Abstract
B-cell clonality detection in whole tissue is considered indicative of B-cell non-Hodgkin lymphoma (NHL). We tested frozen tissue of 24 classical Hodgkin lymphomas (cHL) with a varying tumor cell load with the multiplex polymerase chain reaction (PCR) primer sets for IGH and IGK gene rearrangement (BIOMED-2). A clonal population was found in 13 cases with the IGH FR1 and/or FR2/FR3 PCRs. Using the IGK-VJ and IGK-DE PCRs, an additional six cases had a dominant clonal cell population, resulting in a detection rate of 79% in frozen tissue. Of 12 cases, also the formalin-fixed and paraffin-embedded (FFPE) tissue was tested. Surprisingly, in eight of the 12 FFPE cases with acceptable DNA quality (allowing PCR amplification of >200 nt fragments), the IGK multiplex PCRs performed better in detecting clonality (six out of eight clonal IGK rearrangements) than the IGH PCRs (four out of nine clonal rearrangements), despite a rather large amplicon size. There was no evidence of B-cell lymphoma during follow-up of 1 to 6 years and no correlation was found between the presence of a clonal result and Epstein–Barr virus in the tumor cells. Our results indicate that the present routine PCR methods are sensitive enough to detect small numbers of malignant cells in cHL. Therefore, the presence of a clonal B-cell population does not differentiate between cHL and NHL.
Keywords: B cell, Hodgkin lymphoma, PCR clonality
Introduction
Although classical Hodgkin lymphoma (cHL) is a B-cell lymphoma, due to the scarcity of tumor cells, lack of immunoglobulin expression, and the heavy somatic mutation load of the Ig gene rearrangements [1], the clonal B-cell nature is difficult to detect by standard immunological and molecular techniques. Previously, clonal B-cell receptor rearrangements have therefore been considered more supportive of B-cell non-Hodgkin lymphoma (NHL). Polymerase chain reaction (PCR)-based clonality detection is increasingly used in lymphoma diagnostics. We performed routine PCR-based clonality studies on frozen tissue of 24 lymph nodes involved by cHL. The results were compared to formalin-fixed and paraffin-embedded (FFPE) tissue in 12 of these cases. To cover both productive and non-productive rearrangements that are less prone to somatic mutation, such as IGH-DJ and IGK-DE targets, we used the BIOMED-2 set of multiplex PCR tubes that have been shown to detect 98% of B-NHL [2]. Previous studies using only the IGH primers in a series of 42 samples [3] and IGH and IGK in 12 cases of HL [4] resulted in detection rates of 24–42%.
Materials and methods
Patients
The study was performed on 24 lymph node biopsies with cHL with availability of tissue to perform cytogenetics and immunoflowcytometry and to store frozen tissue. All cases were collected consecutively during the years 2003–2006. Four patients were relapsed cHL after chemotherapy 1–11 years before. This high percentage of relapses is due to an enrichment of patients that are sent to a referral hospital for second-line therapy because of poor outcome. None of the patients had a history of other hematological neoplasm or developed evidence of NHL during follow-up that ranged from 14 to 76 months (median 42). Simultaneous bone marrow biopsy (BMB) for the purpose of staging was insufficient in one patient and showed involvement with cHL in three. The clinical and pathological data are summarized in Table 1.
Table 1.
