Abstract
Background
Normalization is a prerequisite for accurate real time PCR (qPCR) expression analysis and for the validation of microarray profiling data in microbial systems. The choice and use of reference genes that are stably expressed across samples, experimental conditions and designs is a key consideration for the accurate interpretation of gene expression data.
Results
Here, we evaluate a carefully selected set of reference genes derived from previous microarray-based transcriptional profiling experiments performed on Acidithiobacillus ferrooxidans and identify a set of genes with minimal variability under five different experimental conditions that are frequently used in Acidithiobacilli research. Suitability of these and other previously reported reference genes to monitor the expression of four selected target genes from A. ferrooxidans grown with different energy sources was investigated. Utilization of reference genes map, rpoC, alaS and era results in improved interpretation of gene expression profiles in A. ferrooxidans.
Conclusion
This investigation provides a validated set of reference genes for studying A. ferrooxidans gene expression under typical biological conditions and an initial point of departure for exploring new experimental setups in this microorganism and eventually in other closely related Acidithiobacilli. The information could also be of value for future transcriptomic experiments in other bacterial systems.
Background
Gene expression interrogation, both at single-gene (classical gene expression analysis) and genome-wide (global transcriptome analysis) levels, are prominent fields of study. By providing quantitative measures of mRNA changes, it has the extraordinary potential of identifying the functional consequences of genetic variability and environmental influence. Accurate quantification of gene expression is providing great insight into the physiology and metabolic complexity of microbes and their consortia and is thus contributing to our understanding of both fundamental and applied issues of major interest.
Microarray transcript profiling is the most widely used technique to evaluate global gene expression in microbial systems. However, due to methodological uncertainties inherent to the technique, it is imperative to validate the expression of key genes by an alternative procedure and quantitative real-time PCR (qPCR) has become the method of choice. qPCR is accurate, exhibits a broad dynamic range, and is sensitive and reproducible [1-6]. However, when performing qPCR, several parameters need to be controlled in order to obtain accurate and reliable expression measurements; these include variations in the amounts of starting material between samples, RNA extraction efficiency, RNA integrity/quality, efficiency of cDNA synthesis, and differences in the overall transcriptional activity of the cells analyzed [7].
The most frequently used strategy to control for such variations is relative normalization, where the expression of a target gene is measured with respect to total RNA, rRNA or a stably expressed internal reference gene. The use of rRNA or other reference gene has the advantage that their expression permits normalization against the cumulative errors of the entire process [8]. Ideally, the reference gene should be universally valid, expressed stably and at a similar level across all samples, cells, experimental treatments, and designs. Unfortunately, no such reference gene has been identified [9-12] and even widely used control genes have proven unsuitable in certain situations [6,13,14].
Housekeeping genes are usually chosen as reference genes both in eukaryotes [15] and prokaryotes [16] because they are assumed: a) to be essential, b) to be ubiquitous, c) not to be regulated or influenced by the experimental procedure and d) to be expressed at similar levels in different types of cells. However, it is becoming increasingly clear that some commonly chosen housekeeping genes vary considerably across cell types, with time, or due to experimental treatment [13]. This may be explained partially by the fact that some housekeeping proteins participate in other functions as well as in basal cell metabolism [17-19].
Therefore, for an accurate comparison of mRNA transcription in different samples, either validated reference genes are required for normalization or new ones should be determined empirically for each experimental system and condition studied [20,21]. Here, we evaluate a carefully selected set of reference genes for improving the interpretation of gene expression profiles in a model microorganism, Acidithiobacillus ferrooxidans. The implementation of high-throughput microarray analyses of A. ferrooxidans [22,23] and PCR techniques for expression profiling [24-37], have greatly enhanced our understanding of the genetic and physiological potential of this bioleaching bacterium. However, suitable reference genes to evaluate gene expression have not previously been identified in A. ferrooxidans.
In order to address this lacuna, we screened high-density oligonucleotide array-based expression profiles available for A. ferrooxidans and identified a set of nine genes with minimal variability under different experimental setups. Quantitative real-time PCR was then used to determine the mRNA levels of these genes, comparing their transcription in five different experimental conditions. Finally, we evaluated the suitability of these and other previously reported reference genes to monitor the expression of four selected target genes from A. ferrooxidans grown with different energy sources. This study defines reference genes for normalization of gene expression for future research, sparing other researchers in this and related fields from cumbersome and time-consuming screenings for an ideal reference gene, provided that they verify the stability of these candidates under their conditions of study.
Results & Discussion
Selection of candidate reference genes
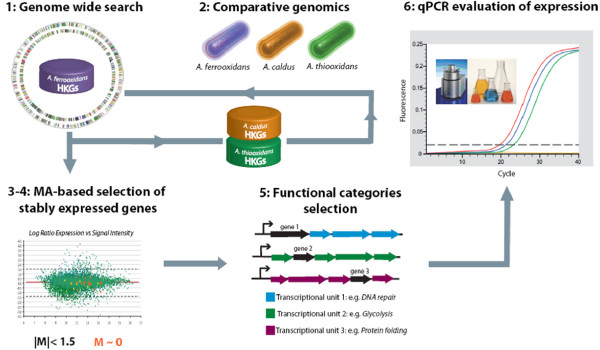
The most useful reference genes for standards in gene expression studies should be stably expressed over a range of experimental conditions and should be of wide phylogenetic distribution. Taking these requirements into account, a combined computational and experimental approach was devised to identify reference genes in the Acidithiobacilli (Figure 1). The strategy involves the following steps:
Figure 1.
Pipeline for the computational and experimental strategy used to identify suitable reference genes for normalization of expression.
1) A genome-wide bioinformatic identification of candidate reference genes
An initial set of candidate reference genes was compiled from a gene list that includes "housekeeping genes" of wide phylogenetic distribution [38]. This set was used to textmine the A. ferrooxidans ATCC 23270 genome (GenBank/EMBL/DDBJ accession number CP001219) [39]. Due to difference in ontological descriptions of gene function, not all genes could be recovered by textmining and additional candidates were identified in the A. ferrooxidans genome by BLASTP and TBLASTX searches using the housekeeping genes as queries. The combined set of candidate reference genes was then used to formulate bidirectional BLASTP and TBLASTX searches of the genomes of A. thiooxidans and A. caldus. This search for well conserved orthologs across Acidithiobacilli was performed in order to better define the set of essential genes for this bacterial genus. Such an approach will promote the ability to carry out future comparative gene expression studies within the Acidithiobacilli. Only genes present in all three genomes were accepted and provided the initial bioinformatic compilation of candidate reference genes (Additional File 1).
