Abstract
One of the long-term goals of synthetic biology is to reliably engineer biological systems that perform human-defined functions. Currently, researchers face several scientific and technical challenges in designing and building biological systems, one of which is associated with our limited ability to access, transmit, and control molecular information through the design of functional biomolecules exhibiting novel properties. The fields of RNA biology and nucleic acid engineering, along with the tremendous interdisciplinary growth of synthetic biology, are fueling advances in the emerging field of RNA programming in living systems. Researchers are designing functional RNA molecules that exhibit increasingly complex functions and integrating these molecules into cellular circuits to program higher-level biological functions. The continued integration and growth of RNA design and synthetic biology presents exciting potential to transform how we interact with and program biology.
Introduction
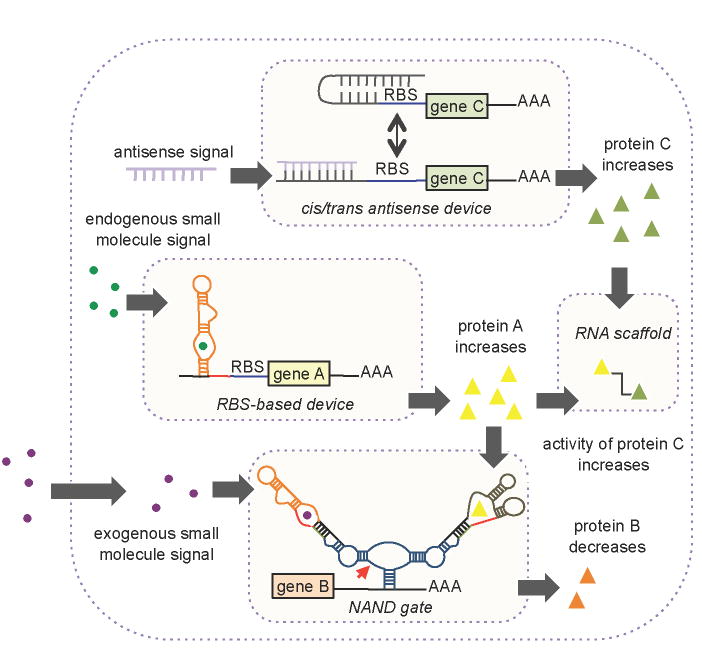
The engineering of biological systems that process information, materials, and energy holds great promise for developing solutions to many global challenges, including renewable energy production, material synthesis, and medical advancement. Our ability to understand and routinely engineer biological systems is limited by the tools available to broadly access, transmit, and control molecular information encoded in the various properties of biomolecules. Synthetic biology is a rapidly growing interdisciplinary field that involves the application of engineering principles to the design and construction of synthetic biological systems. Core objectives of modern synthetic biology have focused on the engineering of complex biological systems (Basu et al., 2005; Ro et al., 2006) and the development of engineering frameworks that support the reliable programming of biological function, including abstraction, standardization, and modularity (Endy, 2005). Within the context of these objectives, we review the emerging field of RNA programming in living systems, which is itself founded on the more mature fields of RNA biology, in vitro nucleic acid engineering and computing (Joyce, 2007; Seeman, 2005). Advances in RNA synthetic biology are providing the foundational tools and knowledge-base that will support broader efforts in encoding cellular information processing and control operations in synthetic RNA molecules and integrating these engineered components with biological networks to program higher-level biological function (Figure 1).
Figure 1.
The Integration of RNA Parts and Devices as Information Processing and Control Components into Engineered Biological Systems
RNA provides a programmable molecular substrate that exhibits diverse function
Biological functions are encoded within the variety of biomolecules present in living organisms. The biological functions of one class of nucleic acids, RNA, can be grouped into protein-coding and nonprotein-coding functions. Noncoding RNA exhibits diverse functional properties, including gene-regulatory, enzymatic, and ligand-binding properties. In addition, RNA molecules can exhibit structural flexibility, which enables them to dynamically adopt different conformations and thereby exhibit allosteric properties. RNA is composed of four nucleotide residues that interact through well-defined hydrogen-bond, base-stacking, and electrostatic interactions. RNA secondary structure is dictated by these hydrogen-bond interactions, allowing the development of folding programs that predict the secondary structures and associated free energies of RNA molecules (Mathews, 2006; Zuker, 2003). The energies involved in RNA folding are contributed to a lesser degree by tertiary structure, such that the relationship between RNA sequence, structure, and function is relatively accessible and predictable. These properties make RNA a powerful design substrate in synthetic biology.
RNA parts
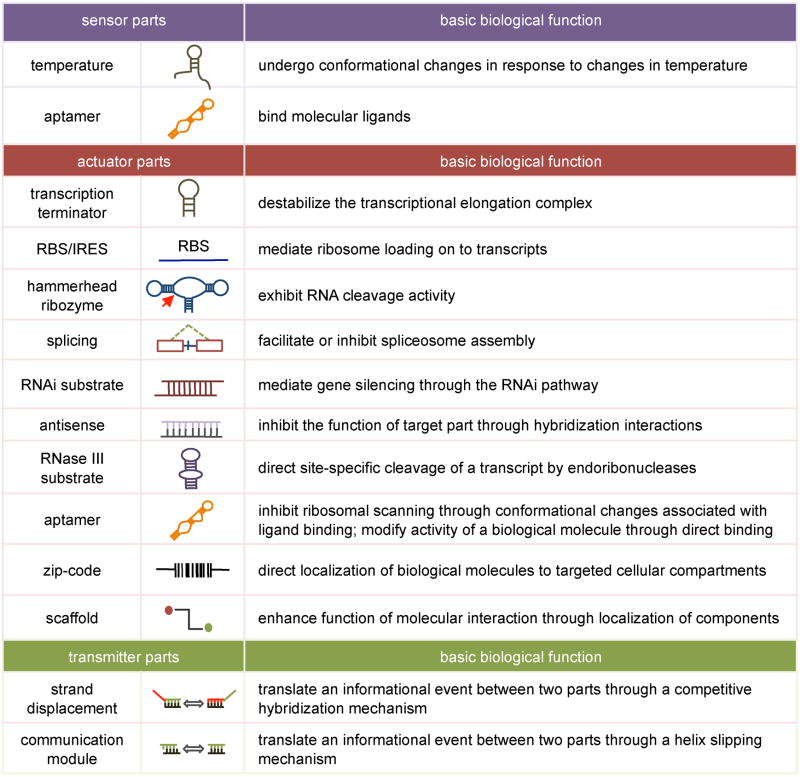
In engineering design, a part is a component that can perform a basic function. RNA parts are genetic components composed of RNA molecules that are capable of performing basic biological functions such as gene regulation, directed conformation change, and ligand binding. RNA parts can be generally grouped into three categories based on function: sensors, actuators, and transmitters (Table 1).
Table 1.
Examples of RNA Parts and their Associated Functions
|
RNA sensors
Sensors are parts that detect signals. RNA sensors detect diverse signals, such as temperature and molecular ligands, through various binding events, including hybridization and tertiary interactions. The binding event encoded in an RNA sensor is generally transduced to an actionable event such that RNA sensors are typically coupled to other RNA parts.
Temperature sensors
RNA senses temperature through the temperature-dependent nature of hybridization interactions. Naturally-occurring RNA sequences can adopt conformations that are sensitive to small temperature changes (Lai, 2003). Generally, temperature-sensitive RNA parts exhibit less structured conformations as a function of increasing temperature (Narberhaus et al., 2006). Effort has been directed toward engineering and refinement of RNA temperature sensors through selection/screening and rational design approaches. RNA temperature sensor parts have been developed in combination with different RNA actuator parts to link changes in temperature to gene-regulatory events (Neupert et al., 2008; Waldminghaus et al., 2008; Yoshimatsu and Nagawa, 1989).