Clinical and pathological data of 24 cHL patients
| Patient | Age/sex | Diagnosis | EBV status | Immunophenotypea | Background B cellsb | BMB |
|---|---|---|---|---|---|---|
| 1 | 15 M | Rel. LRHL | neg | CD30+/−, CD15+/−, CD20+, CD79a+, CD3− | ++ | pos |
| 2 | 38 M | NSHL | pos | CD30+, CD15−, CD20−, CD79a+/−, CD3− | + | neg |
| 3 | 20 M | Rel. NSHL | neg | CD30+, CD15+, CD20−, CD79a−, CD3− | ++ | neg |
| 4 | 14 F | MCHL | na | CD30+, CD15−, CD20+/−, CD79a+/−, CD3− | + | insuff |
| 5 | 47 M | MCHL | neg | CD30+, CD15−, CD20−, CD79a−, CD3− | + | neg |
| 6 | 13 F | NSHL | neg | CD30+, CD15+, CD20−, CD79a−, CD3+/−, CD2+, CD4+, CD5+, CD8− | − | neg |
| 7 | 17 F | NSHL | pos | CD30+/−, CD15+/−, CD20+/−, CD79a−, CD3− | + | insuff |
| 8 | 19 F | NSHL | neg | CD30+, CD15−, CD20−, CD79a+/−, CD3− | + | neg |
| 9 | 22 M | NSHL | neg | CD30+/−, CD15+/−, CD20−, CD79a−, CD3− | +/− | neg |
| 10 | 24 F | NSHL | neg | CD30+, CD15+/−, CD20+/−, CD79a−, CD3− | + | neg |
| 11 | 15 F | NSHL | na | CD30+, CD15+/−, CD20−, CD79a−, CD3− | + | neg |
| 12 | 67 M | NSHL | pos | CD30+, CD15+/−, CD20+/−, CD79a+/−, CD3− | ++ | neg |
| 13 | 27 M | NSHL | neg | CD30+, CD15+/−, CD20−, CD79a−, CD3− | + | pos |
| 14 | 19 F | Rel. NSHL | pos | CD30+, CD15+, CD20−, CD79a−, CD3− | ++ | neg |
| 15 | 14 M | NSHL | neg | CD30+, CD15+, CD20−, CD79a−, CD3− | ++ | neg |
| 16 | 32 M | NSHL | neg | CD30+/−, CD15−, CD20−, CD79a−, CD3− | + | neg |
| 17 | 14 F | NSHL | neg | CD30+, CD15+/−, CD20−, CD79a−, CD3− | + | neg |
| 18 | 61 M | NSHL | neg | CD30+, CD15+, CD20+/−, CD79a+/−, CD3− | + | neg |
| 19 | 76 M | NSHL | neg | CD30+, CD15+/−, CD20−, CD79a−, CD3− | − | neg |
| 20 | 33 M | NSHL | pos | CD30+, CD15+, CD20+/−, CD79a+/−, CD3− | + | neg |
| 21 | 44 M | MCHL | pos | CD30+, CD15−, CD20−, CD79a−, CD3− | + | neg |
| 22 | 43 M | Rel. NSHL | pos | CD30+, CD15−, CD20−, CD79a−, CD3− | ++ | neg |
| 23 | 22 M | iHL | pos | CD30+/−, CD15+/−, CD20−, CD79a−, CD3− | ++ | neg |
| 24 | 14 M | NSHL | neg | CD30+, CD15+, CD20−, CD79a−, CD3− | ++ | neg |
M male, F female, rel. relapsed, BMB bone marrow biopsy, NSHL nodular sclerosing Hodgkin lymphoma, LRHL lymphocyte-rich Hodgkin lymphoma, MCHL mixed cellularity Hodgkin lymphoma, iHL interfollicular Hodgkin lymphoma, neg negative, pos positive, insuff insufficient material
aImmunophenotype of the HRS cells: − <10% positive, +/− 10–50% positive, + >50% positive
bPercentage of background B cells: − <5%, + 5–30%, ++ >30%
Immunophenotypic studies
All lymph node biopsies were received fresh at the Department of Pathology where a part was collected for liquid nitrogen storage. The remaining tissue was formalin-fixed and paraffin-embedded. Four-micrometer tissue sections were stained with hematoxylin–eosin, and with monoclonal antibodies for CD20, CD30, CD79a (all DAKO), CD3 (Neomarkers), and CD15 (BD Diagnostics). A typical example is presented in Fig. 1. Staining was scored on the Hodgkin and Reed–Sternberg cells (HRS) cells: − <10%, +/− 10–50%, and + >50%. Average HRS cell percentages on HE and CD30 stains were independently scored by two observers (KH and HvK). In situ hybridization for Epstein–Barr virus (EBER) was performed on 22 cases according to the instructions of the manufacturer (DAKO). Immunophenotype and Epstein–Barr virus (EBV) status of the HRS cells is shown in Table 1. In addition, the percentage of small B cells in the background was estimated: − <5%, + 5–30%, and ++ >30%.
