Abstract
The well-recognized phospholipids (PLs) of Mycobacterium tuberculosis (Mtb) include several acidic species such as phosphatidylglycerol (PG), cardiolipin, phosphatidylinositol and its mannoside derivatives, in addition to a single basic species, phosphatidylethanolamine. Here we demonstrate that an additional basic PL, lysinylated PG (L-PG), is a component of the PLs of Mtb H37Rv and that the lysX gene encoding the two-domain lysyl-transferase (mprF)-lysyl-tRNA synthetase (lysU) protein is responsible for L-PG production. The Mtb lysX mutant is sensitive to cationic antibiotics and peptides, shows increased association with lysosome-associated membrane protein–positive vesicles, and it exhibits altered membrane potential compared to wild type. A lysX complementing strain expressing the intact lysX gene, but not one expressing mprF alone, restored the production of L-PG and rescued the lysX mutant phenotypes, indicating that the expression of both proteins is required for LysX function. The lysX mutant also showed defective growth in mouse and guinea pig lungs and showed reduced pathology relative to wild type, indicating that LysX activity is required for full virulence. Together, our results suggest that LysX-mediated production of L-PG is necessary for the maintenance of optimal membrane integrity and for survival of the pathogen upon infection.
Author Summary
The human pathogen Mycobacterium tuberculosis (Mtb) survives in the hostile intracellular environment, in part, by withstanding the actions of host-induced cationic antimicrobial peptides (CAMPs). Membrane phospholipid composition and the resultant charge could play an important role in Mtb survival within the host. Acidic phospholipids such as cardiolipin, phosphatidylinositol and its mannoside derivatives, phosphatidylglycerol, and a single basic species, phosphatidylethanolamine, are constituents of the Mtb membrane bilayer. We demonstrate that lysinylated phosphatidylglycerol (L-PG) represents another basic phospholipid and that the lysX gene, which encodes a two-domain protein with lysyl transferase and lysyl-tRNA synthase activities, is necessary for L-PG production. We show that L-PG is required for maintenance of an optimal membrane potential and resistance towards CAMPs. Phagosomes containing the lysX mutant showed an increased association with lysosomes, and the lysX mutant showed growth defects in mouse and guinea pig lungs, indicating that LysX activity is required for full virulence. Collectively, our results suggest that LysX activity, which is responsible for the production of L-PG, is necessary for maintenance of an optimal membrane potential such that the pathogen can grow optimally upon infection, presumably by withstanding the actions of CAMPs.
Introduction
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis, is a successful human pathogen that has infected more than one-third of the world's population. The success of Mtb as an infectious agent relies, in part, on its ability to modulate the expression of bacterial factors in response to infection so that it can successfully multiply within the hostile host environment [1]. The characteristic lipid-rich cell envelope of Mtb is one of the factors believed to contribute to its survival in vivo [2],[3]. It is generally believed that Mtb polar lipids (PoLs) consisting of acidic phospholipids (PL) such as cardiolipin (CL), phosphatidylglycerol (PG), phosphatidylinositol and its mannoside derivatives, in addition to basic PL such as phosphatidylethanolamine, are important constituents of the Mtb membrane [2]. Mtb PLs are known to function as important immune modulators [4] and have been shown to be released within phagosomes and transferred into lysosomes [5],[6]. It is interesting to note that PG, which is an abundant PL in other bacteria, is only a minor species in Mycobacteria, whereas CL is a major species [2],[3] with a high turnover rate [7].
The relative ratio of acidic to basic PLs is one of the determinants of net membrane charge. In some Gram-positive pathogens such as Staphylococcus aureus and Listeria monocytogenes, a fraction of the PG or CL molecules, or both, are lysinylated by the esterification of a glycerol hydroxyl group to lysine. Lysinylation imparts a net positive charge to these acidic PLs. This could, in turn, influence the ratio of acidic to basic PLs, resulting in an altered membrane charge. This could explain the bacterial susceptibility to cationic antibiotics (CAMAs) and peptides (CAMPs) [8],[9]. Although Mtb PLs have been well characterized for more than four decades, it is unknown if lysinylated PLs are a subset of the Mtb PLs and, if so, what the consequences associated with the absence of these lysinylated PLs might be. The present study demonstrates that the Mtb lysX gene, encoding a two-domain protein, is required for the production of lysinylated PG (L-PG), and the absence of L-PG is associated with changes in membrane potential, increased sensitivity to CAMAs and CAMPs, and growth defects in vivo.
Results
Identification of Lysinylated PLs in Mtb
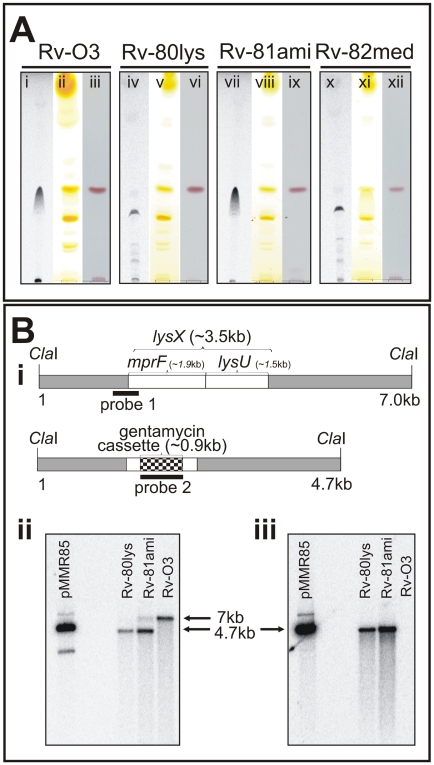
In order to detect lysinylated PLs in Mtb, actively growing cultures were incubated with 14C-lysine for 3 days; total lipids were extracted, and PoLs were resolved by thin layer chromatography (TLC). A distinct radiolabeled lipid was evident, indicating that lysinylated PLs are members of the Mtb PoL pool (Fig. 1A, lane i). In S. aureus, the mprF gene is responsible for L-PG production [8]. Homology searches of the Mtb genome identified Rv1640c as lysX, which encodes an mprF-like gene as a fusion to a lysyl-tRNA synthetase (lysU). The latter gene is distinct from the essential housekeeping tRNA synthetase (Rv3598c). The mprF gene in S. aureus encodes a protein with potential lysyl transferase activity [10]. In order to evaluate the function of lysX, we created a lysX mutant strain, Rv-80lys, by replacing the majority of the coding region comprising the mprF and lysU domains with a gentamycin resistance cassette using homologous recombination (see Methods section). A complementing derivative of this strain, Rv-81ami, was created by integrating a plasmid expressing the intact lysX gene under the control of the amidase promoter [11]. The lysX mutant strain was found to be defective in the production of L-PoLs (Fig. 1A, lane iv compared with lane i). L-PoL production was restored, however, in the lysX complemented strain Rv-81ami (Fig. 1A, see lane vii), confirming that the lysX gene product is responsible for the production of L-PoLs. Staining TLC plates with iodine (lanes ii, v, viii and xi) or ninhydrin (lanes iii, vi, ix and xii), on the other hand, did not detect L-PoLs, indicating that they may not be an abundant lipid species. We cultured Mtb in the presence of 14C-acetic acid and extracted total lipids, followed by TLC separation and subsequent quantification of L-PoL relative to total input radioactivity, and found that L-PoL accounts for approximately 0.3% of the total lipids (data not shown).
Figure 1. Polar lipid and Southern blot analysis of the lysX mutant strain.
A: Mtb strains were grown in the presence and absence of 14C-lysine. Total lipids were extracted in chloroform∶methanol (2∶1 v/v) and resolved by TLC on Silcia Gel 60 (EMD Chemicals, New Jersey) in a solvent system of chloroform∶methanol∶water (65∶25∶4 v/v/v). TLC plates were either visualized by autoradiography (lanes i, iv, vii and x), exposed to iodine vapors (lanes ii, v, viii and xi), or stained with ninhydrin (lanes iii, vi, ix and xii). B: Southern blot analysis of Mtb lysX mutant strains. B-i: The ClaI fragment bearing the wild type lysX gene (3.5 kb) with the locations of the mprF and lysU regions marked. The dark box designated as “probe 1” is an approximately 750 bp fragment that hybridizes with the 5′-end of lysX and 160 bp of the lysX coding region. The ClaI fragment bearing the mutant lysX gene disrupted with the gentamycin cassette (0.9 kb) is also shown. The dark band designated as “probe 2” is the 900 bp gentamycin gene that hybridizes with the mutant lysX gene. B-ii: Southern blot analysis of ClaI-digested Mtb genomic DNA hybridized with probe 1. The 7 kb and 4 kb band positions represent Rv-03 and Rv-80lys, respectively. Note that the complemented copy contains a band corresponding to the integrated copy of lysX gene plus the flanking plasmid sequence. B-iii: Southern blot analysis of ClaI-digested Mtb genomic DNA (see Fig. 1B-iii) hybridized with probe 2. pMMR85 is a positive control plasmid containing the mutant lysX gene plus flanking regions.
