Abstract
Successful cell division requires that chromosomes attach to opposite poles of the mitotic spindle (bi-orientation). Aurora B kinase regulates chromosome-spindle attachments by phosphorylating kinetochore substrates that bind microtubules. Centromere tension stabilizes bi-oriented attachments, but how physical forces are translated into signaling at individual centromeres is unknown. Using FRET-based biosensors to measure localized phosphorylation dynamics in living cells, we found that phosphorylation of an Aurora B substrate at the kinetochore depended on its distance from the kinase at the inner centromere. Furthermore, repositioning Aurora B closer to the kinetochore prevented stabilization of bi-oriented attachments and activated the spindle checkpoint. Thus, centromere tension can be sensed by increased spatial separation of Aurora B from kinetochore substrates, which reduces phosphorylation and stabilizes kinetochore microtubules.
Accurate chromosome segregation during cell division is essential to maintain genome integrity. Prior to segregation, kinetochores of sister chromatids attach to microtubules from opposite spindle poles (bi-orientation). This configuration is achieved through a trial-and-error process in which correct attachments exert tension across the centromere, which stabilizes kinetochore-microtubule interactions. Incorrect attachments, for example if both sister chromatids attach to a single spindle pole, exert less tension and are destabilized, providing a new opportunity to bi-orient (1, 2). How tension is coupled to kinetochore-microtubule stability is not known.
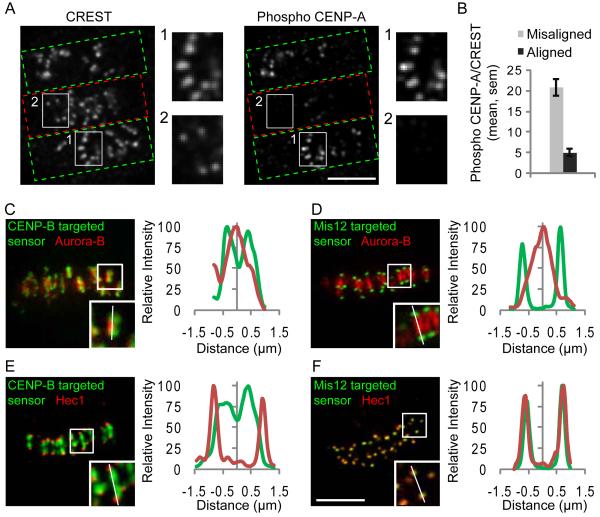
The mitotic kinase Aurora B (Ipl1 in budding yeast) localizes to the inner centromere, between sister kinetochores, and destabilizes microtubule attachments by phosphorylating kinetochore substrates, including Dam1 and the Ndc80 complex (3-10). An appealing model is that Aurora B substrates are selectively phosphorylated at incorrect attachments. To test this model we first examined phosphorylation of CENP-A Ser-7, a known kinetochore substrate (11). We used an assay in which Aurora B inhibition leads to a high frequency of syntelic attachment errors, with sister chromatids connected to a single spindle pole (6) (Fig. S1A). We compared phospho-CENP-A staining at correct and incorrect attachments 10 min after removing the reversible Aurora B kinase inhibitor ZM447439 (12), which re-activates Aurora B. Phospho-CENP-A staining was strongest at incorrect attachments, identified as unaligned kinetochores (Fig. 1A, B), which indicates that phosphorylation of an Aurora B substrate at the kinetochore depends on the microtubule attachment state.
Figure 1.
Phosphorylation of a kinetochore Aurora B substrate depends on the microtubule attachment state. (A-B) Hela cells with both incorrect (green boxes, A) and bi-oriented (red boxes, A) attachments were fixed and stained for kinetochores (CREST) and phospho-CENP-A. Insets show stronger phospho-CENP-A staining on unaligned (1) vs. aligned (2) kinetochores. The phospho-CENP-A/CREST ratio was calculated at individual aligned (N=146) and unaligned (N=89) kinetochores from multiple cells (B). (C-F) Hela cells expressing either the CENP-B-targeted (C,E) or Mis12-targeted (D,F) sensor were fixed and stained for either Aurora B (C,D) or Hec1 (E,F) as markers for the inner centromere and outer kinetochore, respectively. YFP emission (green) shows sensor localization relative to Aurora B or Hec1 (red). Insets show individual centromere pairs used for linescans. Scale bars 5 μm.