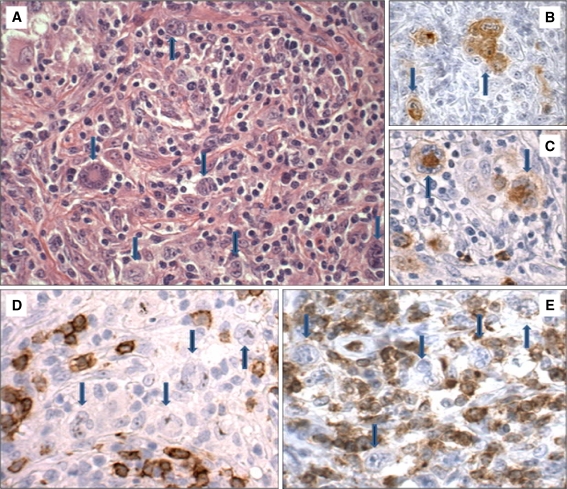
Fig. 1.
Example of nodular sclerosing classical Hodgkin lymphoma (patient 8). a HE stain. The HRS cells, indicated by arrows, are positive for CD30 (b) and CD15 (c). Stains for CD20 (d) and CD3 (e) are negative on the HRS cells (×20)
Clonality analysis
Five- to 10-um frozen tissue sections were used to extract DNA using proteinase K treatment and ethanol precipitation of the genomic DNA. DNA extraction from FFPE tissues was performed using a proteinase K treatment step and subsequent affinity purification of the DNA (QIAamp DNA micro handbook, QIAGEN GmbH, Germany). DNA sample concentration and quality were assessed by spectrophotometry (260/280 nm using the NanoDrop) and by PCR amplification using the BIOMED-2 control gene primer set [5]. All frozen tissues showed amplifiable DNA of 400 nt, which enables molecular clonality assessment. Only eight of 12 FFPE tissues allowed amplification of at least 200 nt amplicons, implying mild fragmentation of DNA and potential risk of lack of detection of clonal rearrangements with a large amplicon size. The quantity of DNA per PCR was 100 ng for DNA from frozen tissue, in duplicate, and 50 and 200 ng per PCR for affinity-purified DNA from paraffin-embedded tissue.
Detection of B-cell targets was performed by amplification of IGH-VJ (FR1, 2, and 3) and incomplete DH-JH gene rearrangements (tube A), IGK-VJ and VK/intron-KDE gene rearrangements using the multiplex PCR protocols developed by BIOMED-2 concerted action BMH4-CT98-3936 [5] in duplicate. Multiplex PCR products of Ig targets were monitored by fluorescent GeneScan analysis on an ABI 3730 platform (Applied Biosystems). GeneScan patterns were processed by the Genemapper (version 3.7) software (ABI prism).
Results
Histology
HE sections showed a picture of cHL in all cases, with scattered large atypical cells that expressed CD30, with variable expression of CD15, CD20, and CD79a, in an inflammatory background. In one case, the HRS cells expressed a T-cell phenotype (CD2+, CD3+/−, CD4+, CD5+, CD8−) but not CD20 or CD79a. Hodgkin lymphoma consisted of nodular sclerosing (n = 19), mixed cellularity (n = 3), lymphocyte-rich (n = 1), and interfollicular (n = 1) types. The histology of case 8, a nodular sclerosing classical Hodgkin lymphoma, is shown in Fig. 1. Most lymph nodes were completely involved by cHL, but some reactive follicles were still present in several cases (3,10,15,16,18, 23,24). The average percentage of HRS cells varied from less than 5% to occasionally 20%, especially in cases with a sclerotic background lacking small lymphocytes (Table 1).
EBER was detected in the HRS cells of eight of 22 patients (Table 1). All were over 15 years of age. One was HIV-positive (case 21), others had no known immune deficiencies. Patient 12 had many slightly enlarged CD30 and EBV-positive B cells in the background in addition to the HRS cells; this was not seen in the other EBV-positive cHL patients. The HRS cells were EBER-negative in patient 19, while sporadic small nuclei were positive.
BMB was involved by cHL in three patients. None of the other patients showed evidence of an abnormal B-cell population in the bone marrow.
Clonality detection
In general, the criteria for evaluating the B-cell clonality results were: (1) sufficient DNA quality, suboptimal samples allowing amplification of only 100 nt of the control gene PCRs cannot be evaluated properly; (2) the clonal results should be detectable in duplicate; and (3) the clonal result should be clearly present almost without polyclonal background, or dominantly present in a Gaussian curve of polyclonal background signals, or should be dominantly present in a region within the expected size range that contains rare gene rearrangements (such as the Vk2/Vk4/Vk5-JK and the Vk2/Vk4/Vk5-Kde rearrangements).