Structural analysis of the lysinylated polar lipid
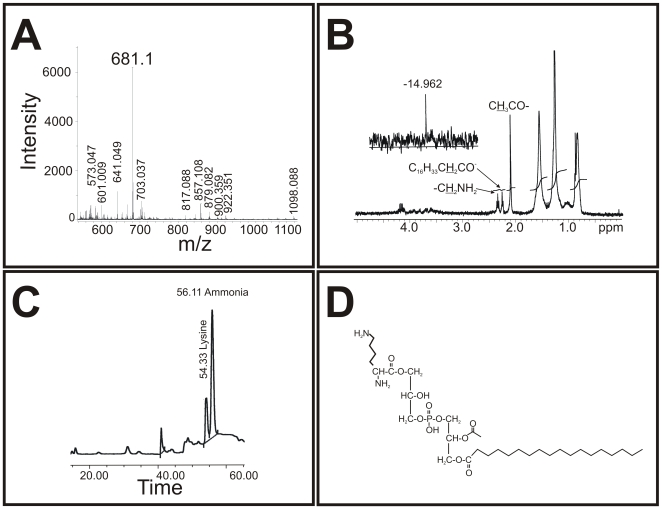
In order to determine the nature of the L-PoL, preparative 2D-TLC was carried out to collect L-PoLs. Structural analysis of the L-PoL was carried out using a combination of MALDI-MS, amino acid analysis and NMR (Fig. 2A–D). The MALDI-MS analysis in negative-ion mode revealed m/z 681 ([M-H]−) to be the molecular ion peak (Fig. 2A). The 1H-NMR results confirmed the presence of an acetyl group at δCH 3 2.1 ppm and δCH 2 from the primary amine in lysine at δ 2.4 ppm (Fig. 2B), whereas the 31P-NMR spectrum showed a shift in the phosphorus resonance spectrum at δ −14.96 ppm (Fig. 2B inset). Fatty acid analysis demonstrated that the molecule was C18 (data not shown), and amino acid analysis following acid hydrolysis confirmed the presence of lysine (Fig. 2C). Together, these data demonstrate that lysine is covalently linked to PG with the predicted structure shown in Fig. 2D. The L-PoL in the text is referred to hereafter as L-PG. Similar structural analyses of the corresponding unlabeled PoL of the slower migrating radioactive lipid species of lysX (Fig. 1A, lane iv) could not be done, given that it was present in negligible quantities (not shown). The thermal decomposition products of lysine are well characterized [12]. We speculate that the PoL accumulation in the lysX strain is a consequence of lysine degradation. Further studies are required to clarify the nature of the lipid species accumulating in the lysX mutant.
Figure 2. Structural analysis of L-PoL.
A: MALDI mass spectrometry analysis of L-PG in negative-ion mode. The m/z 681 [M-H]− represented the molecular ion peak. B: Structural analysis of L-PG. 1H-NMR spectrum of L-PG. The acetyl group can be found at δCH3 2.1 ppm, and δCH2 from the primary amine in lysine was detectable at 2.4 ppm. The inset shows the 31P-NMR spectrum, in which the phosphorus resonance shifted at −14.96 ppm. C: The amino acid profile of L-PG. Pure lipid was hydrolyzed with 6 N HCl for 24 h, and the soluble hydrolytic product was analyzed in an amino acid analyzer. Note that the elution of lysine at 54.33 min coincides with the standard (not shown). The small peak at position 41 min corresponds to a buffer change during the run, and the ammonia peak at 56.11 min is from the buffer used to run the analyzer. D: The proposed L-PG structure. The proposed structure of L-PG with a C18 fatty acid identified from fatty acid analysis (data not shown).
An intact lysX gene containing lysU and mprF domains is necessary for the production of L-PG
As previously noted, the Mtb lysX is a fusion gene encoding both mprF and lysU activities, with mprF located at the 5′ end of the lysX gene (see Fig. 1B). The Gram-positive bacteria that have been shown to produce L-PG, however, contain only mprF. The Mtb MprF and S. aureus MprF share three domains of unknown function, DUF470, DUF471 and DUF472. In order to evaluate whether L-PG production in Mtb requires the activities of both the LysU and MprF domains, we generated Rv-82med, a lysX complemented derivative that produces only the MprF domain (see Methods) and evaluated its ability to produce L-PG following the incubation of actively growing cells with radiolabeled lysine. The Rv-82med strain, much like Rv-80lys, was defective in L-PG production (see Fig. 1A, lane x and compare with lane iv). Quantitative real-time PCR analysis using primers and TaqMan probes targeted to the mprF region of lysX (compared to the 16S rRNA housekeeping gene) revealed that the expression of mprF in Rv-82med was comparable to that in Rv-03 wild type and Rv-81ami (data not shown). Together, these results indicate that Rv-82med expresses mprF and that the MprF domain alone is not sufficient for the production of L-PG in Mtb.
Phenotypes associated with the absence of L-PG production
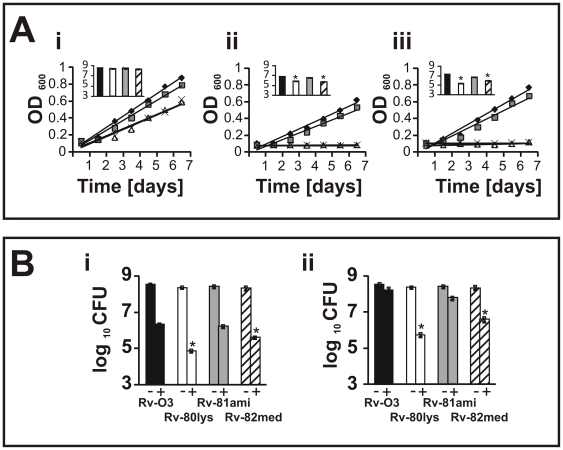
Gram-positive organisms such as S. aureus and B. subtilis are sensitive to cationic antimicrobial antibiotics (CAMAs) such as vancomycin (Van) and polymyxin-B (PMB) and to cationic antimicrobial peptides (CAMPs) such as human neutrophil peptide (HNP-1) and lysozyme. On the other hand, Mtb is generally tolerant to these compounds. HNP-1 and lysozyme are produced in neutrophils and macrophages, respectively. It is generally believed that CAMPs induce cell death by interfering with the integrity of the negatively charged membrane. Furthermore, the ability of intracellular pathogens to resist the action of CAMPs produced by the host is, in part, responsible for pathogen proliferation upon infection [13]. The presence of lysine groups on the acidic PG would impart a net positive charge and, therefore, could affect the net ratio of positively charged to negatively charged PL species. Thus, the absence of L-PG could make the Mtb membrane relatively acidic, thereby sensitizing the bacterium to the action of CAMAs. To test this possibility, actively growing Rv-03, Rv-80lys, Rv-80ami and Rv-82med were exposed to Van and PMB, and growth and viability were measured (Fig. 3A). Van and PMB interfered with the growth and viability of Rv-80lys and Rv-82med (see Fig. 3A-ii and 3A-iii), inset showing viability after 3 days of exposure; and Figure S1 showing viability after 6 days of exposure). Comparisons of growth, measured as the change in optical density (OD), and viability, measured as the change in CFU, revealed that while the lysX mutant was relatively more sensitive to Van and PMB than Rv-03, it was able to recover when grown in the absence of antibiotics, indicating that Van and PMB do not exert potent bactericidal activity. All of the strains grew well in the absence of antibiotics, although the lysX mutant showed a small reduction in growth rate in the absence of antibiotics (Fig. 3A-i), inset shows an approximately 0.3 log reduction in viability). Visualization of Rv-80lys cells following nucleoid staining and bright field or fluorescence microscopy did not reveal any significant differences in cell morphology or nucleoid organization (data not shown).
