Abstract
One of the most fascinating topics in biology is to understand the development of highly differentiated cells such as photoreceptors (PRs). This process involves successive steps, starting with the generation of the eye primordium, recruitment and specification of PRs and finally, expression of the proper rhodopsin, the photopigment that initiates the signaling cascade underlying light input excitation. In this review, we describe the sequential steps that take place in the Drosophila eye, from the initial neuronal specification of PRs through their full maturation, focusing specifically on the transcription factors and signaling pathways involved in controlling the precise expression of different rhodopsins in specialized PRs.
Introduction
Although eyes are morphologically and evolutionarily distinct in different organisms such as the Drosophila compound or the camera-like eye in mammals, the molecular mechanisms that govern their development have shown surprising homologies [1]. In Drosophila, the generation of mature adult photoreceptors (PRs) results from a cascade of regulatory steps that controls the specification and morphological changes that define each class of PRs. Drosophila eye development starts with the determination of the eye-antennal disc during embryogenesis. Then, during the third larval stage of development, a wave of differentiation requiring BMP and Hedgehog signaling (called the morphogenetic furrow, MF) initiates at the posterior margin of the eye disc and progresses anteriorly through undifferentiated cells [2••,3•,4]. This wave leaves patterned clusters of eight PR neurons, called ommatidia. Although this patterning wave was initially thought to be unique to the compound eye of insects, a similar mechanism seems to be used in the zebrafish retina where PR differentiation, mediated by Sonic Hedgehog activity, starts in the center of the prospective eye field and then propagates outward [5•].
Toward the end of pupal development, the eye undergoes profound morphological changes to acquire its final adult shape: PRs form their rhabdomeres (the specialized, membrane-rich, light guiding apical organelle that serves as the site of phototransduction), and the concomitant induction of different wavelength-sensitive rhodopsins, the light receptors, occurs [6]. During this process, two morphologically distinct classes of PRs become distinguished within each ommatidium: outer PRs (R1–6) contain wide rhabdomeres spanning the whole retina and express the broad spectrum opsin Rh1. Inner PRs (R7 and R8) are located in the center of each ommatidium. Their rhabdomeres are smaller and span only half of the retina, with R7 rhabdomeres sitting on top of R8’s. Inner PRs present a complex pattern of rhodopsin expression: R7 express stochastically either of the UV-sensitive opsins, Rh3 or Rh4, whereas R8 express either Rh5 (blue) or Rh6 (green). Interestingly, Rh3 expression in R7 is primarily coupled with Rh5 in R8, and Rh4 is coupled with Rh6. This defines two functional classes of PRs distributed randomly throughout the whole eye: pale (Rh3/Rh5) and yellow subtypes (Rh4/Rh6) [7].
Through their different rhodopsin gene expression, outer and inner PRs mediate different functions. Outer PRs, the fly equivalent of vertebrate rods, are involved in motion detection and image formation. They send their projections to the first optic lobe neuropil, the lamina. Inner PRs, similar to vertebrate cones, are involved in color vision and compare their output in the medulla in a deeper region of the optic lobe [8].
The retinal determination gene network in Drosophila
The specification of the eye primordium requires the expression of a network of ‘retinal determination genes’ [1,9–11], formed by eyeless (ey), eye gone (eyg), sine oculis (so), eyes absent (eya), and dachshund (dac) transcription factor families (Figure 1). Mutations in any of these genes cause reduction or absence of the eye, whereas ectopic expression of these genes, alone or in combinations, is sufficient to induce eye formation in other tissues [12,13,14••,15,16•]. Remarkably, this network has been repetitively used throughout evolution in different organisms for the generation of different organs in a tissue and context-specific manner, integrating multiple signaling pathways [1,17]. ey and its highly related neighboring gene, twin of eyeless, are the homologues of vertebrate Pax6 [18]. Pax6 is essential for eye development in most species where it has been studied, and it is able to induce ectopic eyes when expressed in different tissues, both in flies and vertebrates [13,14••,19,20]. Since the vertebrate and insect eyes are likely to be evolved independently [21••], the common use of Pax6 likely reflects a common ancestral function in regulating PR fate in both lineages [9,22]. ey and toy sit at the top of the transcriptional hierarchy of eye development, while the remaining retinal determination genes so, eya, and dac comprise the downstream regulators that are all required for eye development [1,13,23]. Interestingly, extensive crosstalk occurs between these factors, with multiple feedback loops, thus allowing downstream members of the pathway to retain their ability to induce ectopic eyes [24] (Figure 1).