Molecular sensors
RNA generally senses molecular signals through direct binding interactions with the molecular target. RNA aptamers are the most common class of RNA sensor parts capable of binding ligands with high affinities and specificities (Hermann and Patel, 2000). The flexibility of RNA as a sensor is highlighted by the diverse ligands against which RNA aptamers have been generated, including carbohydrates, proteins, and small molecules (Hermann and Patel, 2000). RNA aptamers are typically generated de novo using an iterative in vitro selection strategy or SELEX (Systematic Evolution of Ligands by EXponential enrichment) (Ellington and Szostak, 1990; Tuerk and Gold, 1990). In addition, RNA can bind nucleic acids through Watson-Crick basepairing, enabling the engineering of RNA sensors to nucleic acid ligands.
RNA actuators
Actuators are parts that control a process or event. RNA actuators control the activity of other biological molecules, thereby affecting responses in living systems, and exhibit a variety of functions such as gene-expression regulation, posttranslational regulation, and directed localization.
Gene-expression actuators
The most predominant class of RNA actuators are parts that regulate gene expression, where actuation can occur through different mechanisms, including transcription, translation, splicing, and stability. These actuators can exhibit their regulatory effects either in cis, when the part is embedded within the target transcript, or in trans, when the part acts on a separate target RNA.
Transcription termination actuators
In prokaryotes, transcription attenuation is actuated by cis-acting RNA parts called transcription terminators (Henkin, 2000). Intrinsic terminators are the most commonly used class of termination parts and are composed of GC-rich stem loops followed by a series of U residues. Transcription termination parts are largely taken from nature and little engineering has been done to refine or enhance their function. These parts are implemented downstream of coding regions and their function is relatively insensitive to the distance from the stop codon. Researchers have implemented multiple transcription termination parts in series to improve the activity exhibited from a single part (Shetty et al., 2008).
Translation initiation actuators
In prokaryotes, translation is initiated at a cis-acting RNA part called a ribosome binding site (RBS) that mediates ribosome loading onto a transcript (Kozak, 1999). RBSes consist of several consensus nucleotides that hybridize with the 3′ end of the 16S rRNA. In contrast, ribosome loading in eukaryotes is initiated at the 5′ end of a transcript through a 7-methyl guanosine cap that recruits the translation initiation machinery. While translation initiation typically occurs through this cap-dependent mechanism, translation can be initiated internally at a cis-acting RNA part called an internal ribosome entry site (IRES) (Hellen and Sarnow, 2001). IRESes range from short unstructured sequences, which facilitate translation initiation through hybridization with 18S rRNA, to large structured elements, which interact with the initiation machinery (Chappell et al., 2000; Kieft et al., 2002). RNA engineers have generated synthetic RBS and IRES sequences with varying activities (Owens et al., 2001; Rackham and Chin, 2005; Zhou et al., 2003). The activity of a RBS is determined by the number of interactions it forms with the 16S rRNA and its distance upstream of the start codon (Chen et al., 1994), whereas the activity of an IRES is less dependent on this distance and can be increased when linked in multiple copies (Chappell et al., 2000).
Catalytic actuators
Ribozymes are catalytic RNA parts that most commonly catalyze the cleavage and/or ligation of RNA molecules via phosphodiester bond rearrangement involving metal ions as cofactors (Doudna and Cech, 2002). The hammerhead ribozyme (hhRz) has been extensively utilized as a gene-regulatory part due to its small size, ease of design, and rapid kinetics. The hhRz motif contains a conserved 11 nt catalytic core encompassed by three stems (I, II, III) (Blount and Uhlenbeck, 2005), where interactions between the nucleotides in stem loops I and II are critical to intracellular activity (Khvorova et al., 2003).
RNA engineers have put significant effort into refining ribozymes for gene-regulatory functions. For example, synthetic trans-acting hhRzs have been constructed such that the trans-ribozyme sequence binds the target transcript through targeting arms to form a catalytically-active structure (Vaish et al., 1998). HhRzs have been implemented to actuate gene expression through directed cleavage in cis or in trans by targeting various regions within a transcript. For example, hhRzs inserted within the 3′ UTR of target transcripts have been shown to effectively down-regulate gene expression in eukaryotes (Khvorova et al., 2003; Meaux and Van Hoof, 2006). HhRz-directed cleavage and gene inhibition has also been shown within the 5′ UTR (Yen et al., 2004), although studies have indicated that implementation in this location in eukaryotes can result in non-specific inhibition of expression due to structural effects (Yim et al., 2000). Trans-acting hhRzs exhibit greater flexibility in targeting both translated and untranslated transcript regions (Kijima et al., 1998; Morino et al., 2000; Sakamoto et al., 1996).
Constitutive/alternative splicing actuators
In eukaryotes, a vast majority of genes consist of protein-coding (exons) and noncoding (introns) regions, in which the exons are joined in processes of constitutive or alternative splicing to produce single or multiple protein isoforms, respectively. Cis-acting RNA sequences direct splicing events through binding components of the spliceosome or other auxiliary protein factors that mediate spliceosome assembly (Maniatis and Tasic, 2002). RNA parts that actuate splicing events include 5′ and 3′ splice sites, branch point, and enhancer and silencer sequences. Little effort has been directed to the engineering, refinement, and utilization of RNA parts that actuate splicing events, as our understanding of the ‘splicing code’ is incomplete (Wang and Burge, 2008). However, researchers have utilized canonical splicing sequences to mediate splicing (Yoshimatsu and Nagawa, 1989) and generated enhancer and silencer sequences that can potentially be used as refined parts for regulating splicing patterns (Wang et al., 2004).
RNA interference-based actuators
Small interfering RNAs (siRNAs) and microRNAs (miRNAs) are gene-regulatory parts that silence gene expression through the RNA interference (RNAi) pathway (Meister and Tuschl, 2004). RNAi-mediated gene silencing is triggered by double-stranded RNAs (dsRNAs) that are introduced exogenously into cells or generated endogenously from primary miRNA transcripts (pri-miRNAs) (Meister and Tuschl, 2004). Pri-miRNAs are processed in the nucleus by the RNase III enzyme Drosha to precursor miRNAs (pre-miRNAs) (Lee et al., 2003), which are exported to the cytoplasm by Exportin-5 (Lund et al., 2004). Exogenously introduced dsRNAs and pre-miRNAs are processed in the cytoplasm by the RNase III enzyme Dicer (Bernstein et al., 2001). The cleaved RNAs are unwound and loaded into multi-protein complexes, where these RNA parts hybridize to target transcripts and direct cleavage or translational repression based on the degree of complementarity (Hammond et al., 2000; Hutvagner and Zamore, 2002).
Due to the efficacy and flexibility of RNAi-mediated gene silencing, RNA engineers have developed various types of synthetic RNAi substrates (Dykxhoorn et al., 2003). RNAi substrates can be expressed from RNA polymerase II or III promoters as siRNAs, short hairpin RNAs (shRNAs), pre-miRNAs, or pri-miRNAs (Brummelkamp et al., 2002; McManus et al., 2002; Miyagishi and Taira, 2002; Zeng et al., 2002). While design rules have been proposed to guide RNAi substrate design (Reynolds et al., 2004), typically many sequences must be screened to generate an effective RNAi substrate. Synthetic RNAi substrates are commonly designed to be fully complementary to their targets to induce cleavage, which is a more effective mechanism of gene silencing. Combinatorial targeting of multiple RNAi substrates to a single transcript has been used to increase gene silencing, detect cellular state, and implement more complex regulatory schemes (Brown et al., 2007; Rinaudo et al., 2007).
Antisense-based actuators
Antisense RNAs are single-stranded RNA parts that control the function of their target RNAs through a hybridization event (Green et al., 1986). Natural antisense RNAs have been characterized that actuate through a variety of mechanisms, including cleavage of antisense-target hybrids by dsRNA-cleaving enzymes and steric inhibition of gene expression machinery by the bound antisense (Crooke, 2004). Due to the simplicity of their design and gene-regulation potency, significant effort has been directed to the engineering and refinement of antisense RNA parts. Trans-acting antisense RNAs vary in length (~20–700 nt) and have been engineered to inhibit the expression of different target genes in various cellular systems (Bonoli et al., 2006; Bunch and Goldstein, 1989; Coleman et al., 1984). Alternatively, cis-acting antisense parts can be implemented to inhibit the function of other RNA parts. For example, an antisense part can be implemented in cis to hybridize to a terminator part to form an antiterminator that inhibits transcription termination (Henkin and Yanofsky, 2002). Cis-acting antisense parts are typically located within close proximity to their targets and are of sufficient length to disrupt target function (~10–20 nt).