Figure 3. Phenotype of lysX strains.
Panel A: The growth and viability of the Mtb lysX strains in 7H9 broth in the absence of antibiotics -i. A-ii: The growth and viability of cultures grown in the presence of 1.0 µg/ml Van. At the indicated times, growth was measured. After 3 days of growth in broth, viability was determined by plating cells on Middlebrook 7H11 agar and determining the CFU. Symbols: Filled diamonds – Rv-03; grey squares – Rv81-ami; white triangles – Rv-80lys; crosses – Rv-82med. The inset shows the viable cell count. Black bars - Rv-03, grey bars - Rv-81ami, white bars - Rv-80lys and dashed bars - Rv82-med. A-iii: The growth and viability of cultures grown in the presence of 100 units/ml PMB. * Represents a P value<0.001 versus Rv-03 and Rv-81ami (Student-Newman-Keuls Method); bars represent means±standard error. Panel B: The growth and viability of Mtb strains in the presence of 1 mg/ml lysozyme -i- or HNP-1 -ii. There was no significant reduction in viability compared to cultures grown without TFA (data not shown). The stars represent P<0.001 versus Rv-03 and Rv-81ami (Student-Newman-Keuls Method). The bars represent the mean±standard error.
The increased sensitivity of the lysX strain to antibiotics suggests an enhanced association between the two. To test this possibility, actively growing lysX and Rv-03 cells were stained with fluorescent-vancomycin (Fl-Van), and the staining patterns were visualized by fluorescence microscopy. Earlier studies revealed that in stained Mtb cells, Fl-Van associates with the nascent growth zones, primarily at the poles and mid-cell septa [14]. These studies also indicated that not all Mtb cells could be stained with Fl-Van [14]. We found that a higher percentage of lysX cells were stained with Fl-Van compared to Rv-03 (see Figure S2). Approximately 52% of lysX cells showed staining patterns not only at the mid-cell and polar septa but also over the entire length; meanwhile, only 32% of wild type cells showed such a staining pattern (see Figure S2 legend for details). These results are consistent with the idea that Van is able to gain better access to Rv-80lys cells compared to Rv-03 cells.
Next, we examined whether the lysX mutant was also sensitive to lysozyme and HNP-1. Similar to the results seen with the CAMAs, HNP-1 and lysozyme significantly reduced the viability of Rv-80lys compared to Rv-03 and Rv-81ami (Fig. 3B-i and 3B-ii), see legends for P values). The phenotype of Rv-82med was found to be similar to that of Rv80lys (Fig. 3B). Together, these results indicate that the absence of L-PG production is associated with increased sensitivity of the bacterium to the actions of CAMPs and CAMAs. Importantly, these experiments also showed that complementation of the lysX mutant restored the wild type growth phenotype under these conditions (Fig. 3A and 3B).
Altered membrane potential of Mtb lysX mutants
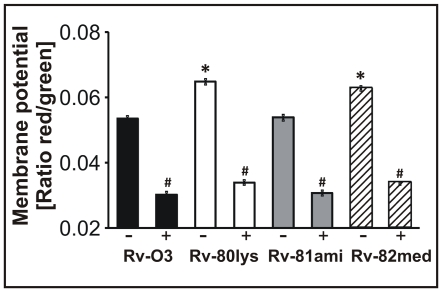
We wished to test whether the absence of L-PG production in Rv-80lys cells was associated with changes in the properties of the PL bilayer (e.g., membrane potential). The membrane potential of the Rv-80lys cells was determined using a slow-response membrane potential-sensitive dye, DiOC2(3), and comparing with Rv-03 cells. This cationic cyanine dye exhibits green fluorescence (Ex = 488 nm and Em = 520 nm) in the monomeric state and red fluorescence (Ex = 488 nm and Em = 620 nm) in the aggregated or oligomeric state. As a negative control, the membrane potential was measured following exposure of the cells to the proton ionophore m-chlorophenylhydrazone (CCCP), which is known to eliminate the proton gradient across the membrane. As seen in Figure 4, the membrane potentials (measured as the ratio of red to green fluorescence) of the lysX mutant Rv-80lys and Rv-82med were 21% and 17%, respectively, higher than that of the Rv-03 and complemented Rv-81ami (P<0.002). The increased ratio of red to green fluorescence observed in lysX mutants suggests accumulation of the positively charged lipophilic dye on the negatively charged membrane. The red to green fluorescence ratio in all strains was decreased to similar levels (∼41%) in the presence of CCCP (P = 0.001). Presumably, this reduction reflects the completely depolarized state of the membrane. Together, these results indicate that the membranes of Rv-80lys and Rv-82med are hyperpolarized relative to Rv-03 and Rv-81ami.
Figure 4. Determination of the membrane potential of Mtb strains.
The relative membrane potential was calculated using the average mean fluorescence intensity (the ratio between the average red fluorescence and the average green fluorescence). The graph shows the average of four experiments. Wild type and lysX mutant strains were incubated with 3 µM DiOC2(3) for 5 h in either the presence (+) or absence (−) of 100 µM CCCP. The bars represent the mean±standard error. * Represents P<0.002 versus Rv-03 and Rv-81ami (Student-Newman-Keuls Method), whereas # refers to P = 0.001 compared to untreated with CCCP.
lysX mutant phenotype in macrophages
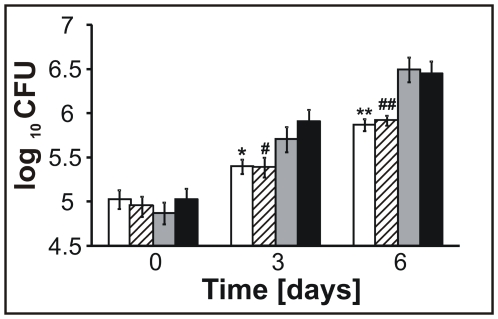
We next examined whether the Mtb lysX mutant Rv-80lys showed proficient growth in macrophages upon infection of the THP-1 macrophage cell line. The Rv-80lys showed a modest growth defect in macrophages compared to Rv-03 and complemented Rv-81ami (see Fig. 5, P = 0.01 for day 3, P = 0.006 for day 6). Similar results were also noted for Rv-82med (Fig. 5, P<0.014 for day 3 and P<0.03 for day 6 compared to Rv-03).
Figure 5. The viability of lysX in macrophages.
THP-1-derived macrophages were infected with Mtb strains; at the indicated times following infection, the macrophages were lysed, and the Mtb viability was determined. The white bars represent Rv-80lys; the dashed bars represent Rv-82med; the grey bars represent Rv-81ami; and the black bars represent Rv-03. Data are mean±standard error from three independent experiments, and the Mann-Whitney Rank Sum Test was used for data analyses.
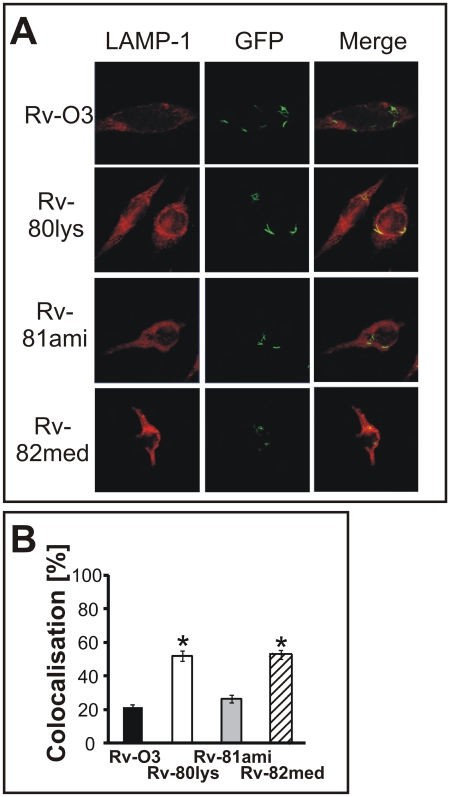
Intracellular replication of Mtb is, in part, due to its ability to resist the delivery of its phagosomes to lysosomes [15]. This process can be visualized by examining the co-localization of Mtb with the lysosome-associated-membrane protein (LAMP-1). In order to address whether the lysX mutant had a phagosome-lysosome fusion defect, we infected macrophages with Mtb strains expressing green-fluorescent protein and visualized co-localization with LAMP-1. Increased association of phagosomes containing Rv-80lys with lysosomes was evident compared to Rv-03 and complemented Rv-81ami (Fig. 6, p<0.001). Rv-82med behaved like Rv-80lys, indicating that the full-length lysX gene is required for functional activity. These results are consistent with the hypothesis that the lysX mutant is not as proficient as the Rv-03 strain in preventing fusion of phagosomes with lysosomes, which could contribute to defects in intramacrophage replication.
