Abstract
Peripheral myelin protein 22 (PMP22) is a dose-sensitive, disease-associated protein primarily expressed in myelinating Schwann cells. Either reduction or overproduction of PMP22 can result in hereditary neuropathy, suggesting a requirement for correct protein expression for peripheral nerve biology. PMP22 is post-transcriptionally regulated and the 3′untranslated region (3′UTR) of the gene exerts a negative effect on translation. MicroRNAs (miRNAs) are small regulatory molecules that function at a post-transcriptional level by targeting the 3′UTR in a reverse complementary manner. We used cultured Schwann cells to demonstrate that alterations in the miRNA biogenesis pathway affect PMP22 levels, and endogenous PMP22 is subjected to miRNA regulation. GW-body formation, the proposed cytoplasmic site for miRNA-mediated repression, and Dicer expression, an RNase III family ribonuclease involved in miRNA biogenesis, are co-regulated with the differentiation state of Schwann cells. Furthermore, the levels of Dicer inversely correlate with PMP22, while the inhibition of Dicer leads to elevated PMP22. Microarray analysis of actively-proliferating and differentiated Schwann cells, in conjunction with bioinformatics programs, identified several candidate PMP22-targeting miRNAs. Here we demonstrate that miR-29a binds and inhibits PMP22 reporter expression through a specific miRNA seed binding region. Over-expression of miR-29a enhances the association of PMP22 RNA with Argonaute 2, a protein involved in miRNA function, and reduces the steady-state levels of PMP22. In contrast, inhibition of endogenous miR-29a relieves the miRNA-mediated repression of PMP22. Correlation analyses of miR-29 and PMP22 in sciatic nerves reveal an inverse relationship, both developmentally and in post-crush injury. These results identify PMP22 as a target of miRNAs and suggest that myelin gene expression by Schwann cells is regulated by miRNAs.
Keywords: Schwann cell, myelin, microRNA, gene regulation, myelination
Introduction
Peripheral myelin protein 22 (PMP22) is a 22 kDa tetraspan glycoprotein that is predominantly expressed by the myelinating Schwann cells (Snipes et al. 1992). PMP22 was first identified as growth arrest-specific 3 (gas-3) gene in NIH 3T3 fibroblasts (Schneider et al. 1988) and its expression increases as cells reach density-dependant inhibition (confluency) (Manfioletti et al. 1990; Zoidl et al. 1995). The significance of the growth arrest-specific expression is still undetermined. Although PMP22 protein expression is highly restricted, the mRNA is present ubiquitously throughout the body, including the CNS, kidney, heart, muscle and lung (Amici et al. 2006; Baechner et al. 1995; Suter et al. 1994). PMP22 is detected in Schwann cells, at epithelial and endothelial cell junctions, and in specific motor and sensory neurons (Baechner et al. 1995; Maier et al. 2003; Notterpek et al. 2001; Roux et al. 2004). In the developing rat sciatic nerve, PMP22 message steadily increases and reaches maximal expression at around postnatal day 21, which correlates with the completion of myelination and Schwann cell differentiation (Garbay et al. 2000). In comparison, PMP22 levels drop significantly post-nerve crush injury (Snipes et al. 1992) in accordance with the de-differentiation of Schwann cells. These findings suggest the involvement of post-transcriptional mechanisms in controlling PMP22 expression.
Point mutations, gene duplication, and deletion of PMP22 are associated with demyelinating neuropathies, including Charcot-Marie-Tooth disease type 1A (CMT1A) (Lupski and Garcia 1992). CMT1A has been linked with a duplication of a 1.5 Mb region on chromosome 17p11.2 (Patel et al. 1992) which includes PMP22. Since the PMP22 gene does not suffer a loss of function with the duplication, the phenotypes are likely the result of altered gene dosage. In hereditary neuropathy pressure palsies (HNPP), one copy of the PMP22 gene is deleted (Chance et al. 1993). The Schwann cells in the neuropathic models and in patients show an impaired ability to myelinate (Nobbio et al. 2004). Therefore, PMP22 levels must be tightly controlled as it is estimated that a 50% reduction in expression will result in HNPP, while a 50% increase leads to CMT1A (Maier et al. 2002). Recent data suggest that the PMP22 transcript is misregulated in a number of diseases, including schizophrenia, depression (Aston et al. 2005; Dracheva et al. 2006) and cancer (van Dartel and Hulsebos 2004). These findings may imply leaky transcription and a requirement to regulate undesired message at a post-transcriptional level.
The 5′-untranslated region (5′UTR) of PMP22 contains two known promoters, which result in two transcripts that differ only in the inclusion of UTRs, primarily untranslated exon 1. The two mRNA transcripts, P1 and P2, appear to have tissue specific functions (Maier et al. 2003; Suter et al. 1994). In addition, the 3′UTR exerts a negative effect on RNA translation, which is observed even after the three AU-rich elements have been removed (Bosse et al. 1999). A recently elucidated mechanism of post-transcriptional gene control involves microRNAs (miRNAs). MiRNAs are small, non-coding regulatory RNA molecules that bind to the 3′UTR of target genes based upon reverse complementarity (Grimson et al. 2007; He et al. 2005; Valencia-Sanchez et al. 2006). MiRNAs are transcribed via RNA polymerase II, cleaved by Drosha, exported into the cytoplasm, and then processed by Dicer to form the mature miRNA. The binding of the miRNA to the target site on mRNA can either signal for the degradation via the RNA induced silencing complex (RISC), which contains the Argonaute proteins, or repress translation without degradation through other less defined mechanisms (Bagga et al. 2005; Pillai et al. 2005). The RISC has been localized to structures termed processing bodies (P-bodies) or GW bodies (GWB). These cytoplasmic foci contain the RNA-binding protein GW182 and serve as the sites where miRNAs are believed to exert the majority of their function (Ding and Han 2007; Liu et al. 2005). Mutations creating or deleting miRNA target sites can result in abnormal phenotypes in vivo (Clop et al. 2006). Recently it was reported that autoimmunity to the GW-bodies is associated with motor and sensory neuropathy in humans (Bhanji et al. 2007) although the histopathology remains undefined. Coincidentally, it has been hypothesized that PMP22 RNA may be degraded by a non-coding RNA molecule (Manfioletti et al. 1990).
In this study we characterize the miRNA expression profile (miRNAome) of Schwann cells in response to different growth conditions and demonstrate that miR-29a represses the expression of both endogenous and reporter PMP22. In addition, we examine the expression of miR-29 during sciatic nerve development and in response to nerve crush injury. The elucidation of the mechanism of post-transcriptional regulation of PMP22 provides novel insight into the etiology of myelin-associated diseases and may identify new therapeutic targets in controlling myelin gene regulation.
Materials and Methods
Plasmids and miRNA Precursors and Inhibitors
The psicheck2 luciferase vector (Promega, Madison, WI) was used for the luciferase assays. The 3′UTR of PMP22 was inserted using the Xho1/Not1 sites. Site directed deletion of the miR-29a seed region was performed using the Genetailor™ site directed Mutagenesis System (Invitrogen, Carlsbad, CA) with specific primers designed using the PrimerX program (http://www.bioinformatics.org/primerx/): 5′-ACAAGCAATCTGTGAAAATAGATTTACCAT-3′ and 5′-TTTCACAGATTGCTTGTCTCTGACGTCT-3′. The c-myc-Ago2 plasmid was a kind gift from Dr. Hannon’s Laboratory (Cold Springs Harbor, NY) (Karginov et al. 2007). Pre-miRNA precursors and anti-miRNA inhibitors were obtained from Ambion (Austin, TX) and used at the indicated concentrations. Fragments of the 3′UTR of PMP22 were obtained by PCR using the rat PMP22 cDNA as template. For PMP400, the primers were 5′-AGGCCTCTCGAGGCGCCCGACGCACCATCCGTCTAGGC-3′ and 5′-GAGCGGCCGCTGAGCAAAACAAAAAGATGA-3′. For PMP800, the primers were 5′-AGGCCTCTCGAGGCGCCCGACGCACCATCCGTCTAGGC-3′ and 5′-GAGCGGCCGCTAAAACTGTTAATTGAGTT-3′. PCR products were digested by Xho I/Not I and cloned into the psicheck2 plasmid (Promega).
Cell Culture and Transfection
Primary Schwann cell cultures were established from newborn rat pups (Ryan et al. 2002). Schwann cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS) (Hyclone, Logan, UT), 5 μM forskolin (Calbiochem, La Jolla, CA) and 10 μg/ml bovine pituitary extract (Biomedical Technologies Inc, Stoughton, MA). To analyze proliferating Schwann cells, the cell were harvested at ~75% confluency. To stimulate differentiation, the cells were subjected to 0.5% FCS/DMEM for 72 h prior to collection (Yang et al. 2004). Alternatively, the cultures were switched to a defined medium (DMEM-F12, 100 U/ml Penicillin, 100 μg/ml Streptomycin, 100 μg/ml BSA, N2 supplement, 38 ng/ml dexamethasone, 50 ng/ml thyroxine, 50 ng/ml tri-iodothyronine) for 48 h to promote growth-arrest and differentiation (Cheng and Mudge 1996). Schwann cells cultured under these conditions are primarily non-dividing, as determined by bromodeoxyuridine incorporation (Cheng and Mudge 1996).
Transient transfections of Schwann cells were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. In brief, the cells were split the day before and then transfected with either RNA and/or DNA at the indicated concentrations. Transfection efficiency was approximately 30 %, as judged by the expression of a plasmid encoding enhanced green fluorescent protein (EmGFP). RNA transfection efficiency was over 90% as determined using a fluorochrome-conjugated scrambled miRNA (Ambion). Cells were harvested for RNA analysis using TRIzol (Invitrogen). Protein samples were obtained by harvesting cells in gel sample buffer and protein concentrations were determined using the BCA assay kit (Pierce, Rockford, IL).