2) The selection of stably expressed candidate reference genes for A. ferrooxidans
The expression profiles for the set of candidate reference genes was then evaluated in three different growth conditions of A. ferrooxidans (iron, pH 1.6 vs. sulfur, pH 3.5, iron-sulfur mixture, pH 1.6 vs. sulfur, pH 1.6 and high iron, pH 1.6 vs. low iron, pH 1.6; see Methods for more details). Candidate reference genes that exhibited non-differential expression (log ratio expression |M| < 1.5) and had the most similar level of expression (log ratio expression M~0) between every pair of conditions in all three experiments were further selected (Additional file 2).
3) Removal of redundant candidate reference genes
The genetic context of these candidate reference genes in the genome of A. ferrooxidans was evaluated using the DNA sequence viewer and annotation tool Artemis v.10 [40]. In the case where more than one candidate gene belonged to the same operon or gene cluster, only one gene was selected for further experimental validation. Also, only one candidate was chosen from genes belonging to the same functional category as defined by TIGRfams [41]. This reduced redundancy in the set.
4) Evaluation of the expression profiles of the candidate reference genes by real time PCR
The expression of these selected candidate reference genes was analyzed by quantitative PCR in order to evaluate if the stability of expression observed in microarray experiments was supported by more sensitive and rigorous evaluation methods. Transcriptional levels were compared by assessing Ct values of each gene for two of the former experimental conditions (iron pH 1.6 and sulfur pH 3.5) and three new ones (sulfur pH 2.5, sulfur pH 4.5 and thiosulfate pH 4.5, see Methods for further detail). These conditions are frequently used in the laboratory to study the biology of Acidthiobacilli because they simulate environmental conditions. Expression values (Ct) of the selected reference genes and their dispersion are plotted in Additional file 3.
The combined bioinformatics and experimental strategy identified nine candidate reference genes (coaE, era, gmk, gyrA, map, nth, rplI, rpoC, trpS).
Expression stability of candidate reference genes
The expression of the 9 selected reference genes was analyzed in the five different experimental conditions described above by the methods of Vandesompele and Andersen as implemented in the Visual Basic Application geNorm [11] and Visual Basic Application NormFinder [7], respectively (Table 1). According to geNorm (which determines a gene expression stability value M to produce a rank where the best genes are those with the lowest M value) map, rpoC, era and gmk showed the least variability in expression in all conditions evaluated (range of expression of 1.029, 1.040, 1.046 and 1.067 fold respectively). The NormFinder approach (which enables identification of the single best genes in a ranking) showed era to be the most stable gene and rpoC and gmk to rank within the four most stably expressed genes. Slight differences observed between the two techniques are to be expected and can be explained by the way in which both methods analyze the data. geNorm selects the gene with the most stable expression independent of the expression of the other genes under analysis. NormFinder instead focuses on the genes with least intra- and inter-group expression variation, thus the selection of the best reference gene is affected by the other genes being analyzed. Similar differences between the results of geNorm and NormFinder have been reported in other studies [42,43]. Taking into account the results from both geNorm and NormFinder, we can conclude that rpoC and era are the most suitable reference genes for studies with the Acidithiobacilli.
Table 1.
Expression stability ranking of candidate reference genes.
| Gene | geNorm | NormFinder |
| map | 0.258 (1)* | 0.311 (6) |
| rpoC | 0.258 (1)* | 0.290 (3) |
| era | 0.416 (2) | 0.201 (1)* |
| gmk | 0.477 (3) | 0.299 (4) |
| trpS | 0.542 (4) | 0.306 (5) |
| nth | 0.581 (5) | 0.265 (2) |
| rplI | 0.616 (6) | 0.425 (8) |
| gyrA | 0.664 (7) | 0.412 (7) |
| coaE | 0.763 (8) | 0.628 (9) |
Ranking of the nine evaluated reference genes according to their expression stability under five different experimental conditions (♣) using geNorm [11] and NormFinder [7] applications.
* Genes with the most stable expression values.
() The rank of the genes is indicated in parentheses.
(♣) Experimental conditions. Condition 1: cells grown in 9 K medium at pH 1.6 containing 200 mM FeSO4; condition 2: cells grown in 9 K medium at pH 2.5 containing 1% elemental sulfur; condition 3: cells grown in 9 K medium at pH 3.5 containing 1% elemental sulfur; condition 4: cells grown in 9 K medium at pH 4.5 containing 1% elemental sulfur; condition 5: cells grown in DSMZ71 medium at pH 4.5 containing 0.5% thiosulfate.
Expression stability of reference genes previously used in studies of A. ferrooxidans
Three genes, recA, alaS and rrs (16S rRNA) have been used in prior studies as internal controls for experimental investigations in A. ferrooxidans [28,30,44,45], but there has been no formal report showing that they are reliable references. Conversely, there is evidence showing differential expression of recA and rrs under cellular stress [46] and starvation [45,47]. In addition, use of rrs as a reference gene has been challenged because it is a very abundant species of RNA present at concentrations outside most calibration ranges [16].
The expression of recA, alaS and rrs was analyzed under the same experimental conditions and their expression values were ranked with respect to the new reference genes derived above using geNorm [11] and NormFinder [7] (Table 2). The variability in expression of rrs and alaS in five different experimental setups shows them to perform well although slightly poorer than rpoC, era and map (Figure 2). Both geNorm and NormFinder identified rrs and alaS among the six more stable genes. On the contrary, recA ranks further down, indicating less stable expression, independently of the ranking method used (Table 2). In addition, the expression of recA varies more than two fold, and together with coaE and trpS exhibits the least suitable expression profile of the genes assessed in this study (Figure 2).
Table 2.