Figure 6. Co-localization of Mtb with LAMP-1-expressing phagosomes.
Panel A: THP-1-derived macrophages were infected with GFP-expressing Mtb strains (Rv-03, Rv-80lys, Rv-81ami and Rv-82med). Bacteria (green spots) inside LAMP-1-positive phagosomes (red) produce a yellow signal indicating co-localization (merged). Panel B: The percent co-localization was determined by visual scoring of yellow spots after 72 h of infection. We analyzed 126 macrophages for Rv-80lys, 164 for wild type, 168 for complemented, and 127 for Rv-82med and scored 534 bacterial cells for Rv-80lys, 1,307 for wild type, 848 for Rv-81ami, and 402 for Rv-82med. Mtb Rv-03 (black bar; 21.2±1.6%), Rv-80lys (white bar; 52.0±3.0%), Rv-81ami (grey bar; 26.4±2.2%) and Rv-82med (dashed bar; 52.6±2.8%). * P<0.001 versus Rv-03 and Rv-81ami using the Student-Newman-Keuls Method; bars represent mean±standard error.
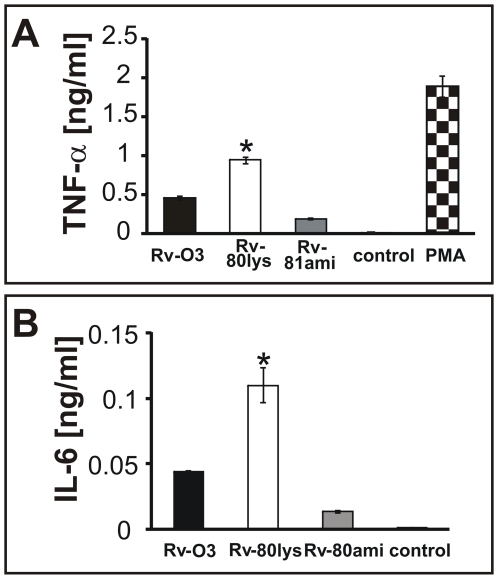
The production of inflammatory cytokines tumor necrosis factor-alpha (TNF-α), IL-6 and IL-10 is necessary to mount a protective immune response against Mtb infection [16],[17],[18]. TNF-alpha restricts the growth of Mtb in alveolar macrophages [18], and the multiplication of virulent Mtb in monocyte-derived macrophages (MDMO) is associated with suppression of TNF-α production during the early periods after infection. To test selected pro-inflammatory cytokine responses of macrophages, MDMO were infected with Rv-80lys and Rv-03 strains, and the production of TNF-α and IL-6 was measured (Fig. 7). As can be seen, the secretion of TNF-α was elevated after infection with the lysX mutant compared to the wild type and complemented strains, similar to MDMO cells exposed to PMA (Fig. 7A). Similarly, the secretion of IL-6 was also increased following infection with the lysX mutant compared to the wild type and complemented strains (Fig. 7B).
Figure 7. Select cytokine responses of macrophages.
Panel A: MDMO were infected with Mtb strains, and TNF-α release was measured by ELISA. PMA was used as a positive control. TNF-α levels in panel A were measured after 24 h. * Represents P<0.05 compared to Rv-03, Rv-81ami, control (untreated cells) and PMA. Panel B: Secretion of IL-6 by MDMO after 48 h of infection. * P<0.05 compared to Rv-03, Rv-81ami and control. The experiments were done in duplicate, and representative results are shown. Data are mean±standard error. Student-Newman-Keuls Method was used.
lysX has growth defects in vivo
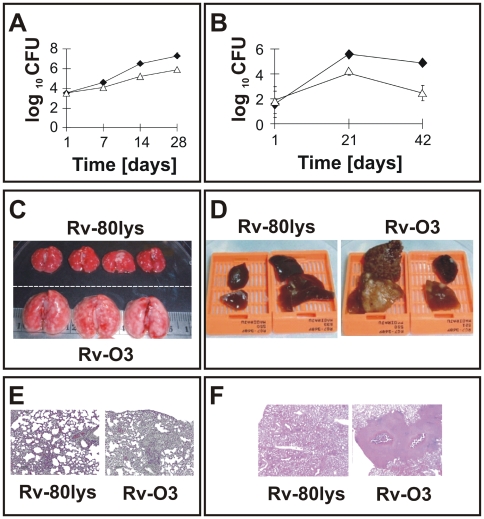
To evaluate the phenotype of lysX in vivo, C57BL/6 mice and Hartley strains of guinea pigs were aerosol infected with lysX, and the viability of the pathogen was measured (Fig. 8). The lysX mutant showed only a modest growth defect in mice (Fig. 8A) but was clearly attenuated in guinea pigs (Fig. 8B) and showed reduced dissemination to the spleen (Figure S4). Gross pathology and histopathology of the lungs of infected mice at 28 days (Fig. 8C and E) and guinea pigs at 42 days (Fig. 8D and F) showed distinct differences between the wild type and the lysX mutant. Hematoxylin-eosin staining confirmed that the lungs infected with Rv-03, but not those infected with Rv-80lys, had extensive inflammation in both species and showed caseating granulomas in guinea pigs. The vast differences in the growth kinetics and pathology between the wild type and lysX mutant pathogens indicate that LysX activity is required for full virulence. Interestingly, the lysX complemented strain Rv-81ami behaved like the lysX mutant with respect to in vivo growth and pathology (data not shown), indicating that the complemented strain is not able to restore lysX function in vivo.
Figure 8. Growth of lysX and WT strains in vivo.
Panel A: C57BL/6 female mouse lungs were infected with lysX (triangles) and WT (dark diamonds) strains via an aerosol route. The mean CFU counts were obtained from lung homogenates of at least three mice per group and plated on Middlebrook 7H10 agar plates. All plates were incubated at 37°C for at least three weeks before colonies were counted. Panel B: Guinea pigs (Hartley strain) were infected with lysX (open symbols) or WT (dark symbols) strains via an aerosol route. Five guinea pigs were sacrificed at days 1, 21 and 42 days after infection, and the survival of Mtb strains in the entire lung homogenate was determined by plating on agar medium as described above. Panels C, D: Gross pathology of the lungs infected with the Rv-80lys and Rv-03 Mtb strains. Lungs from mouse -C- and guinea pig -D- were excised, stored in 10% formalin, embedded, and stained with hematoxylin and eosin for histopathological analysis. Note the presence of tubercles on the surface of the lungs for Rv-03 compared to Rv-80lys. Panels E, F: Histopathology of mouse -E- and guinea pig -F- lungs infected with the Rv-80lys and Rv-03 Mtb strains. Hematoxylin-eosin staining confirmed that the lungs infected with Rv-03, but not those infected with Rv-80lys, had extensive inflammation in both species and showed caseating granulomas in guinea pigs.
The ability of Mtb strains to produce complex cell wall-associated lipids called phthiocerol dimycocerosates (PDIM) and to bind and reduce neutral red dye is associated with virulence. Avirulent and attenuated strains are defective in these processes. Furthermore, virulent Mtb strains propagated in the laboratory often lose these properties [2],[19],[20],[21]. The neutral red reduction and PDIM profiles of the lysX mutant were comparable to those of wild type cells (Figure S5), indicating that the observed in vivo growth defects of the lysX mutant are not due to a loss of PDIM and defect in neutral red reduction.
Discussion
The primary conclusion of our data is that L-PG is one of the basic PLs in Mtb and that the lysX gene, encoding the two-domain LysX protein, is responsible for its production. Although L-PG is a minor PL species of Mtb, its absence has several consequences, one of which is an alteration of the membrane potential. This underscores the role of LysX activity in maintaining optimal membrane function. Presumably, the absence of L-PG in the lysX mutant shifts the ratio of acidic to basic PLs, thereby hyperpolarizing the membrane. A consequence of the absence of L-PG is the increased sensitivity of the pathogen to lipophilic antibiotics such as PMB and Van. It is likely that hyperpolarization of the membrane in the lysX mutants due to its net negative charge promotes interactions with cationic peptides and antibiotics produced by the host immune system, which in turn could lead to the killing of the invading pathogens [13],[22]. It is known that host-induced CAMPs are one of the frontline defenses against invading pathogens. Therefore, sensitization of lysX mutant Mtb cells to the action of CAMPs suggests that maintenance of the optimal membrane potential is necessary for Mtb growth in vivo.
