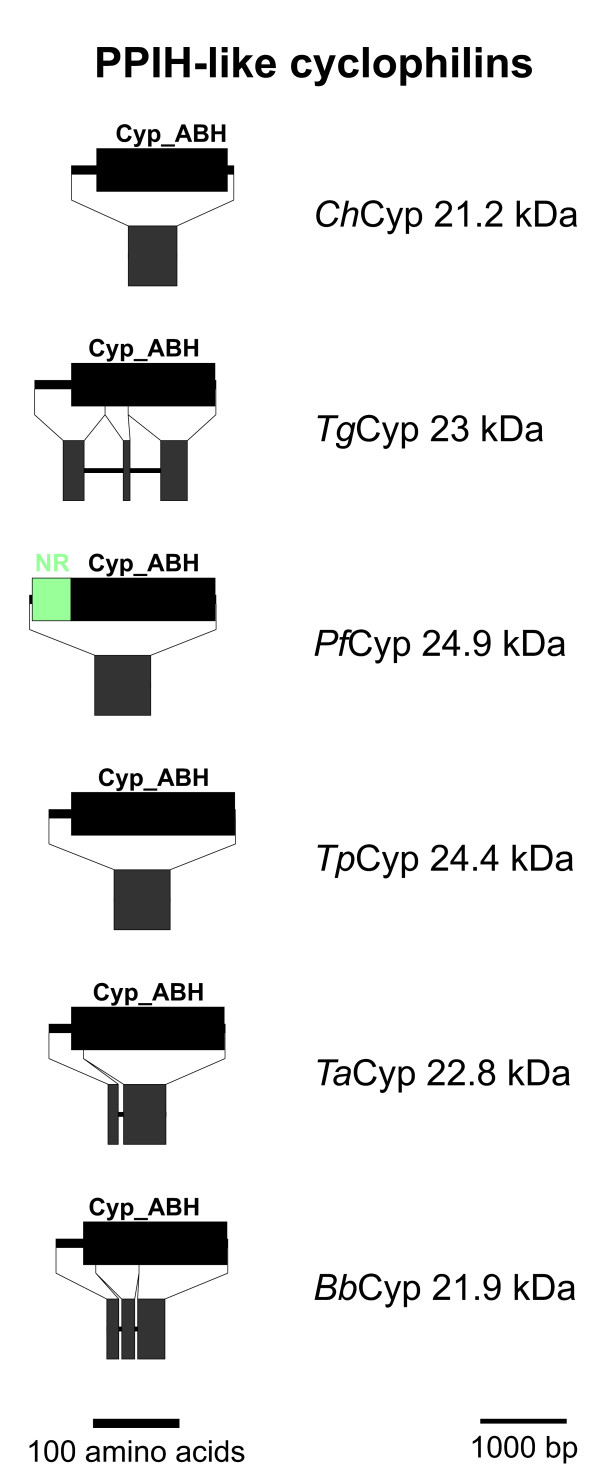
The PPIH-like Cyps represent another subfamily containing a Cyp_ABH domain that is predicted to be present in all analyzed apicomplexan genomes (Figures 1 and 7). In addition to their Cyp domain, these putative proteins have a short NH2-terminal extension which does not contain any recognizable motifs or domains. Only in PfCyp24.9 this NH2-terminal region is characterized by its richness in Asn residues. Though none of the putative apicomplexan PPIH-like Cyps contains any obvious subcellular localization signals, it should be mentioned that their human ortholog has been described to be located in the nucleus and to be associated with the splicing machinery [49,50]. Specifically, HsPPIH is able to interact independently with the factors HsPrp3 and HsPrp4 that both integrate into the U4/U6 di-snRNP particle. The binding-site of HsPrp3 and HsPrp4 for HsPPIH is highly homologous, and binding does not need enzymatic activity of PPIH since it is not impaired by the presence of CsA. PPIH-like Cyps are highly conserved between apicomplexa, fungi and mammals suggesting that the apicomplexan orthologs might carry out similar functions as well.
Figure 7.
PPIH-like Cyps. Domain architecture and genomic organization of PPIH-like Cyps. Species are abbreviated as in Fig. 1. Cyp_ABH, ABH-type Cyp domain (CD accession-no.: [cd01926]); NR-rich, Asn-rich domain.
FCBP proteins
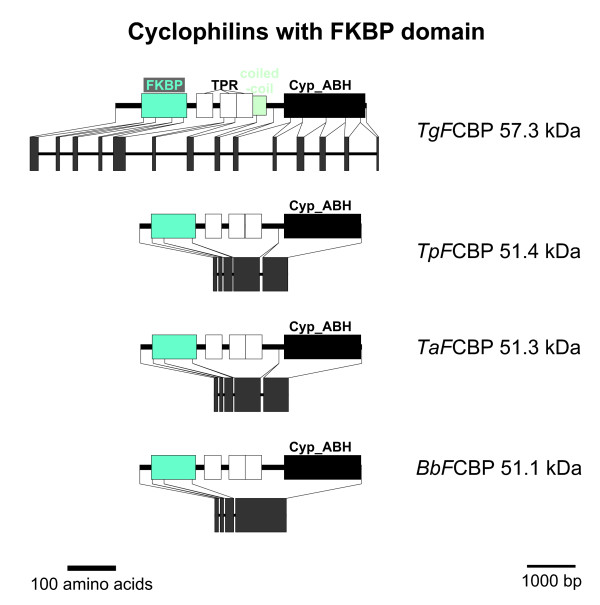
The next multi-domain Cyp subfamily to be described here are the recently identified FCBP proteins [28] which contain two phylogenetically unrelated PPIase domains, i.e. an FK506-binding domain (FKBP) at the NH2-terminus and a Cyp_ABH type domain in the COOH-terminus (Figure 8). Between these two enzymatic domains, there are three tetratricopeptide repeat domains (TRP) [cd00189] which are typically involved in protein/protein interactions and might contribute to recruitment of specific substrates for FCBP proteins.
Figure 8.
FCBP proteins. Domain architecture and genomic organization of FCBPs from apicomplexa. Species are abbreviated as in Fig. 1. Cyp_ABH, ABH-type Cyp domain (CD accession-no.: [cd01926]); FKBP, FK506-binding domain (PFAM accession-no.: [pfam00254]); TPR, Tetratricopeptide repeat (InterProScan accession-no.: [IPR001440]).
Isomerase and chaperone activity have been demonstrated for both PPIase domains of TgFCBP57.3 and the inhibitors FK506 and CsA can suppress activity of the FKBP and Cyp domain, respectively [28]. Moreover, Adams et al. [28] could show that only the complex of the FKBP domain with FK506 but not the complex of the Cyp domain with CsA was able to inhibit T. gondii calcineurin protein phosphatase activity. Although a weakly synergistic inhibitory effect of FK506 and CsA on parasite growth was noted, this must not necessarily be due to action of TgFCBP57.3 but can also involve any of the other Cyp or FKBP proteins expressed by T. gondii. More convincing as a first hint for an important role of FCBPs in the physiology of apicomplexa is the fact that suppression of TgCyp57.3 expression by RNA interference results in severely decreased incorporation of [3H]uracil [28].