Expression stability ranking of previously reported control genes.
| Gene | geNorm | Normfinder |
| rpoC | 0.258 (1) | 0.310 (6) |
| map | 0.258 (1) | 0.331 (8) |
| alaS | 0.299 (2)* | 0.301 (5)* |
| era | 0.411 (3) | 0.229 (1) |
| gmk | 0.487 (4) | 0.295 (4) |
| rrs | 0.522 (5)* | 0.278 (3)* |
| nth | 0.559 (6) | 0.254 (2) |
| trpS | 0.585 (7) | 0.312 (7) |
| rplI | 0.614 (8) | 0.426 (10) |
| gyrA | 0.657 (9) | 0.525 (9) |
| recA | 0.720 (10)* | 0.476 (11)* |
| coaE | 0.786 (11) | 0.641 (12) |
Ranking of the three control genes previously used in A. ferrooxidans gene expression studies according to their expression stability with respect to the nine candidate reference genes under five different experimental condition using geNorm and NormFinder applications. See Table 1 legend for further detail on the ranking methods and experimental conditions.
* Control genes used in previous studies [28,30,45].
() The ranks of the genes are indicated in parentheses.
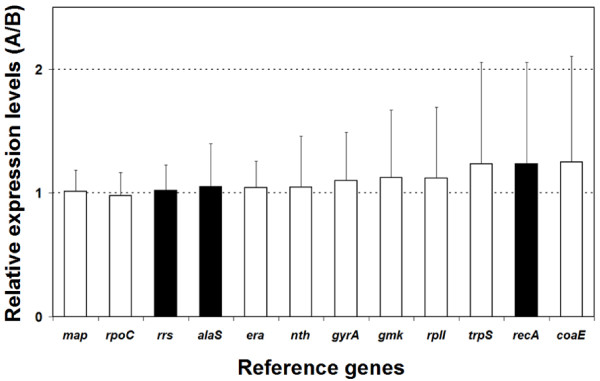
Figure 2.
Relative expression levels of alaS, recA and rrs and the nine candidate reference genes in all possible pairwise combinations (A/B) of five experimental conditions analyzed. Expression levels of the reference genes in each experimental condition (A) was compared to their respective expression levels in all other conditions evaluated (B). Boxes represent the average ratios (A/B) in all possible pairwise combinations of experimental conditions analyzed and bars represent the standard deviations of the average ratios A/B. White boxes correspond to reference genes selected after applying the strategy outlined in Figure 1. Black boxes correspond to reference genes previously used in A. ferrooxidans research [28,30,45].
It can be concluded that, among the previously used reference genes in A. ferrooxidans, only alaS is suitable and can be used with confidence as a normalizer. Given the variability observed in the present study, use of recA as a normalizer is not recommended as it would introduce noise to the analysis and eventually produce misleading results. In addition, use of rrs as a reference gene is not recommended despite its stable expression in the five conditions analyzed because its abundance prejudices the analysis of lowly abundant transcripts.
Use of multiple reference genes for improved normalization
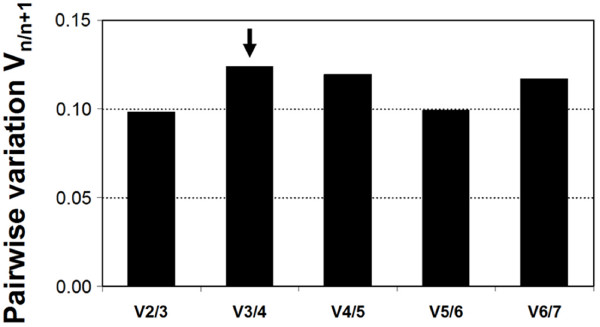
Normalizing gene expression based upon the expression levels of a carefully selected set of reference genes, usually referred to as a normalization factor, performs better than normalizing against any single gene alone [11]. This raises the question as to whether a compromise could be discovered in which a pool of genes, fewer than nine but more than one, would provide greater confidence for use as a reference group in A. ferrooxidans. For this purpose, several normalization factors (NF) were calculated following the criteria defined by Vandesompele et al. [11]. A NF represents the geometric mean of n genes, and the pairwise variation between sequential normalization factors (NFn and NFn+1) gives an idea of how well each of these perform. Geometric means for seven of most stable genes, showing less than 1.5 fold variation in all experimental conditions (map, rpoC, alaS, era, gyrA and nth) were obtained and pairwise variations between any two subsequent values was calculated. As shown in Figure 3, addition of a fourth gene leads to a non-significant change in the average of the gene variance estimates. According to geNorm ranking, it is concluded that the NF derived from the pool of the three genes map, rpoC and alaS (NF1) is suitable for reliable normalization of gene expression of target genes. In spite of this we posit that era, which ranked first according to the Normfinder method, could also be included in the selected reference gene set.
Figure 3.
Pairwise variation (Vn/n+1) analysis between normalization factors (NFn/NFn+1) to determine the optimal number of reference genes required for normalization.
Use of selected reference genes to normalize expression of differentially expressed genes in Fe-S cells
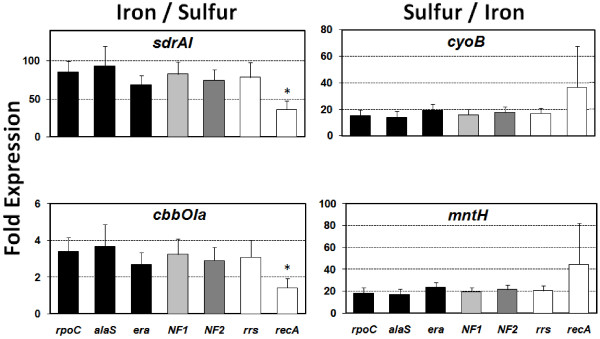
To assess the value of our study, the relative expression levels of selected target genes was analyzed using the following normalization strategies: a) the three best reference genes selected by geNorm and Normfinder rpoC, era and alaS were used individually, b) a NF derived from the combination of the three genes selected by geNorm, rpoC, map and alaS (NF1), c) a NF derived from the combination of the top ranking genes selected by geNorm and Normfinder method rpoC, map and era (NF2) or d) the frequently cited reference genes rrs and recA. For this purpose, four target genes were selected that are known to be differentially expressed in A. ferrooxidans cultures: a) sdrAI (AFE0007) is 95 fold induced in iron [34], b) cyoB (AFE2407) is 12 fold induced in sulfur [22], c) cbbOIa (AFE1408) is 3 fold induced in iron [23] and d) mntH (AFE2920) is 24 fold induced in sulfur (unpublished results).