Analysis was performed in duplicate for frozen and paraffin-embedded tissue, showing consistent results in all PCRs. The expected size ranges of the BIOMED-2 multiplex PCRs are listed in Table 2.
Table 2.
BIOMED-2 PCR product size ranges
| BIOMED-2 PCR | Estimated size range (nt) |
|---|---|
| IGH FR1 (tube A) | 310–360 |
| IGH FR2 (tube B) | 250–295 |
| IGH FR3 (tube C) | 100–170 |
| IGH-DJ (tube D) | 110–390 |
| IGK-VJ (tube A) | 120–300 |
| IGK Vκ/intron-Kde (tube B) | 210–390 |
Frozen tissue
The results of the PCRs for the different target genes on frozen tissue and FFPE are summarized in Table 3. An example of GeneScan results for different targets in a control sample and a patient sample (case 12) is given in Fig. 2. Overall, 19 cases showed clonally rearranged IGH and/or IGK genes, including case 6 with a T-cell phenotype of the HRS cells. This case did not show TCR rearrangement (data not shown). Five cases remained polyclonal for all targets, and these were cases with an estimated HRS cell percentage below 10%, two even below 5%. Ten other cases with an estimated tumor cell percentage from 5% to 10% had a clonal population. Of the five polyclonal cases, four were EBV-positive (cases 20–23). The four clonal EBV-positive cHL (cases 2, 7, 12, and 14) did not show a difference in IGH or IGK detection rate compared to EBV-negative cases. All cases showed a polyclonal background in some targets, confirming the presence of reactive B cells. Analysis of the different targets revealed that 13 of the 19 clonal cases would have been detected by testing only IGH FR1–3, and FR3 was positive in only seven of these cases. Clonal IGK rearrangements were detected in six additional cases, with one clonal IGK-VJ and six clonal IGK-DE rearrangements identified. IGH-DJ showed a clonal rearrangement in two cases but was always accompanied by other rearrangements. The detection rate of clonal rearrangements in this study was 79% providing sufficient DNA quality. In 11 of the 19 clonal cHL cases, multiple (two or more) clonally rearranged targets were found. The eight cases with isolated clonal rearrangements showed either IGH FR2 (two cases), FR3 (one case), or IGK-DE (five cases).
Table 3.
Results of immunoglobulin gene rearrangement studies in frozen tissue (A) and FFPE tissue (B)
| # | Tissue | HRS (%) | DNA (bp) | IGH | IGK | Result | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| FR1 | FR2 | FR3 | DJ | VJ | DE | |||||
| 1 | A | 10 | 400 | C+PCB | C+PCB | C+PCB | p | C+PCB | p | Clonal with PCB |
| B | 200 | ne | ne | p | ne | C+PCB | ne | Clonal with PCB | ||
| 2 | A | 10 | 400 | C+PCB | C | C+PCB | ne | p | C+PCB | Clonal with PCB |
| B | 300 | ne | p | C+PCB | ne | p | C+PCB | Clonal with PCB | ||
| 3 | A | 15 | 400 | C+PCB | C+PCB | p | p | C+PCB | C+PCB | Clonal with PCB |
| 4 | A | <10 | 400 | C+PCB | C+PCB | C+PCB | p | p | p | Clonal with PCB |
| 5 | A | 10 | 400 | C | C | p | p | p | p | Clonal with PCB |
| 6 | A | <10 | 400 | p | C | C+PCB | p | p | p | Clonal with PCB |
| 7 | A | <15 | 400 | C+PCB | p | C+PCB | p | p | p | Clonal with PCB |
| 8 | A | <20 | 400 | C+PCB | p | p | p | p | C+PCB | Clonal with PCB |
| 9 | A | 15 | 400 | p | C+PCB | p | p | p | p | Clonal with PCB |
| B | 200 | ne | C+PCB | ne | ne | p | ne | Clonal with PCB | ||