Bioinformatics
Bioinformatic scans of the rat 3′UTR of PMP22 were conducted using three web based miRNA target prediction programs (Targetscan (http://www.targetscan.org/), miRbase (http://microrna.sanger.ac.uk/targets/v5/), and Pictar (http://pictar.bio.nyu.edu/)). MiRNAs were chosen based upon their prediction by more than one program, conservation of the binding region, and the strength of predicted interaction.
Luciferase and Gel Shift Assays
Luciferase assays were performed using the Dual-Luciferase assay kit (Promega). Schwann cells were co-transfected in 24-well plates with the indicated psicheck2 luciferase construct (0.4 μg/well) and miRNA precursor (10 nM). After 48 h, the cells were harvested in passive lysis buffer and luciferase activities were determined using a Bio-Tek Synergy HT luminometer (Bio-Tek Instruments Inc, Winooski, VT). The luciferase data is expressed as a ratio of Renilla Luciferase (RL) to Firefly Luciferase (FL) to normalize for transfection variability between samples. Luciferase experiments were repeated at least three independent times, in triplicate or greater, as indicated.
Gel shift assays were performed with in vitro transcribed PMP22 RNA using a T7 in vitro transcription kit (Promega). The RNA was incubated for two hours with biotin-labeled miRNAs (1 pmol) at 42° C. The samples were separated on 3% agarose gels, transferred to a nylon membrane, and the complexes revealed using a nucleic acid detection kit HRP-streptavidin substrate (Pierce). Total RNA loading was monitored using SYBR Gold (Invitrogen) staining of the gel.
Primary Antibodies
A previously characterized mouse monoclonal antibody (Chemicon, Temencula, CA), (Notterpek et al. 1999) was used to detect PMP22 in the immunolabeling experiments. For the PMP22 Western blots, we utilized a rabbit polyclonal antibody raised against synthetic peptide of the rat PMP22 (amino acids 117–132) (Pareek et al. 1993). We used a human anti-GWB antibody (Eystathioy et al. 2002) to detect GW bodies in cells (kind gift from the Chan lab, Gainesville, FL) and a mouse anti-GW182 antibody (Abcam, Cambridge, MA) for the Western blot. Antibodies against c-myc (Santa Cruz, CA), phospho-histone H3 (Ser10) (Upstate, Temecula, CA), Dicer (Santa Cruz, CA) and GAPDH (Encore Technologies, Alachua, Florida) were obtained from the indicated suppliers.
Immunoblotting
Equal amounts of protein lysates (40 μg for cells or 5 μg for nerves) were separated on sodium dodecyl sulfate gels and transferred to nitrocellulose membranes. Endoglycosidase digestion with PNGase F (New England Biolabs, Beverly, MA) was performed overnight at 37°C prior to Western blot (Pareek et al. 1997). After transfer, the membranes were incubated with primary and then horseradish peroxidase (HRP) conjugated secondary antibodies (Sigma, St. Louis, MO) (Amici et al. 2006). Films were digitally imaged using a GS-800 densitometer (Bio-Rad Laboratories, Hercules, CA) and figures assembled using Adobe Photoshop 5.5. Quantification of Western blot data was performed using Scion Image (Frederick, MD). The specific band intensities were obtained and the data was normalized for GAPDH to obtain relative protein expression levels.
Immunostaining of Schwann cells
Immunofluorescence experiments were performed as described (Notterpek et al. 1999). Bound primary antibodies were detected using Alexa Fluor 594 (red) anti-mouse and Alexa Fluor 488 (green) anti-human antibodies (Molecular Probes, Eugene, OR; Zymed, San Francisco, CA). Hoechst dye (10 μg/ml, Molecular Probes) was included in the secondary antibody solution to visualize nuclei. Control samples without primary antibodies were processed in parallel. Coverslips were mounted on slides using the Prolong Anti-fade kit (Molecular Probes). Images were acquired with a SPOT digital camera (Diagnostic Instrumentals, Sterling Heights, MI) attached to a Nikon Eclipse E800 microscope (Tokyo, Japan).
Immunoprecipitation
For the c-myc-Ago2 immunoprecipitation experiments, rat Schwann cells were transfected with the c-myc-Ago2 plasmid and either the negative (Neg.) scrambled miR or miR-29a (Karginov et al. 2007). Two days post-transfection, the cells were incubated in lysis buffer (10 mM Tris, pH 7.5, 10 mM KCl, 2 mM MgCl2, 5 mM DTT) supplemented with a mixture of protease inhibitors (Complete™; Roche, Indianapolis, IN) for 15 min on ice and lysed by pipetting. Five-fold concentrated ATP depletion mix (4 units/ml RNaseIn (Promega), 100 mM glucose, 0.5 units/ml hexokinase (Sigma), 1 mg/ml yeast tRNA (Invitrogen), 450 mM KCl) was added and the cell lysates were centrifuged at 16,000×g for 30 min at 4°C. Prior to the immunoprecipitation, anti-c-myc beads (Sigma, St. Louis, MO) were preblocked for 30 min in wash buffer (0.5% Nonidet P-40, 150 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 20 mM Tris, pH 7.5, 5 mM DTT, with EDTA-free protease inhibitors) containing 1 mg/ml yeast tRNA and 1 mg/ml BSA. Wash buffer was added to the lysates and samples were incubated and agitated with the beads for 4 h at 4°C. The beads were washed first in wash buffer and then with wash buffer containing 650 mM NaCl two times. Next, the slurry was transferred to a fresh tube and bound RNA was extracted with TRIzol (Invitrogen) and the RNA concentrations were determined.
Sciatic nerve crush injury
Experiments were performed on 8-weeks old male CD1 mice obtained from Charles River Laboratories (Wilmington, MA, USA) (Islamov et al. 2004). The animal use protocol was approved by Animal Care and Use Committee of East Carolina University. Mice were anesthetized and an incision was made on the right thigh. The right sciatic nerve was exposed and crushed at the level of the sciatic notch for 15 sec with a fine hemostat. The wound was closed and the animals were allowed to recover for 4 or 5 days. After specified time periods, the mice were euthanized and sciatic nerves were quickly removed, snap frozen in liquid nitrogen. The control sample (naïve nerve) was taken from the contralateral side. The excised crush sample was the injury site, plus ~ 4 mm proximal and 4 mm distal from the point of injury.
MiRNA Expression Analysis
Expression of mature miR-29a was verified using real-time PCR and normalized to miR-125a, using the 2−ΔΔCT method (Livak and Schmittgen 2001). MiR-125a is equally expressed in proliferating and non-proliferating Schwann cells (see data in Figure 4). Total RNA was reverse transcribed using the Taqman Reverse Transcription Kit (Applied Biosystems, Foster City, CA) and PCR was performed in triplicate using Taqman primers specific for miR-29a and miR-125a (Applied Biosystems). To analyze RNA levels in rat and mouse sciatic nerves, the animals were sacrificed and the nerves were removed and immediately frozen in liquid nitrogen. To obtain an adequate yield of total RNA from the developing rat nerves, at least two animals were pooled for each sample and three independent samples were analyzed per time point (postnatal days 2, 4, 8, 16 and 21). For RNA analysis of mouse nerves, ten crush sites or control nerve sites were pooled. The nerves were crushed under liquid nitrogen and total RNA was isolated using either the TRIzol (Invitrogen) or the mirVana miRNA Isolation kit (Ambion). For quantitative RT-PCR analysis, 0.1 μg of RNA per reaction was employed with the Quantitech SYBR Green RT-PCR kit (Qiagen) and primers specific for PMP22 (Qiagen). For nerve miRNA analysis, the NCODE miRNA first-strand cDNA synthesis and qRT-PCR kit (Invitrogen) was used with primers specific for miR-29a, miR-29b, and miR-24. Each sample was repeated in triplicate and the results were normalized using primers to GAPDH (Qiagen, for PMP22) or miR-24 (Invitrogen, for miRNA analysis) MiR-24 was used to control for equal RNA input because its expression was not affected by either Schwann cell differentiation or crush nerve injury. The relative expression of each message was determined using the 2−ΔΔCT method (Livak and Schmittgen 2001).
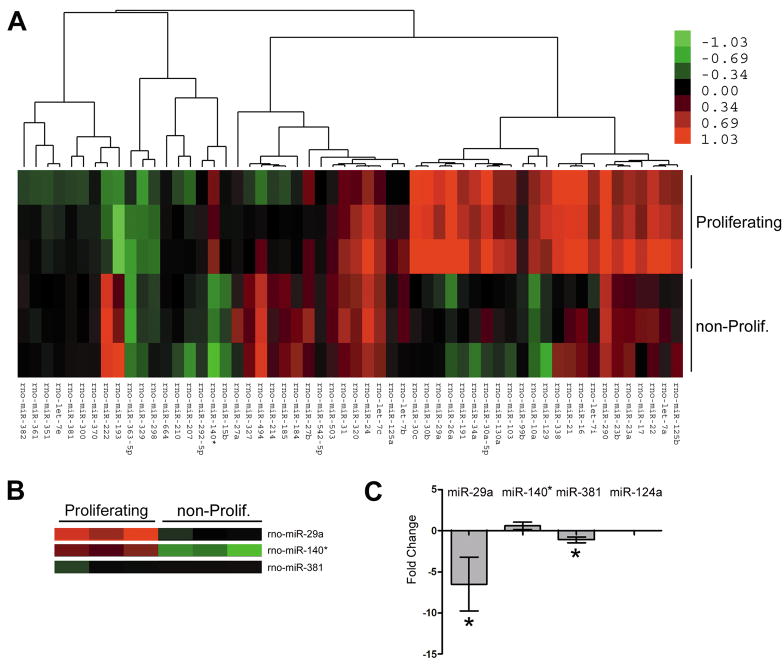
Figure 4. Growth conditions alter the miRNA expression profile of Schwann cells.