Expression of the four genes of interest in iron versus sulfur grown cells was evaluated using the relative expression analysis software qBase v1.3.5 [48]. Figure 4 shows a significant increase in the expression of the sdrA1 and cbbOIa genes in cells grown in ferrous iron and of the cyoB and mntH genes in cells grown in sulfur. In all cases, normalization by individual reference genes outlined here (rpoC, era or alaS), by the geNorm derived NF1 (rpoC plus map plus alaS), by the combined NF2 (rpoC plus map plus era) and by rrs gave similar results. For example, sdrA1 was up-regulated 60–100 fold depending on whether the normalization strategy was a single stable reference gene or the normalization factors. Conversely, normalization by the recA gene dramatically altered the relative expression ratio of the target gene, revealing an up-regulation of less than 50.
Figure 4.
Comparison of different normalization strategies. Mean relative expression levels of four differentially expressed genes using different normalization strategies. Black: stable reference genes used individually, light grey: geometric mean of three stable reference genes selected by geNorm (NF1: rpoC, map and alaS), dark grey: geometric mean of four stable reference genes selected by geNorm (NF2: rpoC, map and era) and white: classical reference genes used individually. (*) Significantly different according to a two tailed unpaired t-test at 95% confidence.
These results demonstrate how the interpretation of bacterial gene expression levels can be affected by the choice of the reference genes in quantitative real-time RT-PCR analysis. If a single gene is to be used, e.g. in studies where only one or a few target genes are being evaluated, the reference gene should be one of the three validated stable reference genes rpoC, era or alaS. In investigations where a larger number of target genes are to be evaluated or a higher degree of confidence is desired, use of the NF derived from the pool of the genes rpoC, map and alaS or era is advisable.
Conclusion
Normalization is a prerequisite for accurate real time PCR expression profiling. Significant random fluctuations or, even worse, directional changes in the expression of chosen reference genes between samples, can lead to the lack of detection of small differences between genes of interest or to erroneous results. Therefore, it is extremely important to find appropriate reference genes with minimal variability. This cumbersome task is often avoided and frequently priorly used reference genes are assumed to be good normalizers without further evaluation in unexplored experimental setups.
The geometric mean of few carefully selected genes, rpoC, map, alaS and/or era, is demonstrated to be the best normalizer for A. ferrooxidans in the diverse experimental conditions used in this study. Use of a single gene for normalization, instead, may result in relatively large variations in target gene expression and significant errors depending on the gene in question and the experimental setup, as showed to be the case when using the recA gene. Conversely, it is suggested that rpoC, era or alaS could be used as normalizers if only one reference gene is strictly necessary. Since ribosomal RNA is much more abundant than most target mRNA transcripts and its quantification falls outside most calibration ranges, the use of rrs is not recommended especially for the measurements of low abundance transcripts.
Whatever strategy is used to normalize for differences in quality and quantity of input RNA it must be validated for a particular experimental model on an individual basis. This investigation provides a validated set of reference genes for those studying A. ferrooxidans gene expression under typical biological conditions and an initial point of departure for those exploring new experimental setups in this microorganism or other closely related Acidithiobacilli or possibly also in other bacterial models.
Methods
Bioinformatic selection of candidate reference genes
An initial set of candidate reference genes was compiled from a gene list that includes "housekeeping genes" of wide phylogenetic distribution [38]. This set was used to textmine the A. ferrooxidans ATCC 23270 genome (GenBank/EMBL/DDBJ accession number CP001219) [39]. Additional candidates were identified in the A. ferrooxidans genome by BLASTP and TBLASTX searches. The combined set of candidate genes was then used to formulate bidirectional BLASTP and TBLASTX searches of the genomes of A. thiooxidans and A. caldus. Genomic context for genes present in all three genomes was analyzed using the DNA sequence viewer and annotation tool Artemis v.10 [40]. Candidate genes belonging to the same predicted operon or gene cluster were excluded from further analysis. Reference genes were classified by function using TIGRfams [41] and one gene per functional category was selected. These last two steps were included to reduce redundancy in the gene set.
Bacterial strains and growth conditions
Gene expression was evaluated under the following experimental conditions: sulfur at pH 2.5, pH 3.5 and pH 4.5; thiosulfate at pH 4.5 and ferrous iron 200 mM pH 1.6. A. ferrooxidans strain ATCC 23270 was grown in modified 9 K basal salt media (0.7 mM (NH4)2SO4, 0.2 mM K2HPO4, 1.6 mM MgSO4.7H2O) containing iron (9 K + Fe: 200 mM FeSO4; adjusted to pH 1.6 with H2SO4) or sulfur (9 K + S: 1% ethanol-sterilized powdered sulfur, adjusted to pH 2.5; 3.5 and 4.5 with H2SO4). DSMZ71 medium was used for thiosulfate growth (20 mM Na2S2O3.5H2O, 22 mM KH2PO4, 2 mM MgSO4.7H2O, 22 mM (NH4)2SO4 and 1.7 mM CaCl2.2H2O). All cultures were incubated at 30°C under aerobic conditions on a rotary shaker at 150 r.p.m.
Cell collection
A. ferrooxidans cultures to be used for nucleic acid purification were harvested at 8000 r.p.m. for 10 min. The cell pellet was washed in 9 K basal salt solution (adjusted at the corresponding pH). Washed cells were collected by centrifugation at 12000 r.p.m. for 10 min.