In partial support of this claim, we found that the lysX mutant showed defects in intracellular replication (Fig. 5) and that infection of macrophages with lysX led to increased production of pro-inflammatory cytokines (Fig. 7). We also found that the lysX mutant showed increased co-localization with LAMP-1 vesicles (Fig. 6). Finally, we showed that the lysX mutant was attenuated in guinea pig lungs and had a modest growth defect in mouse lungs (Fig. 9). Together, these results are consistent with the hypothesis that LysX activity is required to maintain an optimal membrane potential and possibly to promote pathogen survival upon infection. Notably, the gross pathological differences between the lysX mutant and wild type were striking compared to the modest differences in growth in vitro and ex vivo (see Figs. 3, 5 and 8). The reduced bacterial burden and the reduced pathology and size of granulomas in the lungs of guinea pigs clearly suggest that LysX activity is required for bacterial multiplication and virulence. Evaluation of the host-induced cytokine response following different stages of infection with wild type and lysX mutant pathogens could provide valuable insights into lysX function. Our studies also showed that the lysX mutant, like wild type, retained the ability to produce PDIMs and reduce neutral red (Fig. S5). It remains to be evaluated, however, if other membrane and cell wall-associated lipids are modulated in the lysX background.
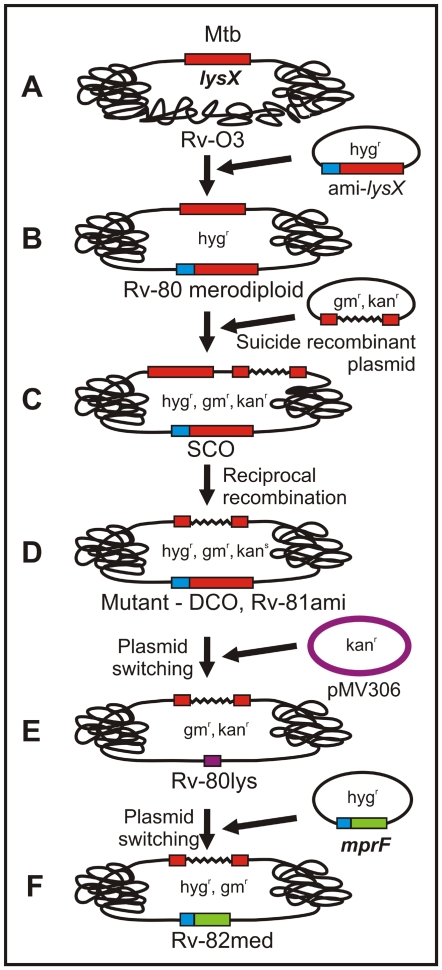
Figure 9. Cartoon showing construction of the lysX mutant and complemented strains.
A- refers to the wild type strain carrying the lysX gene at its native locus. B- refers to the Rv-80 merodiploid (hygr) strain produced by the integration of a plasmid expressing the lysX gene from the amidase promoter. C- refers to a single crossover (SCO, hygr, kanr, gmr) recombinant produced by integration of the suicide recombination plasmid. D- refers to a mutant double crossover (DCO, hygr, kans, gmr) produced following a reciprocal recombination event. E- refers to Rv-80lys (kanr, gmr) produced following switching of the resident integrated plasmid (hygr) with the incoming pMV306 plasmid (kanr). F- refers to Rv-82med (hygr, gmr) produced following switching of the resident pMV306 plasmid (kanr) with the incoming plasmid expressing the mprF fragment from the amidase promoter (hygr).
The production of L-PG is believed to involve two biochemical steps: the generation of lysyl-tRNA by the LysU protein and the transfer of a lysine group from the lysyl-tRNA to PG by MprF, a membrane-bound lysyl-transferase protein [23]. The Gram-positive bacteria shown to produce L-PG carry a single housekeeping lysU gene that encodes a cytosolic LysU protein [8],[9],[24]. E. coli does not contain L-PG, but ectopic expression of the S. aureus mprF gene allows E. coli to accumulate L-PG in their membranes, suggesting that cytosolic LysU and membrane-bound MprF cooperate to produce L-PG [10],[25]. Mtb contains two lysU genes, one encoded by Rv3598c, which is an essential gene, and the other encoded by the lysU domain of lysX [26]. Since expression of the mprF fragment of lysX does not lead to the production of L-PG (Fig. 2), it appears that in Mtb, unlike in other bacteria, the cytosolic LysU and the membrane-bound MprF do not cooperate to produce L-PG.
This raises the question as to why a dedicated lysU gene product, distinct from the housekeeping gene, is required for L-PG production in Mtb. One possibility is that the lysinylation reaction occurs on the membrane, and the local presence of LysU and MprF activities are required to transfer lysine from the lysyl-tRNA to the membrane-bound PG. If the cytosolic lysyl-tRNA could not diffuse through the Mtb plasma membrane, a separate activity would be needed to replace it. Nonetheless, such dedicated activities imply that PG lysinylation in Mtb is a tightly regulated reaction. The temporal expression profile of Mtb genes upon infection in mice shows that lysX is upregulated during acute and chronic infection [27]. Presumably, increased expression levels of lysX would ensure that sufficient levels of L-PG were produced to maintain the optimal ratio of acidic to basic PLs. This would, in turn, ensure that the optimal membrane potential required for Mtb proliferation upon infection is maintained. Another possibility, although unlikely, is that the demand for lysyl-tRNA required for lysinylation and protein synthesis cannot be met by a single housekeeping enzyme. Clearly, however, further studies are required to address this issue.
While this manuscript was in preparation, Vandal et al. reported the characterization of several transposon mutants of Mtb that were hypersensitive to acidic pH, one of which was lysX [28],[29]. Their transposon mutants were hypersensitive to antibiotics and other stressors such as heat, SDS and DETA-NO. Although the lysX mutant was moderately sensitive to DETA-NO, its growth was not attenuated in murine lungs. It is unknown whether L-PG is produced in the lysX transposon mutant and whether the lysX mutant shows any residual activity. As shown in Fig. 1A, our lysX mutant was generated by removing most of the coding sequence responsible for producing the mprF and lysU activities. We demonstrated that L-PG was not produced in the lysX mutant and that maintenance of the membrane potential and resistance to CAMPs were dependent on LysX activity. Importantly, we showed that LysX activity was required for full virulence in mice and guinea pigs. These results underscore the importance of lysX function in Mtb survival upon infection. One limitation of our results, however, is that the complemented Rv-81ami was not able to restore the lysX defect in vivo, although it did restore defects in other assays reported in this study. One possibility is that the expression of lysX in-trans at an attB locus was not sufficient to restore the LysX activity to optimal levels, and small changes in activity could have consequences for the complementation phenotype in vivo. Further studies are required to address this issue.
L-PG appears to be a minor lipid species, yet the loss of L-PG production affected membrane potential and Mtb growth in vitro and in vivo. It is interesting to note that PG, the purported substrate of L-PG, is also a minor lipid species in Mtb and other mycobacterial species [2],[3],[30],[31]. This raises the question of how the lysinylation of a minor PL species contributes to the observed phenotype. It is known that PG is a biosynthetic intermediate of CL, one of the major PL species of mycobacteria. Indeed, the enzymatic activities responsible for CL production from PG pools have been detected in mycobacteria [32]. PG also accumulates as a result of CL catabolism and, if unregulated, could be further processed to produce a diacylglycerol intermediate via the action of phospholipases [33],[34]. Accordingly, we speculate that the lysinylation step helps to prevent PG degradation such that the optimal membrane potential required for Mtb survival upon infection is maintained. Our results also suggest that changes in membrane potential are a potential mechanism for regulating CAMP sensitivity in Mtb and possibly in the mprF mutants of other bacteria; therefore, this could be exploited to develop novel antimicrobial compounds. It is tempting to speculate that by manipulating LysX activity, we could promote the action of other conventional antibiotics against Mtb.
Methods
Ethics statement
Mouse and guinea pig infection protocols were approved by the Animal Care and Use Committee at Johns Hopkins School of Medicine, Baltimore, MD for mice and at Texas A & M University, College Station, TX for guinea pigs, under the NIH contract (Tuberculosis Animal Research and Gene Evaluation Task force).