RNase activity actuators
RNases are RNA processing enzymes that act on cellular RNAs through endoribonucleolytic or exoribonucleolytic activities (Alifano et al., 1994). Specific RNA parts are recognized and processed by cellular RNases. For example, Rnt1p, an RNase III enzyme in Saccharomyces cerevisiae, cleaves hairpins with conserved tetraloops (Chanfreau et al., 2000). As another example, RNase E is an Escherichia coli endoribonuclease that cleaves single-stranded AU-rich sequences (McDowall et al., 1994). Substrates for both RNases have been discovered in cellular transcripts, highlighting their functional roles in processing and degrading mRNA (Ge et al., 2005; Lundberg et al., 1990). Significant effort has been directed toward the engineering, refinement, and utilization of cis-acting RNA parts that actuate RNase activities to regulate gene expression. The length and location of such RNA parts will depend on the associated RNase and its intended gene-regulatory function. For example, stabilizing hairpin parts have been placed at the 5′ and 3′ ends of transcripts to inhibit RNase E and exoribonuclease activities, respectively (Carrier and Keasling, 1997). RNase activity actuation parts exhibiting varying activities have been generated through sequence and structural modification (Carrier and Keasling, 1999; Smolke et al., 2000).
Aptamer-based translational actuators
In addition to acting as sensors, RNA aptamers can function as cis-acting gene-regulatory parts. RNA aptamer-based translational actuators responsive to various small molecules have been embedded within the 5′ UTR of target eukaryotic transcripts (Harvey et al., 2002; Suess et al., 2003; Werstuck and Green, 1998). The aptamer sequence adopts a more structured conformation upon binding its ligand that inhibits ribosomal scanning, thereby reducing expression levels in a ligand-dependent manner. However, the general relationship between the relative location of the aptamer within the 5′ UTR and translational efficacy remains unclear, as other studies have indicated that secondary structures within this region inhibit translation (Pelletier and Sonenberg, 1985). It is possible that the ability of RNA aptamers to function as gene-regulatory parts is dependent on specific properties of the aptamer and its location relative to other RNA parts in the transcript. RNA aptamer-based translational actuators encode two functions, sensing and actuation, and are therefore an example of a double-coding part.
Posttranslational activity actuators
A second class of RNA actuators functions to regulate the activity of biological molecules. RNA aptamers can function as trans-acting RNA parts that actuate the activity of molecules posttranslationally through binding interactions. Binding of a synthetic RNA aptamer to its molecular ligand can interact with chemical moieties important to the functional activity of the ligand, resulting in inhibition (Famulok et al., 2001) or enhancement of activity (Babendure et al., 2003). RNA-based posttranslational activity actuators are another example of a double-coding part. Intramers are a class of RNA aptamer-based posttranslational activity actuators that are expressed intracellularly to inhibit the activity of various molecular ligands (Famulok et al., 2001). For example, intramers against viral proteins (Kim and Jeong, 2004; Nishikawa et al., 2003) have been shown to inhibit the activities of the target viral proteins in human cells, thereby reducing or inhibiting viral infectivity.
Localization actuators
A third class of RNA actuators functions to direct localization of biological molecules to different molecules, cellular compartments, or specific cells. Localization of mRNA is used to achieve higher local protein concentrations (Hazelrigg, 1998), whereas localization of gene-regulatory RNAs and transcripts can be used to achieve conditional or enhanced control of gene-regulatory functions (Lee et al., 1999). Sequences that bind protein factors responsible for localization effects, termed zip codes, can be used as cis-acting localization parts (Bassell et al., 1999). Localization strategies can also be performed by scaffold parts that bring molecular targets close together to enhance function associated with their interactions. Although such scaffolding functions are typically performed by proteins in nature (Pawson and Scott, 1997), the ability of RNA to bind molecular ligands supports its potential use as a scaffold part.
RNA localization parts have either been adapted from nature or generated as RNA aptamers to specific molecular targets. For example, heterologous transcripts harboring RNA zip-code parts within their 3′ UTRs have been localized to targeted compartments within the cytoplasm (Kislauskis et al., 1993). In addition, such RNA zip-code parts have been integrated with other RNA actuation parts to enhance gene-regulatory activities through colocalization. For example, the human α- and β-actin zip codes were engineered into trans-acting hhRzs and target genes, and the constructs harboring matched zip codes (α-α, β-β) exhibited enhanced colocalization and ribozyme-mediated inhibition of target expression (Lee et al., 1999). As another example, synthetic RNA scaffolds have been built linking multiple RNA aptamers to localize DNA-binding and transcriptional activation proteins to allow transcription initiation in yeast (Cassiday and Maher, 2001).
RNA transmitters
Transmitters are parts that send or convert signals between different components. RNA transmitters are RNA sequences that translate an informational event (such as signal detection by a sensor) from one RNA part to another through conformational changes, and thus are typically coupled to other RNA parts.
Communication module parts
Communication module parts are based on a single sequence that can bind to a partially complementary target strand in more than one way, where the energetics of strand binding allow the part to move between different bound states through a helix slipping event (Soukup and Breaker, 1999).
Strand displacement parts
Strand displacement parts are based on two sequences that compete for binding to a complementary target strand, where the energetics of each binding event allow the part to move between different bound states through a competitive hybridization event (Mandal and Breaker, 2004).
RNA devices
A device in engineering design is a combination of refined parts that can perform a human-defined function (Endy, 2005). RNA devices are composed of more than one functionally distinct RNA parts and are capable of performing programmed biological functions. Riboswitches are naturally-occurring counterparts of RNA devices that link molecular sensing and gene-regulatory functions (Mandal and Breaker, 2004). Riboswitches control gene expression in response to specific molecular inputs and act through diverse gene expression mechanisms (Breaker, 2008). While the majority of riboswitches respond to cellular metabolites, riboswitches that incorporate temperature sensors have also been characterized (Narberhaus et al., 2006). Riboswitches that incorporate multiple sensor and actuator domains through more complex architectures to perform higher-order regulatory functions, such as cooperativity and multi-input signal integration, have also been identified (Breaker, 2008).
Building on these natural examples, engineers have built a variety of RNA devices that process and transmit molecular inputs to targeted gene expression outputs in different cellular systems (Isaacs et al., 2006; Suess and Weigand, 2008). Engineers have adopted various design strategies to build RNA devices from combinations of diverse parts and have employed both screening/selection and rational design approaches.
RNA devices based on the direct coupling of sensor and actuator parts
A direct coupling device design strategy is one that links the sensor and actuator parts directly without the inclusion of a distinct transmitter part (Figure 2A). In this strategy, information (in the form of input binding to the sensor) is typically transmitted between the two parts through conformational changes associated with ligand binding to the aptamer that are specific to each aptamer-ligand pair. As a result, the coupling of parts into a functional RNA device in this strategy can be dependent on mechanistic properties of the sensor and actuator.