DNA isolation
A. ferrooxidans cultures were grown for 72 h until stationary phase. DNA isolation was carried out by phenol-chloroform extraction. Briefly, cells were collected and resuspended in buffer TE (25:10) pH 8.0 with 5 mg/ml lysozyme, and incubated at 37°C for 30 minutes, followed by another hour of incubation at the same temperature with 1% SDS and 0.2 mg/ml proteinase K. Cell lysis was completed by alternate shifting of the suspension from 80°C to -80°C. DNA extraction was performed twice with a mixture of phenol:chloroform:isoamylic alcohol (25:24:1). Removal of the residual phenol was accomplished by one treatment with a mixture of chloroform:isoamylic alcohol (24:1). The DNA contained in the final aqueous phase was precipitated overnight at -20°C with absolute ethanol, washed with 70% ethanol, and finally resuspended in sterilized water. Genomic DNA quality and integrity were assessed by 1% (w/v) agarose gel electrophoresis and standard PCR, and concentration was determined by absorbance at 260 nm.
Total RNA isolation
RNA was isolated from A. ferrooxidans mid-logarithmic cultures grown in modified 9 K basal salt medium in the presence of iron 200 mM, sulfur (1%, pH 2.5, 3.5 and 4.5) or 0.5% thiosulfate. Briefly, cells were collected and resuspended in ice-cold buffer TE (25:10) pH 8.0 with 1× Extraction Buffer (per liter: 1% SDS, 50 mM Tris-HCl pH 8.0, and 2 mM EDTA). Cell lysis was accomplished by incubation at 100°C for 5 minutes. The suspension was treated with TRIzol (Invitrogen), and the recovered aqueous phase was treated with chloroform followed by two extractions with acid phenol and chloroform. RNA was precipitated with absolute ethanol overnight at -20°C, washed with 70% ethanol, and finally resuspended in sterilized water. Samples were treated with DNase and purified with the Roche High Pure RNA Isolation Kit, following the manufacturer's recommendations and checked for DNA contamination by standard PCR, including a genomic DNA positive control. RNA quality was evaluated by 1.0% agarose gel electrophoresis and its concentration was measured by absorbance at 260 nm.
Reverse transcription
cDNA was prepared from 1 μg total RNA using random hexamers and Superscript II reverse transcriptase (Invitrogen) according to manufacturer instructions. The resulting cDNA was diluted 1:10 in distilled water and stored in aliquots at -20°C until further use.
Real-time PCR
Primers for real-time PCR assays and amplification efficiencies (E) are shown in Table 3. The real-time PCR reactions were performed in the Mx3000P QPCR System (Stratagene) using the SYBR GreenER qPCR SuperMix Universal Kit (Invitrogen). The 20 μl PCR reactions contained 2 μl of a 1:100 diluted cDNA sample; 200 nM of each primer and 1× SYBR GreenER qPCR SuperMix Universal (Invitrogen). The reference dye ROX was included at a final concentration of 5 nM. The cycling protocol was as follows: initial denaturation for 10 min at 95°C followed by 40 cycles of 30 s at 95°C, 15 s at 52°C; 30 s at 72°C. Fluorescence was measured after the extension phase at 72°C. The PCR products were subjected to a melting curve analysis, that commenced at 52°C and increased at 0.5°C s-1 up to 95°C, with a continuous fluorescent measurement. Specific amplification was confirmed by a single peak in the melting curve. For each experimental condition total RNA was extracted from two independent A. ferrooxidans cultures. Each RNA sample was retro-transcribed and the expression of all genes was assessed on the same cDNA sample. The reactions for each target gene were performed in triplicate and in the same PCR run. Thus, data sets consist of 6 values per gene per experimental set-up generated under standardized PCR cycling conditions. Stationary phase genomic DNA 10-fold dilutions (ranging from 10 ng to 1 pg) were used to generate a 5-point standard curve for every gene by using the Cycle Threshold (Ct) value versus the logarithm of each dilution factor. Reaction efficiency (E = [10(-1/slope)]-1) for every gene was derived from the slope of the corresponding standard curves. Transcript quantities were calculated from the standard curve by the software accompanying the MxPro3000P QPCR System (Stratagene) set with default parameters. Each experiment included a no template control.
Table 3.
Reference genes, qPCR primers and reaction parameters.
| Gene |
Forward primer (5'-3') |
Reverse primer (5'-3') |
Amplicon size |
Q-PCR Linear correlation coefficient |
Q-PCR Efficiency (%) |
Average Ct ± SD |
| Candidate reference genes | ||||||
| coaE | ACT ATC GCC CAT TGC TGG AT | TTG GTA ATG ATC CAG TCG GC | 169 | 0.999 | 93 | 25.50 ± 0.69 |
| era | CAT GGA TGA GAT CAA GAG CG | GTA TCC CGA GAA TCT GAT CC | 174 | 0.999 | 94 | 22.56 ± 0.21 |
| gmk | ATG GCA CTA GTG AAC CTT GG | AAT GAC CGT CTC CGA ATC CT | 197 | 0.998 | 92 | 24.49 ± 0.50 |
| gyrA | TAC CTC GAT TAC GCC ATG AG | TGT CAT AGA CAG CGG TAT CG | 202 | 0.998 | 95 | 26.24 ± 0.45 |
| map | TTA CCA CCG ATG AAC TGG AC | AGC CAT CCT TGA TAA CCG TG | 222 | 0.999 | 94 | 20.64 ± 0.29 |
| nth | ATC GTC TGG GAC TGT TCA AC | TAT TGA GGA CGA CAT TGG CG | 153 | 0.997 | 94 | 25.31 ± 0.40 |
| rplI | GAT GCC ATG GTT ACG ATT GC | GAC CAT CAC ATC CAG TTC GA | 210 | 1.000 | 92 | 21.68 ± 0.51 |
| rpoC | AAT GCG GTG TTG AGG TAA CC | AGG TAC TGG TCT TCG GTA AG | 238 | 0.997 | 95 | 20.79 ± 0.23 |
| trpS | TCT GCT CAT CGA ATG GTT GG | GAA TGT CTG CCG TCA TCA AC | 230 | 0.999 | 89 | 23.86 ± 0.70 |
| Reference genes used in prior studies | ||||||
| alaS | CTG GAA TCT GGT CTT CAT GC | GCT TGA AGA GAT CGG TGT CA | 152 | 1.000 | 94 | 22.35 ± 0.39 |
| recA | CCG CCA ACA TTT CCC GGA CC | ACG CCG CGG TCC ACC AGT TC | 301 | 0.998 | 98 | 29.41 ± 0.88 |
| rrs | ACA CTG GGA CTG AGA CAC GG | ACC GCC TAC GCA CCC TTT AC | 277 | 0.999 | 95 | 8.93 ± 0.27 |
| Differentially expressed target genes | ||||||
| cbbOIa | ACG GCA TTG AGC TAT ACC G | TCC GGG TCT AGT AGT GCA T | 265 | 0.996 | 97 | 21.55 ± 0.72 |
| cyoB | CAA TTA CAT GGT CCA CAA CA | AGC GTA TAC ACC ACG ATA CC | 300 | 0.995 | 92 | 28.11 ± 2.17 |
| mntH | ATA TCG GTG CCG TCA TCA TG | GAG AGC TGA TGC ACC GTA TT | 217 | 0.996 | 97 | 21.92 ± 2.02 |
| sdrAI | GTT TGG GTG CTG AAG TAG TG | GCA ACA GTG GCA AAC AGG C | 255 | 0.995 | 93 | 23.61 ± 2.82 |
Primer sequences and reaction parameters used for quantitative real time PCR expression analysis.