In addition to TgFCBP57.3, putative FCBP proteins can be found only in the genomes of T. parva, T. annulata, and B. bovis but not in any of the Plasmodium or Cryptosporidium species (Figures 1 and 8). In all four apicomplexan FCBP proteins, the enzymatically active domains are separated by TRP repeats. Conspicuously, BLASTp and tBLASTn analyses of protein and nucleic acid databases as well as the CDART tool reveal that putative proteins containing both a Cyp and a FKBP domain are present even in very distantly related organisms such as bacteria [5,28] (see Tables S2 and S3 in Additional file 4 in the supplemental online material for accession-no.). Furthermore, putative FCBP proteins can also be identified in the ciliophora T. thermophila and P. tetraurelia (Figure S3 in Additional file 5). Since ciliophora and apicomplexa are considered to be phylogenetically related and are usually placed together with dinoflagellates in the infrakingdom alveolata [51], this finding suggests that FCBP proteins were already present in their common ancestors. This hypothesis is also supported by the fact that the deduced FCBPs of ciliophora are also separated by TRP repeats. However, at least the putative TtFCBP131.6 appears to have evolved new or additional functions, since this protein exhibits the presence of an additional (though incomplete) NTPase domain in its very long NH2-terminus. Such an NTPase domain can be found neither in its homologs in Paramecium nor in the apicomplexan FCBPs. An alternative explanation for the large NH2-terminus might be an incorrect prediction of the intron/exon structure resulting in fusion of two adjacent but distinct genes in the database entry. An important argument for the latter hypothesis is the incompleteness and therefore presumably non-functionality of the NTPase domain in the predicted sequence of TtFCBP131.6.
Putative FCBPs can also be identified in the oomycete Phytophora capsici, the green algae O. tauri (chlorophyta) and in archaebacteria (Figure S3 in Additional file 5). Whereas PcaFCBP52.5 also contains a Cyp_ABH domain, the Cyp domains in O. tauri CPR7 is truncated and therefore only recognized as Cyp superfamily (accession-no.: [cl00197]). In both predicted archaebacterial FCBPs, CD-BLAST identifies only a Cyp domain without further specification (accession-no.: [cd00317]). In contrast to PcaCyp52.5, neither OtCPR7 nor the archaebacterial FKBPs do contain TRP repeats separating the two PPIase domains (Figure S3). Finally, it should be mentioned that the OtCPR7 sequence might be COOH-terminally truncated since the Cyp domain itself is truncated. In contrast to all other FCBP proteins identified here, OtCPR7 contains an NH2-terminal mitochondrial localization signal as predicted with high significance by both PSORT II [52] and MitoProt II.
There are also several putative dual-family immunophilins with an NH2-terminal Cyp and a COOH-terminal FKBP domain in proteo- and flavobacteria as well as in spirochaeta (Figure S3). Here, these proteins are called CFBPs, and they do not contain any TRP repeats. All these putative bacterial CFBPs are very similar in size and domain architecture, however, Borrellia hermsii CFBP38 has a prokaryotic membrane lipoprotein lipid attachment site (Prosite accession-no.: [PS51257]) at its immediate NH2-terminus as identified by InterProScan suggesting that BhCFBP38 is exported by the bacterium. The domain architecture of all non-apicomplexan FCBPs and some representative CFBPs are shown in Figure S3.
The discontinuous distribution pattern of FCBPs and CFBPs in phylogenetically unrelated clades raises the question whether these proteins evolved multiple times independently. Alternatively, a common evolutionary origin of proteins with this domain architecture might be assumed followed by either loss from most genomes or horizontal gene transfer. In order to address this question, BLAST analyses were used to identify those Cyps and FKBPs in archaebacteria, eubacteria, and eukaryotes that show the highest similarity to the diverse FCBPs and CFBPs. All proteins used for these analyses are listed in Tables S2 and S3 in Additional file 4. Then, maximum likelihood analyses were performed independently on ClustalW2-built alignments of Cyp and FKBP domains. Results of these phylogenetic analyses are presented in Figure 9. The cyclophilin domains of all eukaryotic FCBPs are closely related (i.e. most of them are recognized as Cyp_ABH domain by CD-BLAST) and therefore form a highly significant cluster in Figure 9A (group in the dendrogram with blue background). However, they are clearly not monophyletic as there are several non-FCBP Cyps within this group and FCBP proteins have apparently evolved at least three times independently – i.e. in chlorophyta, oomycetes and alveolata. For OtCPR7 this conclusion is further supported by the fact that this FCBP does not contain any TRP repeats. Cyp domains of the putative archaebacterial FCBPs are not even closely related to this group and form a completely independent cluster. The Cyp domains of proteo-/flavobacterial CFBP proteins are monophyletic – in contrast to those of spirochaetes. However, for the latter group there are currently only members known from Treponema denticula and four Borrelia species. It is for instance possible that one of these two proteins is highly divergent from the average spirochaete CFBP due to secondary evolutionary changes. In particular, the presence of a lipoprotein anchor at the NH2-terminus of BhCFBP38 suggests an extracellular localization of the mature protein and therefore a significantly altered function.
Figure 9.
Phylogram showing evolutionary relationships for Cyp and FKBP domains of FCBPs and CFBPs. Cyp domains (A) and FKBP domains (B) of FCBPs and CFBPs were aligned with related domains identified by BLAST analyses in archaebacteria, eubacteria and eukaryotes. Unrooted maximum likelihood phylograms were calculated using PhyML [34]. Statistical support for branches is given as approximate likelihood ratio at the nodes. Only likelihoods of at least 70% are presented. FCBPs of apicomplexa, ciliophora, oomyceta, chlorophyta, and archaebacteria are highlighted in red, orange, yellow, green, and purple, respectively. CFBP of spirochaetes and flavo-/proteobacteria are marked in different blue tones. Species abbreviations: Ta, Theileria annulata; Tp, Theileria parva; Tg, Toxoplasma gondii; Bb, Babesia bovis; Gj, Griffithsia japonica; Pt, Paramecium tetraurelia; Tt, Tetrahymena thermophila; Cw, Crocosphaera watsonii; Pca, Phytophora capsici; Mm, Mus musculus; Eh, Entamoeba histolytica; Ot, Ostreococcus tauri; Tc, Trypanosoma cruzi; Ss, Synechocystis spec.; Cl, Codonopsis lanceolata; Cb, Caenorhabditis briggsae; Bm, Blastupirellula marina; Sa, Stigmatella aurantiaca; Ar, uncultured archaeon GZfos18C8; Cbe, Clostridium beijerincki; Mb, Methanococcoides burtonii; Gf, Gramella forsetii; Ca, Croceibacter atlanticus; Fb, Flavobacteriales bacterium; Fba, Flavobacteria bacterium; Cs, Celluphaga spec. MED134; Lb, Leeuwenhoekiella blandensis; Dp, Desulfotalea psychrophilia; Td, Treponema denticulata; Bh, Borrelia hermsii; Hm, Haloarcula marismortui; Haloquadrantum walsbyi; Mg, Magnaporthe grisea; Pn, Phaeosphaeria nodorum; Aa, Aedes aegyptii; Lm, Leishmania major; Tn, Tetraodon nigroviridis; Py, Plasmodium yoelii; Pc, Plasmodium chabaudi; Pb, Plasmodium berghei; Pf, Plasmodium falciparum; Ec, Entodinium caudatum; Te, Trichodesmium erythraeum; No, Nitrosococcus oceani; Ps, Polaromonas spec. Js666; Yl, Yarrowia lipolytica; Gs, Geobacter spec. FRC-32; Mba, Methanosarcina barkeri; Mbu, Methanococcoides burtonii; Mt, Methanotherococcus thermolithotrophicus; Ma, Methanosarcina acetivorans; Mma, Methanoculleus marisnigri; Cf, Chlorobium ferrooxidans.
The Cyp domains of FCBPs of ciliophora and apicomplexa are closely related, surprisingly, however, a non-FKBP Cyp from the rhodophyte Griffithsia japonica is proposed to be a member of the same cluster as revealed by maximum likelihood analysis. Since red algae are frequently supposed to be the evolutionary origin of the apicoplast, one explanation for this result may be that the Cyp domain of FKBPs in alveolata was derived from the genome of a rhodophyte-related secondary endosymbiont.