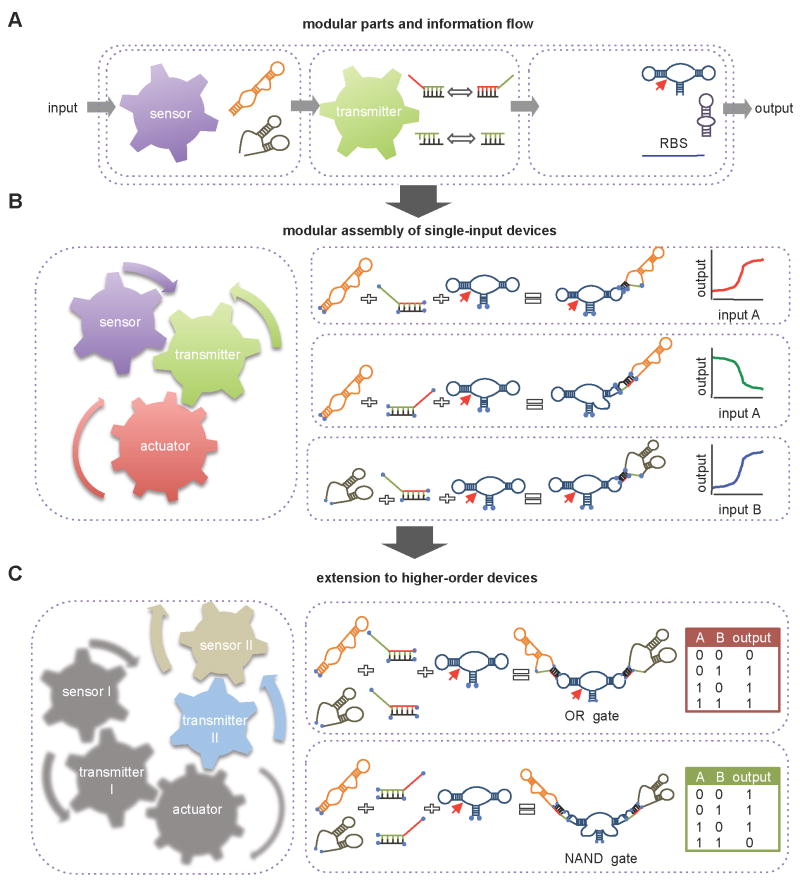
Figure 2. RNA Device Design Strategies.
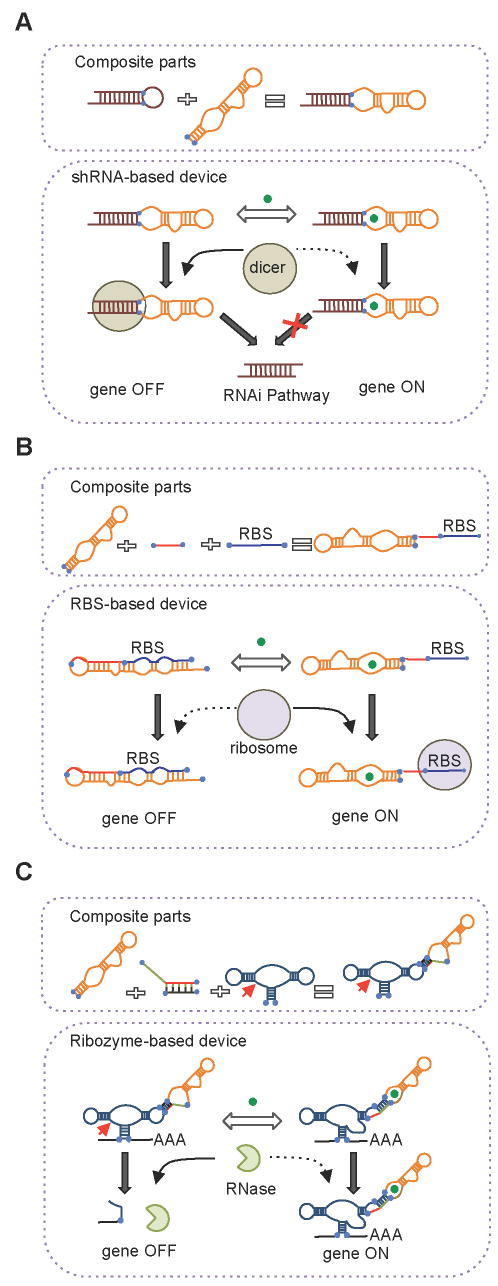
(A) A device design strategy based on the direct coupling of sensor and actuator parts.
(B) A device design strategy based on the integration of a transmitter part between the sensor and actuator parts.
(C) A device design strategy based on the integration of a transmitter part that enables the modular assembly of sensor, actuator, and transmitter parts.
Early examples of the direct coupling strategy were implemented based on the replacement of a portion of the actuator with the sensor. For example, a ribozyme-based RNA device was built by replacing a stem in the group I self-splicing intron from the bacteriophage T4 thymidylate synthase gene with the theophylline aptamer (Thompson et al., 2002). The device was integrated into its native gene and demonstrated theophylline-dependent splicing and regulation of cell growth in an E. coli thymidine auxotroph strain. In another example (Buskirk et al., 2004), an input-dependent transcriptional activator activity was built by replacing a stem in a RNA part that functions as a transcriptional activator (Buskirk et al., 2003) with the tetramethylrosamine (TMR) aptamer (Grate and Wilson, 1999). A functional TMR-responsive device was generated by randomizing several nucleotides in the region connecting the two parts and selecting for TMR-responsive sequences, which allowed the conformational change associated with TMR binding to activate the transcriptional activator.
Other examples are based on the direct coupling of the sensor and actuator sequences. In one example, the theophylline aptamer was directly coupled to a shRNA actuator (An et al., 2006) (Figure 2A). Dicer cleavage of the shRNA sequence was inhibited by theophylline, resulting in small molecule regulation of RNAi-based gene silencing. The device function was shown to be very sensitive to the distance between the Dicer cleavage site and ligand binding site, as changes of even one basepair abolished function. RNA devices have also been shown to effectively regulate splicing and alternative splicing through this strategy by coupling small molecule aptamers to 5′ splice site and branch point sequences (Kim et al., 2008; Weigand and Suess, 2007).
RNA devices based on integration of a distinct information transmission function
A second type of device design strategy is one that links the sensor and actuator parts through a separate transmitter part (Figure 2B). The transmitter part typically translates information between the sensor and actuator through a change in secondary structure. In this way information is transmitted through a mechanism that is itself independent of input binding to the sensor, such that the molecule adopts at least two different conformations associated with an active or inactive sensor. Input binding to the conformation harboring an active sensor shifts the distribution between these conformations to favor the input-bound state. By introducing an information transmission function that is distinct from the input binding event, devices can achieve greater flexibility in the coupling of actuator and sensor parts.
Several examples of RNA devices that link aptamers to translation initiation parts through linker sequences that perform information transmission functions have been described. The actuator of these devices is regulated by modulating its accessibility to translational machinery through hybridization interactions with the actuator sequence. For example, RBS-based devices were developed by coupling the theophylline aptamer to a RBS through a randomized linker region (Figure 2B). The linker region was varied in length and theophylline-responsive devices were screened through colorimetric, flow cytometry, and cell motility assays in E. coli by altering the gene regulatory output (Desai and Gallivan, 2004; Lynch and Gallivan, 2008; Topp and Gallivan, 2008). The devices function through modulating ribosomal accessibility to the RBS via a strand displacement mechanism, where the RBS was partially basepaired with the aptamer in a gene OFF state or released from basepairing in the gene ON state in the input-bound conformation. The aptamer sequence in this device design strategy encodes for both ligand binding and antisense functions such that the sequences of the sensor and actuator are not independent of one another and cannot be independently modified while maintaining device function. A similar RBS-based device was built through the coupling of the theophylline aptamer to a RBS through a helix slipping-based transmitter part, or communication module (Suess et al., 2004), where small nucleotide shifts led to changes in the accessibility of the RBS and active state of the aptamer. Similar selection strategies have been applied to generate RBS-based devices that exhibit logic operations (AND, NAND) by coupling the theophylline aptamer to a natural riboswitch element through a randomized linker region (Sharma et al., 2008).