(A) The miRNA expression profiles of Schwann cells grown in serum-containing (proliferating) or defined medium (non-proliferating) were determined using an Exiqon microarray (n=3 for each condition, color scale bar indicates fold-change). (B) Three miRNAs predicted to target the 3′UTR of PMP22 are expressed in Schwann cells and show unique profiles upon growth condition. (C) The fold-change in the expression of predicted PMP22 regulating miRNAs in proliferating, as compared to non-proliferating Schwann cells (*p<0.05, n=3, 2−ΔΔCT method). MiR-124a serves as a negative control.
MiRNA microarrays were performed by isolating total RNA from Schwann cells in proliferating and defined media (triplicate samples per condition), purified using the mirVana miRNA Isolation Kit (Ambion, Austin, TX) and analyzed with an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA). Total RNA (5 μg) was labeled with a Cy3-conjugated RNA-linker and hybridized to Locked Nucleic Acid (LNA) based miRCURY™ arrays (Exiqon, Woburn, MA). Images were acquired using an Axon scanner (4000B) and processed with Genepix 6 (Molecular Devices, Sunnyvale, CA).
RNA Expression Analysis
Real-time reverse-transcriptase PCR was performed as described (Notterpek et al. 2001). RNA was isolated from Schwann cells transfected with the indicated plasmid, miRNA-precursor or inhibitor, or from sciatic nerves using TRIzol (Invitrogen). QuantiTech primers specific for PMP22 RNA were obtained from Qiagen (catalog number QT00175938). Control primers for GAPDH (Qiagen QuantiTech assay catalog number QT00199633) were included. Each sample was analyzed in triplicate (0.2 μg of RNA per reaction) using the Applied Biosystems 7300 real-time PCR system. The SYBR green QuantiTech kit was obtained from Qiagen. Data was normalized to GAPDH using the 2−ΔΔCT method (Livak and Schmittgen 2001).
Statistics
Data from multiple independent experiments were analyzed using Microsoft Excel 2007 and Graphpad Prism v5.0. For analysis of two independent groups, Student’s t-test was used with significance at p<0.05. For determination of significance between three or more groups, one-way ANOVA and post-hoc Tukey’s t-tests were utilized with significance at p<0.05. All graphs represent the means and the error bars represent the standard deviation of the mean. For correlation studies, a linear regression analysis was performed and the r2 and p-values were calculated using Graphpad Prism v5.0.
Results
PMP22 levels inversely correlate with GW-body formation and Dicer expression
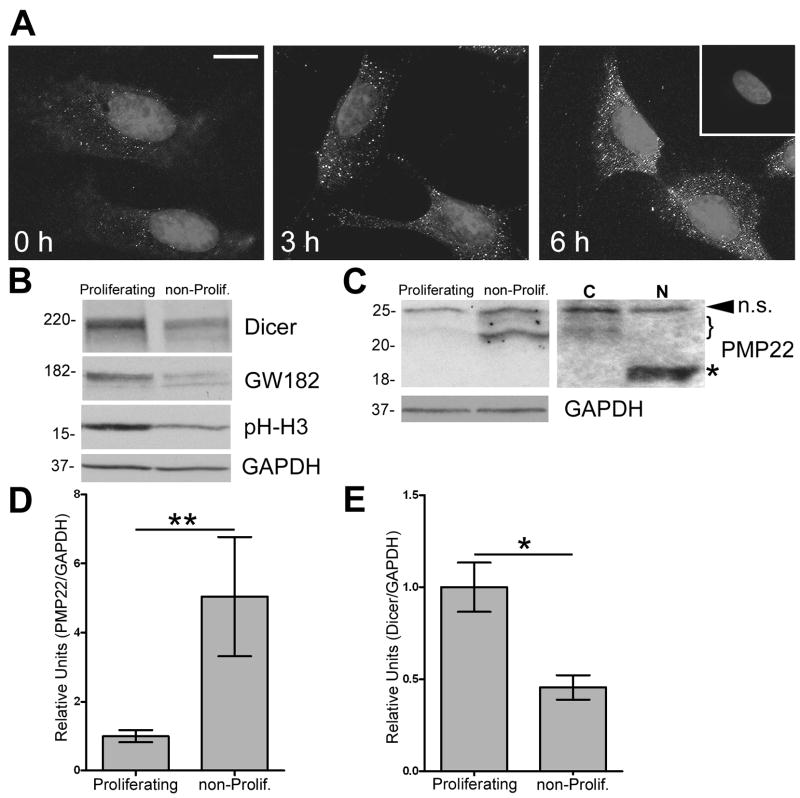
To investigate whether PMP22 is regulated by miRNAs, we first determined the formation of GW-bodies (GWBs) during different growth conditions in rat Schwann cells, where PMP22 is most expressed (Pareek et al. 1997). Schwann cells were subjected to growth-arrest by serum starvation, and then stimulated to proliferate by the addition of 10% serum (Zoidl et al. 1995). At the indicated time points, the cells were fixed and processed for immunostaining using an anti-GWB antibody (Fig. 1A). Increases in both the size and abundance of GWBs occur in a time dependent manner upon release of the cells from growth-arrest. Western blot analysis on whole cell lysates demonstrates greater GW182 protein expression in actively proliferating, when compared to non-proliferating, differentiated Schwann cells (Fig. 1B). In addition, we investigated another miRNA associated protein, Dicer, which is required for mature miRNA biogenesis for differential expression (Valencia-Sanchez et al. 2006). Similar to GW182, we observe the highest expression of Dicer when the cells are actively growing. The reduction in the steady-state levels of phospho-histone H3, a mitotic marker, in samples from the defined medium confirms that there are fewer cells in division (Fig. 1B). In comparison to the miRNA pathway associated proteins, the expression of PMP22 is low in proliferating and high in differentiating Schwann cells (Fig. 1C). To demonstrate that the detected heterogeneous bands at around ~22 kDa are differentially glycosylated forms of PMP22, we performed N-glycosidase reactions (Pareek et al. 1997). Upon incubation of the cell lysates with PNGase F, which completely removes the carbohydrate moiety of PMP22, all the detected bands, except the top band, shift to the core 18 kDa protein (Pareek et al. 1993). Quantification of the PMP22 protein bands in three independent experiments reveals that PMP22 expression is significantly (**p<0.01) elevated in non-proliferating, differentiated cells (Fig. 1D). In comparison, Dicer expression is high in proliferating, as compared to differentiating Schwann cells (Fig. 1E). Taken together, these data indicate that the expression of PMP22 inversely correlates with both Dicer and GW182, two essential proteins for miRNA biogenesis.
Figure 1. GW body formation and Dicer expression are enhanced in actively-proliferating Schwann cells.
(A) Rat Schwann cells were subjected to growth arrest, followed by the addition of serum (for time-points indicated) to induce cellular growth. Cells were fixed and labeled with a human anti-GWB antibody. Hoechst dye is used to visualize nuclei. The inset in the upper right corner represents a no primary antibody control. Scale bar, 10 μm. (B) Western blot analysis on total lysates (40 μg/lane) of proliferating and non-proliferating (non-prolif.) cells are shown using the indicated antibodies. Phospho-Histone H3 (pH-H3) serves as a mitotic marker. (C) On the anti-PMP22 Western blot, the arrowhead indicates a non-specific (n.s.) band whereas the bracket denotes differentially glycosylated isoforms of PMP22. Upon incubation of the cell lysates with PNGase F (N), the indicated ~22 kDa PMP22 bands shift to the core 18 kDa core protein (*). C: no enzyme control. (B, C) GAPDH serves as a loading control. Molecular mass in kDa. Quantification of three independent experiments reveals increased PMP22 (D) and decreased Dicer (E) expression in non-proliferating, as compared to proliferating cells (**p<0.01; *p<0.05). (D, E) Values are expressed as relative units normalized for GAPDH.
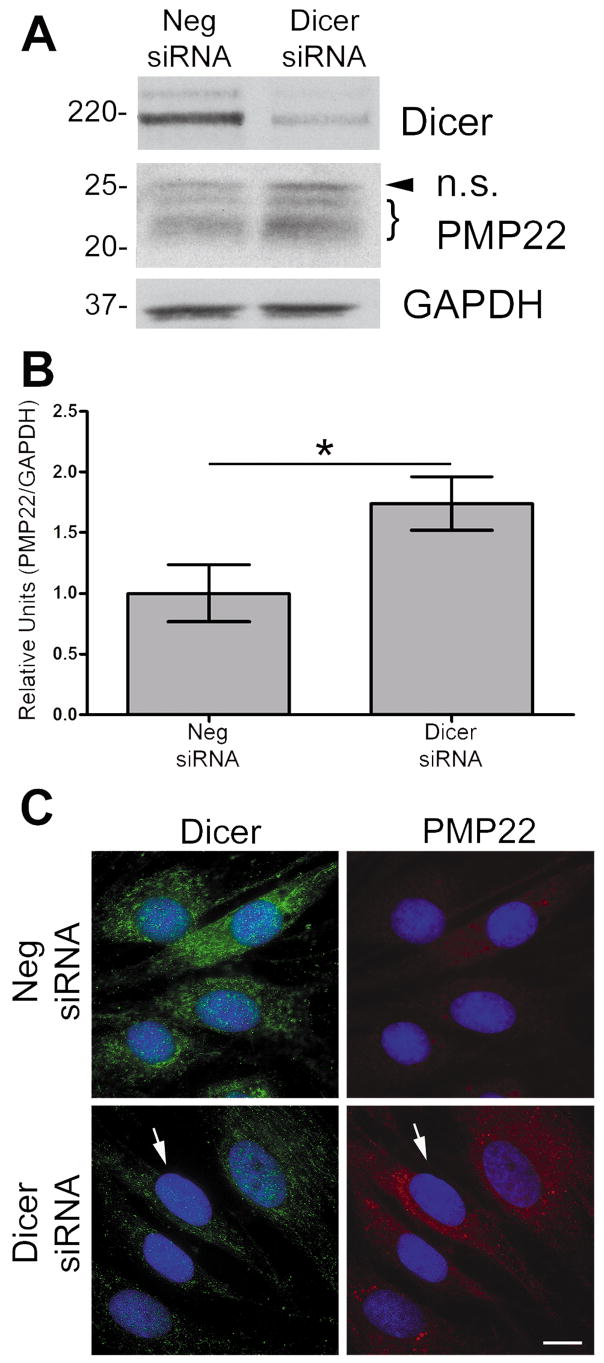
To further establish a functional relationship between PMP22 levels and Dicer, we inhibited Dicer expression in Schwann cells using siRNA (Fig. 2A). We used a negative (Neg.) scrambled siRNA to control for any non-specific effects of transfection. In the Dicer inhibited cells, we observe an increase in PMP22 protein levels at 72 h post-transfection (Fig. 2A), as compared to the Neg. siRNA transfected cells. Densitometric analysis of three independent experiments indicates an ~60 % increase in PMP22 protein upon the suppression of Dicer (Fig. 2B). The biochemical results were reinforced by immunostaining Schwann cells transfected with either the Neg. or Dicer siRNA (Fig. 2C). Cells with Dicer expression inhibited show less Dicer-like immunoreactivity (green) and an increase in PMP22-like staining (red) (Notterpek et al. 1999). Therefore, the inhibition of Dicer enhances PMP22 levels and indicates that mature miRNAs are regulating PMP22 expression in Schwann cells.