We considered two models to explain how selective phosphorylation at incorrect attachments might be achieved. First, tension could directly regulate kinase activity by inducing a conformational change either in the kinase itself or in its associated regulatory proteins (13). Second, kinase activity could be constant, but tension pulls kinetochores away from Aurora B at the inner centromere (4, 14). This spatial separation might lead to decreased phosphorylation of Aurora B substrates at the kinetochore. To distinguish between these models, we measured phosphorylation of an Aurora B substrate localized to either the centromere or the kinetochore, using biosensors that report quantitative changes in phosphorylation by Aurora B in living cells through changes in fluorescence resonance energy transfer (FRET) (15). Sensors were targeted either to the centromere using the centromere-targeting domain of CENP-B (16) or to the kinetochore using Mis12 (17), (Fig. S1A), and their localization was verified (Fig. 1C-F). Both sensors respond to Aurora B inhibition, consistent with previous findings (15) (Fig. S1B-C).
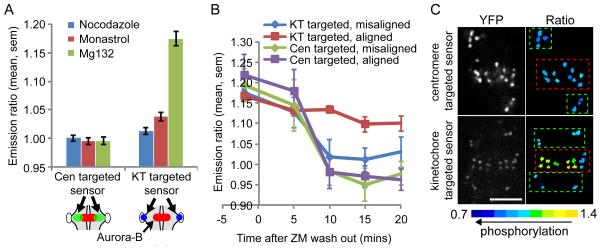
We first examined steady-state phosphorylation levels in cells treated with either nocodazole to depolymerize microtubules, the kinesin-5 inhibitor monastrol to create monopolar spindles and syntelic attachments (18), or the proteasome inhibitor MG132 to arrest cells at metaphase with bi-oriented attachments. Measurements of inter-kinetochore distances (19) confirmed that tension was low in cells treated with nocodazole (0.6±0.1 μm, mean±stdev) or monastrol (0.7±0.1 μm) and high in cells treated with MG132 (1.4±0.3 μm, Fig. S1D-E). The centromere-targeted sensor was phosphorylated under all conditions examined, independent of tension, while the kinetochore-targeted sensor was phosphorylated when tension was low and dephosphorylated when tension was high (Fig. 2A). Thus phosphorylation depends on spatial separation of Aurora B from its substrate, rather than on direct regulation of Aurora B kinase activity.
Figure 2.
Phosphorylation of Aurora B substrates depends on spatial separation of kinase from substrate. Hela cells expressing either the kinetochore (KT)-targeted or centromere (Cen)-targeted sensors were treated as indicated and imaged live. (A) Cells were treated with nocodazole, monastrol, or MG132, and the YFP/TFP emission ratio was calculated. An increase in emission ratio indicates dephosphorylation. Each bar represents an average over multiple cells, ≥30 kinetochores analyzed per cell. Note that y-axis is scaled to include the range of the data. Schematic shows sensor localization relative to Aurora B, as in Fig. S1A. (B-C) Cells were treated as in Fig. 1A, then imaged live before and after ZM447439 washout. The emission ratio was calculated at aligned and unaligned centromeres/kinetochores at the indicated timepoints (B). Each point represents ≥14 kinetochores/centromeres analyzed over multiple cells. Images (C) show sensor localization (YFP) and a color-coded representation of the emission ratio 10 min after ZM447439 washout. Boxes indicate aligned (red) and unaligned (green) centromeres/kinetochores. Scale bar 5 μm.