Phylogenetic analysis of the deduced FCBP domains does also not support a monophylic origin of alveolate FCBPs. Overall, phylogenetic distances between FKBP domains are much larger than for Cyp domains (compare scale bar between Figures 9A and 9B) indicating that the latter are far better conserved. Moreover, the phylogram reveals poor sequence conservation even within groups containing a well conserved Cyp domain. For instance, the CFBPs of proteo-/flavobacteria do not form a monophyletic group when FKBP domains are analyzed (Figure 9B). The two spirochaete CFBPs are clearly separated and the green algal OtCPR7 does not show any close relationship with FKBP domains from other eukaryotic FCBPs but appears to be closely related to bacterial FKBPs suggesting that it might have been acquired from a cyanobacterial endosymbiont. Though the FKBP domains of all alveolat FCBPs can be found in the same highly significant cluster (group with blue background), this group also contains non-FCBP FKBPs. Conspicuously, however, all FKBP proteins within this group also contain TRP repeats (compare Figures 8, S3, 9B, and Table S3). In contrast to the results obtained for Cyp domains, not even the FKBP domains of FCBPs from ciliophora and apicomplexa appear to be monophyletic. On one hand, it is quite unlikely that the same structure of FKBP domain and Cyp domain connected by TRP repeats arose multiple times independently and it can therefore be suspected that this result is due to high and diversifying evolutionary pressure on FKBP domains in this protein family. On the other hand, there is a widely distributed monophyletic family of FKBP proteins with TRP repeats. These proteins might indeed have captured a Cyp domain several times independently. The strongest argument for the latter evolutionary pathway is the presence of putative FKBP proteins with TRP repeats and a concurrent absence of predicted FCBP proteins in all Plasmodium species. If FCBP proteins would be a monophyletic, ancient group in alveolata or at least in apicomplexa, secondary loss of the Cyp domain from FCBPs restoring the ancient FKBP/TRP protein has to be postulated for Plasmodium.
In summary, additional data from more species are needed to draw a conclusive pattern of evolution for FCBP proteins in protists. Currently, there are hints supporting either a monophyletic origin or multiple independent origins though the latter option appears to be slightly more likely.
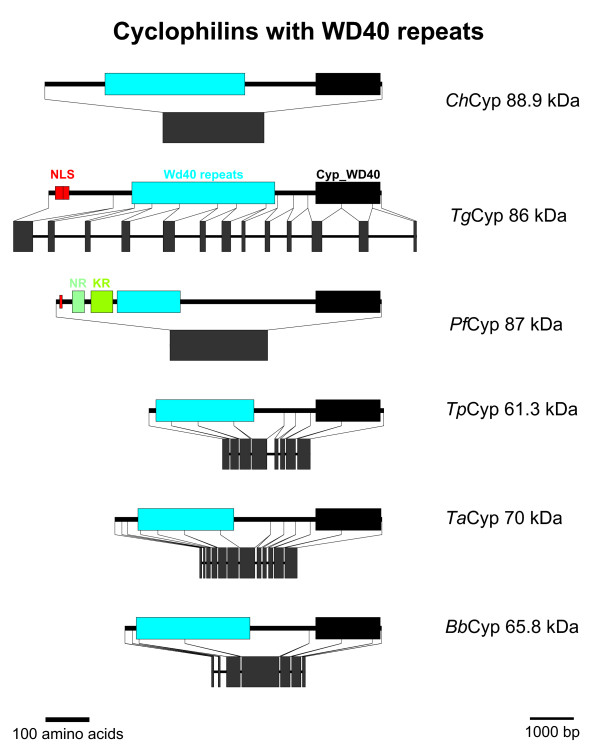
Cyps with WD40 repeats
The first group of putative Cyps with a non-Cyp_ABH domain is represented by the WD40 repeat-containing Cyps which are encoded in all apicomplexan genomes analyzed here (Figure 1). This subfamily contains multi-domain Cyps with a Cyp_WD40 domain (CD accession-no: [cd01927]) in its COOH-terminus (Figure 10). WD40 domains (CD accession no.: [cl02567]) are characterized by repeats of about 40 amino acids containing a characteristic Trp/Asp dipeptide. The length of the deduced WD40 domain ranges from 145 amino acids in P. falciparum to 321 and 328 amino acids in C. hominis and T. gondii, respectively. The WD40 repeat domain is either located close to the NH2-terminus as in Theileria species and in B. bovis, or is preceded by a longer NH2-terminal extension without identifiable domains or motifs as in T. gondii, P. falciparum, and C. hominis. There is no sequence similarity between the orthologs within this region. In T. gondii, however, a nuclear localization signal can be found here, and in P. falciparum there are two short stretches rich in Asn and Lys, respectively. An orthologous protein from E. tenella has recently been described to contain a very Ser- and His-rich NH2-terminus [29].
Figure 10.
Cyps with WD40 repeats. Domain architecture and genomic organization of Cyps with WD40 repeats. Species are abbreviated as in Fig. 1. NLS, nuclear localization signal; WD40 repeat (CD accession-no.: [cl02567]), Cyp_ABH, ABH-type Cyp domain (CD accession-no.: [cd01926]); NR, Asp-rich region; KR, Lys-rich region.
The genomic organization differs largely between species with intronless genes in C. hominis and P. falciparum while there are between 6 (B. bovis) and 12 introns (T. gondii) in the other species.
Deckert et al. [53] showed that the human WD40-repeat Cyp is a component of the spliceosomal B complex which contains the complete set of U snRNAs in a precatalytic state. However, its precise role in splicing or regulation of splicing has not been addressed yet. Recent structural analyses show that – in the crystal – the NH2-terminus of the protein binds to the active site of a neighboring molecule in a substrate-analogous manner [54]. Binding of this sequence to the active center without subsequent isomerization was also confirmed by NMR solution studies.
PPIL1-like Cyps
For Cyps of the PPIL1/SpCyp2 subfamily, CD-BLAST does not recognize any special Cyp domain but only the Cyp superfamily in general (accession-no.: [cl00197]). Only two putative members of this subfamily can be found in the apicomplexan genomes analyzed here, i.e. TgCyp21 and PfCyp23.2 (Figure 1 and Figure S4 in Additional file 6). PfCyp23.2 is also predicted to have an NH2-terminal coiled-coil region. However, there is no hint for such a domain in other PPIL1-like Cyps such as TgCyp21 or SpCyp2.
HsPPIL1 has been demonstrated to be a part of the spliceosomal machinery [38] and to directly interact with the highly conserved transcriptional cofactor SKIP [55]. Although PPIL1-like Cyps are widely spread, they are for instance missing in a large number of fungal genomes [9] indicating that the function of this subfamily is not essential. Since SKIP is also involved in splicing and remains bound to the spliceosome throughout both trans-esterification steps [55], PPIL1-like Cyps and SKIP might be involved in the complex linkage of transcription and splicing during mRNA processing.