Figure 2. Suppression of Dicer increases PMP22 levels.
(A) In whole cell lysates (40 μg/lane), the suppression of Dicer by siRNA is confirmed using an anti-Dicer antibody, as compared to scrambled Neg. siRNA. Increased PMP22 protein is observed in cells transfected with Dicer siRNA. The arrowhead corresponds to a non-specific (n.s.) band and the bracket indicates PMP22. GAPDH serves as a loading control. Molecular mass in kDa. (B) Quantification of three independent experiments reveals that inhibition of Dicer expression results in an increase in PMP22 levels, as compared cells transfected with Neg. siRNA (*p<0.05). PMP22 levels were normalized to GAPDH as a loading control. (C) Dicer suppression is confirmed using an anti-Dicer antibody (green), as compared to Neg. siRNA transfected cells. Increased PMP22 in cells treated with Dicer siRNA is detected using an anti-PMP22 antibody (red). The arrow signifies the same cell in both panels for comparison. Hoechst dye is used to visualize nuclei. Scale bar, 10 μm.
MicroRNAs are predicted to target PMP22 and regulate reporter expression
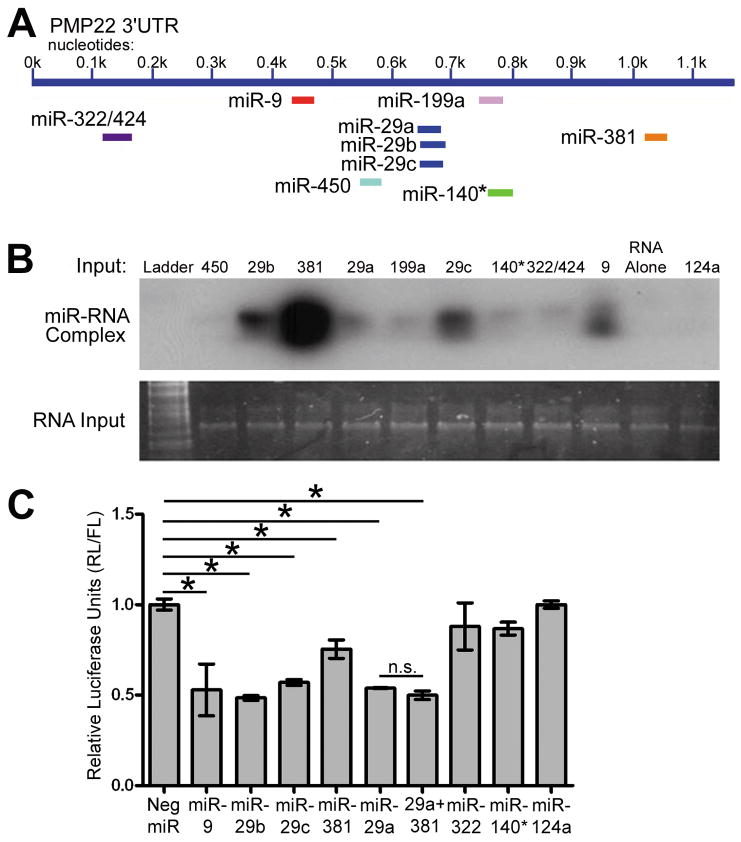
Bioinformatic scans are the “in silico” standard for assembling a list of candidate miRNAs predicted to target the 3′UTR of a given RNA. We used three programs (Targetscan, miRbase and Pictar) to generate a list of ten miRNAs with high probability to bind to the 3′UTR of PMP22. A schematic of where these miRNAs are predicted to bind is shown (Fig. 3A). Since RNA is known to have significant secondary structure and this can affect miRNA binding (Kertesz et al. 2007), we evaluated the binding ability of the predicted miRNAs to the 3′UTR of PMP22 using a gel shift assay (Fig. 3B). The presence of intact RNA and relative loading in each lane is confirmed using SYBR gold staining of the gel. The miRNA/RNA complexes were resolved using a HRP-conjugated streptavidin. MiR-29a-c, -381, and -9 all demonstrate strong binding ability, with miRs-199a, -140*, and -322/424 showing weaker binding. MiR-450 does not possess any detectable binding (Fig. 3B). MiR-124a is not predicted to target the 3′UTR of PMP22 and is used as a negative control. RNA alone lane only contains the PMP22 RNA with no labeled miRNA probe.
Figure 3. The binding and regulatory ability of predicted PMP22 targeting miRNAs.
(A) MiRNAs that are predicted to bind to the 3′UTR of PMP22 are shown. (B) Binding of candidate miRNAs are detected as miRNA/PMP22 RNA complexes by a gel shift assay. The RNA alone lane contains PMP22 RNA only, whereas miR-124 serves as an additional negative control. Total RNA input is shown using SYBR gold staining. (C) Luciferase assays were performed after co-transfection of the PMP22 3′UTR luciferase reporter and the indicated miRNA-precursors. Luciferase signal is shown after Renilla Luciferase (RL) readings were normalized to Firefly Luciferase (FL). Neg. miR does not code any known miRNA, and miR-124 is used as a non-PMP22 targeting control. (*p<0.05, n=3), n.s.: not significant.
To investigate which miRNAs may be regulating PMP22 expression in Schwann cells, we established a PMP22 3′UTR-luciferase construct with the 3′UTR of PMP22 inserted downstream and in frame with the Renilla Luciferase (RL) gene. This construct allows us to quickly and quantitatively evaluate miRNA effects on the 3′UTR of PMP22. We utilized the psicheck2 dual luciferase vector that contains a separate Firefly Luciferase (FL) gene to normalize for transfection efficiency. The 3′UTR-luciferase construct was co-transfected in Schwann cells with 10 nM miRNAs and the cells were harvested at 48 h post-transfection. MiR-29a, miR-29b, miR-29c, miR-9, and miR-381 all significantly (*p<0.05) reduce luciferase activity when compared to the Neg. scrambled miRNA, while miR-322/424 and miR-140* do not (Fig. 3C). As miR-381 and miR-29a are expressed endogenously in Schwann cells (see below in Fig. 4A), we examined if co-transfection of these two miRs may have an additive effect on reporter expression. As shown in Figure 3C, the co-transfection of miR-381 with miR-29a does not significantly enhance the inhibitory activity of miR-29a. MiR-124a serves as a non-PMP22 targeting control to ensure that activation of the miRNA pathway alone is not affecting reporter expression. These data demonstrate that specific PMP22 targeting miRNAs reduce reporter expression in Schwann cells.
MicroRNAs are differentially expressed in Schwann cells upon growth condition
To substantiate the potential functional significance of PMP22 targeting miRNAs, we next determined the miRNA expression profile (microRNAome) of actively proliferating compared to non-proliferating Schwann cells. Several miRNAs demonstrate differential expression based upon growth condition (Fig. 4A). In addition, miRNAs predicted to target PMP22 are expressed by Schwann cells, including miR-29a, miR-381 and miR-140* (Fig. 3A). The relative expression of these particular miRNAs is consistent among the independent triplicate samples (Fig. 4B). MiR-9 was not detected in this microarray, nor was it found to be expressed in Schwann cells by RT-PCR or Northern blot (data not shown) indicating that this miRNA likely to regulate PMP22 expression in other cell types. We validated the microarray data for miR-29a, miR-381 and miR140* using RT-PCR (Fig. 4C). MiR-124a is included as a negative control since it was not detected by the microarray, or by RT-PCR. In agreement with the microarray data, miR-29a is notably down-regulated when the cells are promoted to differentiate. We observed a similar repression of miR-29a when Schwann cells are cultured in a reduced serum medium (data not shown). Based on the observed inverse correlation of miR-29a and PMP22, we decided to further characterize this specific miRNA in Schwann cells.
MicroRNA-29a specifically regulates PMP22 reporter expression
To demonstrate that the endogenous Schwann cell miR-29a is regulating the expression of PMP22, we employed miRNA inhibitors (anti-miRs). As shown in Fig. 3C, miR-29a reduces the expression of the 3′UTR-luciferase construct (Fig. 5A). More importantly, the anti-miR-29a relieves the repression by the endogenous miR-29a, as compared to the Neg. control. In comparison, inhibition of other predicted PMP22 targeting miRNAs, including miR-381, miR-322/424 and miR-140*, does not (Fig. 5B). These results indicate that although both miR-29a and miR-381 can reduce luciferase signal when their levels are elevated via transfection, only endogenous miR-29a is actively repressing PMP22 expression.