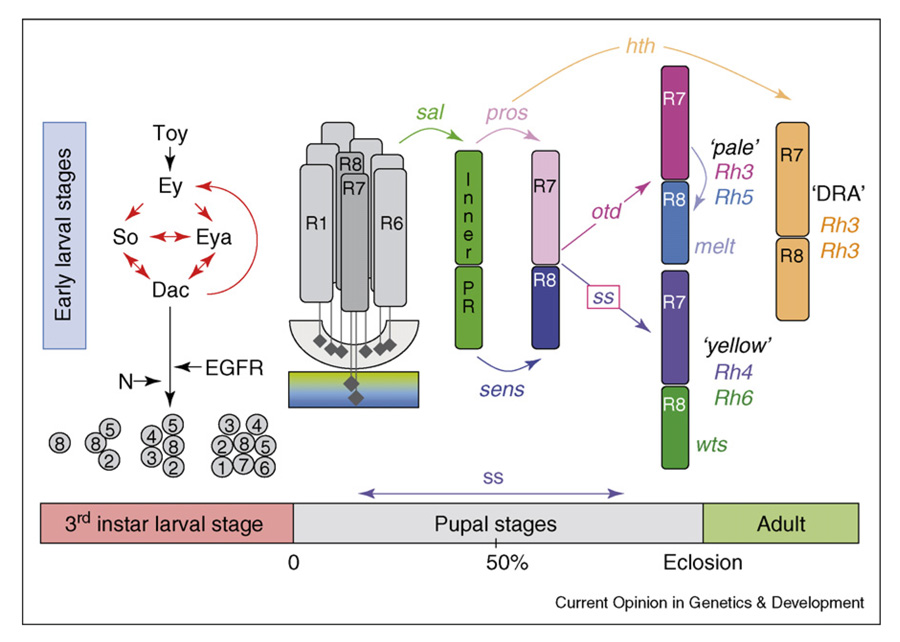
Figure 1.
Developmental timetable of the Drosophila retinal mosaic. After the eye-antennal disc is specified by the eye determination genes, PRs are recruited sequentially into an ommatidium (first R8, then R2 and R5, later R3 and R4, and finally R1 and R6). In the late larval and early pupal stages initial cell-fate decisions are made. First, a generic color PR fate is established through the action of sal. Then R7 and R8 cells are distinguished from each other by the specific expression of pros and sens in R7 and R8, respectively. Hth commits inner PRs to the DRA fate; while otd generates the pale fate, ss patterns, the yellow ommatidial subset. Finally, the subset-specific expression of wts and melt in yellow and pale R8 cells, respectively, enforces the decision that has been communicated by the R7 cell.
Photoreceptor determination in Drosophila
Once cells in the eye imaginal disc have been selected during embryogenesis to participate in eye development, PR differentiation initiates during larval stages at the morphogenetic furrow (Figure 1). At least five different signaling pathways are involved in this process. The hedgehog (hh) and dpp pathways are involved in the initiation and progression of the MF [3•,4,25]. Conversely, wg acts as a negative regulator of the MF, preventing inaccurate retinal specification [26,27••].