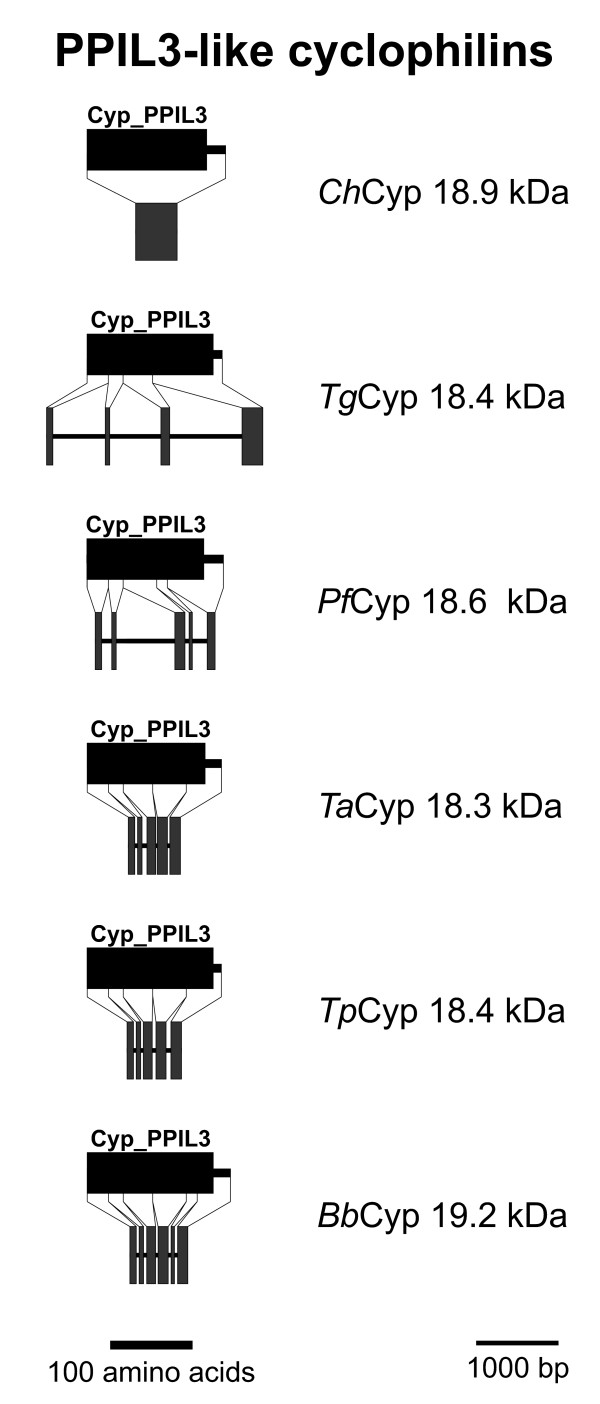
PPIL3-like Cyps
Within the non Cyp_ABH group, the PPIL3-like Cyps are the only subfamily of small single domain Cyps that is widely distributed among apicomplexa. PPIL3-like Cyps can be predicted in all the apicomplexan genomes and consist of little more than the Cyp_PPIL3 domain (accession-no.: [cd01928]) itself (Figures 1 and 11). Since apparent localization signals are missing, a predominant cytosolic localization of the putative proteins might be assumed. However, at least one splice form of human PPIL3 has been identified as part of the B complex of the spliceosome in the nucleus [38] though its precise role in the spliceosome B complex has not been analyzed yet.
Figure 11.
PPIL3-like Cyps. Domain architecture and genomic organization of PPIL3-like Cyps. Species are abbreviated as in Fig. 1. Cyp_PPIL3, PPIL3-type Cyp domain (CD accession-no: [cd01928]).
In contrast to the PPIA-like subfamily, the major subfamily of small cytosolic Cyps, most of the PPIL3-like Cyps are encoded by genes consisting of four (P. falciparum) to six (B. bovis) exons (Figure 11). In this case, the only exception is ChCyp18.9, which has an intronless coding sequence.
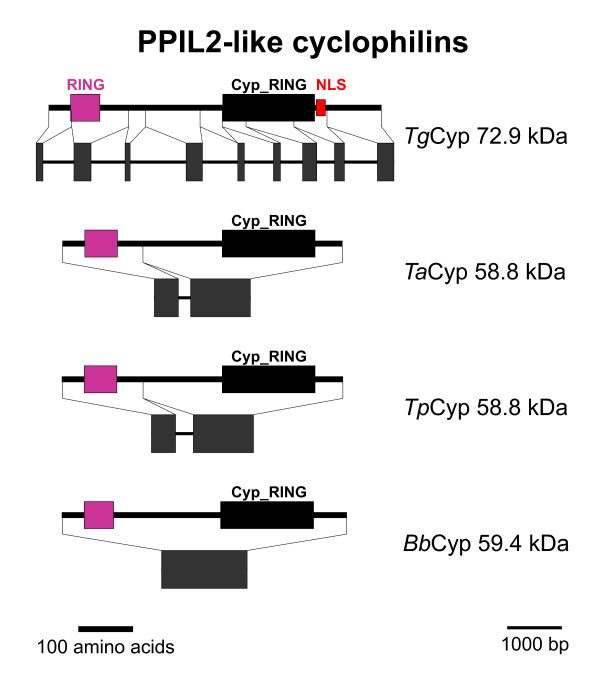
PPIL2-like Cyps
Putative PPIL2-like Cyps form a very robust phylogenetic cluster (Figure 1) and are characterized by the presence of a so-called RING finger domain that has been reported to facilitate E3 ubiquitin-ligase activity [56]. Cyps with RING finger motif in their NH2-terminus are widely spread among different organisms including fungi, plants and mammals, and the Arabidopsis thaliana ortholog, AtPUB49, has been shown to be an active E3 ubiquitin-ligase and to exhibit PPIase and chaperone activity suggesting that it is involved in protein folding and degradation processes. The Cyp_RING domain (accession-no: [cd01923]) is present in the vicinity of the COOH-terminus (Figure 12). This subfamily is apparently missing in the genomes of P. falciparum and C. hominis. The deduced sequence of TgCyp72.9 is somewhat larger than its orthologs in Theileria and Babesia (between 58.8 and 59.4 kDa) due to a longer COOH-terminus. Moreover, TgCyp72.9 contains a putative nuclear localization signal which is not present in its haemosporidian orthologs.
Figure 12.
PPIL2-like Cyps. Domain architecture and genomic organization of Cyps with RING finger domain. Species are abbreviated as in Fig. 1. RING, RING finger domain (Interpro accession-no.: IPR003613); Cyp_RING, RING-type Cyp domain (CD accession-no: [cl00197]); NLS, nuclear localization signal.
The genomic organization of Cyps with RING finger domain again shows signs of intron-loss during evolution with eight introns in T. gondii, one intron in both Theileria species and no intron left in B. bovis.
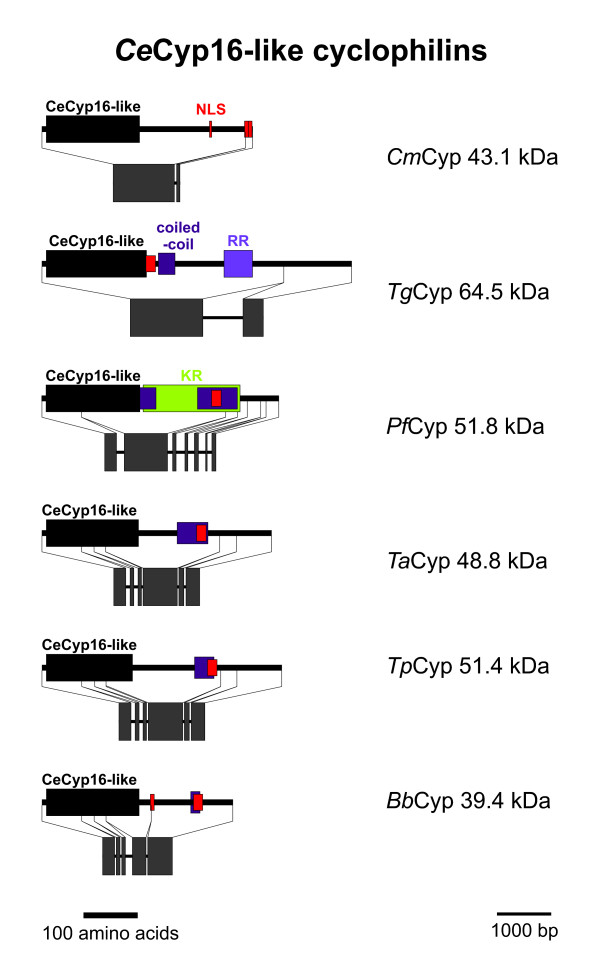
CeCyp16-like Cyps
In contrast to most other moderate to large size Cyps, the subfamily containing a Cyp-CeCyp16-like domain (accession number: [cd01925]) does not contain any additional domain that could be identified by CD-BLAST or InterProScan (Figure 13). However, there is a nuclear localization signal detectable in all putative apicomplexan CeCyp16-like Cyps, which is located in approximately the same distance from the Cyp domain in all subfamily members with the exception of TgCyp64.5 where it immediately follows the Cyp domain. Moreover, using PSORTII at least one coiled-coil protein-protein interaction domain can be identified in all these proteins but CmCyp43.1 (for CmCyp43.1 the score for a coiled-coil region is only slightly too small to be judged as significant by PSORTII). PfCyp51.8 is even predicted to contain two coiled-coil regions. Moreover, PfCyp51.8 has a large Lys-rich region encompassing both coiled-coil domains. A comparable but much smaller positively charged region consisting of a large number of Arg residues is present in TgCyp64.5.