Other examples of RNA devices built with separate transmitter parts couple multiple actuators into a single device. For example, two recent RBS-based devices were designed to regulate ribosome access to the RBS through ribozyme cleavage (Ogawa and Maeda, 2008; Wieland and Hartig, 2008). The RBS and hhRz actuators were coupled such that the RBS was sequestered within the ribozyme structure and cleavage of the ribozyme resulted in separation of the two parts, thereby increasing ribosome access and expression levels. Ribozyme cleavage was in turn modulated by binding of theophylline to its aptamer, which was linked through a communication module to the ribozyme. In another example, a RBS-based device was designed to regulate ribosome access to the RBS through cis- and trans-acting antisense parts (Isaacs et al., 2004). A cis-acting antisense was implemented to inhibit ribosome access to a RBS and a trans-acting antisense was employed to activate expression by hybridizing with the cis-acting antisense. Although this device design offers a greater level of flexibility in tailoring parts due to the isolation of binding events to hybridization interactions, the sequence of the cis-acting antisense is dependent on both the RBS and the trans-acting antisense, thereby rendering interdependence among the parts.
Functional composition frameworks in device design that support modular assembly strategies
One of the goals of synthetic biology is to develop foundational technologies that make the engineering of biology easier and more reliable (Endy, 2005). Efforts are being directed to the development of functional composition frameworks, which are a type of device design strategy that supports the construction of devices through modular assembly of distinct parts and are therefore characterized by functional modularity. In engineering design, such frameworks support the efficient and reliable engineering of diverse device functions from a smaller number of refined parts through a plug-and-play type strategy without complex device redesign.
Functional composition frameworks have recently been proposed for single-input single-output RNA device design. In the proposed design strategies functional modularity is achieved through the separation of functions (sensing, actuation, information transmission) into distinct and independent parts (Figures 3A). A common design approach is the encoding of the information transmission function in a distinct transmitter part that employs a strand displacement event solely with the sequences of that part, such that the sequences of the sensor and actuator do not depend on one another and can be changed independently. Therefore, the integration of sensor-actuation functions is simplified via a distinct transmitter part that insulates part functions and controls the interactions between parts through predictive hybridization interactions.
Figure 3. A Functional Composition Framework that Supports Modular Assembly of Single-Input Single-Output RNA Devices and Extension to Higher-Order Devices.
(A) The flow of information through the modular parts of an RNA device.
(B) Modular assembly of single-input single-output RNA devices that exhibit different information processing functions.
(C) Extension to the modular assembly of RNA devices that exhibit higher-order information processing functions.
In one example, a functional composition framework for RNA device design was proposed based on the assembly of three parts (Win and Smolke, 2007): a sensor, made of an RNA aptamer; an actuator, made of a self-cleaving hhRz; and a strand displacement-based transmitter part (Figure 2C). Several design strategies were employed in the implementation of this device platform to support engineering properties such as portability, utility, and composability. For example, the RNA device was integrated into the 3′ UTR of the target gene, which is a flexible and extensible sequence space where the integration of RNA devices is anticipated to have minimal non-specific effects on expression. In addition, ribozyme self-cleavage in the 3′ UTR inactivates a transcript and thereby lowers gene expression independent of cell-specific machinery, thus enabling portability across organisms. Flanking sequences were also included to insulate the device from surrounding sequences that may disrupt its structure and therefore activity to support reliable coupling to other parts and devices.
Ribozyme-based devices were built in which the input-bound conformation was associated with the disruption or restoration of the ribozyme catalytic core and thus converted a molecular input to increased (ON switch) or decreased (OFF switch) expression, respectively, based on the transmitter part (Figure 3B). The device response properties were programmed by altering sequences within the transmitter part, which alters the energetic and kinetic properties of the strand displacement event and therefore the ability of the device to access the different conformational states. The functional modularity of the framework was shown to support direct sensor replacement strategies, such that aptamer sequences to different molecular inputs (theophylline and tetracycline) were swapped into the framework and the function of the device was maintained while being made responsive to a new input (Figure 3B).
A similar framework was used to build RNA devices that actuate through the RNAi pathway by coupling an aptamer to a shRNA through a distinct strand displacement-based transmitter part (Beisel et al., 2008). shRNA-based devices were built in which the input-bound conformation was associated with the disruption of the shRNA stem, thereby inhibiting Dicer processing and subsequent gene silencing, converting a molecular input to increased (ON switch) expression of reporter and endogenous gene targets. The functional modularity of both the sensor and actuator parts in this framework was further highlighted in this example.
Composition frameworks that support extensions to higher-order device functions
One of the important properties of a composition framework is its extensibility to the assembly of more complex devices that enable sophisticated programmed information processing and control functions from basic sensing, actuation, and information transmission parts. Such frameworks will provide general approaches for the forward engineering of multi-input, higher-order devices and support the combinatorial assembly of many information processing, transduction, and control devices from a smaller number of refined parts.
Recent work has described the extension of the functional composition framework described above for single-input single-output ribozyme-based devices to the modular assembly of devices that perform higher-order functions from a small set of refined sensor, actuator, and transmitter parts (Win and Smolke, 2008) (Figure 3C). Three signal integration schemes based on single layer device architectures were developed that involve the assembly of multiple single-input single-output RNA devices and the coupling of multiple sensor-transmitter components to single ribozyme actuators. RNA devices that function as logic gates (AND, NOR, NAND, OR gates), signal and bandpass filters, and cooperative control devices were built on the extended framework.
RNA device performance characterization and measurement standards
In support of the reuse of refined components and their efficient integration into engineered biological systems, synthetic biologists are fueling discussions on technical standards in part and device characterization (Canton et al., 2008). Different metrics have been used to report the performance of RNA parts and devices to-date, making performance comparisons and the downstream integration of these components into different application-specific systems challenging.
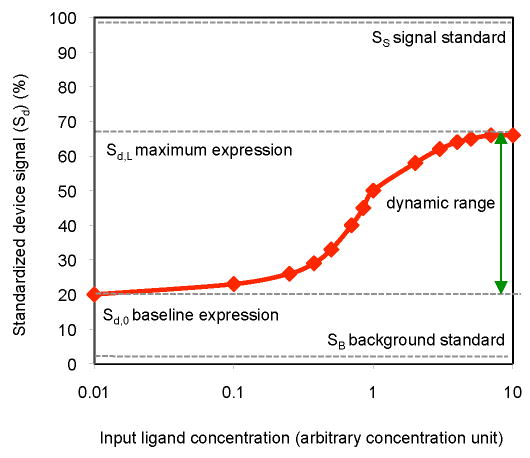
The properties that characterize the performance of a RNA device based on gene expression actuation include output dynamic range (reported as either a difference, Sd,L-Sd,0, or ratio, Sd,L/Sd,0), baseline expression (Sd,0, expression activity in the absence of input), and input response range (input concentration over which device output changes) (Figure 4). Generally, these performance descriptors cannot be compared across different genetic systems, such that reporting properties relative to standards is critical to enabling device comparison. The use of two standards in RNA device characterization was recently proposed (Win and Smolke, 2008): (i) the gene expression activity from the genetic construct in the absence of the RNA device (100%; signal standard, SS); and (ii) the gene expression activity in the absence of the genetic construct (0%; background standard, SB). These proposed standards determine the RNA device performance across the full transcriptional range of a specified promoter, without any non-specific effects that a device may exhibit due to its location relative to other components in the construct.
Figure 4.
RNA Device Performance Characterization
With the goal of integrating RNA devices into genetic circuits composed of diverse biological components, such standardized characterization information is critical to match component properties to achieve the desired system response. For example, proteins can exhibit very different thresholds of titratable function depending on their activities and the properties of the other components comprising the genetic circuit. Such system properties will guide the design of the RNA device properties to match those of the components in the circuit by optimizing for different design criteria. The demonstrated ability to tune RNA device response to fit performance requirements of specific applications using (1) energetic tuning strategies (Beisel et al., 2008; Win and Smolke, 2007); (2) higher-order architectures (Win and Smolke, 2008); and (3) component matching strategies (Yokobayashi et al., 2002) is a critical property for their utility in downstream applications.