Strains, media and culture conditions
The Mtb strains were cultured in Middlebrook 7H9 broth supplemented with 10% OADC (oleic acid, albumin, dextrose, catalase) and 0.05% Tween-80. As needed, 50 µg/ml hygromycin (hyg), 10 µg/ml kanamycin (kan) or 50 µg/ml gentamycin (gm) was added. For the determination of viable colonies and the scoring of recombinants, bacterial cells were plated on Middlebrook 7H11 agar plates containing the appropriate antibiotics. In some experiments, cultures were grown in the presence of L-[U-14C]-lysine (300 mCi/mmol, Amersham Pharmacia Biotech) or [1,2-14C] acetic acid (46 mCi/mmol, PerkinElmer), and total lipids were extracted and resolved by TLC. The radioactivity present in the L-PG spot was determined and normalized relative to total in put radioactivity.
Construction of the lysX deletion and complemented derivative strains
The lysX coding region was cloned downstream of the amidase promoter in an integration-proficient, hygromycin-resistant plasmid and electrotransformed into Mtb in order to generate the lysX merodiploid strain. The chromosomal copy of the lysX gene was disrupted in the lysX merodiploid background by homologous recombination as described previously [35]. Using this approach, 90% of the lysX coding region was replaced with a 900-bp gentamycin resistance cassette. This strain, designated as Rv-81ami, was the lysX complemented strain. Next, the resident integrated plasmid encoding the functional lysX gene was replaced with an empty kanamycin-resistant plasmid to generate the lysX mutant strain, designated as Rv-80lys, as described [14],[35],[36],[37]. A cartoon depicting the lysX mutant and complemented strain construction is shown (Fig. 9). All strains were confirmed by PCR and Southern blot analysis.
For the generation of the lysX complemented strain expressing the mprF domain, a 1,950 bp lysX gene encoding the mprF domain was amplified by PCR using the primers MVM530lysF (5′-GGCGAATTCCATATGGGACTCCACTTAACTG-3′) and lys650MM606R (5′-AGC AGCAAGCTTCTAGAATCACGCCAACCGCTCGGGACTGC-3′) and cloned into the pJFR19 vector under the control of the amidase promoter [38]. The integrity of cloned insert was verified by DNA sequencing. This recombinant plasmid was used to replace the resident empty plasmid in the Rv-80lys mutant to generate Rv82-med. This strain was confirmed by PCR and Southern blot analysis.
Intracellular growth measurements
The human monocyte cell line THP-1 (American Type Culture Collection, Rockville, Maryland) was used. Cells were grown in RPMI 1640 (Invitrogen, CA) supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, 10% fetal bovine serum (Invitrogen) and 100 U/ml penicillin G (Sigma, MO). The viability of the macrophages was determined using trypan blue staining. Monocytes were differentiated into macrophages by exposure to 50 nM PMA (phorbol 12-myristate 13-acetate; Sigma) and 7.5 ng/ml IFN-γ (human interferon-gamma, Peprotech) for 24 h, followed by a 24 h incubation with 50 nM PMA alone. The macrophages were washed three times with RPMI 1640 medium and incubated in medium that was not supplemented with PMA or IFN- γ for the next 24 h. Single cell suspensions of Mtb strains in RPMI 1640 media were used to infect 4.5×105 macrophages in triplicate in a 24-well plate at a multiplicity of infection of 1∶2–4. After 3 hours of infection, macrophages were lysed in 0.09% SDS, and viability was determined to get a t0 count. No statistical differences in viability among these strains were noted at t0. Subsequently, macrophages at 3 and 6 days post-infection were also processed in order to determine Mtb viability. The lysX strains were no more sensitive than the other strains in terms of the concentrations of SDS used to lyse the macrophages and process them for viability determination (see Figure S3).
Co-localization of M. tuberculosis with phagosomes expressing LAMP-1
THP-1 macrophages (5×105) attached to glass coverslips were infected with GFP-producing Mtb (Rv-03, Mtb Rv-80lys, Rv-81ami and Rv-82med). The macrophages were fixed, blocked and incubated with H4A3 monoclonal antibodies to LAMP-1, followed by a rhodamine-conjugated goat anti-mouse IgG, as described [38]. Bacterial co-localization with LAMP-1-positive vesicles appeared as yellow spots. The experiments were done in duplicate, and representative images are shown.
Cytokine measurement in MDMO
Peripheral blood mononuclear cells (PBMC) were isolated from healthy volunteers by differential gradient centrifugation on Ficoll-Paque Plus (Amersham Biosciences). Adherent monocytes were isolated by seeding 5×106 cells in 24-well plates in MDMO-media (RPMI 1640 supplemented with 10% heat-inactivated human serum) and incubating for 90 min at 37°C in 5% CO2. Following the removal of non-adherent cells, MDMO-media was added, and cells were incubated at 37°C for 4 days to mature into macrophages and then used for infection with Mtb strains. 5×105 macrophages were infected in triplicate in 24-well plates at a multiplicity of infection of 1∶5 as described previously [38]. At indicated periods of infection, supernatants were removed, and the TNF-α and IL-6 levels were measured using ELISA assays (eBioscience, Inc., CA) according to the manufacturer's instructions. In some experiments, MDMO were stimulated with 150 nM PMA, and the secretion of TNF-α was measured.
Isolation of mycobacterial lipids
The extraction of total lipids from whole cells using chloroform∶methanol (2∶1 v/v) and the separation of polar lipids in a solvent system containing chloroform∶methanol∶water (65∶25∶4) in the first dimension and chloroform∶methanol∶acetic acid∶water (80∶12∶16∶4) in the second dimension were performed as described previously [7],[32],[39]. Polar lipids were visualized by exposing the plates to iodine vapors or staining them with ninhydrin in order to detect amino acid-containing lipids. In some experiments, autoradiography was used to detect radiolabeled lipids. For PDIMs analysis hexane∶diethylether∶acetic acid (80∶20∶1, vol/vol/vol) solvent system was used.
MALDI-MS, ESI-MS and ESI-MS/MS
MALDI-MS was performed using an UltraFlex TOF/TOF (Bruker Daltonics, Billurica, CA) as described previously [39]. The L-PG sample in acetonitrile was mixed 1∶1 with 2,5-dihydroxylbenzoic acid matrix for spotting onto the target plate.
Fatty acid analysis
The L-PG was hydrolyzed with 3 N HCl in methanol for 4 h at 80°C. The sample was dried and treated with silylation reagent (TRI-SIL, Pierce Biotechnology, Rockford, IL) for 30 min at room temperature. The trimethylsilylated derivatives were analyzed by GC/MS. Specifically, the sample was applied to a DB-5 column at an initial temperature of 60°C for 1 min, then increased to 130°C at a rate of 30°C/min, and finally increased to 280°C at a rate of 5°C/min.
NMR spectroscopy
1H and 31PNMR were performed at a concentration of 2 mg L-PG sample per 0.6 mL of CDCl3 on a Varian Inova 400 MHz instrument.
Amino acid analysis
The purified L-PG was incubated overnight at 100°C in 6 N HCl in a heat-block. Samples were cooled, evaporated to dryness, resuspended in water and subjected to amino acid analysis.
Neutral red assay
Neutral red chemical staining of Mtb wild type and lysX mutants was carried out following the protocol described by Soto et al. [40].
Experiments evaluating the lysX phenotype
In order to evaluate the growth inhibitory effects of cationic compounds, Van (1 ug/mL), PMB (100 units/uL), human neutrophil peptide-1 (25 ug/mL) or lysozyme (0.5 mg/mL) was added to the growth media. The human neutrophil peptide stock was dissolved in 0.1% tri-fluoro acetic acid (TFA), and the cultures contained 0.025% TFA. No growth inhibition was noted at this concentration of TFA. The cultures were initially diluted to an OD600 of 0.05, dispensed into a 96-well microplate (100 uL per well) and incubated at 37°C with rotation at 60 rpm. At the indicated time periods, the change in the optical density (A600) was measured, and viability was determined. Low dose aerosol infection experiments of mice (C57BL/6 female mice) and guinea pigs (Hartely strains) for evaluating the growth and viability of lysX strain were essentially as described previously [41].