As Hh drives the progression of the MF across the eye disc, it induces the expression of the transcription factor Atonal [28], which becomes expressed in regularly spaced precursors that are specified as R8 in each cluster [29•]. The recruitment of the other PRs from the surrounding pool of undifferentiated cells is mediated by the epidermal growth factor receptor (EGFR) and Notch (N). First, an EGFR ligand, Spitz, is secreted by R8 [30••]. As a result of this process, five PRs (R8 plus R2–5) are initially recruited in each ommatidium, forming a five-cell pre-cluster. These five PRs maintain their G1 arrest and differentiate in response to EGFR. The surrounding cells that did not respond to EGFR signaling, however, enter the cell cycle one last time in a second mitotic wave [31•,32] before R1 and R6 are finally recruited by EGFR and become differentiated. The R7 cell fate is acquired through the activation of two signals: Bride-of-Sevenless (Boss), which is expressed by R8, activates the Sevenless receptor on the surface of the presumptive R7 cell [33,34,35••]. A second signal involves the Delta ligand expressed by R1 and R6, which directs the R7 cell fate through activation of the Notch receptor [35••,36•]. The two signaling pathways might interact in a combinatorial fashion in R7 or they might act sequentially with activation of the EGFR pathway first recruiting R7, followed by Notch inhibiting the Ras pathway [35••,37]. Finally, accessory cells are recruited by EGFR activity to the ommatidium, which then comprises 20 cells.
The initial patterning of PRs in the eye disc is followed by PR maturation that happens much later, during late pupal stages. This maturation involves sequential steps, which restrict individual cells to a particular fate and finally results in the expression of genes specific for each class of PRs. Several transcription factors that act sequentially in these processes have recently been identified (Figure 1).
Establishment of a generic ‘color PR’ fate
The first cell-fate decision necessary for generating different functional classes of PRs appears to be the distinction between inner and outer PRs. The spalt genes (sal) encode two zinc-finger transcription factors and are specifically expressed in both R7 and R8. They are essential to commit these cells to an inner color PR cell fate (Figure 1). As loss-of-function of sal results in the transformation of inner PRs into outer PRs, their rhabdomeres become longer and broader and they express Rh1. Projections of these PRs are, however, not redirected toward the lamina, maintaining their overall projection pattern to the medulla [38•]. Unlike vertebrate PRs that are dedicated to light perception, with the retinal ganglion cells being in charge of transmitting the information to higher brain center, fly PRs are both neurons and PRs: sal is only involved in the differentiation of PRs and does not affect their neuronal fate.
Distinguishing between R7 and R8
sal determines the fate of R7 and R8 as ‘generic’ inner/color PRs. Further specification is, however, required to determine the specific differentiation into R7 (long rhabdomere on top, expressing Rh3 or Rh4) or R8 (rhabdomere at the bottom of the retina, expressing Rh5 or Rh6). Indeed, the next cell-fate decision involves the distinction of R7 cells from R8 cells. prospero, which encodes a transcription factor, is specifically expressed in R7 cells where it represses R8 opsins (Rh5 and Rh6) by directly binding their promoters [39•] (Figure 1). Furthermore, prospero is required for correct nuclear positioning in the R7 cell. In the absence of prospero, R7 cells take on the ‘generic’ inner PR fate, which appears to be close to the R8 fate. R8 specification away from the generic fate, however, requires a different cell-specific transcription factor, senseless (sens). Sens is expressed specifically in R8 cells and has been shown to be able to induce Rh6 expression when mis-expressed in outer PRs [40] (T. Cook, submitted) (Figure 1). Therefore, sal creates a generic inner PR fate, while prospero and senseless push these cells toward the R7 and R8 fates, respectively.