Figure 5. miR-29a regulates PMP22 3′UTR-luciferase reporter expression through one specific binding site.
(A) Schwann cells were co-transfected with the 3′UTR luciferase reporter and either miR-29a or anti-miR-29a, and luciferase activity was quantified (**p<0.01, n=4). (B) Inhibition of miR-322/424, miR-381, and miR-140* does not significantly increase PMP22 3′UTR reporter expression when compared to Neg. control (n=3, p>0.05). (C) Schematic representation of PMP22 3′UTR truncation constructs. The predicted miR-29a binding site is at 0.66 kb after the stop codon (grey line). (D) Quantification of luciferase assays after transfection with the indicated constructs (**p<0.01; *p<0.05; ***p<0.001, n=4). (E) A schematic depicting the deletion of the predicted 7 nt seed region for the miR-29a binding site. (F) Luciferase assays were performed after transfection with either the full length or the 29a seed Del PMP22 3′UTR construct in the presence of miR-29a (*p<0.05) (n=3).
To confirm the binding sites for miR-29a in the 3′UTR of PMP22 and demonstrate the specificity of the interaction, we established constructs with or without the predicted binding region at 0.66 kb past the stop codon (Fig. 5C). Mir-29a and the Neg. miRNA were transfected with the base psicheck2 vector which did not contain the 3′UTR of PMP22. Under these conditions, miR-29a does not reduce reporter signal thus eliminating off-target effects. However, in agreement with previous studies (Bosse et al. 1999), the 3′UTR of PMP22 does instill a notable reduction on reporter activity compared to the base vector. When the PMP800 construct, which contains the predicted miR-29a site, is co-transfected with miR-29a, a further reduction in luciferase activity is seen. In comparison, the PMP400 construct, in which the predicted miR-29a site is removed, demonstrates significantly greater luciferase activity than the full length 3′UTR construct (Fig. 5D). These data indicate that the endogenous Schwann cell miR-29a regulates PMP22 expression. To demonstrate the specificity of miR-29a on the 3′UTR of PMP22, we deleted the 7 nt seed region in the predicted miR-29a binding site (Fig. 5E), which eliminated the reduction in luciferase activity (Fig. 5F). However, experiments with the non-mutated 3′UTR performed in parallel still retain the miR-29a associated repression of reporter signal (Fig. 5F). These results validate the specificity of the interaction between miR-29a and the 3′UTR of PMP22.
Endogenous Schwann cell PMP22 is regulated by miR-29a
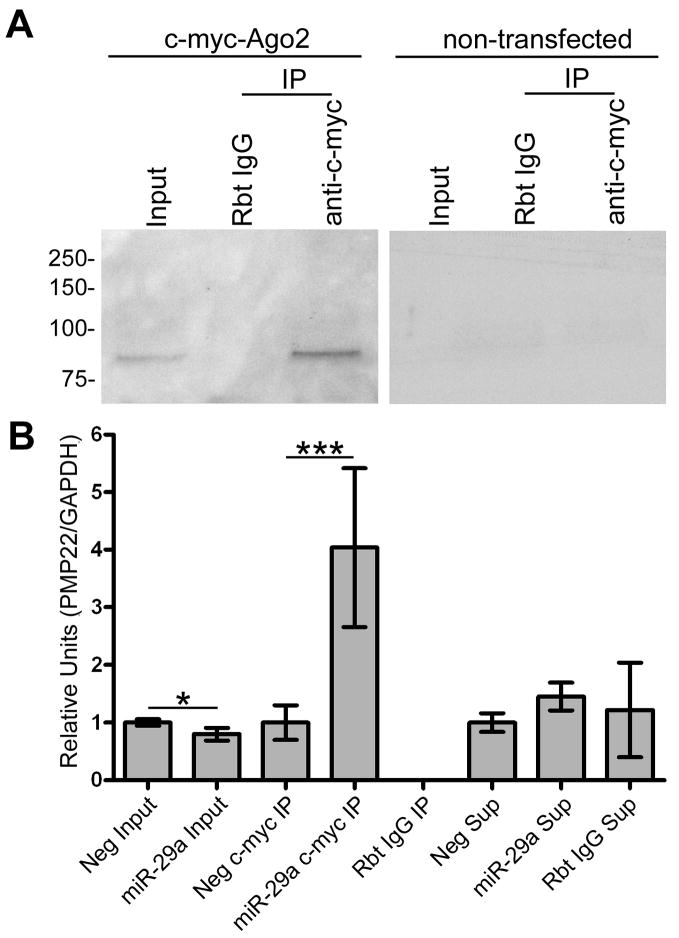
A recently developed biochemical approach to validate miRNA targets (Karginov et al. 2007) utilizes the knowledge that the Argonaute proteins of the RISC complex bind to target RNAs to exert repression (Meister et al. 2004). Thus if the Argonaute (Ago2 in these experiments) protein is immunoprecipitated ‘primed’ with exogenous miR-29a, the precipitated protein should show an enhanced association with the PMP22 RNA. To test this, we co-transfected c-myc-Ago2 with miR-29a, or the scrambled Neg. miRNA. Western blot analysis with anti-c-myc antibody shows efficient and specific immunoprecipitation (IP) of the c-myc-Ago2, as compared to beads conjugated to non-specific rabbit IgG (Fig. 6A). The same experiment performed with non-transfected cells is shown on the right, and serves as a negative control. Next, RNA was isolated from all the IP fractions and semi-quantitative RT-PCR was performed on equal amounts of RNA. In the input samples, co-transfection of miR-29a and c-myc-Ago2 significantly reduces the levels of steady-state PMP22 RNA when compared to the Neg. miRNA (Fig. 6B). In addition, cells with c-myc-Ago2 primed with miR-29a contain the majority of the PMP22 RNA associated with the Ago2 protein, detected in the c-myc IP fraction (Fig. 6B). Non-specific rabbit IgG conjugated beads do not precipitate any detectable PMP22 RNA, which remained in the post-precipitation supernatant fraction. These data indicate that even in the Neg. miRNA transfected cells, endogenous PMP22 RNA is associated with Ago2. Significantly, when the abundance of miR-29a is elevated, the amount of PMP22 in complex with Ago2 further increases. Therefore, the endogenous PMP22 RNA is regulated via the RISC complex in Schwann cells and when the levels of miR-29a are increased, this interaction is enhanced.
Figure 6. Endogenous PMP22 RNA associates with Ago2 and the interaction is enhanced by miR-29a.
(A) C-myc-Ago2 transfected and non-transfected Schwann cells were processed for immunoprecipitation (IP) using either a non-specific rabbit IgG (Rbt IgG) or rabbit anti-c-myc conjugated (c-myc IP) agarose beads. Western blot using an anti-c-myc antibody is shown. The lack of signal in the non-transfected cells confirms the specificity of the IP. (B) RNA was isolated from total cell lysates (input), IP fractions (IP) and post-immunoprecipitation supernatants (Sup), from the samples analyzed in panel A. Semi-quantitative RT-PCR was performed using primers specific for PMP22 RNA. GAPDH was used as a control for equal total RNA input (*p<0.05; ***p<0.001; n=4).
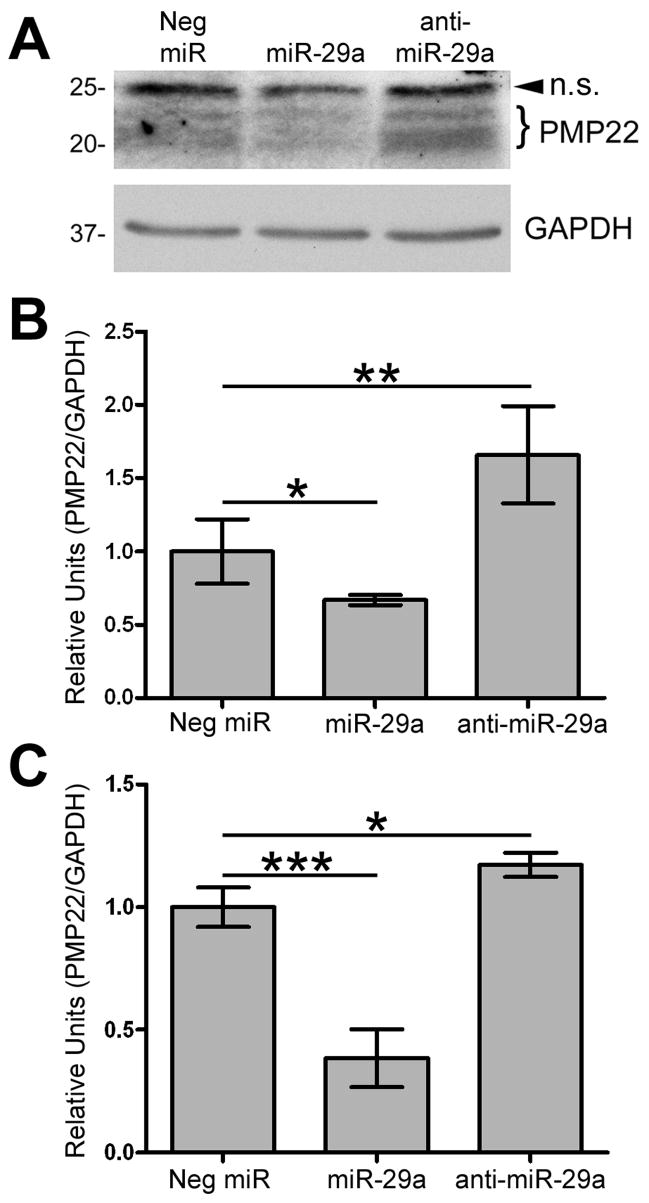
To further show that miR-29a regulates endogenous PMP22 in Schwann cells, we transfected the cells with miR-29a, anti-miR-29a or Neg. miR, followed by protein analysis (Fig. 7A). While miR-29a reduces steady-state PMP22 protein levels within Schwann, when endogenous miR-29a is inhibited, the steady-state levels of PMP22 are greater than in Neg. control miRNA cells (Fig. 7A). Quantification of four independent experiments reveals that miR-29a reduces PMP22 protein levels by approximately 45%, when compared to Neg. controls. On the other hand, inhibition of endogenous miR-29a results in a significant increase in steady-state PMP22 (Fig. 7B). Since miRNA-mediated gene regulation can ultimately reduce the steady-state levels of the target RNA (Wu and Belasco 2008), we used real-time RT-PCR to determine the effect of miR-29a on PMP22 RNA. In cells with elevated miR-29a, the levels of PMP22 message are reduced, as compared to control (Fig. 7C). In comparison, when endogenous miR-29a is inhibited, the steady-state levels of PMP22 RNA are elevated. Taken together, these results demonstrate that miR-29a actively modulates PMP22 protein and RNA expression within Schwann cells.