Figure 13.
CeCyp16-like Cyps. Domain architecture and genomic organization of CeCyp16-like Cyps. Species are abbreviated as in Fig. 1. Cyp_CeCyp16, CeCyp16-type Cyp domain (CD accession-no: [cd01925]); NLS, nuclear localization signal; coiled-coil, coiled-coil protein interaction region; RR, Arg-rich region; KR, Lys-rich region.
The C. elegans ortholog CeCyp16 has been shown to be expressed predominantly in the intestine [57] and high-troughput RNAi screening experiments revealed strong phenotyps for CeCyp16 including lethality, embryonic lethality, morphological abnormalities and maternal sterility [58,59] indicating that this subfamily of Cyps is very important at least in multi-cellular organisms. However, since CeCyp16 is much smaller than its apicomplexan orthologs and most of the latter miss a clearly definable positively charged domain in their COOH-terminus that can be found in nematode CeCyp16-like proteins [57], it is not yet possible to draw any conclusions regarding the function of CeCyp16-like Cyps in apicomplexa. Even certain biochemical observations made on recombinant CeCyp16-like proteins from the nematodes C. elegans and Onchocerca volvolus cannot simply be extended to their ortologs in apicomplexa. In particular, although both nematode and apicomplaxan CeCyp16-like Cyps reveal absence of an otherwise highly conserved Trp residue in the active center of the enzyme, there are acidic residues in this position in nematode CeCyp16-like proteins but a wide variety of different amino acids residues in CeCyp16-like Cyps of apicomplexa including Gln, Val, Tyr, Cys, and Phe. Since the conserved Trp residue has been shown to be crucial for CsA binding/sensitivity, it can therefore be assumed that CeCyp16-like proteins of apicomplexa are in general rather resistant to CsA. However, predictions about changes in substrate specificity cannot be made using the results of biochemical analyses made on C. elegans and O. volvolus orthologs.
PPIL4-like Cyps
Putative PPIL4-like Cyps are only identifiable in the genomes of C. hominis and T. gondii (Figures 1 and Figure S5 in Additional file 7). Although orthologs are present in other Cryptosporidium species (data not shown) and many but not all genomes of fungi [9], this subfamily is apparently absent from all other apicomplexan genomes analyzed. ChCyp34.5, just like its orthologs SpCyp6 and HsPPIL4, contains an RRM (RNA recognition motif) closer to its COOH-terminus and a Cyp_RRM domain (accession-no.: [cd01921]) in its immediate NH2-terminus (Figure S5). In contrast to other PPIL4-like Cyps, the deduced sequence of ChCyp34.5 is missing a Ser/Arg-rich SR domain in its COOH-terminus. The predicted sequence of TgCyp66.3 is very unusual since it contains a Cyp domain that is interrupted by a large insertion which remains to be confirmed experimentally. However, in contrast to ChCyp34.5, TgCyp66.2 is more typical for PPIL2-like Cyps since it possesses a Ser-rich and highly positively charged domain in its COOH-terminus. Although it also contains multiple Lys in addition to Arg residues, it can be assumed that this domain fulfills a function similar to that of the SR domain of mammalian and fungal PPIL4-like Cyps. It is not unlikely that both ChCyp34.5 and TgCyp66.3 are not yet predicted accurately (e.g. a missing exon in ChCyp34.5 and a missing intron in TgCyp66.3 would explain the current results) and it will finally turn out that both possess normal Cyp domains and an SR domain.
PPIL4-like Cyps should not be confused with the PPIE-like Cyps, a subfamily that is missing in all apicomplexan genomes. PPIE-like Cyps contain an RRM motif in the NH2-terminus and a Cyp_ABH domain in their COOH-terminus.
ChCyp34.5 contains a nuclear localization signal within its Cyp domain and PSORTII predicts a nuclear localization. Due to its high content of positively charged amino acid residues, the putative TgCyp66.3 is predicted to have a multitude of overlapping nuclear localization signals in its COOH terminus in addition to one signal about 100 amino acids away from its NH2-terminus. Indeed, the orthologous AtCyp59 protein from A. thaliana has been described to be localized in the nucleus but outside of those nuclear speckles rich in SR domain proteins [60]. Although interaction with other SR domain proteins implicated in RNA splicing could be demonstrated using yeast-two-hybrid and pull-down assays, the punctuate nuclear localization pattern and a measurable interaction with the COOH-terminal domain of RNA polymerase II suggest that AtCyp59 predominantly participates in transcriptional processes and that it is only marginally involved in splicing [60]. It is still too early to speculate whether PPIL2-like Cyps of apicomplexa have similar functions as AtCyp59 or other PPIL2-like Cyps since the SR domain responsible for all known AtCyp59 interactions is missing in ChCyp34.5 and the Cyp domain of TgCyp66.3 might well be non-functional. In this context it is also noteworthy that the Cyp domain of this protein is less conserved than that of other PPIL4-like Cyps. Whereas this domain in SpCyp6 and HsPPIL4 is recognized as Cyp_RRM domain (accession no.: [cd01921]), CD-Blast only recognizes a domain belonging to the Cyp superfamily (accession no.: [cd00197]). It is also possible that the truncated or disrupted PPIL4-like Cyps in Cryptosporidia and Toxoplasma containing a slightly degenerated Cyp domain represent transition states that ultimately led to complete loss of this gene in other apicomplexa.
Genomic organization of Cyp genes
Loss of introns during evolution of organisms exhibiting a parasitic live mode such as Giardia [61], Trypanosoma [62], Trichomonas [63], and Encephalitozoon [64] has been recognized previously and has also been described for apicomplexan parasites on a genome wide scale [65]. While Toxoplasma is known to have a genomic organization with a very high number of about five introns on average per gene, Theileria and Plasmodium species have approximately two and one intron on average per gene. Only a very small number of introns can be found in the genomes of Cryptosporidium species which have introns in only about 5% of their genes. However, most of the few remaining introns in Cryptosporidium genomes are not conserved to T. gondii introns and are therefore supposed to be not of ancient origin [65]. Apparently, loss of introns occurred independently in the lineages leading to crypto- and haemosporidia.