Color versus polarization detection
Subsequent steps involve the commitment of inner PRs to fates that provide color-sensitive versus polarization-sensitive properties to different ommatidia. Dorsal rim area (DRA) ommatidia are located in one to two rows of ommatidia in the dorsal edge of the retina and function in the detection of the e-vector of polarized light. Their inner rhabdomeres are larger and contain stacks of microvilli that are perpendicular to each other in R7 and R8, thus serving as polarizing filters. Interestingly, both R7 and R8 cells in the DRA express the same rhodopsin, Rh3. The commitment to the DRA fate is achieved by a localized expression of homothorax (hth), which encodes a homeoprotein (Figure 1). hth is expressed specifically in one row of ommatidia at the dorsal edge of the eye in response to two dorsal cues: wingless signaling from the head cuticle and the specification of dorsal tissue by IroC genes [41•,42••]. Loss of hth leads to the loss of the typical DRA morphology. Additionally, the specific expression of Rh3 in both R7 and R8 is lost, and an unusual coupling of Rh3 in R7 with Rh6 in R8 is observed. Mis-expression of hth is also sufficient to transform all ommatidia into DRA ommatidia with increased rhabdomere diameter and expression of Rh3 in both R7 and R8. Thus, hth is the key factor for determining the DRA fate and provides the fly with polarized light vision for navigation [42••].
pale versus yellow stochastic ommatidial choice
Inner PRs in non-DRA ommatidia develop for color vision and express Rh4/Rh6 in 70% of (yellow) ommatidia and Rh3/Rh5 in the remaining 30% of (pale) ommatidia. How the stochastic choice of pale versus yellow ommatidia is made and how a specific and highly conserved ratio is achieved remain important developmental questions. Two factors have been implicated in this choice: a permissive factor required for the pale choice and an instructive factor that specifies the yellow ommatidial subtype. The K50 homeodomain transcription factor encoded by orthodenticle (otd) was identified as a key factor for the generation of the pale subtype (Figure 1). Otd positively regulates the expression of Rh3 and Rh5 by directly binding to their promoter [43]. Although loss of otd leads to the loss of Rh3 and Rh5 in the pale subtype, the yellow subtype (Rh4 and Rh6) does not expand into pale ommatidia, leading to 30% of ommatidia that are not committed to expressing any given opsin. Thus, ommatidial subtype specification and rhodopsin expression seem to be two separable processes [6].
Although otd is necessary for pale ommatidia rhodopsin expression, mis-expression of otd to all cells is not sufficient to induce this cell fate. In contrast, recent studies have identified the bHLH-PAS transcription factor encoded by spineless (ss) as both necessary and sufficient in patterning yellow ommatidia. ss is expressed in a large subset of R7 cells during pupation where it leads to the expression of Rh4 in adult R7 cells [44••] (Figure 1). The remaining R7 cells, which lack ss, express Rh3 by default. Mis-expression of ss activates rh4 in all PRs and prevents expression of rh3, indicating that ss lies upstream of otd-mediated activation of the pale-specific opsin. Although ss encodes a transcription factor, it is not expressed in adult and therefore does not act directly on the rh4 promoter. Rh expression starts at 78% after puparium formation (APF) with the expression of Rh1 and is followed by Rh3, Rh4, Rh5, and Rh6 shortly after [45]. This means that other factors downstream of ss are responsible for controlling the expression of Rh4 and antagonizing the signal generated by the R7 cell. ss is the first factor other than Rh that is expressed in a subset of PR cells. It will be very interesting to identify the factors upstream of ss that lead to its remarkable stochastic expression pattern.
Signaling from R7 to R8 to coordinate subtype choice
The choice of becoming a pale versus a yellow ommatidium is first made by ss in the R7 cell. Once this decision is made, this information is communicated to the underlying R8. When the signal from R7 is absent (e.g. in sevenless mutants), all R8 express Rh6 [46••,47]. While the exact nature of this signal emanating from pR7 remains elusive, downstream events in R8 are well characterized. Two genes warts and melted play opposite roles to maintain the decision that has been communicated by R7, but they do not affect the normal distribution of the pale and yellow R7 (Figure 1). warts encodes a Ser/Thr kinase similar to human large tumor suppressor (Lats). It is specifically expressed in yellow R8. Loss-of-function of warts leads to the transformation of all yellow (Rh6) R8 into pale (Rh5) R8, as if all R8 were receiving the R7 signal. melted, which encodes a Pleckstrin Homology (PH) domain protein, is expressed in a complementary fashion in pale R8 [48••]. Loss of melted results in the transformation of all pale (Rh5) R8 into yellow (Rh6) R8, as if no R7 signal was received. Additionally, generalized expression of warts or melted induces generalized Rh6 or Rh5 expression in R8, respectively, again without affecting R7 [48••]. These two proteins interact in a negative interaction loop, where melted responds to the signal coming from R7 while warts regulates the output of the loop. This bi-stable loop ensures that a robust decision is made in R8 to express the correct Rh. warts/Lats is part of the Lats/Hippo/Salvador tumor suppressor pathway that normally prevents proliferation and promotes cell death. Melt is a member of the Tor/Insulin growth control pathway. It appears that the entire Lats/Hippo pathway (but no other member of the Tor/Insulin pathway) becomes re-utilized in R8 specification after it is no longer required when PRs have exited cell cycle. Since warts and melted do not encode transcription factors, although they are regulated at the transcriptional level, it will be very interesting to identify the transcriptional effectors of this loop. R8 specification thus provides an excellent system to identify both upstream and downstream regulators that control tumor suppression.