| 10 | A | 5 | 400 | p | C+PCB | p | p | p | p | Clonal with PCB |
| 11 | A | 10 | 400 | p | C | p | C | p | C | Clonal with PCB |
| 12 | A | 20 | 400 | p | p | C+PCB | C | p | C+PCB | Clonal with PCB |
| B | 200 | ne | ne | C+PCB | C | p | C+PCB | Clonal with PCB | ||
| 13 | A | 20 | 400 | p | p | C+PCB | p | p | p | Clonal with PCB |
| 14 | A | 20 | 400 | p | p | p | p | C+PCB | C+PCB | Clonal with PCB |
| B | 200 | ne | p | p | ne | C+PCB | C+PCB | Clonal with PCB | ||
| 15 | A | <15 | 400 | p | p | p | p | p | C | Clonal with PCB |
| B | 200 | ne | p | p | p | p | ne | Polyclonal | ||
| 16 | A | <10 | 400 | p | p | p | p | p | C | Clonal with PCB |
| 17 | A | 5 | 400 | p | p | p | p | p | C+PCB | Clonal with PCB |
| 18 | A | <15 | 400 | p | p | p | p | p | C+PCB | Clonal with PCB |
| B | 200 | ne | ne | ne | ne | p | ne | Polyclonal | ||
| 19 | A | 5 | 400 | p | p | p | p | p | C+PCB | Clonal with PCB |
| B | 300 | ne | p | ne | p | p | C+PCB | Clonal with PCB | ||
| 20 | A | 10 | 400 | p | p | p | p | p | p | Polyclonal |
| 21 | A | 10 | 400 | p | p | p | p | p | p | Polyclonal |
| 22 | A | <5 | 400 | p | p | p | p | p | p | Polyclonal |
| 23 | A | <5 | 400 | p | p | p | p | p | p | Polyclonal |
| 24 | A | 5 | 400 | p | p | p | p | p | p | Polyclonal |
Note that six of the eight FFPE tissues give concordant results to the analysis of the frozen tissues
HRS Hodgkin and Reed–Sternberg cells, bp base pairs, FR framework, DE kappa deleting element, C clonal, p polyclonal, PCB polyclonal background, ne not evaluable
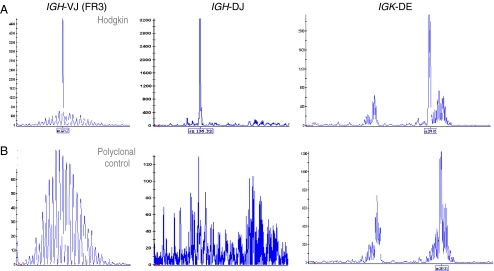
Fig. 2.
Clonality assessment. GeneScan results of the gene rearrangement profile of a cHL frozen biopsy sample (patient 12) showing clonal Ig rearrangements with multiple primer sets in a polyclonal background of B cells (a) and a polyclonal tonsil biopsy control (b)
Paraffin-embedded tissue
Sufficient FFPE tissue was available from 12 of the cases. From these 12 cases, the DNA was seriously fragmented in four cases, which showed amplification of only the 100-nt product in the Gene Control PCR. Clonality assessment was not successful in three of these although in some targets a weak polyclonal signal was seen. Surprisingly, however, one case (3) allowed detection of a weak clonal product of 145 nt in the IGK-VJ PCR. Of the eight cases that showed amplification of 200 nt or even 300 nt in the control gene PCR, six showed concordant results for the final interpretation of the clonality pattern. The targets that could be amplified in FFPE tissue showed identical sizes as in the corresponding frozen tissue.
From the IGH multiplex PCRs, four out of nine clonal rearrangements were identified in the FFPE tissues, notably one out of three IGH FR2, two out of three FR3, and one IGH-DJ. None of the two clonal IGH FR1 was detected in the corresponding FFPE samples. Of the clonal IGK rearrangements, both IGK-VJ rearrangements of the FFPE tissues (detected product sizes 138 and 136 nt) were detected and four out of six IGK-DE rearrangements. Note that the four clonal IGK-DE rearrangements that were detected in FFPE tissue had amplicon sizes of 281, 274, 283, and 352 nt, and the two that were undetected fell into the same range with 280 and 274 nt.