As expectable from the differences in the general genomic organization, the exon/intron structure of Cyp genes varies widely between species. The number of introns per Cyp gene, i.e. 0.5 for C. hominis, 4.5 for T. gondii, 2.0 for P. falciparum, 3.1 for T. parva, 3.5 for T. annulata, and 2.8 for B. bovis is very similar to that observed on the genome scale. Despite this overall similarity, however, there are some remarkable exeptions from the general pattern: First, the putative ChCyp22.9 with SP in C. hominis shows a conserved position of introns in comparison to its ortholog in T. gondii. Secondly, some Cyp subfamilies have a high number of introns in both haemosporidians and T. gondii although only minimal conservation of introns can be observed. This group includes the small apicomplexa-specific cyclophilins, the Cyps with WD40 repeats (with exception of PfCyp87), and the PPIL3-like Cyps. Finally, there is one Cyp gene subfamily, the CeCyp16-like Cyps, showing a completely reversed trend with more introns in the haemosporidian genomes than in T. gondii. Since several of the introns in haemosporidia appear to be of ancient origin, it is most likely that TgCyp65.5 has lost its introns since divergence from the last common anchestor with the haemosporidia.
In summary, though the trend of moderate and nearly complete loss of introns observed on a genome-wide scale for haemo- and cryptosporidia, respectively, could also be observed for Cyp genes in general, there are exceptions to this rule in certain subfamilies that might be exploited in the future to decipher the selection forces that contribute to conservation of certain introns despite high overall frequency of intron loss. For instance, it would be highly intriguing to look for any functional roles for the three introns in ChCyp22.9 (e.g. on regulation of gene expression) that might explain counterselection against their deletion during evolution.
Conclusion
The present study was able to identify 16 different Cyp subfamilies in apicomplexa. While some of these subfamilies can be found in the genomes of all species analyzed, there are also two small subfamilies, that can only be found in the genus Cryptosporidium and Toxoplasma (PPIL4-like Cyps) or even only in Toxoplasma (PPIL6-like Cyps), respectively. Six out of these 16 subfamilies (i.e. PPIH-like, SYF2-containing, WD40-containing, PPIL-3-like, PPIL-4-like, and PPIL-1-like Cyps) have been described to be a part of the extremely complex transcription and/or splicing machinery in mammalian or yeast cells indicating that regulation of protein conformation in these very large protein or ribonucleoprotein complexes catalyzing RNA processing is a highly conserved major function of eukaryotic Cyps.
While most apicomplexa are predicted to posses typical cytoplasmic PPIA-like Cyps, these putative proteins in both Theileria species are predicted to have an NH2-terminal apicoplast localization signal. Surprisingly, these are the only Cyps that are predicted to be transported to the apicoplast. Apicomplexa might be more easily able to cope with loss of cytosolic PPIA-like proteins than other eukaryota since members of the apicomplexa-specific group of relatively small Cyps with Cyp_ABH domain might be able to functionally replace PPIA-like cytosolic Cyps. Moreover, at least one member of the Cyp subfamily with signal peptides has been reported not to be confined to the secretory pathway but to be present in the cytosol as well [24]. This Cyp subfamily is very closely related to cytosolic PPIA-like Cyps and therefore unique in so far as it does not represent orthologs of the PPIB-like subfamily that is present in the secretory pathway of other eukaryotes.
Since the Cyp antagonist CsA has been shown to have anti-parasitc activity against a wide variety of apicomplexa [13,16,17,19,20], Cyps represent an attractive target for the identification of new drugs against this important group of pathogens. These might either include non-immunosuppressive CsA derivatives or completely new, structurally unrelated agents. Systematic identification and characterization of the apicomplexan Cyp repertoire as commenced in this bioinformatic survey will enable future analysis of suitable drug targets in more detail. The encouraging fact that there are Cyp subfamilies that are absent from their mammalian hosts, such as Cyps with signal peptides, small apicomplexa-specific Cyps, Plasmodium-specific Cyps, and Cyps with SYF2 domain, already points out obvious drug target candidates.
Methods
Identification of Cyp genes
Initially, putative apicomplexan Cyps were identified using BLASTp and tBLASTn algorithms to search in GenBank® protein and nucleic acid databases as well as in PlasmoDB, ToxoDB, CryptoDB, and in the Theileria parva genome database of TIGR. S. pombe Cyp1 and Cyp2 were used as query sequences. These Cyps were chosen because they are not closely related. If a Cyp subfamily member was not identified in one of the apicomplexan organisms, a Cyp of the same subfamily from a closely related apicomplexan parasite was used as query to search in protein, cDNA, EST and genome databases. This method ensures that no Cyps are missed in any of the taxa. In order to prevent that no complete subfamilies was overseen, BLAST analyses were also performed using the complete T. gondii Cyp repertoire as a query. However, no additional Cyp sequences could be identified.
In contrast to conventional nomenclature for many Cyps, molecular mass suffixes in the names were given with one position after the decimal point since otherwise identical names would have resulted in a few cases. It was decided not to use suffix letters to avoid a possible confusion with mammalian Cyps. For instance, a Cyp19A might have been confused with a human CypA/PPIA. In addition it should be mentioned that all molecular mass suffixes used have been derived from the predicted sequence of unprocessed proteins. Although this can currently be only a provisional nomenclature, consecutive naming with numbers or letters would result in different names for orthologues Cyps and identical names for unrelated Cyps of different apicomplexa. A more function based nomenclature of apicomplexan Cyps should be introduced later, when at least for one apicomplexan genome all Cyps have been verified experimentally. For human and S. pombe Cyps, names according to the entries in the ENSEMBL database were used.
Phylogenetic analyses
Homologous putative protein sequences were aligned using ClustalW2 [33]. Maximum likelihood phylogenetic trees were then calculated with PhyML [34] using the approximate likelihood ratio test option and the JTT model [66] for amino acid substitution. The program was set to estimate the proportion of invariable sites and the gamma distribution parameter, while the number of substitution rate categories was set to four. The input tree was built using the BIONJ algorithm implemented in PhyML. The resulting trees in Newick format were visualized and processed using MEGA4 [67,68].
Identification of protein domains
For identification of protein domains, CD-BLAST [31,32] and InterPro Scan [69] were used. Moreover, protein sequences were scanned for subcellular localization signals with PSORT, SignalP [70], PATS [40], PlasMit [45,71], and Mitoprot [44].
Abbreviations
ABC: ATP-binding casette; AP: apicoplast transit signal; CFBP: cyclosporin A- and FK506-binding proteins; CsA: cyclosporin A; Cyp: cyclophilin; ER: endoplasmic reticulum; FCBP: FK506- and cyclosporin A-binding proteins; FK506: tacrolimus; FKBP: FK506-binding proteins; ORF: open reading frames; PPIase: peptidyl-prolyl cis/trans isomerase; snRNP: small nuclear ribonucleoprotein particle; SP: signal peptide; SR domain: Ser- and Arg-rich domain.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JK, GG, and GvSH designed the study and wrote the final manuscript. JK performed bioinformatic analyses and drafted the manuscript. All authors approved the final version.
Supplementary Material
Table S1: Cyclophilins from H. sapiens and S. pombe. Listing of the human and fission yeast Cyp repertoire used for comparison with apicomplexan Cyps. Accession-no., protein size and domain architecture are summarized.
Figure S1 – Plasmodium-specific Cyps. Domain architecture and genomic organization of Plasmodium-specific Cyps.
Figure S2 – PPIL6-like Cyp TgCyp36.7. Domain architecture and genomic organization of TgCyp36.7.
Tables S2 and S3: Protein sequences used to analyse evolution of Cyp and FKBP domains in dual class immunophilins. Listing of the genes used for comparison with apicomplaxan FKBPs in Figures 5 and 6.
Figure S3 – FCBP and CFBP proteins in non-apicomplexa. Domain architecture of FCBPs and CFBPs from non-apicomplexan organisms.