Figure 7. miR-29a regulates endogenous PMP22 levels in Schwann cells.
(A) Schwann cells were transfected with Neg. miR, miR-29a, or anti-miR-29a and processed for Western blotting (40 μg/lane) with the indicated antibodies. Arrowhead corresponds to a non-specific (n.s.) band and the bracket indicates differentially glycosylated forms of PMP22. GAPDH is shown as a loading control. (B) Quantification of PMP22 protein levels, after normalization to GAPDH, in cells transfected with the indicated constructs (*p<0.05; **p<0.01; n=4). (C) Quantification using the 2−ΔΔCT method of real-time RT-PCR experiments on RNA from Schwann cells transfected with Neg. miR, miR-29a, or anti-miR-29a (***p<0.001; *p<0.05; n=4). Data is normalized to GAPDH and expressed in relative units.
PMP22 and miR-29 expression inversely correlate in vivo
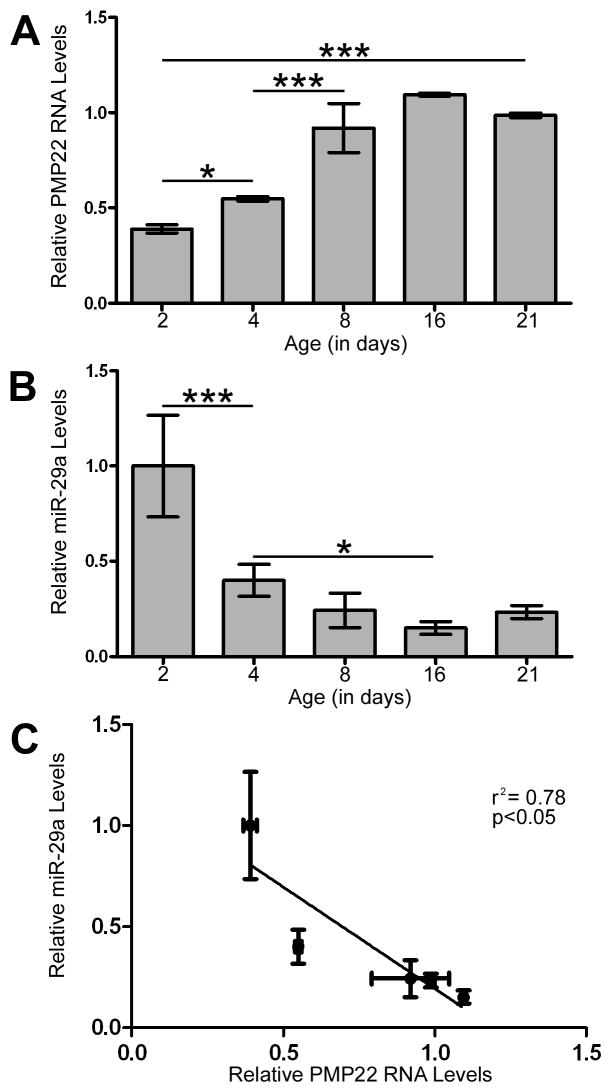
The expression of PMP22 RNA and protein increase as peripheral nerves develop and the Schwann cells differentiate into myelin forming cells (Garbay et al. 2000; Snipes et al. 1992). To investigate a potential relationship between PMP22 and miR-29a, we isolated sciatic nerves from rat pups at postnatal days 2, 4, 8, 16 and 21. Quantitative RT-PCR reveals that PMP22 RNA expression steadily increases during early postnatal nerve development, reaching maximal level at around P16 (Fig. 8A). Analyses of the same samples for miR-29a shows the highest levels at P2, followed by reduced expression already at P4 (Fig. 8B). Linear regression analysis between the two RNAs detected a statistically significant correlation, with increased miR-29a levels being associated with decreased PMP22 expression (r2=0.78, p<0.05) (Fig. 8C). These data indicate an inverse relationship between PMP22 and miR-29a in vivo, supporting that Schwann cell differentiation state affects the miRNA profile during sciatic nerve development.
Figure 8. PMP22 and miR-29 expression are inversely correlated in the developing rat sciatic nerve.
(A) Total RNA was isolated from rat sciatic nerves at the indicated time points and the expression of PMP22 was quantified by RT-PCR. GAPDH was used to normalize for loading and relative change in expression was determined using the 2−ΔΔCT method (n=3, *p<0.05, ***p<0.001). (B) RT-PCR on the same RNA samples determines the expression of miR-29a in sciatic nerves during development. MiR-24 was used to normalize for equal loading (*p<0.05, ***p<0.001; n=3). (C) Data from the relative expression of PMP22 (x-axis) and miR-29a (y-axis), as determined in panels A and B, were analyzed by linear regression (p<0.05, r2 = 0.078).
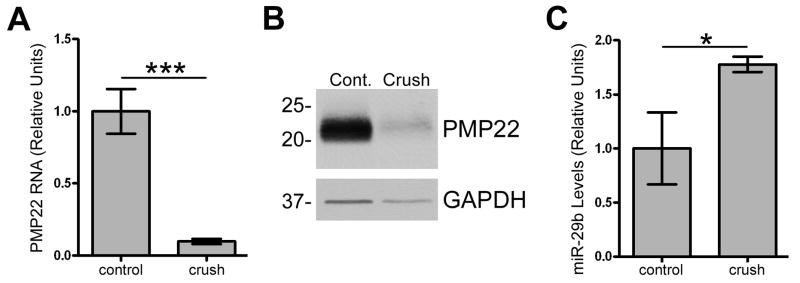
To further investigate a functional relationship between PMP22 and miR-29, we analyzed RNA and protein from control and injured mouse sciatic nerves. Post-crush injury, myelin genes, including PMP22, are down regulated and the Schwann cells de-differentiate and proliferate allowing the axons to heal (Bosse et al. 2006; Snipes et al. 1992). In agreement, we found that PMP22 RNA (Fig. 9A) and protein (Fig. 9B) are reduced post-injury when compared to control. MiRNA microarray analyses of control and crush injured mouse nerves identified miR-29b as the predominate form of miR-29 expressed in response to injury (data not shown). MiR-29 levels are low in mature myelinated nerves, while there is an approximately 2-fold increase in miR-29b expression at four day post-injury (data not shown). To validate the microarray results, miR-29b levels were determined using quantitative RT-PCR on RNA isolated from crush and control nerves (Fig. 9C). In agreement with the microarray data, miR-29b levels are elevated in samples subjected to crush injury when compared to control. MiR-29a and miR-29b are located within the same miRNA cluster in the mouse, rat and human genome, and miR-29b has the same predicted binding site in the 3′UTR of PMP22 as miR-29a (Fig. 3C). These data further support a functional relationship between miR-29 and PMP22 in the PNS and implicate the miRNA pathway in peripheral nerve development and repair.
Figure 9. Nerve crush injury reduces PMP22 expression and elevates miR-29 levels.
(A) Total RNA was isolated from control or crush injured mouse sciatic nerves and relative expression of PMP22 was determined using quantitative RT-PCR. GAPDH was used to normalize for RNA loading (n=10 nerves, triplicates used for each run; ***p<0.001). Data are expressed as relative units. (B) Western blot analysis of total nerve lysates (5 μg/lane) at 5 d post-crush injury. GAPDH is shown as a loading control. Molecular mass, in kDa. (C) Relative quantitative RT-PCR was used to determine the levels of miR-29b at 4 d post-injury in mouse nerves. MiR-24 was used to normalize for equal loading (n=3 for each run and n=10 nerves for each condition, *p<0.05).
Discussion
Myelin gene expression is regulated by transcriptional and post-transcriptional events (Svaren and Meijer 2008; Wegner 2000) and here we show that functional RNA molecules within glia are involved in this process. Specifically, we found differential miRNA expression profiles based upon Schwann cell phenotype, which also correlates with an induction of Dicer and GWB formation. The expression of PMP22 inversely correlates with Dicer, and steady-state PMP22 levels can be increased by the inhibition of Dicer. In addition, we demonstrate that several miRNAs present in Schwann cells bind to the 3′UTR of PMP22 and are able to reduce the expression of a luciferase reporter. Although a number of miRNAs had negative effects on reporter expression, only the inhibition of endogenous miR-29a relieved the miRNA-mediated repression, supporting a functional relationship between miR-29a and PMP22. MiR-29a specifically interacts with the 3′UTR of PMP22 and regulates the expression of the endogenous PMP22 protein and RNA, while the levels of miR-29 and PMP22 inversely correlate in vivo.
