Abstract
An increase in extracellular Ca2+ induces the nuclear localization of the Crz1 transcription factor and the activation of target genes in yeast. A recent study indicates that nuclear entry occurs in short stochastic bursts that are unsynchronized within the population of cells. The frequency but not the amplitude of the bursts is controlled by Ca2+. Modulation of the frequency of the burst coordinates aspects of expression of Crz target genes.
Signaling pathways must coordinate the activities of dozens or even hundreds of genes in response to extrinsic stimuli. Activation of these genes, in turn, often sets off cascades of downstream gene induction, which can give rise to permanent changes, such as cell fate determination, or transient accommodation to some external stimulus. Keeping such signal transduction cascades advancing in a predictable way in the face of a host of concurrent influences must be difficult. What mechanisms assure the coordinate transcription of large groups of genes that are regulated by a specific signaling pathway?
A recent study in yeast suggests that one way of coordinating the early events in these gene induction cascades is by modulating the frequency of small bursts of nuclear localization of transcription factors. By visualizing the nuclear localization of green fluorescent protein (GFP)–linked Crz1 (Fig. 1A) in single budding yeast cells, Cai et al. found that this calcium- and calcineurin-sensitive transcription factor moved into the nucleus in a synchronized manner about 15 to 20 min (τdelay) after extracellular addition of Ca2+ (1). Subsequently, Crz1 exited and moved back into the nucleus in stochastic, short, un-synchronized bursts of nuclear translocation events, each lasting about 2 min, during the time that extracellular Ca2+ was increased. Crz1 exits the nucleus by some undefined but rapid mechanism. The frequency, but not the amplitude, of these short nuclear bursts of Crz1 was responsive to the concentration of extracellular Ca2+, such that burst frequency increased with increased extracellular Ca2+. These bursts were independent of other cellular processes, such as cell cycle, and occurred without directly preceding spikes in intracellular Ca2+ concentration. The authors also examined localization of two other yeast transcription factors, the stress-responsive yeast transcription factor Msn2 and the glucose responsive repressor Mig1, and found similar bursts of nuclear localization. Both of these transcription factors had previously shown nuclear localization bursts (2, 3); however, in the Cai et al. study, Msn2 and Mig1 bursts were found to be Ca2+-independent and un-correlated with Crz1 bursts.
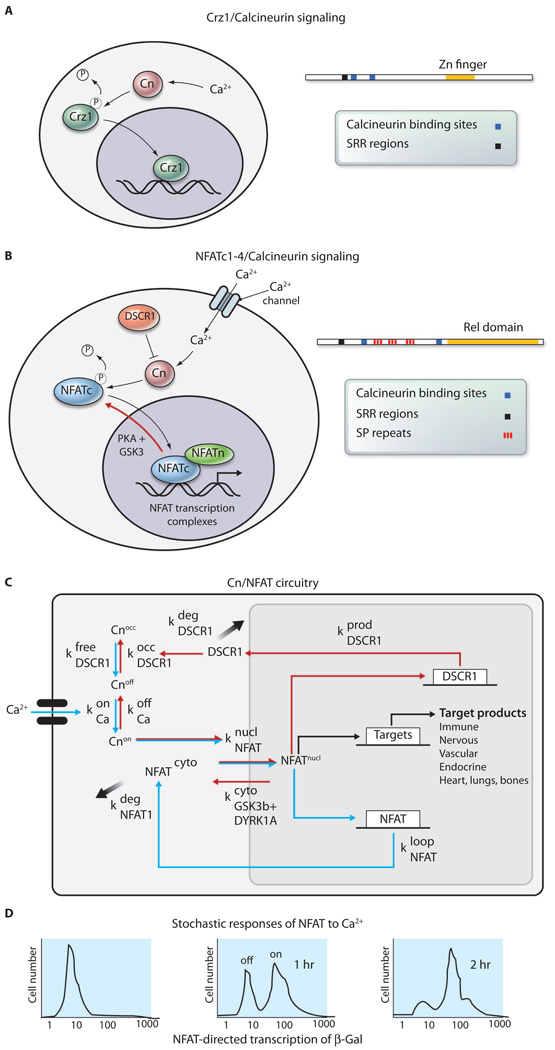
Fig. 1.