Figure S4 – PPIL1-like Cyps. Domain architecture and genomic organization of PPIL1-like Cyps.
Figure S5 – PPIL4-like Cyps. Domain architecture and genomic organization of ChCyp34.5, the only apicomplexan PPIL4-like Cyp which contains an RNA recognition motif.
Contributor Information
Jürgen Krücken, Email: juergen.kruecken@tiho-hannover.de.
Gisela Greif, Email: gisela.greif@bayerhealthcare.com.
Georg von Samson-Himmelstjerna, Email: Georg.von.Samson-Himmelstjerna@tiho-hannover.de.
References
- Wang P, Heitman J. The cyclophilins. Genome biology. 2005;6:226. doi: 10.1186/gb-2005-6-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Furutani M. Archaeal peptidyl prolyl cis-trans isomerases (PPIases) Front Biosci. 2000;5:D821–836. doi: 10.2741/maruyama. [DOI] [PubMed] [Google Scholar]
- Fischer G, Aumüller T. Regulation of peptide bond cis/trans isomerization by enzyme catalysis and its implication in physiological processes. Reviews of physiology, biochemistry and pharmacology. 2003;148:105–150. doi: 10.1007/s10254-003-0011-3. [DOI] [PubMed] [Google Scholar]
- Gothel SF, Marahiel MA. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol Life Sci. 1999;55:423–436. doi: 10.1007/s000180050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S. Immunophilins: for the love of proteins. Cell Mol Life Sci. 2006;63:2889–2900. doi: 10.1007/s00018-006-6215-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ, Kay JE. The cyclophilin repertoire of the fission yeast Schizosaccharomyces pombe. Yeast (Chichester, England) 2005;22:927–945. doi: 10.1002/yea.1288. [DOI] [PubMed] [Google Scholar]
- Arevalo-Rodriguez M, Wu X, Hanes SD, Heitman J. Prolyl isomerases in yeast. Front Biosci. 2004;9:2420–2446. doi: 10.2741/1405. [DOI] [PubMed] [Google Scholar]
- Pemberton TJ, Kay JE. Identification and Comparative Analysis of the Peptidyl-Prolyl cis/trans Isomerase Repertoires of H. sapiens, D. melanogaster, C. elegans, S. cerevisiae and Sz. pombe. Comp Funct Genomics. 2005;6:277–300. doi: 10.1002/cfg.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ. Identification and comparative analysis of sixteen fungal peptidyl-prolyl cis/trans isomerase repertoires. BMC genomics. 2006;7:244. doi: 10.1186/1471-2164-7-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinski K, Muir S, Cardenas M, Heitman J. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13093–13098. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell A, Monaghan P, Page AP. Peptidyl-prolyl cis-trans isomerases (immunophilins) and their roles in parasite biochemistry, host-parasite interaction and antiparasitic drug action. International journal for parasitology. 2006;36:261–276. doi: 10.1016/j.ijpara.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Bueding E, Hawkins J, Cha YN. Antischistosomal effects of cyclosporin A. Agents Actions. 1981;11:380–383. doi: 10.1007/BF01982474. [DOI] [PubMed] [Google Scholar]
- Thommen-Scott K. Antimalarial activity of cyclosporin A. Agents and actions. 1981;11:770–773. doi: 10.1007/BF01978803. [DOI] [PubMed] [Google Scholar]
- Chappell LH, Wastling JM. Cyclosporin A: antiparasite drug, modulator of the host-parasite relationship and immunosuppressant. Parasitology. 1992;105:S25–40. doi: 10.1017/s0031182000075338. [DOI] [PubMed] [Google Scholar]
- Silverman JA, Hayes ML, Luft BJ, Joiner KA. Characterization of anti-Toxoplasma activity of SDZ 215–918, a cyclosporin derivative lacking immunosuppressive and peptidyl-prolyl-isomerase-inhibiting activity: possible role of a P glycoprotein in Toxoplasma physiology. Antimicrob Agents Chemother . 1997;41:1859–1866. doi: 10.1128/aac.41.9.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell SP, Scheibel LW, Cole GA. Inhibition by cyclosporin A of rodent malaria in vivo and human malaria in vitro. Infect Immun . 1982;37:1093–1100. doi: 10.1128/iai.37.3.1093-1100.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack DG, McLeod R. New micromethod to study the effect of antimicrobial agents on Toxoplasma gondii: comparison of sulfadoxine and sulfadiazine individually and in combination with pyrimethamine and study of clindamycin, metronidazole, and cyclosporin A. Antimicrob Agents Chemother. 1984;26:26–30. doi: 10.1128/aac.26.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillehoj HS. Effects of immunosuppression on avian coccidiosis: cyclosporin A but not hormonal bursectomy abrogates host protective immunity. Infection and immunity. 1987;55:1616–1621. doi: 10.1128/iai.55.7.1616-1621.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose ME, Hesketh P. Eimeria vermiformis and E. mitis: inhibition of development in vivo by cyclosporin A. Experimental parasitology. 1989;68:289–296. doi: 10.1016/0014-4894(89)90111-2. [DOI] [PubMed] [Google Scholar]
- Perkins ME, Wu TW, Le Blancq SM. Cyclosporin analogs inhibit in vitro growth of Cryptosporidium parvum. Antimicrob Agents Chemother . 1998;42:843–848. doi: 10.1128/aac.42.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed JS, Rintelen M, Hartwig H, Schein E. Effect of cyclosporin A on the proliferation of bovine lymphocytes to concanavalin A and on the growth of Theileria annulata-infected bovine cells. Trop Med Parasitol. 1991;42:375–380. [PubMed] [Google Scholar]
- Bell A, Wernli B, Franklin RM. Roles of peptidyl-prolyl cis-trans isomerase and calcineurin in the mechanisms of antimalarial action of cyclosporin A, FK506, and rapamycin. Biochemical pharmacology. 1994;48:495–503. doi: 10.1016/0006-2952(94)90279-8. [DOI] [PubMed] [Google Scholar]
- Hirtzlin J, Farber PM, Franklin RM, Bell A. Molecular and biochemical characterization of a Plasmodium falciparum cyclophilin containing a cleavable signal sequence. European journal of biochemistry/FEBS. 1995;232:765–772. [PubMed] [Google Scholar]
- Gavigan CS, Kiely SP, Hirtzlin J, Bell A. Cyclosporin-binding proteins of Plasmodium falciparum. International journal for parasitology. 2003;33:987–996. doi: 10.1016/s0020-7519(03)00125-5. [DOI] [PubMed] [Google Scholar]
- Kumar R, Musiyenko A, Barik S. Plasmodium falciparum calcineurin and its association with heat shock protein 90: mechanisms for the antimalarial activity of cyclosporin A and synergism with geldanamycin. Mol Biochem Parasitol . 2005;141:29–37. doi: 10.1016/j.molbiopara.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Dobson S, May T, Berriman M, Del Vecchio C, Fairlamb AH, Chakrabarti D, Barik S. Characterization of protein Ser/Thr phosphatases of the malaria parasite, Plasmodium falciparum: inhibition of the parasitic calcineurin by cyclophilin-cyclosporin complex. Mol Biochem Parasitol. 1999;99:167–181. doi: 10.1016/s0166-6851(99)00010-9. [DOI] [PubMed] [Google Scholar]
- Gavigan CS, Shen M, Machado SG, Bell A. Influence of the Plasmodium falciparum P-glycoprotein homologue 1 (pfmdr1 gene product) on the antimalarial action of cyclosporin. The Journal of antimicrobial chemotherapy. 2007;59:197–203. doi: 10.1093/jac/dkl461. [DOI] [PubMed] [Google Scholar]