To determine whether phosphorylation dynamics occur on relevant timescales for regulation of kinetochore-microtubule attachments, we followed kinase activity during the correction of attachment errors (6). Cells were treated as in Fig. 1A and imaged live after Aurora B re-activation, and we measured phosphorylation dynamics at aligned and unaligned kinetochores and centromeres. The centromere-targeted sensor was rapidly phosphorylated at both aligned and unaligned centromeres within 10 min of removing the Aurora B inhibitor (Fig 2B). The kinetochore-targeted sensor was also rapidly phosphorylated at unaligned kinetochores, but remained dephosphorylated at aligned kinetochores (Fig. 2B). Color-coded images of the YFP/TFP emission ratio showed that the kinetochore-targeted sensor was selectively phosphorylated at unaligned kinetochores10 min after inhibitor washout, while the centromere-targeted sensor was phosphorylated at all centromeres (Fig. 2C and Fig. S1F). Thus Aurora B is active at both correct and incorrect attachments, and differential phosphorylation of kinetochore substrates may depend on distance from the kinase.
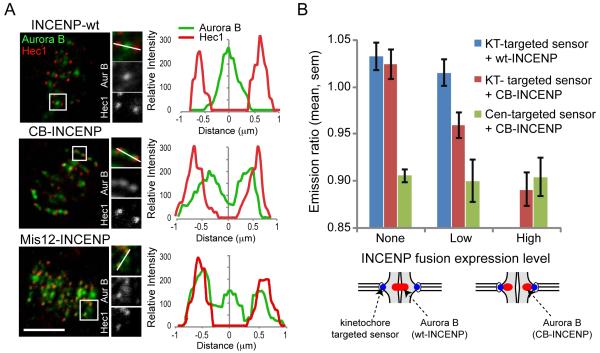
The spatial separation model predicts that positioning Aurora B closer to the kinetochore should lead to increased phosphorylation of kinetochore substrates in metaphase. Aurora B localizes to the inner centromere through interactions with INCENP (20). We manipulated the position of Aurora B by creating INCENP fusion proteins in which the inner centromere-targeting domain of INCENP is replaced either with the centromere-targeting domain of CENP-B (CB-INCENP) (16, 21) or with full-length Mis12 (Mis12-INCENP) (Fig. S2A). Endogenous Aurora B localized closer to or at the kinetochore in cells expressing CB-INCENP or Mis12-INCENP, respectively (Fig 3A). The extent of Aurora B redistribution increased with the expression level of CB-INCENP (Fig. S2C), which suggests that CB-INCENP competes with endogenous INCENP for Aurora B binding. To determine the relationship between kinase position and phosphorylation in cells expressing CB-INCENP, we measured phosphorylation of the kinetochore-targeted sensor at chromosomes aligned on the metaphase plate. Phosphorylation increased as the expression level of CB-INCENP increased (Fig. 3B), while the average inter-kinetochore distances (1.4±0.3 μm, mean±stdev) remained constant (Fig. S2 D-E), which indicates that the increased phosphorylation was not due to reduced tension. Thus phosphorylation of a kinetochore substrate depends on the relative position of the kinase.
Figure 3.
Positioning Aurora B closer to the kinetochore leads to increased phosphorylation of kinetochore substrates. (A) U2OS cells expressing the indicated vsv-tagged INCENP fusion proteins were fixed and stained for Aurora B (green) and Hec1 (red). Scale bars 5 μm. (B) Hela cells were transfected with a phosphorylation sensor together with mCherry-tagged wt-INCENP or CB-INCENP as indicated, imaged live, and the YFP/TFP emission ratio was calculated. Cells were grouped based on levels of the INCENP fusion proteins at centromeres, calculated based on mCherry fluorescence intensity (wt-INCENP was not present at as high levels as CB-INCENP). Each bar represents an average over multiple cells, ≥30 kinetochores analyzed per cell.