(A) Crz1 enters the nucleus in short stochastic bursts lasting about 2 min after undergoing dephosphorylation in the cytoplasm by the calcium-activated phosphatase calcineurin (Cn) (1, 19). The mechanism through which Crz1 exits the nucleus is not clear. (B) NFATc proteins are similar to Crz1 in that they are dephosphorylated by calcineurin on similar motifs and enter the nucleus, where they combine with other proteins to form complexes on the regulatory regions of target genes (16). The SRR is the serine-rich region, which is multiply phosphorylated. (C) Feedback loops in the calcineurin/NFAT signaling pathway predict oscillation with certain parameters (such as concentrations of the proteins) of the pathway that might be achieved in specific tissues. k, reaction constant. Occ refers to occluded or inactive state for calcineurin. (D) NFAT target genes are activated in a stochastic manner with the probability of being in the on state determined by time after stimulation with Ca2+ (9).
The expression of Crz1 target genes (both a synthetic Crz1-responsive reporter construct as well as an endogenous Crz1 target gene) was proportional to the burst frequency of Crz1 nuclear translocation, and protein expression followed the exposure to high extracellular Ca2+ concentrations with a 1-and-1/2-hour delay. Removal of Crz1 from the nucleus between bursts appeared to be incomplete: The total cellular distribution went from highly nuclear to diffuse. However, the changes were large enough that a substantial number of target promoters must have been stripped of the transcription factor yet seemed to “remember” that it was there. The mechanisms involved in this memory mechanism are not clear, but could involve such processes as the formation of partially assembled preinitiation complexes or chromatin remodeling.
Modeling this system with no information about the mechanisms producing rapid exit of Crz1 or the nature of the events taking place during the delay until Crz1 target genes are activated is challenging. Nevertheless, the authors then mathematically modeled the transcriptional response to the changes in nuclear burst frequency and found that frequency modulation (FM) regulation of nuclear translocation bursts would permit coordinated transcriptional regulation of target genes of a signaling pathway in a manner that is independent of the input function of the target gene promoter.
To test their prediction, the authors built synthetic reporter genes, which were regulated by increasing numbers of calcineurin-dependent response elements (CDREs), and also studied the behavior of endogenous fluo-rescently tagged calcineurin and Crz1 target genes. They found that the synthetic reporter genes as well as many of the tested endogenous target genes are coordinated with each other by FM regulation of Crz1 nuclear bursts, despite the fact that these genes must be regulated by several transcription factors acting in an enhancesome as well as being controlled by a number of different pathways in addition to calcium-calcineurin. The 90-min delay before target gene activation is perplexing because genes such as fos are activated and protein appears within 15 min after serum stimulation. Does this long delay imply that brief intracellular Ca2+ bursts produce an accumulation of some critical intermediate at the promoter that must achieve a specific level before firing? Does it reflect a higher-order assembly that would critically affect the mathematical modeling? Or could the long delay be due to the requirement for another transcription factor at the promoter? If the delay simply reflects the insensitivity of the GFP assay that the authors used, they will need to adapt a different assay for accurate modeling.
Nonetheless, frequency modulation could provide a way of coordinating the first wave of gene expression after an environmental stimulus (Fig. 2). Despite the large number of vertebrate or insect transcription factors that have been tagged with GFP and observed over long and short time courses, there seem to be few if any that show these rapid bursts of nuclear localization. However, some studies using cultured mammalian cell lines have found asynchronous oscillations in nuclear localization of three transcription factors [p53, nuclear factor κB (NF-κB), and Hes1] with a much longer time period (2 to 3 hours) (4–8). In the case of NF-κB, inhibition of oscillatory nuclear shuttling blocked transcription from a NF-κB–dependent promoter (6).