Conclusions
Retinal patterning involves many different specification strategies that ultimately give rise to the highly organized adult retina. Most of these events are regulated by coordinated actions of many transcription factors and signaling pathways with highly restricted functions. Early patterning events involve sequential recruitment of PRs into preclusters. Later events lead to the precise specification of localized PR subtypes (e.g. DRA ommatidia) or ommatidia exhibiting stochastic distribution (p versus y). Once a choice is made in R7, it is communicated to R8 where it is maintained by a highly coordinated loop. A more detailed understanding of pattern formation in the Drosophila retina will help us understand more general problems in neurobiology.
Acknowledgements
The authors would like to thank Tiffany Cook for helpful comments on the manuscript. This work was supported by Grant NIH R01 EY013010 to C.D.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Silver SJ, Rebay I. Signaling circuitries in development: insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- 2. Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. This is the classical paper that is basis of most work on the fly retina.
- 3. Heberlein U, Wolff T, Rubin GM. The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell. 1993;75:913–926. doi: 10.1016/0092-8674(93)90535-x. The role of signaling pathways in the morphogenetic furrow.
- 4.Dominguez M, Hafen E. Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev. 1997;11:3254–3264. doi: 10.1101/gad.11.23.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neumann CJ, Nuesslein-Volhard C. Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science. 2000;289:2137–2139. doi: 10.1126/science.289.5487.2137. An interesting parallel between the morphogenetic furrow in flies and the wave of Shh that sweeps through the zebrafish retina, inducing its own expression and neurogenesis.
- 6.Wernet MF, Desplan C. Building a retinal mosaic: cell-fate decision in the fly eye. Trends Cell Biol. 2004;14:576–584. doi: 10.1016/j.tcb.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Franceschini N, Kirschfeld K, Minke B. Fluorescence of photoreceptor cells observed in vivo. Science. 1981;213:1264–1267. doi: 10.1126/science.7268434. [DOI] [PubMed] [Google Scholar]
- 8.Morante J, Desplan C. Building a projection map for photoreceptor neurons in the Drosophila optic lobes. Semin Cell Dev Biol. 2004;15:137–143. doi: 10.1016/j.semcdb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Desplan C. Eye development: governed by a dictator or a junta? Cell. 1997;91:861–864. doi: 10.1016/s0092-8674(00)80475-4. [DOI] [PubMed] [Google Scholar]
- 10.Pappu KS, Mardon G. Genetic control of retinal specification and determination in Drosophila. Int J Dev Biol. 2004;48:913–924. doi: 10.1387/ijdb.041875kp. [DOI] [PubMed] [Google Scholar]
- 11.Chen R, Halder G, Zhang Z, Mardon G. Signaling by the TGF-beta homolog decapentaplegic functions reiteratively within the network of genes controlling retinal cell fate determination in Drosophila. Development. 1999;126:935–943. doi: 10.1242/dev.126.5.935. [DOI] [PubMed] [Google Scholar]
- 12.Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- 13.Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- 14. Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. A dramatic paper that reported the ability of Pax6/eyeless to induce ectopic eyes.