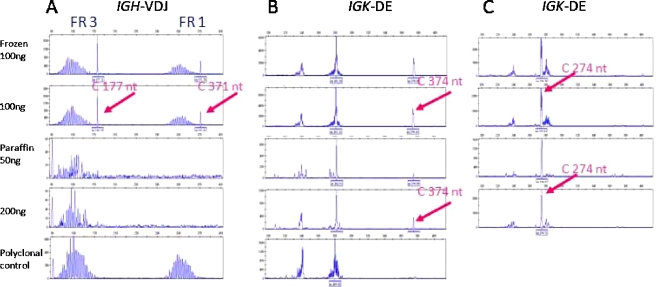
The clonal products amplified from paraffin-embedded tissue, even from samples with suboptimal DNA quality (showing amplification of 200 nt, weakly), were clearly detectable, as shown in Fig. 3 for the clonal IGK-DE rearrangement of case 12. In case 14, also having suboptimal DNA quality (200 nt), the clonal IGK-DE rearrangement of 374 nt can be detected in duplicate. Detection of this particular rearrangement is possible because Vk2/Vk4/Vk5-Kde gene rearrangements (size 350–390 nt) are rare rearrangements and hardly present in the tonsil DNA, which is used as polyclonal control. Therefore, detection of peaks in duplicate in this size range, even when these are small peaks, is highly suspicious for a true clonal rearrangement, in this case in a polyclonal background. The IGH-V(D)J rearrangement of case 1 with the FR3 and FR1 amplicon sizes of 177 and 371 nt is clearly detectable in the DNA from frozen tissue, but even the smaller FR3 rearrangement is not detected in the DNA sample from paraffin-embedded tissue, despite amplification of a weak 300-nt band in the control gene PCR.
Fig. 3.
Examples of PCR on DNA samples from corresponding frozen and paraffin-embedded tissue. All examples show clonal products and a polyclonal background. (a) Case 1: the IGH-V(D)J rearrangement with the FR3 and FR1 amplicon sizes of 177 and 371 nt is clearly detectable in the DNA from frozen tissue, but not in the DNA sample from paraffin-embedded tissue. Case 14 (b) and case 12 (c), IGK-DE amplicon sizes of 374 and 274 nt are detected in frozen and paraffin-embedded tissue. DNA obtained from a tonsil is used as the polyclonal control sample in each PCR experiment
Comparing the EBV status, relapse or primary biopsy, the percentage of small B cells in the background, the expression of CD20 and/or CD79a on the HRS cells, and the clonality status did not result in a correlation, except for a higher number of cases with CD20 and/or CD79a expression in the group with detectable clonal IGH rearrangement. Seven of 13 IGH rearranged cases showed B-cell antigen expression compared to two of 11 cases without detectable clonal IGH rearrangement.
Discussion
Clonality testing for complete and incomplete immunoglobulin (Ig) heavy chain and kappa light chain using the recently developed primer sets of the Biomed consortium of a series of 24 cHL revealed a clonally rearranged B-cell population in 79% of the cases, including a case with a T-cell phenotype.
Former attempts of clonality analysis by Southern blot analysis failed to show rearranged Ig genes in most cases of cHL because of the insensitivity of the technique and the large amount of DNA needed [6, 7]. PCR on tumor DNA with primers for IGH FR2/Jh and FR3/Jh improved sensitivity of clonality detection to 3–40% in different studies [1, 6–9]. Single-cell PCR of laser-dissected HRS cells further increased the detection rate of clonal Ig gene rearrangements [8, 9]. With the current BIOMED-PCR, not only complete IGH gene rearrangements (FR1-3) are detected, but also incomplete IGH-DJ and light chain gene rearrangements, including IGK-DE. This is an important addition, since the variable regions of the IGH genes are frequently somatically mutated in cHL [1, 10, 11], hampering clonality detection by IGH-VJ PCR. Light chain rearrangements, particularly IGK-DE, and IGH-DJ have been proven less prone to somatic mutation [11].
In agreement with the reported high frequency of somatic hypermutation in cHL, FR3 detected clonality in only seven out of our 19 clonal cases (37%), stressing the importance of multiple target testing in these lymphomas. This percentage is comparable to the reported (borderline) monoclonal result in 40% of cHL in a recent study limited to the use of BIOMED-2 IGH primers [3].