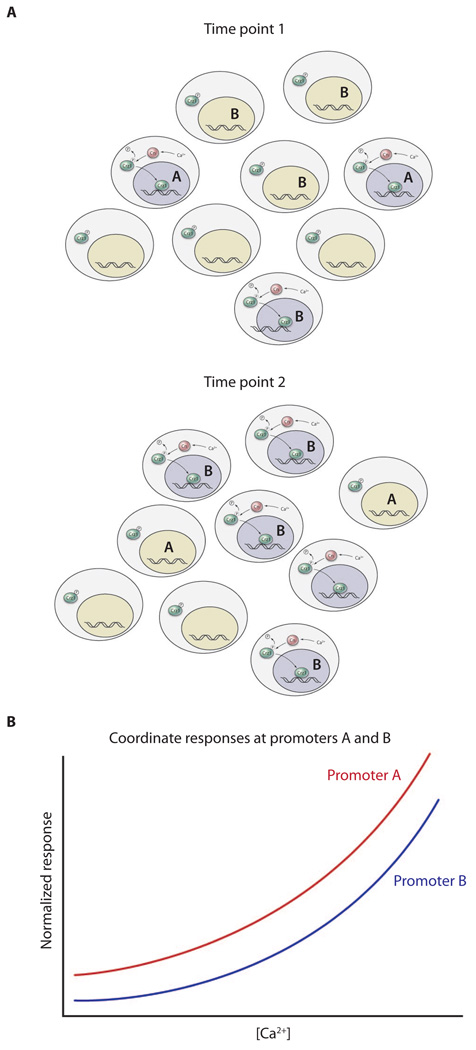
Fig. 2.
FM coordination. A field of cells undergoing brief stochastic bursts of Ca2+ entry (A) are brought to coordinated behavior with proportional target gene expression. Gray nuclei in (A) represent bursts of Crz1 nuclear localization; green nuclei are those in which Crz1 bursts are not taking place. (B) Both promoters show similar responses to Ca2+ concentration.
The first transcription factors shown to function in a quantal or stochastic manner, in that they are either in an “on” or “off” state, were the nuclear factor of activated T cells (NFAT) (9) and the glucocorticoid receptor (10). The NFATc family of transcription factors in vertebrates, which are necessary for lymphocyte activation as well as neural, cardiovascular, bone, epithelial, neural crest, lymphocyte, muscle, and lung development, are regulated by calcineurin activity (11–15) like yeast Crz1. Calcium influx in response to numerous receptors and ligands activates calcineurin, leading to the dephosphorylation and nuclear entry of the NFATc proteins (16). There, they assemble into NFAT transcription complexes on the promoters of target genes (Fig. 1B). Although Crz1 is not homologous to NFATc, the calcineurin binding site and some of the motifs that are dephosphorylated by calcineurin in Crz1 and the NFATc proteins are similar (Fig. 1, A and B) (17–19).
Activation of NFAT-dependent transcription in lymphocytes is sensitive to Ca2+ oscillation frequencies (20–22). In neurons, NFATc translocation to the nucleus is greater in response to low-frequency stimuli (5 Hz) than in response to high-frequency stimulation (23). During muscle development, NFAT activity is induced by electrostimulation with a tonic low-frequency impulse pattern, mimicking the firing pattern of slow motor neurons, but not with a phasic high-frequency pattern typical of fast motor neurons (24–27). NFATc proteins move into the nucleus quickly (τdelay = 2 to 10 min) after a Ca2+ stimulus, and single-cell assays in lymphocytes have shown that promoters are stochastically distributed in either off or on status, with the intensity of the Ca2+ signal determining the probability of being on (Fig. 1D) (9).
The NFATc pathway also involves negative feedback control at the level of the calcineurin inhibitor, DSCR1 (Down syndrome critical region 1), also called MCIP1 (modulatory calcineurin-interacting protein-1) or RCAN (regulator of calcineurin) (28–30), and positive feedback control on the NFATc1 and c4 promoters (Fig. 1, B and C) (31). The interplay of these feedback loops could produce oscillations in nuclear localization that depend on the concentrations of the proteins in the circuit in a specific cell type (Fig. 1C). However, GFP-tagged NFATc proteins in lymphocytes and neurons accumulate continuously in the nucleus over 2 to 30 min in response to an increase of intracellular Ca2+ concentration and do not show bursts of nuclear localization or detectable oscilliations (23, 32).