- 15.Bonini NM, Bui QT, Gray-Board GL, Warrick JM. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- 16. Shen W, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;124:45–52. doi: 10.1242/dev.124.1.45. A follow-up on the ability of retinal determination genes to induce ectopic eyes: this interesting paper shows that this does not happen anywhere but only at specific locations.
- 17.Hanson IM. Mammalian homologues of the Drosophila eye specification genes. Semin Cell Dev Biol. 2001;12:475–484. doi: 10.1006/scdb.2001.0271. [DOI] [PubMed] [Google Scholar]
- 18.Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 19.Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- 20.Onuma Y, Takahashi S, Asashima M, Kurata S, Gehring WJ. Conservation of Pax 6 function and upstream activation by Notch signaling in eye development of frogs and flies. Proc Natl Acad Sci USA. 2002;99:2020–2025. doi: 10.1073/pnas.022626999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salvini-Plawen LV, Mayr E. On the evolution of photoreceptors and eyes. Evol. Biol. 1977;10:207–263. This paper is a very thorough survey of eyes in a large number of animal phyla and concludes that eyes have evolved 40 times independently through evolution. This is a ‘classic’.
- 22.Zuker CS. Cell signalling. A taste of things to come. Nature. 1995;376:22–23. doi: 10.1038/376022a0. [DOI] [PubMed] [Google Scholar]
- 23.Ostrin EJ, Li Y, Hoffman K, Liu J, Wang K, Zhang L, Mardon G, Chen R. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16:466–476. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedrich M. Continuity versus split and reconstitution: exploring the molecular developmental corollaries of insect eye primordium evolution. Dev Biol. 2006;299:310–329. doi: 10.1016/j.ydbio.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 25.Curtiss J, Mlodzik M. Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development. 2000;127:1325–1336. doi: 10.1242/dev.127.6.1325. [DOI] [PubMed] [Google Scholar]
- 26.Baonza A, Freeman M. Control of Drosophila eye specification by wingless signaling. Development. 2002;129:5313–5322. doi: 10.1242/dev.00096. [DOI] [PubMed] [Google Scholar]
- 27. Treisman JE, Rubin GM. Wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development. 1995;121:3519–3527. doi: 10.1242/dev.121.11.3519. The role of wingless in morphogenetic furrow progression. It acts at the edges of the eye disc, preventing neuronal differentiation through inhibition of hh and dpp expression.
- 28.Dominguez M. Dual role for Hedgehog in the regulation of the proneural gene atonal during ommatidia development. Development. 1999;126:2345–2353. doi: 10.1242/dev.126.11.2345. [DOI] [PubMed] [Google Scholar]
- 29. Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. Atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. Atonal specifies R8, the first photoreceptors to be recruited to the ommatidium and the only one that is independent of the EGFR pathway.
- 30. Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. The first thorough report of the multiple functions of the EGFR pathway for the recruitment of all cell types in the retina.
- 31. Baker NE, Yu SY. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell. 2001;104:699–708. doi: 10.1016/s0092-8674(01)00266-5. The authors describe the role of the EGFR pathway in the second mitotic wave to recruit R1, R6 and R7 PRs.
- 32.Yang L, Baker NE. Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev Cell. 2003;4:359–369. doi: 10.1016/s1534-5807(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 33.Simon MA, Bowtell DD, Dodson GS, Laverty TR, Rubin GM. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 34.Kramer H, Cagan RL, Zipursky SL. Interaction of bride of sevenless membrane-bound ligand and the sevenless tyrosine-kinase receptor. Nature. 1991;352:207–212. doi: 10.1038/352207a0. [DOI] [PubMed] [Google Scholar]
- 35. Tomlinson A, Struhl G. Delta/Notch and Boss/Sevenless signals act combinatorially to specify the Drosophila R7 photoreceptor. Mol Cell. 2001;7:487–495. doi: 10.1016/s1097-2765(01)00196-4. One of two reports on the role of Notch signaling for R7 specification.