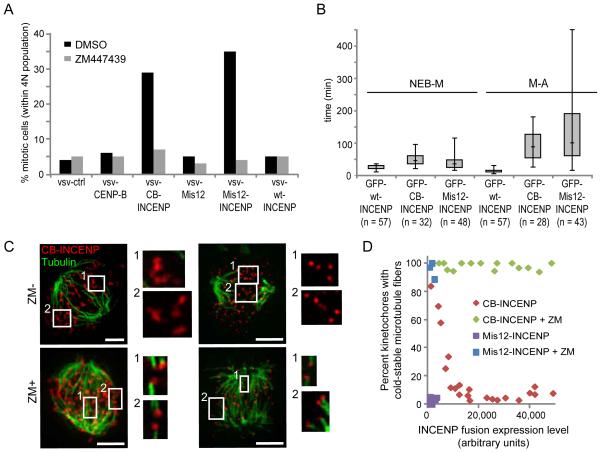
Because phosphorylation of endogenous Aurora B substrates at the kinetochore reduces microtubule binding (7-10), the spatial separation model predicts that repositioning the kinase closer to the kinetochore should impair the stabilization of bi-oriented attachments and activate the spindle checkpoint (2). Indeed, expression of either CB-INCENP or Mis12-INCENP induced a dramatic mitotic arrest, with a mitotic index >25% vs. ∼5% in control cells (Fig. 4A and Fig. S3A-B). The mitotic arrest was dependent on Aurora B kinase activity (Fig. 4A and Fig. S3A) and on the spindle checkpoint (Fig. S3B). Live imaging revealed that the timing of chromosome alignment increased from 20 min (median) in cells expressing wt-INCENP to 45 or 35 min for CB-INCENP or Mis12-INCENP, respectively. Moreover, the time spent in metaphase increased substantially from 10 min for wt-INCENP to 87.5 min or 100 min for CB-INCENP or Mis12-INCENP, respectively (Fig. 4B, Fig. S3C-D and movies S1-S3). Paired sister-chromatids frequently exited and re-entered the metaphase plate during the metaphase arrest (Fig. S3E, movie S4), followed by aberrant anaphases with lagging chromosomes (Fig. S3D-E, movies S1-S4). In addition, most metaphase cells contained at least one kinetochore positive for Mad1 (Fig. S4). Thus microtubules can sufficiently attach to allow congression, but stabilization of bi-oriented attachments is impaired when Aurora B is positioned close to the kinetochore.
Figure. 4.
Positioning Aurora B closer to the kinetochore activates the spindle checkpoint and destabilizes kinetochore-microtubule attachments. (A) U2OS cells expressing the indicated proteins were released from a G1/S block with or without ZM447439 for 14 hrs. The mitotic index was determined by propidium iodide/MPM2 mAb labeling and FACS analysis. One representative experiment out of 3 is shown. (B) Box-and-whisker plots (median, interquartile range, full range) showing time from nuclear envelope breakdown to metaphase (NEB-M) and from metaphase to anaphase (M-A) of U2OS cells expressing the indicated INCENP fusion proteins. (C-D) Hela cells expressing vsv-tagged CB-INCENP or Mis12-INCENP were treated with or without ZM447439, then analyzed for cold-stable microtubules (green) and vsv immunofluorescence (red). Brightness of vsv-Mis12-INCENP staining is enhanced relative to vsv-CB-INCENP so that it is visible. The percent of kinetochores with cold-stable microtubules was determined and plotted vs. expression level of the INCENP fusion proteins (D). Each data point represents >80 kinetochores from one cell.
To test directly whether repositioning Aurora B prevents the stabilization of kinetochore microtubules, we examined cold-stable microtubules. Kinetochore fibers remain intact at 4 °C, while most other microtubules in the cell depolymerize (22). The number of kinetochores with cold-stable attachments decreased as CB-INCENP expression increased, and at high CB-INCENP levels, where the kinetochore-targeted sensor was highly phosphorylated (Fig. 3B), very few kinetochores had cold-stable microtubule fibers (Fig. 4C-D). Kinetochores also lacked cold-stable microtubule fibers in cells expressing detectable Mis12-INCENP (Fig. 4C-D), while fibers were stable in cells expressing wt-INCENP (Fig. S3F). Aurora B inhibition stabilized kinetochore microtubules in cells expressing either CB-INCENP or Mis12-INCENP (Fig. 4C-D), which indicates that the destabilization depends on increased kinase activity at the kinetochore. Thus repositioning Aurora B closer to the kinetochore leads to both increased phosphorylation of kinetochore substrates (Fig. 3B) and destabilization of kinetochore microtubules (Fig. 4).