NFATc proteins are rapidly excluded from the nucleus through sequential phosphorylation by DYRK1a [dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A] and GSK3 (glycogen synthase kinase 3) (23, 31, 33, 34). Mathematical modeling of the NFAT genetic regulatory circuit indicated that calcineurin-NFAT signaling is robust under most conditions encountered during development.However, it is particularly sensitive to the gene dosage of two negative regulators of this signaling pathway, DSCR1 (RCAN1) and DYRK1a, which are located on chromosome 21 (31). Increased expression of DYRK1a and DSCR1, which cooperatively reduce nuclear occupancy of the NFATc proteins, destabilizes the NFAT regulatory circuit and leads to the development of many of the phenotypic features of human Down syndrome (31).
The advantage of FM regulation of nuclear localization of transcription factors appears to be in enabling coordination of the activation of large numbers of target genes after a stimulus (Fig. 2). The disadvantage might be that it reduces the information that could be encoded in the frequency of Ca2+ spikes, which has, for example, been shown to be important in the development of some NFAT-dependent cell types. It is surprising that similar rapid bursts of nuclear localization have not been seen in mammal, fly, or worm cells. Could it be that they have been missed? The studies of Cai et al. indicate that a careful reinspection is worthwhile. Another avenue for addressing this question would be “reverse mathematical modeling”: beginning by assuming that there are advantages to coordinating the first wave of gene activation after nuclear entry of a transcription factor, dreaming up mathematical circuits for achieving this, and then testing each dreamed-up possibility by searching for corresponding biological systems and observations to support it. Critical to validation of FM coordination is whether mutations that affect the oscillatory dynamics of transcription factors underlie developmental defects, human disease, or both. Perhaps Down syndrome, in which the balance of nuclear localization of the NFATc family members is tipped toward the cytoplasm by the increased dosage of the calcineurin inhibitor DSCR1and the NFAT exporter Dyrk1a (31), is worth careful study in this regard.
References
- 1.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485–490. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacquet M, Renault G, Lallet S, De Mey J, Goldbeter A. Oscillatory behavior of the nuclear localization of the transcription factors Msn2 and Msn4 in response to stress in yeast. Sci. World J. 2003;3:609–612. doi: 10.1100/tsw.2003.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeVit MJ, Waddle JA, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lev Bar-Or R, Maya R, Segel LA, Alon U, Levine AJ, Oren M. Generation of oscillations by the p53-Mdm2 feedback loop: A theoretical and experimental study. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11250–11255. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IκB-NF-κB signaling module: Temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 6.Nelson DE, Ihekwaba AEC, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MRH. Oscillations in NF-κB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 7.Hirata H, Yoshiura S, Ohtsuka T, Bessho Y, Harada T, Yoshikawa K, Kageyama R. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- 8.Monk NA. Oscillatory expression of Hes1, p53, and NF-κB driven by transcriptional time delays. Curr. Biol. 2003;13:1409–1413. doi: 10.1016/s0960-9822(03)00494-9. [DOI] [PubMed] [Google Scholar]
- 9.Fiering S, Northrop JP, Nolan GP, Mattila PS, Crabtree GR, Herzenberg LA. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Dev. 1990;4:1823–1834. doi: 10.1101/gad.4.10.1823. [DOI] [PubMed] [Google Scholar]
- 10.Ko MS, Nakauchi H, Takahashi N. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 1990;9:2835–2842. doi: 10.1002/j.1460-2075.1990.tb07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. Identification of a putative regulator of early T cell activation genes. Science. 1988;241:202–205. [PubMed] [Google Scholar]