Stability of gene expression and relative quantification
The stability of gene expression was evaluated using the Excel-based applications geNorm [11] and Normfinder [7] and the relative expression was calculated with qBase 1.3.5 [48]. Briefly, the geNorm method is based on a pairwise comparison approach and depends on the calculation of an M value, defined as the average pairwise variation of a particular gene with all others. The NormFinder method is based on a different mathematic model that considers the intra- and inter-treatment variation in expression for gene ranking. The normalization factors were calculated following the criteria defined by Vandesompele [11]. These include: a) to use the geometric mean (n numbers are multiplied and then the nth root of the resulting product is taken) as this controls better for possible outliers and abundance differences between genes and b) to define the minimum number of reference genes needed for a reliable calculation by evaluating the pairwise variation between sequential normalization factors including three, four, five or more stable reference genes (NFn/NFn+1).
Evaluation of reference gene expression by microarray transcript profiling
A. ferrooxidans gene expression was evaluated under three experimental conditions: (1) cells were grown in 9 K medium containing 62 mM FeSO4 at pH 1.6 versus cells grown in 9 K medium 1% elemental sulfur at pH 3.5 containing; (2) cells were grown in 9 K medium containing 62 mM FeSO4 plus 1% elemental sulfur at pH 1.6 versus cells grown in 9 K medium containing 1% elemental sulfur at pH 1.6 and (3) cells were grown in 9 K medium containing 200 mM FeSO4 at pH 1.6 versus cells grown in 9 K medium containing 62 mM FeSO4 at pH 1.6. Construction, experimental and data analysis protocols for A. ferrooxidans type strain specific oligonucleotide microarrays have been previously described [22] and deposited in the ArrayExpress database under the following accession numbers (A-MEXP-1478, A-MEXP-1479).
Authors' contributions
PAN carried out the real time PCR standardization and analysis; PCC was responsible for the culture handling and preparation of the genomic DNA and total RNA samples. PAN and RQ conceived the study; EJ tutored PAN; RQ and DSH helped in the biological interpretation, and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Initial compilation of candidate reference genes. Selection of conserved candidate reference genes belonging to different functional categories derived from textmining and blast searches in the genomes of three related Acidithiobacilli, A. ferrooxidans ATCC 23270, A. thiooxidans ATCC 19377 and A. caldus ATCC 51756.
Candidate reference genes that survive culling after microarray transcript profiling. Selection of conserved, non-differentially (|M| < 1.5) and stably (M~0) expressed reference genes belonging to different functional categories and different transcriptional units derived from a microarray dataset built upon 3 different experimental conditions. Condition A: cells grown in 9 K medium at pH 1.6 containing 62 mM FeSO4 (treatment) versus cells grown in 9 K medium at pH 3.5 containing 1% elemental sulfur (control); Condition B: cells grown in 9 K medium at pH 1.6 containing 62 mM FeSO4 plus 1% elemental sulfur (treatment) versus cells grown in 9 K medium at pH 3.5 containing 1% elemental sulfur (control); Condition C: cells grown in 9 K medium at pH 1.6 containing 200 mM FeSO4 (treatment) versus cells grown in 9 K medium at pH 1.6 containing 62 mM FeSO4 (control). Candidate reference genes are indicated in blue.
Boxplot graph for the expression levels of candidate reference genes. Comparison of the transcriptional expression levels of the nine candidate reference genes by direct plotting of the Ct values (number of cycles needed for the fluorescence signal to reach a specific threshold level of detection). The Ct median values for 5 different experimental setups are shown as lines, 25th and 75th percentile as boxes and ranges as bars. Condition 1: cells grown in 9 K medium at pH 1.6 containing 200 mM FeSO4, condition 2: cells grown in 9 K medium at pH 2.5 containing 1% elemental sulfur; condition 3: cells grown in 9 K medium at pH 3.5 containing 1% elemental sulfur; condition 4: cells grown in 9 K medium at pH 4.5 containing 1% elemental sulfur; condition 5: cells grown in DSMZ71 medium at pH 4.5 containing 0.5% thiosulfate. Candidate reference genes include: gyrA, DNA gyrase subunit A; coaE, dephospho-CoA kinase; nth, endonuclease III; gmk, guanylate kinase; trpS, tryptophanyl-tRNA synthetase; era, GTP-binding protein; rplI, ribosomal protein L9; rpoC, DNA-directed RNA polymerase subunit β and map, type I methionine aminopeptidase.
Acknowledgments
Acknowledgements
Work supported by Fondecyt 11060164, Fondecyt 1050063, Conicyt Basal CCTE PFB16, UNAB DI-3406-R, Innova 08CM01-03 and a Microsoft Sponsored Research Award.
Contributor Information
Pamela A Nieto, Email: pamelanietop@gmail.com.
Paulo C Covarrubias, Email: paulo.covarrubias@gmail.com.
Eugenia Jedlicki, Email: ejedlick@med.uchile.cl.