Membrane potential analysis
Cytoplasmic membrane potential changes were determined using the slow response, membrane potential-sensitive cyanine dye DiOC2(3) (Sigma). Briefly, actively growing cultures of Mtb strains (OD600 = 0.8) were incubated with 3 µM DiOC2(3) for 5 h. Spectrofluorometry was used to detect the red fluorescence (488 nm/620 nm) associated with aggregates of DiOC2(3), which exhibits green fluorescence (488 nm/520 nm) in the monomeric state. The assay was performed using white 96-well microtiter plates (Perkin Elmer; Waltham, MA) and a Cary Eclipse spectrofluorometer (Varian; Palo Alto, CA). A negative (depolarized) control of 100 µM m-chlorophenylhydrazone CCCP (Sigma), a proton ionophore that destroys the proton gradient and eliminates the bacterial membrane potential, was included. The membrane potential was measured as the ratio of red fluorescence (associated with membrane potential changes) to green fluorescence (a cell size-dependent, membrane potential-independent signal). Preliminary optimization studies revealed that 5 h incubation was optimal for the measurements.
Statistical analysis
Differences between groups were analyzed by multiple comparison procedures (Student-Newman-Keuls Method) with a simple one-way ANOVA or Mann Whitney Rank Sum Test, using SIGMASTAT (SPSS Science, Inc., Chicago, IL). A P value of less than 0.05 was considered significant.
Supporting Information
Viability of Mtb strains in the absence -A- and presence -B- of Van 1.0 µg/ml. At day six of growth, viability was determined by plating cells on Middlebrook 7H11 agar and counting. The bars represent mean±standard error.
(1.57 MB TIF)
Visualization of Fl-Van stained cells. Actively growing cultures (optical density ∼0.01 to 0.04) were grown with fluorescent vancomycin BODIPY (Invitrogen) at a final concentration of 1 µg/mL for 20 hours. The cells were harvested by low speed centrifugation, fixed in 4% paraformaldehyde for 24 h, and imaged on a Nikon Eclipse microscope with a CCD camera; bright field (BF) and fluorescent images (Van) were acquired. Magnification was 100×, and data were analyzed using Metamorph software. At least 100 cells were imaged for each strain; the images were scored for staining patterns and the presence or absence of fluorescent staining. Approximately 35% of wild type and 25% of lysX cells were not stained under these conditions. For clarity, only select staining patterns are shown: defined dye accumulation at the cell poles -A, D- and mid-cell (as in D) or diffuse accumulation throughout the cell - B, E. In some cells, both types of accumulation were observed -C, F. For the lysX mutant, about 52% of cells showed diffuse staining, but this was only seen in about 32% of wild type cells.
(2.97 MB TIF)
Effect of SDS on the viability of the lysX mutant: The viability of Mtb strains was examined under the same conditions used for macrophage infection and lysis in the presence of 0.09% SDS. Actively growing cultures of Mtb strains were harvested, exposed to 0.09% SDS for 3 min, diluted and spread on Middlebrook 7H11 agar plates. Cells untreated with SDS were processed similarly. All plates were incubated at 37°C, and colonies were counted. No statistically significant differences between the SDS treated and untreated groups were noted. Data shown are mean ± standard error.
(1.05 MB TIF)
Growth of the lysX mutant in the spleen of mice and guinea pigs. Growth of the lysX mutant and Rv-03 strain in the spleens of mice -A- and guinea pig -B. Following aerosol infection of mice and guinea pigs, spleens were harvested at the indicated time points. Homogenates were prepared, and viability was determined on agar plates. The lysX mutant showed reduced and/or delayed dissemination compared to wild type in both animal models.
(1.64 MB TIF)
Neutral red staining and PDIM analysis: Panel A: Wild type and lysX mutant cells were stained with neutral red and photographed. As a control, the attenuated strain Mtb H37ra was used. Panel B: The lipids were separated by silica thin-layer chromatography (TLC) with hexane:diethylether:acetic acid (80∶20∶1, vol/vol/vol) as solvent system. The lipids were visualized by spraying with 10% phosphormolybdate in ethanol followed by heating to about 110°C for 15 min.
(5.65 MB TIF)
Acknowledgments
We thank Dr. Barry Starcher for performing the amino acid analysis; Renata Dziedzic for help with the microscopy experiments; Dr. Paul Converse for his interest and helpful discussions at various stages of this work; Dr. Buka Samten for advice and help with the monocyte isolation protocols; and Dr. Chris Fancklyn for discussions during earlier stages of this work. The H4A3 hybridomas used in the present study were obtained from the Developmental Studies Hybridoma Bank under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242.
Footnotes
The authors have declared that no competing interests exist.
We gratefully acknowledge the support of National Institutes of Health (www.nih.niaid.gov) grants and contracts [AI41406, AI73966 (MM); AI48417 (MR); AI37139 (DC); AI 30036, AI 36973, and AI 37856 (WB)]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Smith I. Mycobacterium tuberculosis: pathogenesis and molecular determinants of virulence. Clin Micro Rev. 2003;16:463–496. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan PJ, Nikaido H. The envelope of mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 3.Goren MB. Mycobacterial lipids: selected topics. Bacteriol Rev. 1972;36:33–64. doi: 10.1128/br.36.1.33-64.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant EP, Degano M, Rosat JP, Stenger S, Modlin RL, et al. Molecular recognition of lipid antigens by T cell receptors. J Exp Med. 1999;189:195–205. doi: 10.1084/jem.189.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer K, Chatterjee D, Torrelles J, Brennan PJ, Kaufmann SH, et al. Mycobacterial lysocardiolipin is exported from phagosomes upon cleavage of cardiolipin by a macrophage-derived lysosomal phospholipase A2. J Immunol. 2001;167:2187–2192. doi: 10.4049/jimmunol.167.4.2187. [DOI] [PubMed] [Google Scholar]
- 6.Geisel RE, Sakamoto K, Russell DG, Rhoades ER. In vivo activity of released cell wall lipids of Mycobacterium bovis bacillus Calmette-Guerin is due principally to trehalose mycolates. J Immunol. 2005;174:5007–5015. doi: 10.4049/jimmunol.174.8.5007. [DOI] [PubMed] [Google Scholar]
- 7.Dhariwal KR, Chander A, Venkitasubramanian TA. Turnover of lipids in Mycobacterium smegmatis CDC 46 and Mycobacterium phlei ATCC 354. Arch Microbiol. 1978;116:69–75. doi: 10.1007/BF00408735. [DOI] [PubMed] [Google Scholar]
- 8.Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, et al. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol. 2006;62:1325–1339. doi: 10.1111/j.1365-2958.2006.05452.x. [DOI] [PubMed] [Google Scholar]
- 10.Staubitz P, Neumann H, Schneider T, Wiedemann I, Peschel A. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol Lett. 2004;231:67–71. doi: 10.1016/S0378-1097(03)00921-2. [DOI] [PubMed] [Google Scholar]
- 11.Parish T, Mahenthiralingam E, Draper P, Davis EO, Colston MJ. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology. 1997;143:2267–2276. doi: 10.1099/00221287-143-7-2267. [DOI] [PubMed] [Google Scholar]
- 12.Breitbart DJ, Nawar WW. Thermal decomposition of lysine. J Agric Food Chem. 1979;27:511–513. [Google Scholar]
- 13.Peschel A. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 2002;10:179–186. doi: 10.1016/s0966-842x(02)02333-8. [DOI] [PubMed] [Google Scholar]
- 14.Chauhan A, Lofton H, Maloney E, Moore J, Fol M, et al. Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol Microbiol. 2006;62:132–147. doi: 10.1111/j.1365-2958.2006.05333.x. [DOI] [PubMed] [Google Scholar]
- 15.Roberts EA, Deretic V. The Mycobacterium tuberculosis phagosome. Methods Mol Biol. 2008;445:439–449. doi: 10.1007/978-1-59745-157-4_28. [DOI] [PubMed] [Google Scholar]
- 16.Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer K, et al. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity. 1995;2:561–572. doi: 10.1016/1074-7613(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 17.Ladel CH, Blum C, Dreher A, Reifenberg K, Kopf M, et al. Lethal tuberculosis in interleukin-6-deficient mutant mice. Infect Immun. 1997;65:4843–4849. doi: 10.1128/iai.65.11.4843-4849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch CS, Ellner JJ, Russell DG, Rich EA. Complement receptor-mediated uptake and tumor necrosis factor-alpha-mediated growth inhibition of Mycobacterium tuberculosis by human alveolar macrophages. J Immunol. 1994;152:743–753. [PubMed] [Google Scholar]