- 12.Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T cell activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- 13.Northrop JP, Ho SN, Chen L, Thomas DJ, Timmerman LA, Nolan GP, Admon A, Crabtree GR. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature. 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- 14.Crabtree GR. Generic signals and specific outcomes: Signaling through Ca2+, calcineurin, and NF-AT. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 15.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: Regulation and function. Annu. Rev. Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 16.Flanagan WM, Corthésy B, Bram RJ, Crabtree GR. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. 1991;352:803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- 17.Ho SN, Thomas DJ, Timmerman LA, Li X, Francke U, Crabtree GR. NFATc3, a lymphoid-specific NFATc family member that is calcium- regulated and exhibits distinct DNA binding specificity. J. Biol. Chem. 1995;270:19898–19907. doi: 10.1074/jbc.270.34.19898. [DOI] [PubMed] [Google Scholar]
- 18.Beals CR, Clipstone NA, Ho SN, Crabtree GR. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- 19.Stathopoulos AM, Cyert MS. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 1997;11:3432–3444. doi: 10.1101/gad.11.24.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 21.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 22.Li W-H, Llopis J, Whitney M, Zlokarnik G, Tsien RY. Cell-permeant caged InsP3 ester shows that Ca2+ spike frequency can optimize gene expression. Nature. 1998;392:936–941. doi: 10.1038/31965. [DOI] [PubMed] [Google Scholar]
- 23.Graef IA, Mermelstein PG, Stankunas K, Neilson JR, Deisseroth K, Tsien RW, Crabtree GR. L-type calcium channels and GSK-3 regulate the activity of NF-ATc4 in hippocampal neurons. Nature. 1999;401:703–708. doi: 10.1038/44378. [DOI] [PubMed] [Google Scholar]
- 24.McCullagh KJ, Calabria E, Pallafacchina G, Ci-ciliot S, Serrano AL, Argentini C, Kalhovde JM, Lømo T, Schiaffino S. NFAT is a nerve activity sensor in skeletal muscle and controls activity-dependent myosin switching. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10590–10595. doi: 10.1073/pnas.0308035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, Olson EN. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J. Biol. Chem. 2000;275:4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- 27.Serrano AL, Murgia M, Pallafacchina G, Calabria E, Coniglio P, Lømo T, Schiaffino S. Calcineurin controls nerve activity-dependent specification of slow skeletal muscle fibers but not muscle growth. Proc. Natl. Acad. Sci. U.S.A. 2001;98:13108–13113. doi: 10.1073/pnas.231148598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Rothermel B, Vega RB, Frey N, McKinsey TA, Olson EN, Bassel-Duby R, Williams RS. Independent signals control expression of the calcineurin inhibitory proteins MCIP1 and MCIP2 in striated muscles. Circ. Res. 2000;87:E61–E68. doi: 10.1161/01.res.87.12.e61. [DOI] [PubMed] [Google Scholar]
- 29.Kingsbury TJ, Cunningham KW. A conserved family of calcineurin regulators. Genes Dev. 2000;14:1595–1604. [PMC free article] [PubMed] [Google Scholar]
- 30.Rothermel BA, McKinsey TA, Vega RB, Nicol RL, Mammen P, Yang J, Antos CL, Shelton JM, Bassel-Duby R, Olson EN, Williams RS. Myocyte-enriched calcineurin-interacting protein, MCIP1, inhibits cardiac hypertrophy in vivo. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3328–3333. doi: 10.1073/pnas.041614798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arron JR, Winslow MM, Polleri A, Chang C-P, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, Yamasaki N, Miyakawa T, Francke U, Graef IA, Crabtree GR. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- 32.Timmerman LA, Clipstone NA, Ho SN, Northrop JP, Crabtree GR. Rapid shuttling of NF-AT in discrimination of Ca2+ signals and immunosuppression. Nature. 1996;383:837–840. doi: 10.1038/383837a0. [DOI] [PubMed] [Google Scholar]
- 33.Beals CR, Sheridan CM, Turck CW, Gardner P, Crabtree GR. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 34.Gwack Y, Sharma S, Nardone J, Tanasa B, Iuga A, Srikanth S, Okamura H, Bolton D, Feske S, Hogan PG, Rao A. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature. 2006;441:646–650. doi: 10.1038/nature04631. [DOI] [PubMed] [Google Scholar]