GWBs are the main sites of miRNA-mediated gene regulation in the cell and the formation of these structures appears to be regulated with the cell cycle (Lian et al. 2006; Yang et al. 2004). We detected GWBs in Schwann cells, and in agreement with previous reports (Lian et al. 2006; Yang et al. 2004), their formation is increased when cellular division is stimulated (Fig. 1). These data indicate that although GWBs are present in non-proliferating Schwann cells, they may be more important during cellular division. It will be of interest to determine if Schwann cells require GWBs and/or miRNA regulation to retain their mitotic ability in the mature peripheral nerve, which is necessary for repair upon axonal injury (Clemence et al. 1989). An impairment of miRNA processing may reduce the ability of the cells to divide and remyelinate post-injury. Dicer, an essential miRNA biogenesis protein, is required for developmental processes and has been implicated in disease states, including retinal degeneration, abnormal neuronal spine length, heart failure, and skeletal muscle development (Chen et al. 2008; Damiani et al. 2008; Davis et al. 2008; O’Rourke et al. 2007). Our data indicate that Dicer expression in Schwann cells is differentially regulated depending on the growth condition. In agreement with the growth regulatory activity of PMP22 (Zoidl et al. 1995), Dicer and PMP22 protein levels have an inverse relationship, which can be utilized to modify PMP22 expression (Figs. 1 & 2). Our data suggest that loss of Dicer expression or function could be detrimental to Schwann cell biology by leading to alteration in PMP22 levels.
Using biochemical and molecular approaches, we have identified miR-29a as a regulator of PMP22 in Schwann cells. We employed current prediction programs to assemble a list of potential PMP22-targeting miRNAs (Fig. 3) and compared their relative expression to PMP22 using microarrays (Fig. 4). MiR-29a expression inversely correlates with PMP22, which may signal a functional relationship. Although three miRNAs are able to down regulate the 3′UTR-luciferase reporter (Fig. 3C), miR-29a is the only tested endogenous miRNA whose inhibition leads to increased luciferase activity (Fig. 5B). While miR-381 is also present in Schwann cells and can reduce reporter expression (Figs. 3 and 4), inhibition of endogenous miR-381 does not relieve the repression (Fig. 5B). Therefore, mere binding ability alone does not necessarily dictate functionality in vitro. The de-repression observed with miR-29a anti-miRNA and the maximal effects on the luciferase assays identify endogenous miR-29a as a critical miRNA governing PMP22 expression in Schwann cells. In addition, deletion of the predicted miR-29a binding site on the 3′UTR of PMP22, which is conserved between rat, mouse, human, and chicken (http://www.targetscan.org/), abolishes the miR-29a dependent repression. However, since the miR-29a seed deletion construct did not have greater luciferase signal then the intact 3′UTR (Fig. 5F), we cannot eliminate the contribution of other regulatory domains in the 3′UTR of PMP22. AU-rich elements have been implicated in miRNA-mediated repression (Jing et al. 2005) and the 3′UTR of PMP22 contains three such regions (Bosse et al. 1999), including one in close proximity to the predicted miR-29a seed target site. We validated a functional role for miR-29a in regulating endogenous PMP22 by co-immunoprecipitating Ago2 ‘primed’ with exogenous miR-29a, and PMP22 RNA (Fig. 6). Future studies will examine if other myelin gene RNAs are associated with this complex in Schwann cells.
Schwann cells exist in at least two different phenotypes in the adult nervous system, myelinating and non-myelinating, and can transition between the two states upon injury (Garbay et al. 2000; Jessen and Mirsky 2008). Both of these processes, namely myelination and cellular division, require a rapid and extensive change in gene expression (Scherer 1997). Here we characterize the miRNAome of actively-dividing Schwann cells and compare it to non-proliferating cells cultured in defined media (Cheng and Mudge 1996). There is unique expression of miRNAs based upon growth condition (Fig. 4), suggesting there are different subsets of genes that are post-transcriptionally regulated depending on the cells phenotype. It is also possible that certain genes are preferentially regulated by miRNAs in one of these growth conditions. Our data suggest that PMP22 is regulated by miRNAs primarily in proliferating cells where the expression of miR-29a is highest (Fig. 4). It will be of interest to determine the target messages of the additional differentially regulated miRNAs in Schwann cells and search for their possible roles in neuropathic states or in developmental abnormalities. Other PMP22 binding miRNAs that did not change their relative expression levels might be implicated in conferring cellular identity and controlling leaky transcription (Mattick and Makunin 2005). Such mechanism would support the detection of PMP22 RNA throughout the body and the restricted distribution of the protein (Amici et al. 2006; Baechner et al. 1995).
MiRNAs have been implicated in several disease phenotypes, including Alzheimer’s disease, cancer and heart disease (Blenkiron and Miska 2007; Hebert et al. 2008; van Rooij and Olson 2007), which quickly led to novel therapeutic approaches utilizing artificial miRNAs for treatment (Hammond 2006; McBride et al. 2008). Although no reports have directly associated miRNA regulation with peripheral nerve health, recent observations show that PNS axons contain functional effector complexes (Murashov et al. 2007) as transfection with siRNAs into distal axons selectively downregulated the target and abolished Sema3A-dependent growth cone collapse (Hengst et al. 2006). Since siRNA and miRNA pathways share common effector proteins, these observations indicate that peripheral nerve function may be regulated by the miRNA biosynthetic pathway. The identification of neuropathic patients with auto-immunity to GWBs supports this concept (Bhanji et al. 2007). Although it is not known whether the neuropathy is axonal or glial in origin, it is tempting to hypothesize that any impairment in the miRNA machinery in Schwann cells may alter myelin gene expression and lead to neuropathy. Since certain myelin genes, including PMP22, are dose-sensitive (Berger et al. 2006), loss of a required regulatory process could result in abnormal gene dosages, a mechanism known to lead to disease (Roa et al. 1991). The elevated levels of miR-29 in response to sciatic nerve crush injury and the inverse relationship to PMP22 (Fig. 9) suggest that the miRNA pathway responds to nerve injury and may be involved in regulating the demyelination process.
MiRNAs have been shown to exert their function by either signaling for mRNA degradation via siRNA-like mechanisms, or inhibiting translation (Bagga et al. 2005; Pillai et al. 2005). We provide evidence that miR-29a ultimately does lead to reduced steady-state PMP22 RNA levels (Fig. 7C). It is currently unknown if other myelin genes, such as myelin basic protein or myelin protein zero, are co-regulated by the same miRNAs. Myelin basic protein message has been shown to be translationally repressed and transported for local synthesis in oligodendrocytes (Gould et al. 2000), a mechanism that may involve miRNAs. Nevertheless, the miRNA regulation of PMP22 appears to be cell-type specific, as in oligodendrocytes we identified additional brain-enriched miRNAs that target PMP22 (Lau et al. 2008). Together, these data show that the individual miRNAome of the cell may contribute to refining the genetic profile, whereas more than one miRNA may target a specific RNA dependent on cell type.
In conclusion, here we demonstrate that a disease-associated peripheral myelin gene is regulated by miRNAs. Elucidating this pathway will provide novel insights to the understanding of the molecular signals and mechanisms required for myelination. In addition, miRNAs may be therapeutic tools for diseases associated with altered gene dose in glial cells, such as in demyelinating neuropathies. Future studies will seek to characterize the role of miRNAs in the myelination process, and peripheral nerve development and injury.
Acknowledgments
We thank and acknowledge the Intramural Program of National Institute of Neurological Disease and Stroke (LH), National Research Service Award 1F31NS061465 (JDV), the National Muscular Dystrophy Association, the National Institutes of Health NS041012, the McKnight Brain Institute of the University of Florida, and the Joshua Benjamin Weitzel Fund for Developmental Neurobiology (LN) for partial support of this project. We thank the Chan laboratory (University of Florida, Gainesville, FL) for the human anti-GWB antibody and the Hannon laboratory (Cold Springs Harbor, NY) for providing the c-myc-Ago2 expression plasmid. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disease and Stroke or the National Institutes of Health.