How bi-oriented attachments are selectively stabilized by tension across the centromere is a long-standing question. Our findings support a mechanism linking tension to phosphorylation of Aurora B substrates at the kinetochore and regulation of kinetochore-microtubule stability. In the absence of tension, kinetochore substrates are phosphorylated because they are in close proximity to Aurora B at the inner centromere. Phosphorylation reduces the affinity of these substrates for microtubules (7-10), so attachments are destabilized. Tension is exerted when spindle microtubules pull bi-oriented sister kinetochores in opposite directions, away from the inner centromere, so that kinetochore substrates are dephosphorylated, which increases affinity for microtubules and stabilizes attachments. Many biological systems that respond to mechanical forces do so through protein conformational changes, such as those induced in mechanosensitive ion channels (23). We present another strategy by which physical forces can be converted into biochemical changes in the cell, through modulation of the spatial separation of a kinase from its substrate.
Supplementary Material
Acknowledgments
We thank T.M. Kapoor (Rockefeller University) and R.H. Medema (UMC Utrecht) for their support during the initiation of these studies. We also thank B.E Black, G. Kops, and R.H. Medema for critical reading of the manuscript and helpful comments, and K.V. Le and T. Chiang for assistance with preparing reagents. This work was supported by grants from the American Cancer Society (IRG-78-002-30, MAL), the National Institute of Health (GM083988, MAL), the Searle Scholars Program (MAL), and the Netherlands Organization for Scientific Research (Vidi 917.66.332, SMAL).
Footnotes
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and notes
- 1.Tanaka TU. Chromosoma. 2008;117:521. doi: 10.1007/s00412-008-0173-5. [DOI] [PubMed] [Google Scholar]
- 2.Nicklas RB. Science. 1997;275:632. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 3.Biggins S, Murray AW. Genes Dev. 2001;15:3118. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka TU, et al. Cell. 2002;108:317. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 5.Hauf S, et al. J Cell Biol. 2003;161:281. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Nat Cell Biol. 2004;6:232. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 7.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. Cell. 2006;127:983. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 8.DeLuca JG, et al. Cell. 2006;127:969. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 9.Ciferri C, et al. Cell. 2008;133:427. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gestaut DR, et al. Nat Cell Biol. 2008;10:407. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeitlin SG, Shelby RD, Sullivan KF. J Cell Biol. 2001;155:1147. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ditchfield C, et al. J Cell Biol. 2003;161:267. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruchaud S, Carmena M, Earnshaw WC. Nat Rev Mol Cell Biol. 2007;8:798. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 14.Andrews PD, et al. Dev Cell. 2004;6:253. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 15.Fuller BG, et al. Nature. 2008;453:1132. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pluta AF, Saitoh N, Goldberg I, Earnshaw WC. J Cell Biol. 1992;116:1081. doi: 10.1083/jcb.116.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheeseman IM, Desai A. Nat Rev Mol Cell Biol. 2008;9:33. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 18.Kapoor TM, Mayer TU, Coughlin ML, Mitchison TJ. J Cell Biol. 2000;150:975. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waters JC, Chen RH, Murray AW, Salmon ED. J Cell Biol. 1998;141:1181. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams RR, et al. Curr Biol. 2000;10:1075. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- 21.Eckley DM, Ainsztein AM, Mackay AM, Goldberg IG, Earnshaw WC. J Cell Biol. 1997;136:1169. doi: 10.1083/jcb.136.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rieder CL. Chromosoma. 1981;84:145. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- 23.Orr AW, Helmke BP, Blackman BR, Schwartz MA. Dev Cell. 2006;10:11. doi: 10.1016/j.devcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.