David S Holmes, Email: dsholmes2000@yahoo.com.
Raquel Quatrini, Email: rquatrini@yahoo.com.ar.
References
- Klein D. Quantification using real-time PCR technology: applications and limitations. Trends Mol Med. 2002;8:257–260. doi: 10.1016/S1471-4914(02)02355-9. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol. 2002;30:503–512. doi: 10.1016/S0301-472X(02)00806-8. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Gibson UE, Heid CA, Williams PM. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Zakrajsek BA. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods. 2000;46:69–81. doi: 10.1016/S0165-022X(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Vandecasteele SJ, Peetermans WE, Merckx R, Van Eldere J. Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J Bacteriol. 2001;183:7094–7101. doi: 10.1128/JB.183.24.7094-7101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van RN, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radonic A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- Sturzenbaum SR, Kille P. Control genes in quantitative molecular biological techniques: the variability of invariance. Comp Biochem Physiol B Biochem Mol Biol. 2001;130:281–289. doi: 10.1016/S1096-4959(01)00440-7. [DOI] [PubMed] [Google Scholar]
- Takle GW, Toth IK, Brurberg MB. Evaluation of reference genes for real-time RT-PCR expression studies in the plant pathogen Pectobacterium atrosepticum. BMC Plant Biol. 2007;7:50. doi: 10.1186/1471-2229-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Simons JW. Direct comparison of GAPDH, beta-actin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem Biophys Res Commun. 1999;259:523–526. doi: 10.1006/bbrc.1999.0815. [DOI] [PubMed] [Google Scholar]
- Theis T, Skurray RA, Brown MH. Identification of suitable internal controls to study expression of a Staphylococcus aureus multidrug resistance system by quantitative real-time PCR. J Microbiol Methods. 2007;70:355–362. doi: 10.1016/j.mimet.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Petersen BH, Rapaport R, Henry DP, Huseman C, Moore WV. Effect of treatment with biosynthetic human growth hormone (GH) on peripheral blood lymphocyte populations and function in growth hormone-deficient children. J Clin Endocrinol Metab. 1990;70:1756–1760. doi: 10.1210/jcem-70-6-1756. [DOI] [PubMed] [Google Scholar]
- Singh R, Green MR. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993;259:365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- Ishitani R, Sunaga K, Hirano A, Saunders P, Katsube N, Chuang DM. Evidence that glyceraldehyde-3-phosphate dehydrogenase is involved in age-induced apoptosis in mature cerebellar neurons in culture. J Neurochem. 1996;66:928–935. doi: 10.1046/j.1471-4159.1996.66030928.x. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta. 2008;227:1343–9. doi: 10.1007/s00425-008-0706-4. [DOI] [PubMed] [Google Scholar]
- Quatrini R, Appia-Ayme C, Denis C, Ratouchniak J, Veloso F, Valdes J, Lefimil C, Silver S, Roberto F, Orellana O, et al. Insights into the iron and sulfur energetic metabolism of Acidthiobacillus ferrooxidans by microarray transcriptome profiling. Hydrometallurgy. 2006;83:263–272. doi: 10.1016/j.hydromet.2006.03.030. [DOI] [Google Scholar]
- Appia-Ayme C, Quatrini R, Dennis Y, Denizot F, Silver S, Roberto F, Veloso F, Valdes J, Cardenas J, Esparza M, et al. Microarray and bioinformatic analyses suggest models for carbon metabolism in the autotroph Acidithiobacillus ferrooxidans. Hydrometallurgy. 2006;83:273–280. doi: 10.1016/j.hydromet.2006.03.029. [DOI] [Google Scholar]
- Appia-Ayme C, Guiliani N, Ratouchniak J, Bonnefoy V. Characterization of an operon encoding two c-type cytochromes, an aa(3)-type cytochrome oxidase, and rusticyanin in Thiobacillus ferrooxidans ATCC 33020. Appl Environ Microbiol. 1999;65:4781–4787. doi: 10.1128/aem.65.11.4781-4787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiliani N, Jerez CA. Molecular cloning, sequencing, and expression of omp-40, the gene coding for the major outer membrane protein from the acidophilic bacterium Thiobacillus ferrooxidans. Appl Environ Microbiol. 2000;66:2318–2324. doi: 10.1128/AEM.66.6.2318-2324.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher BG, Rawlings DE. The divergent chromosomal ars operon of Acidithiobacillus ferrooxidans is regulated by an atypical ArsR protein. Microbiology. 2002;148:3983–3992. doi: 10.1099/00221287-148-12-3983. [DOI] [PubMed] [Google Scholar]
- Levican G, Bruscella P, Guacunano M, Inostroza C, Bonnefoy V, Holmes DS, Jedlicki E. Characterization of the pet I and res operons of Acidithiobacillus ferrooxidans. J Bacteriol. 2002;184:1498–1501. doi: 10.1128/JB.184.5.1498-1501.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarzabal A, Appia-Ayme C, Ratouchniak J, Bonnefoy V. Regulation of the expression of the Acidithiobacillus ferrooxidans rus operon encoding two cytochromes c, a cytochrome oxidase and rusticyanin. Microbiology. 2004;150:2113–2123. doi: 10.1099/mic.0.26966-0. [DOI] [PubMed] [Google Scholar]
- Ramirez P, Guiliani N, Valenzuela L, Beard S, Jerez CA. Differential protein expression during growth of Acidithiobacillus ferrooxidans on ferrous iron, sulfur compounds, or metal sulfides. Appl Environ Microbiol. 2004;70:4491–4498. doi: 10.1128/AEM.70.8.4491-4498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas M, Seeger M, Holmes DS, Jedlicki E. A Lux-like quorum sensing system in the extreme acidophile Acidithiobacillus ferrooxidans. Biol Res. 2005;38:283–297. doi: 10.4067/s0716-97602005000200018. [DOI] [PubMed] [Google Scholar]
- Quatrini R, Lefimil C, Holmes DS, Jedlicki E. The ferric iron uptake regulator (Fur) from the extreme acidophile Acidithiobacillus ferrooxidans. Microbiology. 2005;151:2005–2015. doi: 10.1099/mic.0.27581-0. [DOI] [PubMed] [Google Scholar]