- Adams B, Musiyenko A, Kumar R, Barik S. A novel class of dual-family immunophilins. The Journal of biological chemistry. 2005;280:24308–24314. doi: 10.1074/jbc.M500990200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosse RJ, Krücken J, Bierbaum S, Greif G, Wunderlich F. Eimeria tenella: genomic organization and expression of an 89 kDa cyclophilin. Experimental parasitology. 2008;118:275–279. doi: 10.1016/j.exppara.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Potenza M, Galat A, Minning TA, Ruiz AM, Duran R, Tarleton RL, Marin M, Fichera LE, Bua J. Analysis of the Trypanosoma cruzi cyclophilin gene family and identification of Cyclosporin A binding proteins. Parasitology. 2006;132:867–882. doi: 10.1017/S0031182005009558. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic acids research. 2009:D205–210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic acids research. 2004:W327–331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics (Oxford, England) 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic biology. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Dix I, Russell CS, McGarvey M, Beggs JD, Kupiec M. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics. 2000;156:1503–1517. doi: 10.1093/genetics/156.4.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, Brunskill EW, Sayen MR, Gottlieb RA, Dorn GW, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- Schinzel AC, Takeuchi O, Huang Z, Fisher JK, Zhou Z, Rubens J, Hetz C, Danial NN, Moskowitz MA, Korsmeyer SJ. Cyclophilin D is a component of mitochondrial permeability transition and mediates neuronal cell death after focal cerebral ischemia. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12005–12010. doi: 10.1073/pnas.0505294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa A, Somarelli JA, Herrera RJ. Spliceosomal immunophilins. FEBS letters. 2008;582:2345–2351. doi: 10.1016/j.febslet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxwell BM, Woerly G, Husi H, Mackie A, Quesniaux VF, Hiestand PC, Wenger RM, Ryffel B. Identification of several cyclosporine binding proteins in lymphoid and non-lymphoid cells in vivo. Biochimica et biophysica acta. 1992;1138:115–121. doi: 10.1016/0925-4439(92)90050-w. [DOI] [PubMed] [Google Scholar]
- Zuegge J, Ralph S, Schmuker M, McFadden GI, Schneider G. Deciphering apicoplast targeting signals – feature extraction from nuclear-encoded precursors of Plasmodium falciparum apicoplast proteins. Gene. 2001;280:19–26. doi: 10.1016/s0378-1119(01)00776-4. [DOI] [PubMed] [Google Scholar]
- Tonkin CJ, Kalanon M, McFadden GI. Protein targeting to the malaria parasite plastid. Traffic (Copenhagen, Denmark) 2008;9:166–175. doi: 10.1111/j.1600-0854.2007.00660.x. [DOI] [PubMed] [Google Scholar]
- Ebel T, Pelle R, Janoo R, Lipp J, Bishop R. A membrane-anchored Theileria parva cyclophilin with a non-cleaved amino-terminal signal peptide for entry into the endoplasmic reticulum. Veterinary parasitology. 2004;121:65–77. doi: 10.1016/j.vetpar.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Tuo W, Fetterer R, Jenkins M, Dubey JP. Identification and characterization of Neospora caninum cyclophilin that elicits gamma interferon production. Infect Immun . 2005;73:5093–5100. doi: 10.1128/IAI.73.8.5093-5100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. European journal of biochemistry/FEBS. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Bender A, van Dooren GG, Ralph SA, McFadden GI, Schneider G. Properties and prediction of mitochondrial transit peptides from Plasmodium falciparum. Mol Biochem Parasitol . 2003;132:59–66. doi: 10.1016/j.molbiopara.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Haynes SR. The RNP motif protein family. The New biologist. 1992;4:421–429. [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome research. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I, Flicek P, Duan D, Brent MR. Integrating genomic homology into gene structure prediction. Bioinformatics (Oxford, England) 2001;17:S140–148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- Horowitz DS, Kobayashi R, Krainer AR. A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. RNA. 1997;3:1374–1387. [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S, Achsel T, Mundt C, Gothel SF, Cronshagen U, Lane WS, Marahiel M, Luhrmann R. The 20 kD protein of human [U4/U6.U5] tri-snRNPs is a novel cyclophilin that forms a complex with the U4/U6-specific 60 kD and 90 kD proteins. RNA (New York, NY) 1998;4:127–141. [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Kingdom protozoa and its 18 phyla. Microbiological reviews. 1993;57:953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends in biochemical sciences. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol Cell Biol . 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TL, Walker JR, Ouyang H, MacKenzie F, Butler-Cole C, Newman EM, Eisenmesser EZ, Dhe-Paganon S. The crystal structure of human WD40 repeat-containing peptidylprolyl isomerase (PPWD1) The FEBS journal. 2008;275:2283–2295. doi: 10.1111/j.1742-4658.2008.06381.x. [DOI] [PubMed] [Google Scholar]
- Xu C, Zhang J, Huang X, Sun J, Xu Y, Tang Y, Wu J, Shi Y, Huang Q, Zhang Q. Solution structure of human peptidyl prolyl isomerase-like protein 1 and insights into its interaction with SKIP. The Journal of biological chemistry. 2006;281:15900–15908. doi: 10.1074/jbc.M511155200. [DOI] [PubMed] [Google Scholar]
- Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays in biochemistry. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- Ma D, Nelson LS, LeCoz K, Poole C, Carlow CK. A novel cyclophilin from parasitic and free-living nematodes with a unique substrate- and drug-binding domain. The Journal of biological chemistry. 2002;277:14925–14932. doi: 10.1074/jbc.M112293200. [DOI] [PubMed] [Google Scholar]
- Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, Heuvel S van den, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome research. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Gullerova M, Barta A, Lorkovic ZJ. AtCyp59 is a multidomain cyclophilin from Arabidopsis thaliana that interacts with SR proteins and the C-terminal domain of the RNA polymerase II. RNA . 2006;12:631–643. doi: 10.1261/rna.2226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science . 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Simpson AG, Stevens JR, Lukes J. The evolution and diversity of kinetoplastid flagellates. Trends in parasitology. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Vanacova S, Yan W, Carlton JM, Johnson PJ. Spliceosomal introns in the deep-branching eukaryote Trichomonas vaginalis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4430–4435. doi: 10.1073/pnas.0407500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Roy SW, Penny D. Widespread intron loss suggests retrotransposon activity in ancient apicomplexans. Mol Biol Evol. 2007;24:1926–1933. doi: 10.1093/molbev/msm102. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol . 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in bioinformatics. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan – an integration platform for the signature-recognition methods in InterPro. Bioinformatics (Oxford, England) 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nature protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Small I, Peeters N, Legeai F, Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Cyclophilins from H. sapiens and S. pombe. Listing of the human and fission yeast Cyp repertoire used for comparison with apicomplexan Cyps. Accession-no., protein size and domain architecture are summarized.
Figure S1 – Plasmodium-specific Cyps. Domain architecture and genomic organization of Plasmodium-specific Cyps.
Figure S2 – PPIL6-like Cyp TgCyp36.7. Domain architecture and genomic organization of TgCyp36.7.
Tables S2 and S3: Protein sequences used to analyse evolution of Cyp and FKBP domains in dual class immunophilins. Listing of the genes used for comparison with apicomplaxan FKBPs in Figures 5 and 6.
Figure S3 – FCBP and CFBP proteins in non-apicomplexa. Domain architecture of FCBPs and CFBPs from non-apicomplexan organisms.
Figure S4 – PPIL1-like Cyps. Domain architecture and genomic organization of PPIL1-like Cyps.
Figure S5 – PPIL4-like Cyps. Domain architecture and genomic organization of ChCyp34.5, the only apicomplexan PPIL4-like Cyp which contains an RNA recognition motif.