- 19.Cox JS, Chen B, McNeil M, Jacobs WR., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 20.Dubos RJ, Middlebrook G. Cytochemical reaction of virulent tubercle bacilli. Am Rev Tuberc. 1948;58:698. doi: 10.1164/art.1948.58.6.698. [DOI] [PubMed] [Google Scholar]
- 21.Kolattukudy PE, Fernandes ND, Azad AK, Fitzmaurice AM, Sirakova TD. Biochemistry and molecular genetics of cell-wall lipid biosynthesis in mycobacteria. Mol Microbiol. 1997;24:263–270. doi: 10.1046/j.1365-2958.1997.3361705.x. [DOI] [PubMed] [Google Scholar]
- 22.Kraus D, Peschel A. Molecular mechanisms of bacterial resistance to antimicrobial peptides. Curr Top Microbiol Immunol. 2006;306:231–250. doi: 10.1007/3-540-29916-5_9. [DOI] [PubMed] [Google Scholar]
- 23.Lennarz WJ, Bonsen PP, van Deenen LL. Substrate specificity of O-L-lysylphosphatidylglycerol synthetase. Enzymatic studies on the structure of O-L-lysylphosphatidylglycerol. Biochemistry. 1967;6:2307–2312. doi: 10.1021/bi00860a005. [DOI] [PubMed] [Google Scholar]
- 24.Samant S, Hsu FF, Lee H. The Bacillus anthracis MprF is required for synthesis of lysylphosphatidylglycerols and for resistance to cationic antimicrobial peptides. J Bacteriol. 2008 doi: 10.1128/JB.01345-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oku Y, Kurokawa K, Ichihashi N, Sekimizu K. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology. 2004;150:45–51. doi: 10.1099/mic.0.26706-0. [DOI] [PubMed] [Google Scholar]
- 26.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 27.Talaat AM, Lyons R, Howard ST, Johnston SA. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc Natl Acad Sci U S A. 2004;101:4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vandal OH, Roberts JA, Odaira T, Schnappinger D, Nathan CF, et al. Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. J Bacteriol. 2009;191:625–631. doi: 10.1128/JB.00932-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandal OH, Pierini LM, Schnappinger D, Nathan CF, Ehrt S. A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat Med. 2008;14:849–854. doi: 10.1038/nmXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell DG, Mwandumba HC, Rhoades EE. Mycobacterium and the coat of many lipids. J Cell Biol. 2002;158:421–426. doi: 10.1083/jcb.200205034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brennan PJ, Rooney SA, Winder FG. The lipids of Mycobacterium tuberculosis BCG: fractionation, composition, turnover and the effects of isoniazid. Ir J Med Sci. 1970;3:269–284. doi: 10.1007/BF02958861. [DOI] [PubMed] [Google Scholar]
- 32.Mathur AK, Murthy PS, Saharia GS, Venkitasubramanian TA. Studies on cardiolipin biosynthesis in Mycobacterium smegmatis. Can J Microbiol. 1976;22:354–358. doi: 10.1139/m76-054. [DOI] [PubMed] [Google Scholar]
- 33.Bright-Gaertner E, Proulx P. Metabolism of phosphoglycerides in Escherichia coli during growth at 37 degrees C and during a cold-induced lag phase. Biochim Biophys Acta. 1972;270:40–49. doi: 10.1016/0005-2760(72)90175-0. [DOI] [PubMed] [Google Scholar]
- 34.Short SA, White DC. Metabolism of phosphatidylglycerol, lysylphosphatidylglycerol, and cardiolipin of Staphylococcus aureus. J Bacteriol. 1971;108:219–226. doi: 10.1128/jb.108.1.219-226.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chauhan A, Madiraju MV, Fol M, Lofton H, Maloney E, et al. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J Bacteriol. 2006;188:1856–1865. doi: 10.1128/JB.188.5.1856-1865.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair N, Dziedzic R, Greendyke R, Muniruzzaman S, Rajagopalan M, et al. Synchronous replication initiation in novel Mycobacterium tuberculosis dnaA cold-sensitive mutants. Mol Microbiol. 2008 doi: 10.1111/j.1365-2958.2008.06523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajagopalan M, Atkinson MA, Lofton H, Chauhan A, Madiraju MV. Mutations in the GTP-binding and synergy loop domains of Mycobacterium tuberculosis ftsZ compromise its function in vitro and in vivo. Biochem Biophys Res Commun. 2005;331:1171–1177. doi: 10.1016/j.bbrc.2005.03.239. [DOI] [PubMed] [Google Scholar]
- 38.Fol M, Chauhan A, Nair NK, Maloney E, Moomey M, et al. Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol Microbiol. 2006;60:643–657. doi: 10.1111/j.1365-2958.2006.05137.x. [DOI] [PubMed] [Google Scholar]
- 39.Kaur D, Berg S, Dinadayala P, Gicquel B, Chatterjee D, et al. Biosynthesis of mycobacterial lipoarabinomannan: role of a branching mannosyltransferase. Proc Natl Acad Sci U S A. 2006;103:13664–13669. doi: 10.1073/pnas.0603049103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soto CY, Andreu N, Gibert I, Luquin M. Simple and rapid differentiation of Mycobacterium tuberculosis H37Ra from M. tuberculosis clinical isolates through two cytochemical tests using neutral red and nile blue stains. J Clin Microbiol. 2002;40:3021–3024. doi: 10.1128/JCM.40.8.3021-3024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain SK, Hernandez-Abanto SM, Cheng QJ, Singh P, Ly LH, et al. Accelerated detection of Mycobacterium tuberculosis genes essential for bacterial survival in guinea pigs, compared with mice. J Infect Dis. 2007;195:1634–1642. doi: 10.1086/517526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Viability of Mtb strains in the absence -A- and presence -B- of Van 1.0 µg/ml. At day six of growth, viability was determined by plating cells on Middlebrook 7H11 agar and counting. The bars represent mean±standard error.
(1.57 MB TIF)
Visualization of Fl-Van stained cells. Actively growing cultures (optical density ∼0.01 to 0.04) were grown with fluorescent vancomycin BODIPY (Invitrogen) at a final concentration of 1 µg/mL for 20 hours. The cells were harvested by low speed centrifugation, fixed in 4% paraformaldehyde for 24 h, and imaged on a Nikon Eclipse microscope with a CCD camera; bright field (BF) and fluorescent images (Van) were acquired. Magnification was 100×, and data were analyzed using Metamorph software. At least 100 cells were imaged for each strain; the images were scored for staining patterns and the presence or absence of fluorescent staining. Approximately 35% of wild type and 25% of lysX cells were not stained under these conditions. For clarity, only select staining patterns are shown: defined dye accumulation at the cell poles -A, D- and mid-cell (as in D) or diffuse accumulation throughout the cell - B, E. In some cells, both types of accumulation were observed -C, F. For the lysX mutant, about 52% of cells showed diffuse staining, but this was only seen in about 32% of wild type cells.
(2.97 MB TIF)
Effect of SDS on the viability of the lysX mutant: The viability of Mtb strains was examined under the same conditions used for macrophage infection and lysis in the presence of 0.09% SDS. Actively growing cultures of Mtb strains were harvested, exposed to 0.09% SDS for 3 min, diluted and spread on Middlebrook 7H11 agar plates. Cells untreated with SDS were processed similarly. All plates were incubated at 37°C, and colonies were counted. No statistically significant differences between the SDS treated and untreated groups were noted. Data shown are mean ± standard error.
(1.05 MB TIF)
Growth of the lysX mutant in the spleen of mice and guinea pigs. Growth of the lysX mutant and Rv-03 strain in the spleens of mice -A- and guinea pig -B. Following aerosol infection of mice and guinea pigs, spleens were harvested at the indicated time points. Homogenates were prepared, and viability was determined on agar plates. The lysX mutant showed reduced and/or delayed dissemination compared to wild type in both animal models.
(1.64 MB TIF)
Neutral red staining and PDIM analysis: Panel A: Wild type and lysX mutant cells were stained with neutral red and photographed. As a control, the attenuated strain Mtb H37ra was used. Panel B: The lipids were separated by silica thin-layer chromatography (TLC) with hexane:diethylether:acetic acid (80∶20∶1, vol/vol/vol) as solvent system. The lipids were visualized by spraying with 10% phosphormolybdate in ethanol followed by heating to about 110°C for 15 min.
(5.65 MB TIF)