- 36. Cooper MT, Bray SJ. R7 photoreceptor specification requires Notch activity. Curr Biol. 2000;10:1507–1510. doi: 10.1016/s0960-9822(00)00826-5. This report also reports on the role of the Notch pathway for the specification of the R7 cell.
- 37.Yang L, Baker NE. Notch activity opposes Ras-induced differentiation during the second mitotic wave of the developing Drosophila eye. BMC Dev Biol. 2006;6:8. doi: 10.1186/1471-213X-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mollereau B, Dominguez M, Webel R, Colley NJ, Keung B, de Celis JF, Desplan C. Two-step process for photoreceptor formation in Drosophila. Nature. 2001;412:911–913. doi: 10.1038/35091076. Photoreceptors in Drosophila are also neurons; spalt is a key factor for the differentiation of color photoreceptors R7 and R8 but not for their specification as neurons. spalt distinguishes color from black and white (outer) photoreceptors.
- 39. Cook T, Pichaud F, Sonneville R, Papatsenko D, Desplan C. Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev Cell. 2003;4:853–864. doi: 10.1016/s1534-5807(03)00156-4. Identification of prospero as a regulator of R7 cell fate. Prospero is able to directly repress R8 opsins in R7 cells, where it is specifically expressed.
- 40.Domingos PM, Brown S, Barrio R, Ratnakumar K, Frankfort BJ, Mardon G, Steller H, Mollereau B. Regulation of R7 and R8 differentiation by the spalt genes. Dev Biol. 2004;273:121–133. doi: 10.1016/j.ydbio.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 41. Tomlinson A. Patterning the peripheral retina of the fly: decoding a gradient. Dev Cell. 2003;5:799–809. doi: 10.1016/s1534-5807(03)00326-5. The role of wingless and the iroquois gene complex for the determination of polarization-sensitive photoreceptors.
- 42. Wernet MF, Labhart T, Baumann F, Mazzoni EO, Pichaud F, Desplan C. Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell. 2003;115:267–279. doi: 10.1016/s0092-8674(03)00848-1. Homothorax is the key regulator that specifies the polarization-sensitive region of the retina and distinguishes it from the main part of the retina that is involved in color discrimination.
- 43.Tahayato A, Sonneville R, Pichaud F, Wernet MF, Papatsenko D, Beaufils P, Cook T, Desplan C. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev Cell. 2003;5:391–402. doi: 10.1016/s1534-5807(03)00239-9. [DOI] [PubMed] [Google Scholar]
- 44. Wernet MF, Mazzoni EO, Celik A, Duncan DM, Duncan I, Desplan C. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. Spineless in the gene that makes the pale/yellow subset choice. Spineless is expressed in 70% of R7 cells before rhodopsin expression, and it is necessary and sufficient to determine the entire ommatidial fate.
- 45.Earl JB, Britt SG. Expression of Drosophila rhodopsins during photoreceptor cell differentiation: insights into R7 and R8 cell subtype commitment. Gene Expr Patterns. 2006;6:687–694. doi: 10.1016/j.modgep.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 46. Chou WH, Hall KJ, Wilson DB, Wideman CL, Townson SM, Chadwell LV, Britt SG. Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron. 1996;17:1101–1115. doi: 10.1016/s0896-6273(00)80243-3. The first complete report of the stochastic expression of R7 and R8 opsins with the coupling between these two cells in ‘pale’ versus ‘yellow’ ommatidia.
- 47.Papatsenko D, Sheng G, Desplan C. A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development. 1997;124:1665–1673. doi: 10.1242/dev.124.9.1665. [DOI] [PubMed] [Google Scholar]
- 48. Mikeladze-Dvali T, Wernet MF, Pistillo D, Mazzoni EO, Teleman AA, Chen YW, Cohen S, Desplan C. The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell. 2005;122:775–787. doi: 10.1016/j.cell.2005.07.026. How tumor suppressor genes are re-utilized after they are no longer used in post-mitotic cells for the formation of a bi-stable loop that ensures a robust expression of opsins in R8.