Enabling technologies that support the engineering of RNA devices
The engineering of RNA devices requires the integration of several enabling technologies (Figure 5). As a basic enabling technology, strategies that support high-throughput and reliable RNA part generation, refinement, and characterization are critical to RNA device engineering. Generally, RNA gene-actuator parts are adapted from nature with little modification or designed from relatively simple hybridization-based design rules, although some work has focused on the in vitro evolution of RNA actuators exhibiting enhanced function (Conaty et al., 1999; Vaish et al., 1997). RNA sensor parts have been adapted from naturally-occurring elements (Nomura and Yokobayashi, 2007), but are more often generated through in vitro selection strategies. The generation of RNA aptamers exhibiting desired in vivo affinities and specificities is currently a limiting step in device design. In vitro selection strategies for DNA aptamers to proteins have been adapted to high-throughput formats (Cox et al., 2002) and higher efficiency partitioning schemes such as capillary electrophoresis (Berezovski et al., 2005; Drabovich et al., 2005). However, improved aptamer generation schemes have not yet been demonstrated for RNA aptamers and other groups of targets such as small molecules.
Figure 5.
Process Flow of Enabling Technologies Supporting Device Design and Implementation into Engineered Systems
Composition frameworks that support the rapid assembly of RNA devices from parts exhibiting more basic functions are another enabling technology in support of more efficient and reliable device design. Although first-generation frameworks have recently been demonstrated that highlight the design advantages afforded through such engineering design principles (Beisel et al., 2008; Win and Smolke, 2007; Win and Smolke, 2008), much work remains in optimizing and refining RNA device frameworks. For example, effective composition frameworks require the careful refinement of parts such that the parts are more reliably integrated into the proposed framework. Therefore, libraries of well-characterized, refined RNA parts that are compatible with device frameworks are needed to support this enabling technology. In addition, frameworks that address existing challenges in enhancing functional device performance (Beisel et al., 2008; Win and Smolke, 2007), such as dynamic range and insulation of part functions are needed. Such improved frameworks will likely require the integration of new technologies that allow the measurement of RNA folding and switching rates (Greenleaf et al., 2008), thereby advancing our understanding of RNA structure-function relationships. Finally, extended architectures (Deans et al., 2007; Rinaudo et al., 2007; Win and Smolke, 2008), including single- and multi-layered systems composed of heterogeneous components, are important to support the engineering of more complex device functions such as signal restoration and amplification.
Strategies that support the engineering of parts within an RNA device platform can increase the efficiency of new device construction by removing the additional step of part integration and functional optimization. For example, transmitter parts have been generated by applying in vitro (Soukup and Breaker, 1999) and in vivo screening strategies to small libraries within functional device platforms (Lynch et al., 2007; Wieland and Hartig, 2008; Win and Smolke, 2007). The latter strategy benefits from selecting for function within the desired cellular context, as parts that have been generated in vitro do not necessarily translate to functional elements within the cellular environment (Link et al., 2007; Win and Smolke, 2007). More recently, cell-based screening strategies for transmitter parts have been adapted to higher-throughput methods based on fluorescence activated cell sorting (FACS) (Fowler et al., 2008; Lynch and Gallivan, 2008). Researchers have also demonstrated the application of such higher-throughput screening methods to RNA gene-actuation parts in the device platform by extending the randomized region into the actuator (Lynch and Gallivan, 2008). An important area of future research, will be to direct such high-throughput, cell-based screening strategies to the generation of new sensor parts within device platforms.
Finally, computational tools that support the in silico design and programming of device function will provide a critical enabling technology to matching quantitative device response properties to application-specific performance requirements. Recent modeling tools have been described that provide early sequence-to-function computational frameworks for guiding and optimizing RNA device design (Beisel et al., 2008). The refinement of such in silico device design tools will require further insight into RNA structure-function relationships (Martick and Scott, 2006), kinetic and thermodynamic properties of RNA folding (Greenleaf et al., 2008), and improved predictions of RNA secondary and tertiary structures (Parisien and Major, 2008).
The implementation of RNA parts and devices in engineered systems
Within living systems, RNA parts and devices are integrated into genetic circuits composed of heterogeneous parts, including proteins and small molecules, which comprise devices and systems exhibiting human-defined functions. Numerous examples of the integration of RNA parts into heterogeneous systems have been described, while more recent examples have focused on the integration of RNA devices into circuits encoding functions relevant for biotechnological or medical applications, including metabolic engineering, cellular biosensing, phenotype generation, and therapeutics.
In the rapidly growing field of metabolic engineering, RNA parts have been implemented as tools to tune enzyme levels, regulate pathway flux, and optimize product yields. In one example, RNase activity actuators were integrated in intergenic regions of a polycistronic transcript to direct its segmental processing and stability, resulting in differential expression of multiple target genes in E. coli (Smolke et al., 2000). This engineering strategy was applied to control the flux through a synthetic carotenoid pathway, where accumulation levels of the intermediate pathway metabolites were varied through implementation of RNase activity actuators exhibiting different regulatory strengths (Smolke et al., 2001). A modified strategy was developed based on the generation of libraries of combinations of RNase activity actuators, that were screened through FACS for combinations that resulted in desired expression ratios (Pfleger et al., 2006). This strategy was applied to a synthetic mevalonate pathway using a biosensor assay to screen for mevalonate production to optimize the relative expression of three pathway enzymes, resulting in a 7-fold increase in mevalonate levels.
RNA parts have also been implemented as regulatory components in circuits that have potential application in gene therapy. In a recent example, RNAi-based actuators were integrated as modular components into a genetic circuit that was employed as a tight and tunable transgenic regulatory system (Deans et al., 2007). The gene circuit coupled heterogeneous actuator parts, repressor proteins and shRNAs, to significantly reduce the basal expression of the regulated gene from any single actuator. The response of the genetic circuit was shown to be tunable and applied to the small molecule-responsive regulation of mammalian cell growth.
Recent work has also demonstrated the implementation of RNA devices in genetic circuits that are relevant to biotechnological applications. For example, the application of RNA devices to the non-invasive detection of intracellular metabolite levels was recently demonstrated (Win and Smolke, 2007). Ribozyme-based devices were implemented as in vivo biosensors for the detection of xanthine accumulation in S. cerevisiae by transmitting the metabolite binding event to a change in fluorescent protein levels. Genetic circuits that integrate RNA devices toward the regulation of cellular phenotypes and traits have also been demonstrated. In one example, an RNA device was integrated into a genetic circuit in E. coli that enabled cells to detect, follow, and precisely locate a molecular signal (Topp and Gallivan, 2007). The genetic circuit implemented a theophylline-responsive RBS-based device for the regulation of the cheZ gene encoding a chemotaxis protein responsible for cell motility.
RNA devices have also been implemented in potential therapeutic applications. In one example, an RNA device composed of aptamer and antisense parts that interact in trans through hybridization was applied as a controlled therapeutic agent (Rusconi et al., 2002). The device implements an aptamer to coagulant factor IXa as an antagonist and an antisense RNA as an antidote to the function encoded in the aptamer. The aptamer-antisense device was shown to function as a potent anticoagulant drug and antidote combination in plasma and animal models (Rusconi et al., 2004). In another example, an RNA device combining zip-code and gene-regulatory parts was implemented for targeted delivery of gene-actuation parts to silence the expression of survival genes (McNamara et al., 2006). The device was composed of an RNA aptamer that binds to the prostate-specific membrane antigen (PSMA), which is overexpressed in prostate cancer cells, and a siRNA that silences a target gene. The device was demonstrated to trigger targeted gene silencing and apoptosis in cell culture and animal models.
Conclusion
The thoughtful combination of scientific research and engineering theory has resulted in significant advances in the design of functional RNA molecules. Importantly, the field of RNA engineering has been fueled, influenced, and inspired by discoveries and advances in the areas of RNA biology and in vitro nucleic acid engineering. The recent application of synthetic biology principles has resulted in more efficient and effective design strategies supporting the programming of RNA devices that perform complex information processing, transduction, communication, and control functions in living systems. The integration of future scientific and technological advances will lead to enabling technologies supporting the more reliable and robust programming of RNA parts and devices and their integration as functional components into biological networks and systems. The resulting improvements in our ability to transmit information to and from living systems, and implement control within cells themselves, will transform how we interact with and program biology.