Abbreviations
- PMP22
Peripheral myelin protein 22
- DMEM
Dulbecco’s Modified Eagle’s Medium
- FCS
Fetal calf serum
- 3′UTR
3′untranslated region
- RL
Renilla luciferase
- FL
Firefly luciferase
- GWB
GW-bodies
- RISC
RNA-induced silencing complex
- Neg
negative
References
- Amici SA, Dunn WA, Jr, Murphy AJ, Adams NC, Gale NW, Valenzuela DM, Yancopoulos GD, Notterpek L. Peripheral myelin protein 22 is in complex with alpha6beta4 integrin, and its absence alters the Schwann cell basal lamina. J Neurosci. 2006;26(4):1179–89. doi: 10.1523/JNEUROSCI.2618-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. 2005;10(3):309–22. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- Baechner D, Liehr T, Hameister H, Altenberger H, Grehl H, Suter U, Rautenstrauss B. Widespread expression of the peripheral myelin protein-22 gene (PMP22) in neural and non-neural tissues during murine development. J Neurosci Res. 1995;42(6):733–41. doi: 10.1002/jnr.490420602. [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122(4):553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Berger P, Niemann A, Suter U. Schwann cells and the pathogenesis of inherited motor and sensory neuropathies (Charcot-Marie-Tooth disease) Glia. 2006;54(4):243–57. doi: 10.1002/glia.20386. [DOI] [PubMed] [Google Scholar]
- Bhanji RA, Eystathioy T, Chan EK, Bloch DB, Fritzler MJ. Clinical and serological features of patients with autoantibodies to GW/P bodies. Clin Immunol. 2007;125(3):247–56. doi: 10.1016/j.clim.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron C, Miska EA. miRNAs in cancer: approaches, aetiology, diagnostics and therapy. Hum Mol Genet. 2007;16:R106–13. doi: 10.1093/hmg/ddm056. Spec No 1. [DOI] [PubMed] [Google Scholar]
- Bosse F, Brodbeck J, Muller HW. Post-transcriptional regulation of the peripheral myelin protein gene PMP22/gas3. J Neurosci Res. 1999;55(2):164–77. doi: 10.1002/(SICI)1097-4547(19990115)55:2<164::AID-JNR4>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Bosse F, Hasenpusch-Theil K, Kury P, Muller HW. Gene expression profiling reveals that peripheral nerve regeneration is a consequence of both novel injury-dependent and reactivated developmental processes. J Neurochem. 2006;96(5):1441–57. doi: 10.1111/j.1471-4159.2005.03635.x. [DOI] [PubMed] [Google Scholar]
- Chance PF, Alderson MK, Leppig KA, Lensch MW, Matsunami N, Smith B, Swanson PD, Odelberg SJ, Disteche CM, Bird TD. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72(1):143–51. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105(6):2111–6. doi: 10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Mudge AW. Cultured Schwann cells constitutively express the myelin protein P0. Neuron. 1996;16(2):309–19. doi: 10.1016/s0896-6273(00)80049-5. [DOI] [PubMed] [Google Scholar]
- Clemence A, Mirsky R, Jessen KR. Non-myelin-forming Schwann cells proliferate rapidly during Wallerian degeneration in the rat sciatic nerve. J Neurocytol. 1989;18(2):185–92. doi: 10.1007/BF01206661. [DOI] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38(7):813–8. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- Damiani D, Alexander JJ, O’Rourke JR, McManus M, Jadhav AP, Cepko CL, Hauswirth WW, Harfe BD, Strettoi E. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J Neurosci. 2008;28(19):4878–87. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28(17):4322–30. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17(8):411–6. doi: 10.1016/j.tcb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Dracheva S, Davis KL, Chin B, Woo DA, Schmeidler J, Haroutunian V. Myelin-associated mRNA and protein expression deficits in the anterior cingulate cortex and hippocampus in elderly schizophrenia patients. Neurobiol Dis. 2006;21(3):531–40. doi: 10.1016/j.nbd.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13(4):1338–51. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbay B, Heape AM, Sargueil F, Cassagne C. Myelin synthesis in the peripheral nervous system. Prog Neurobiol. 2000;61(3):267–304. doi: 10.1016/s0301-0082(99)00049-0. [DOI] [PubMed] [Google Scholar]
- Gould RM, Freund CM, Palmer F, Feinstein DL. Messenger RNAs located in myelin sheath assembly sites. J Neurochem. 2000;75(5):1834–44. doi: 10.1046/j.1471-4159.2000.0751834.x. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM. MicroRNA therapeutics: a new niche for antisense nucleic acids. Trends Mol Med. 2006;12(3):99–101. doi: 10.1016/j.molmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102(52):19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105(17):6415–20. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst U, Cox LJ, Macosko EZ, Jaffrey SR. Functional and selective RNA interference in developing axons and growth cones. J Neurosci. 2006;26(21):5727–32. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islamov RR, Chintalgattu V, Pak ES, Katwa LC, Murashov AK. Induction of VEGF and its Flt-1 receptor after sciatic nerve crush injury. Neuroreport. 2004;15(13):2117–21. doi: 10.1097/00001756-200409150-00024. [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. Negative regulation of myelination: relevance for development, injury, and demyelinating disease. Glia. 2008;56(14):1552–65. doi: 10.1002/glia.20761. [DOI] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120(5):623–34. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A. 2007;104(49):19291–6. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39(10):1278–84. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28(45):11720–30. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian S, Jakymiw A, Eystathioy T, Hamel JC, Fritzler MJ, Chan EK. GW bodies, microRNAs and the cell cycle. Cell Cycle. 2006;5(3):242–5. doi: 10.4161/cc.5.3.2410. [DOI] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7(7):719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Garcia CA. Molecular genetics and neuropathology of Charcot-Marie-Tooth disease type 1A. Brain Pathol. 1992;2(4):337–49. doi: 10.1111/j.1750-3639.1992.tb00710.x. [DOI] [PubMed] [Google Scholar]
- Maier M, Berger P, Nave KA, Suter U. Identification of the regulatory region of the peripheral myelin protein 22 (PMP22) gene that directs temporal and spatial expression in development and regeneration of peripheral nerves. Mol Cell Neurosci. 2002;20(1):93–109. doi: 10.1006/mcne.2002.1116. [DOI] [PubMed] [Google Scholar]
- Maier M, Castagner F, Berger P, Suter U. Distinct elements of the peripheral myelin protein 22 (PMP22) promoter regulate expression in Schwann cells and sensory neurons. Mol Cell Neurosci. 2003;24(3):803–17. doi: 10.1016/s1044-7431(03)00246-x. [DOI] [PubMed] [Google Scholar]
- Manfioletti G, Ruaro ME, Del Sal G, Philipson L, Schneider C. A growth arrest-specific (gas) gene codes for a membrane protein. Mol Cell Biol. 1990;10(6):2924–30. doi: 10.1128/mcb.10.6.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14(Spec No 1):R121–32. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105(15):5868–73. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185–97. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Murashov AK, Chintalgattu V, Islamov RR, Lever TE, Pak ES, Sierpinski PL, Katwa LC, Van Scott MR. RNAi pathway is functional in peripheral nerve axons. Faseb J. 2007;21(3):656–70. doi: 10.1096/fj.06-6155com. [DOI] [PubMed] [Google Scholar]
- Nobbio L, Vigo T, Abbruzzese M, Levi G, Brancolini C, Mantero S, Grandis M, Benedetti L, Mancardi G, Schenone A. Impairment of PMP22 transgenic Schwann cells differentiation in culture: implications for Charcot-Marie-Tooth type 1A disease. Neurobiol Dis. 2004;16(1):263–73. doi: 10.1016/j.nbd.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Notterpek L, Roux KJ, Amici SA, Yazdanpour A, Rahner C, Fletcher BS. Peripheral myelin protein 22 is a constituent of intercellular junctions in epithelia. Proc Natl Acad Sci U S A. 2001;98(25):14404–9. doi: 10.1073/pnas.251548398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notterpek L, Ryan MC, Tobler AR, Shooter EM. PMP22 accumulation in aggresomes: implications for CMT1A pathology. Neurobiol Dis. 1999;6(5):450–60. doi: 10.1006/nbdi.1999.0274. [DOI] [PubMed] [Google Scholar]
- O’Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, Harfe BD. Essential role for Dicer during skeletal muscle development. Dev Biol. 2007;311(2):359–68. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek S, Notterpek L, Snipes GJ, Naef R, Sossin W, Laliberte J, Iacampo S, Suter U, Shooter EM, Murphy RA. Neurons promote the translocation of peripheral myelin protein 22 into myelin. J Neurosci. 1997;17(20):7754–62. doi: 10.1523/JNEUROSCI.17-20-07754.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek S, Suter U, Snipes GJ, Welcher AA, Shooter EM, Murphy RA. Detection and processing of peripheral myelin protein PMP22 in cultured Schwann cells. J Biol Chem. 1993;268(14):10372–9. [PubMed] [Google Scholar]
- Patel PI, Roa BB, Welcher AA, Schoener-Scott R, Trask BJ, Pentao L, Snipes GJ, Garcia CA, Francke U, Shooter EM, et al. The gene for the peripheral myelin protein PMP-22 is a candidate for Charcot-Marie-Tooth disease type 1A. Nat Genet. 1992;1(3):159–65. doi: 10.1038/ng0692-159. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309(5740):1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Roa BB, Garcia CA, Lupski JR. Charcot-Marie-Tooth disease type 1A: molecular mechanisms of gene dosage and point mutation underlying a common inherited peripheral neuropathy. Int J Neurol. 1991;25–26:97–107. [PubMed] [Google Scholar]
- Roux KJ, Amici SA, Notterpek L. The temporospatial expression of peripheral myelin protein 22 at the developing blood-nerve and blood-brain barriers. J Comp Neurol. 2004;474(4):578–88. doi: 10.1002/cne.20154. [DOI] [PubMed] [Google Scholar]
- Ryan MC, Shooter EM, Notterpek L. Aggresome formation in neuropathy models based on peripheral myelin protein 22 mutations. Neurobiol Dis. 2002;10(2):109–18. doi: 10.1006/nbdi.2002.0500. [DOI] [PubMed] [Google Scholar]
- Scherer SS. The biology and pathobiology of Schwann cells. Curr Opin Neurol. 1997;10(5):386–97. doi: 10.1097/00019052-199710000-00006. [DOI] [PubMed] [Google Scholar]
- Schneider C, King RM, Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54(6):787–93. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- Snipes GJ, Suter U, Welcher AA, Shooter EM. Characterization of a novel peripheral nervous system myelin protein (PMP-22/SR13) J Cell Biol. 1992;117(1):225–38. doi: 10.1083/jcb.117.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter U, Snipes GJ, Schoener-Scott R, Welcher AA, Pareek S, Lupski JR, Murphy RA, Shooter EM, Patel PI. Regulation of tissue-specific expression of alternative peripheral myelin protein-22 (PMP22) gene transcripts by two promoters. J Biol Chem. 1994;269(41):25795–808. [PubMed] [Google Scholar]
- Svaren J, Meijer D. The molecular machinery of myelin gene transcription in Schwann cells. Glia. 2008;56(14):1541–51. doi: 10.1002/glia.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- van Dartel M, Hulsebos TJ. Characterization of PMP22 expression in osteosarcoma. Cancer Genet Cytogenet. 2004;152(2):113–8. doi: 10.1016/j.cancergencyto.2003.11.003. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117(9):2369–76. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M. Transcriptional control in myelinating glia: the basic recipe. Glia. 2000;29(2):118–23. [PubMed] [Google Scholar]
- Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell. 2008;29(1):1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EK. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci. 2004;117(Pt 23):5567–78. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- Zoidl G, Blass-Kampmann S, D’Urso D, Schmalenbach C, Muller HW. Retroviral-mediated gene transfer of the peripheral myelin protein PMP22 in Schwann cells: modulation of cell growth. Embo J. 1995;14(6):1122–8. doi: 10.1002/j.1460-2075.1995.tb07095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]