- Barreto M, Jedlicki E, Holmes DS. Identification of a gene cluster for the formation of extracellular polysaccharide precursors in the chemolithoautotroph Acidithiobacillus ferrooxidans. Appl Environ Microbiol. 2005;71:2902–2909. doi: 10.1128/AEM.71.6.2902-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta M, Beard S, Ponce J, Vera M, Mobarec JC, Jerez CA. Identification of putative sulfurtransferase genes in the extremophilic Acidithiobacillus ferrooxidans ATCC 23270 genome: structural and functional characterization of the proteins. OMICS. 2005;9:13–29. doi: 10.1089/omi.2005.9.13. [DOI] [PubMed] [Google Scholar]
- Bruscella P, Appia-Ayme C, Levican G, Ratouchniak J, Jedlicki E, Holmes DS, Bonnefoy V. Differential expression of two bc1 complexes in the strict acidophilic chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans suggests a model for their respective roles in iron or sulfur oxidation. Microbiology. 2007;153:102–110. doi: 10.1099/mic.0.2006/000067-0. [DOI] [PubMed] [Google Scholar]
- Rivas M, Seeger M, Jedlicki E, Holmes DS. Second acyl homoserine lactone production system in the extreme acidophile Acidithiobacillus ferrooxidans. Appl Environ Microbiol. 2007;73:3225–3231. doi: 10.1128/AEM.02948-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levican G, Katz A, de Armas M, Nunez H, Orellana O. Regulation of a glutamyl-tRNA synthetase by the heme status. Proc Natl Acad Sci USA. 2007;104:3135–3140. doi: 10.1073/pnas.0611611104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera M, Pagliai F, Guiliani N, Jerez CA. The chemolithoautotroph Acidithiobacillus ferrooxidans can survive under phosphate-limiting conditions by expressing a C-P lyase operon that allows it to grow on phosphonates. Appl Environ Microbiol. 2008;74:1829–1835. doi: 10.1128/AEM.02101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R, Silva FJ, Peretó J, Moya A. Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev. 2004;68:518–37. doi: 10.1128/MMBR.68.3.518-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes J, Pedroso I, Quatrini R, Dodson RJ, Tettelin H, Blake R, 2nd, Eisen JA, Holmes DS. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics. 2008;9:597. doi: 10.1186/1471-2164-9-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–5. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol. 2009;10:11. doi: 10.1186/1471-2199-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbeler S, Scharsack JP, Becker S. Housekeeping genes for quantitative expression studies in the three-spined stickleback Gasterosteus aculeatus. BMC Mol Biol. 2008;9:18. doi: 10.1186/1471-2199-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew DA, Knight KL. Molecular design and functional organization of the RecA protein. Crit Rev Biochem Mol Biol. 2003;38:385–432. doi: 10.1080/10409230390242489. [DOI] [PubMed] [Google Scholar]
- He Z, Zhong H, Hu Y, Xiao S, Liu J, Xu J, Li G. Analysis of differential-expressed proteins of Acidithiobacillus ferrooxidans grown under phosphate starvation. J Biochem Mol Biol. 2005;38:545–549. doi: 10.5483/bmbrep.2005.38.5.545. [DOI] [PubMed] [Google Scholar]
- Stevenson DM, Weimer PJ. Expression of 17 genes in Clostridium thermocellum ATCC 27405 during fermentation of cellulose or cellobiose in continuous culture. Appl Environ Microbiol. 2005;71:4672–4678. doi: 10.1128/AEM.71.8.4672-4678.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. Regulation of ribosomal RNA synthesis in E. coli : effects of the global regulator guanosine tetraphosphate (ppGpp) J Mol Microbiol Biotechnol. 2002;4:331–340. [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Initial compilation of candidate reference genes. Selection of conserved candidate reference genes belonging to different functional categories derived from textmining and blast searches in the genomes of three related Acidithiobacilli, A. ferrooxidans ATCC 23270, A. thiooxidans ATCC 19377 and A. caldus ATCC 51756.
Candidate reference genes that survive culling after microarray transcript profiling. Selection of conserved, non-differentially (|M| < 1.5) and stably (M~0) expressed reference genes belonging to different functional categories and different transcriptional units derived from a microarray dataset built upon 3 different experimental conditions. Condition A: cells grown in 9 K medium at pH 1.6 containing 62 mM FeSO4 (treatment) versus cells grown in 9 K medium at pH 3.5 containing 1% elemental sulfur (control); Condition B: cells grown in 9 K medium at pH 1.6 containing 62 mM FeSO4 plus 1% elemental sulfur (treatment) versus cells grown in 9 K medium at pH 3.5 containing 1% elemental sulfur (control); Condition C: cells grown in 9 K medium at pH 1.6 containing 200 mM FeSO4 (treatment) versus cells grown in 9 K medium at pH 1.6 containing 62 mM FeSO4 (control). Candidate reference genes are indicated in blue.
Boxplot graph for the expression levels of candidate reference genes. Comparison of the transcriptional expression levels of the nine candidate reference genes by direct plotting of the Ct values (number of cycles needed for the fluorescence signal to reach a specific threshold level of detection). The Ct median values for 5 different experimental setups are shown as lines, 25th and 75th percentile as boxes and ranges as bars. Condition 1: cells grown in 9 K medium at pH 1.6 containing 200 mM FeSO4, condition 2: cells grown in 9 K medium at pH 2.5 containing 1% elemental sulfur; condition 3: cells grown in 9 K medium at pH 3.5 containing 1% elemental sulfur; condition 4: cells grown in 9 K medium at pH 4.5 containing 1% elemental sulfur; condition 5: cells grown in DSMZ71 medium at pH 4.5 containing 0.5% thiosulfate. Candidate reference genes include: gyrA, DNA gyrase subunit A; coaE, dephospho-CoA kinase; nth, endonuclease III; gmk, guanylate kinase; trpS, tryptophanyl-tRNA synthetase; era, GTP-binding protein; rplI, ribosomal protein L9; rpoC, DNA-directed RNA polymerase subunit β and map, type I methionine aminopeptidase.