Acknowledgments
RNA programming research in the Smolke Lab is supported by the NIH, NSF, DOD, and the Bill and Melinda Gates Foundation. C.D.S. is an Alfred P. Sloan Research Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alifano P, Bruni CB, Carlomagno MS. Control of mRNA processing and decay in prokaryotes. Genetica. 1994;94:157–172. doi: 10.1007/BF01443430. [DOI] [PubMed] [Google Scholar]
- An CI, Trinh VB, Yokobayashi Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. Rna. 2006;12:710–716. doi: 10.1261/rna.2299306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J Am Chem Soc. 2003;125:14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Oleynikov Y, Singer RH. The travels of mRNAs through all cells large and small. Faseb J. 1999;13:447–454. doi: 10.1096/fasebj.13.3.447. [DOI] [PubMed] [Google Scholar]
- Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- Beisel CL, Bayer TS, Hoff KG, Smolke CD. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol Syst Biol. 2008;4:224. doi: 10.1038/msb.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovski M, Drabovich A, Krylova SM, Musheev M, Okhonin V, Petrov A, Krylov SN. Nonequilibrium capillary electrophoresis of equilibrium mixtures: a universal tool for development of aptamers. J Am Chem Soc. 2005;127:3165–3171. doi: 10.1021/ja042394q. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Blount KF, Uhlenbeck OC. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- Bonoli M, Graziola M, Poggi V, Hochkoeppler A. RNA complementary to the 5′ UTR of mRNA triggers effective silencing in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2006;339:1224–1231. doi: 10.1016/j.bbrc.2005.11.137. [DOI] [PubMed] [Google Scholar]
- Breaker RR. Complex riboswitches. Science. 2008;319:1795–1797. doi: 10.1126/science.1152621. [DOI] [PubMed] [Google Scholar]
- Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C, Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Bunch TA, Goldstein LS. The conditional inhibition of gene expression in cultured Drosophila cells by antisense RNA. Nucleic Acids Res. 1989;17:9761–9782. doi: 10.1093/nar/17.23.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk AR, Kehayova PD, Landrigan A, Liu DR. In vivo evolution of an RNA-based transcriptional activator. Chem Biol. 2003;10:533–540. doi: 10.1016/s1074-5521(03)00109-1. [DOI] [PubMed] [Google Scholar]
- Buskirk AR, Landrigan A, Liu DR. Engineering a ligand-dependent RNA transcriptional activator. Chem Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- Carrier TA, Keasling JD. Controlling messenger RNA stability in bacteria: strategies for engineering gene expression. Biotechnol Prog. 1997;13:699–708. doi: 10.1021/bp970095h. [DOI] [PubMed] [Google Scholar]
- Carrier TA, Keasling JD. Library of synthetic 5′ secondary structures to manipulate mRNA stability in Escherichia coli. Biotechnol Prog. 1999;15:58–64. doi: 10.1021/bp9801143. [DOI] [PubMed] [Google Scholar]
- Cassiday LA, Maher LJ., 3rd In vivo recognition of an RNA aptamer by its transcription factor target. Biochemistry. 2001;40:2433–2438. doi: 10.1021/bi002376v. [DOI] [PubMed] [Google Scholar]
- Chanfreau G, Buckle M, Jacquier A. Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc Natl Acad Sci U S A. 2000;97:3142–3147. doi: 10.1073/pnas.070043997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci U S A. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman J, Green PJ, Inouye M. The use of RNAs complementary to specific mRNAs to regulate the expression of individual bacterial genes. Cell. 1984;37:429–436. doi: 10.1016/0092-8674(84)90373-8. [DOI] [PubMed] [Google Scholar]
- Conaty J, Hendry P, Lockett T. Selected classes of minimised hammerhead ribozyme have very high cleavage rates at low Mg2+ concentration. Nucleic Acids Res. 1999;27:2400–2407. doi: 10.1093/nar/27.11.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JC, Rajendran M, Riedel T, Davidson EA, Sooter LJ, Bayer TS, Schmitz-Brown M, Ellington AD. Automated acquisition of aptamer sequences. Comb Chem High Throughput Screen. 2002;5:289–299. doi: 10.2174/1386207023330291. [DOI] [PubMed] [Google Scholar]
- Crooke ST. Progress in antisense technology. Annu Rev Med. 2004;55:61–95. doi: 10.1146/annurev.med.55.091902.104408. [DOI] [PubMed] [Google Scholar]
- Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J Am Chem Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- Drabovich A, Berezovski M, Krylov SN. Selection of smart aptamers by equilibrium capillary electrophoresis of equilibrium mixtures (ECEEM) J Am Chem Soc. 2005;127:11224–11225. doi: 10.1021/ja0530016. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- Famulok M, Blind M, Mayer G. Intramers as promising new tools in functional proteomics. Chem Biol. 2001;8:931–939. doi: 10.1016/s1074-5521(01)00070-9. [DOI] [PubMed] [Google Scholar]
- Fowler CC, Brown ED, Li Y. A FACS-based approach to engineering artificial riboswitches. Chembiochem. 2008;9:1906–1911. doi: 10.1002/cbic.200700713. [DOI] [PubMed] [Google Scholar]
- Ge D, Lamontagne B, Elela SA. RNase III-mediated silencing of a glucose-dependent repressor in yeast. Curr Biol. 2005;15:140–145. doi: 10.1016/j.cub.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Grate D, Wilson C. Laser-mediated, site-specific inactivation of RNA transcripts. Proc Natl Acad Sci U S A. 1999;96:6131–6136. doi: 10.1073/pnas.96.11.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ, Pines O, Inouye M. The role of antisense RNA in gene regulation. Annu Rev Biochem. 1986;55:569–597. doi: 10.1146/annurev.bi.55.070186.003033. [DOI] [PubMed] [Google Scholar]
- Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Harvey I, Garneau P, Pelletier J. Inhibition of translation by RNA-small molecule interactions. Rna. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelrigg T. The destinies and destinations of RNAs. Cell. 1998;95:451–460. doi: 10.1016/s0092-8674(00)81613-x. [DOI] [PubMed] [Google Scholar]
- Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Henkin TM. Transcription termination control in bacteria. Curr Opin Microbiol. 2000;3:149–153. doi: 10.1016/s1369-5274(00)00067-9. [DOI] [PubMed] [Google Scholar]
- Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: how RNA provides instructions for transcription termination/antitermination decisions. Bioessays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- Joyce GF. Forty years of in vitro evolution. Angew Chem Int Ed Engl. 2007;46:6420–6436. doi: 10.1002/anie.200701369. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat Struct Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- Kijima H, Tsuchida T, Kondo H, Iida T, Oshika Y, Nakamura M, Scanlon KJ, Kondo T, Tamaoki N. Hammerhead ribozymes against gamma-glutamylcysteine synthetase mRNA down-regulate intracellular glutathione concentration of mouse islet cells. Biochem Biophys Res Commun. 1998;247:697–703. doi: 10.1006/bbrc.1998.8878. [DOI] [PubMed] [Google Scholar]
- Kim DS, Gusti V, Dery KJ, Gaur RK. Ligand-induced sequestering of branchpoint sequence allows conditional control of splicing. BMC Mol Biol. 2008;9:23. doi: 10.1186/1471-2199-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Jeong S. Inhibition of the functions of the nucleocapsid protein of human immunodeficiency virus-1 by an RNA aptamer. Biochem Biophys Res Commun. 2004;320:1181–1186. doi: 10.1016/j.bbrc.2004.06.077. [DOI] [PubMed] [Google Scholar]
- Kislauskis EH, Li Z, Singer RH, Taneja KL. Isoform-specific 3′-untranslated sequences sort alpha-cardiac and beta-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Lai EC. RNA sensors and riboswitches: self-regulating messages. Curr Biol. 2003;13:R285–291. doi: 10.1016/s0960-9822(03)00203-3. [DOI] [PubMed] [Google Scholar]
- Lee NS, Bertrand E, Rossi J. mRNA localization signals can enhance the intracellular effectiveness of hammerhead ribozymes. Rna. 1999;5:1200–1209. doi: 10.1017/s1355838299990246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Link KH, Guo L, Ames TD, Yen L, Mulligan RC, Breaker RR. Engineering high-speed allosteric hammerhead ribozymes. Biol Chem. 2007;388:779–786. doi: 10.1515/BC.2007.105. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Lundberg U, von Gabain A, Melefors O. Cleavages in the 5′ region of the ompA and bla mRNA control stability: studies with an E. coli mutant altering mRNA stability and a novel endoribonuclease. Embo J. 1990;9:2731–2741. doi: 10.1002/j.1460-2075.1990.tb07460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SA, Desai SK, Sajja HK, Gallivan JP. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem Biol. 2007;14:173–184. doi: 10.1016/j.chembiol.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SA, Gallivan JP. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH. Revolutions in RNA secondary structure prediction. J Mol Biol. 2006;359:526–532. doi: 10.1016/j.jmb.2006.01.067. [DOI] [PubMed] [Google Scholar]
- McDowall KJ, Lin-Chao S, Cohen SN. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J Biol Chem. 1994;269:10790–10796. [PubMed] [Google Scholar]
- McManus MT, Petersen CP, Haines BB, Chen J, Sharp PA. Gene silencing using micro-RNA designed hairpins. Rna. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- Meaux S, Van Hoof A. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. Rna. 2006;12:1323–1337. doi: 10.1261/rna.46306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Morino F, Tokunaga T, Tsuchida T, Handa A, Nagata J, Tomii Y, Kijima H, Yamazaki H, Watanabe N, Matsuzaki S, et al. Hammerhead ribozyme specifically inhibits vascular endothelial growth factor gene expression in a human hepatocellular carcinoma cell line. Int J Oncol. 2000;17:495–499. doi: 10.3892/ijo.17.3.495. [DOI] [PubMed] [Google Scholar]
- Narberhaus F, Waldminghaus T, Chowdhury S. RNA thermometers. FEMS Microbiol Rev. 2006;30:3–16. doi: 10.1111/j.1574-6976.2005.004.x. [DOI] [PubMed] [Google Scholar]
- Neupert J, Karcher D, Bock R. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res. 2008;36:e124. doi: 10.1093/nar/gkn545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa F, Kakiuchi N, Funaji K, Fukuda K, Sekiya S, Nishikawa S. Inhibition of HCV NS3 protease by RNA aptamers in cells. Nucleic Acids Res. 2003;31:1935–1943. doi: 10.1093/nar/gkg291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y, Yokobayashi Y. Reengineering a natural riboswitch by dual genetic selection. J Am Chem Soc. 2007;129:13814–13815. doi: 10.1021/ja076298b. [DOI] [PubMed] [Google Scholar]
- Ogawa A, Maeda M. An artificial aptazyme-based riboswitch and its cascading system in E. coli. Chembiochem. 2008;9:206–209. doi: 10.1002/cbic.200700478. [DOI] [PubMed] [Google Scholar]
- Owens GC, Chappell SA, Mauro VP, Edelman GM. Identification of two short internal ribosome entry sites selected from libraries of random oligonucleotides. Proc Natl Acad Sci U S A. 2001;98:1471–1476. doi: 10.1073/pnas.98.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- Rackham O, Chin JW. A network of orthogonal ribosome x mRNA pairs. Nat Chem Biol. 2005;1:159–166. doi: 10.1038/nchembio719. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Rusconi CP, Roberts JD, Pitoc GA, Nimjee SM, White RR, Quick G, Jr, Scardino E, Fay WP, Sullenger BA. Antidote-mediated control of an anticoagulant aptamer in vivo. Nat Biotechnol. 2004;22:1423–1428. doi: 10.1038/nbt1023. [DOI] [PubMed] [Google Scholar]
- Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, Sullenger BA. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- Sakamoto N, Wu CH, Wu GY. Intracellular cleavage of hepatitis C virus RNA and inhibition of viral protein translation by hammerhead ribozymes. J Clin Invest. 1996;98:2720–2728. doi: 10.1172/JCI119097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman NC. From genes to machines: DNA nanomechanical devices. Trends Biochem Sci. 2005;30:119–125. doi: 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Nomura Y, Yokobayashi Y. Engineering Complex Riboswitch Regulation by Dual Genetic Selection. J Am Chem Soc. 2008 doi: 10.1021/ja805203w. [DOI] [PubMed] [Google Scholar]
- Shetty RP, Endy D, Knight TF., Jr Engineering BioBrick vectors from BioBrick parts. J Biol Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolke CD, Carrier TA, Keasling JD. Coordinated, differential expression of two genes through directed mRNA cleavage and stabilization by secondary structures. Appl Environ Microbiol. 2000;66:5399–5405. doi: 10.1128/aem.66.12.5399-5405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolke CD, Martin VJ, Keasling JD. Controlling the metabolic flux through the carotenoid pathway using directed mRNA processing and stabilization. Metab Eng. 2001;3:313–321. doi: 10.1006/mben.2001.0194. [DOI] [PubMed] [Google Scholar]
- Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc Natl Acad Sci U S A. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suess B, Hanson S, Berens C, Fink B, Schroeder R, Hillen W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003;31:1853–1858. doi: 10.1093/nar/gkg285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suess B, Weigand JE. Engineered riboswitches - Overview, Problems and Trends. RNA Biol . 2008;5 doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- Thompson KM, Syrett HA, Knudsen SM, Ellington AD. Group I aptazymes as genetic regulatory switches. BMC Biotechnol. 2002;2:21. doi: 10.1186/1472-6750-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp S, Gallivan JP. Guiding bacteria with small molecules and RNA. J Am Chem Soc. 2007;129:6807–6811. doi: 10.1021/ja0692480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp S, Gallivan JP. Random walks to synthetic riboswitches--a high-throughput selection based on cell motility. Chembiochem. 2008;9:210–213. doi: 10.1002/cbic.200700546. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Vaish NK, Heaton PA, Eckstein F. Isolation of hammerhead ribozymes with altered core sequences by in vitro selection. Biochemistry. 1997;36:6495–6501. doi: 10.1021/bi963134r. [DOI] [PubMed] [Google Scholar]
- Vaish NK, Kore AR, Eckstein F. Recent developments in the hammerhead ribozyme field. Nucleic Acids Res. 1998;26:5237–5242. doi: 10.1093/nar/26.23.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldminghaus T, Kortmann J, Gesing S, Narberhaus F. Generation of synthetic RNA-based thermosensors. Biol Chem. 2008;389:1319–1326. doi: 10.1515/BC.2008.150. [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. Rna. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Weigand JE, Suess B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Res. 2007;35:4179–4185. doi: 10.1093/nar/gkm425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- Wieland M, Hartig JS. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew Chem Int Ed Engl. 2008;47:2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- Win MN, Smolke CD. From the Cover: A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc Natl Acad Sci U S A. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen L, Svendsen J, Lee JS, Gray JT, Magnier M, Baba T, D’Amato RJ, Mulligan RC. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- Yim SH, Park I, Ahn JK, Kang C. Translational suppression by hammerhead ribozymes and inactive variants in S. cerevisiae. Biomol Eng. 2000;16:183–189. doi: 10.1016/s1389-0344(99)00052-0. [DOI] [PubMed] [Google Scholar]
- Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc Natl Acad Sci U S A. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimatsu T, Nagawa F. Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science. 1989;244:1346–1348. doi: 10.1126/science.2544026. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- Zhou W, Edelman GM, Mauro VP. Isolation and identification of short nucleotide sequences that affect translation initiation in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100:4457–4462. doi: 10.1073/pnas.0437993100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]