Indeed, all three IGH frameworks were negative in six cases that proved clonal by IGK PCR. Especially IGK-DE was a valuable addition, being the second most frequent target with a clonal rearrangement in 11 of 24 tested cases, although its frequency (45%) was somewhat lower than reported in B-cell NHL (53–75%) [2]. This was also seen in 12 cHL tested in the series of McClure et al., where additional IGK testing added an extra clonal case, increasing detection of clonality from 33% to 42% [4]. Clonal incomplete IGH-DJ rearrangement was detected in only two of our cases (8%), and never as the sole target, and seems therefore of no additional use for this lymphoma type.
That the overall detection rate of clonality in cHL is a little less than reported for B-cell NHL is probably due to the often low number of tumor cells in a background that usually harbors significant numbers of reactive B cells, since all cases without clonal rearrangements had a very low tumor load. The lower sensitivity in FFPE tissue can be mainly attributed to DNA fragmentation, allowing only smaller DNA fragments to be amplified. Although the BIOMED-2 primers have not been universally accepted for FFPE tissues, many laboratories use the BIOMED-2 primers for FFPE samples as well. However, when the DNA is fragmented due to tissue fixation and processing, clonal targets may not efficiently be amplified and therefore potentially yield false-negative results (as shown for cases 15 and 18 of our series), Therefore, we do not recommend clonality testing in cases showing severely fragmented DNA (amplification of only the 100-nt product of the BIOMED-2 control gene PCR). The relative high percentage of clonality in the study of Chute et al. using FFPE tissues will thus also be due to the good quality of the DNA samples, showing amplification of a 389-nt-sized control gene in all cases [3]. In agreement with the results of Halldórsdóttir et al. in NHL [12], the IGK multiplex PCRs, most particularly the IGK-DE PCR, performed better in detecting clonality than the IGH PCRs, despite a rather large amplicon size (up to 300 to 390 nt).
Since no correlation was found between the rearrangement pattern and the EBV status of the tumor cells, tumor percentage, relapse, background B cells, or bone marrow involvement, these factors cannot explain our findings.
Our data show that the present standard PCR-based method for clonality detection is sensitive enough to detect clonal rearrangements of the malignant cells in 79% of cHL, if PCR for IGH FR1–3 is complemented with IGK-DE.
Acknowledgments
Conflict of interest statement The authors declare that they have no conflict of interest.
References
- 1.Tamaru J, Hummel M, Zemlin M, Kalvelage B, Stein H. Hodgkin's disease with a B-cell phenotype often shows a VDJ rearrangement and somatic mutations in the VH genes. Blood. 1994;84(3):708–15. [PubMed] [Google Scholar]
- 2.Evans PA, Ch Pott, Groenen PJ, Salles G, Davi F, Berger F, Garcia JF, Krieken JH, Pals S, Kluin P, Schuuring E, Spaargaren M, Boone E, Gonzalez D, Martinez B, Villuendas R, Gameiro P, Diss TC, Mills K, Morgan GJ, Carter GI, Milner BJ, Pearson D, Hummel M, Jung W, Ott M, Canioni D, Beldjord K, Bastard C, Delfau-Larue MH, Dongen JJ, Molina TJ, Cabecadas J. Significantly improved PCR-based clonality testing in B-cell malignancies by use of multiple immunoglobulin gene targets. Report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia. 2007;21(2):207–14. doi: 10.1038/sj.leu.2404479. [DOI] [PubMed] [Google Scholar]
- 3.Chute DJ, Cousar JB, Mahadevan MS, Siegrist KA, Silverman LM, Stoler MH. Detection of immunoglobulin heavy chain gene rearrangements in classic Hodgkin lymphoma using commercially available BIOMED-2 primers. Diagn Mol Pathol. 2008;17(2):65–72. doi: 10.1097/PDM.0b013e318150d695. [DOI] [PubMed] [Google Scholar]
- 4.McClure RF, Kaur P, Pagel E, Ouillette PD, Holtegaard CE, Treptow CL, Kurtin PJ. Validation of immunoglobulin gene rearrangement detection by PCR using commercially available BIOMED-2 primers. Leukemia. 2006;20(1):176–9. doi: 10.1038/sj.leu.2404049. [DOI] [PubMed] [Google Scholar]
- 5.Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 6.Angel CA, Pringle JH, Primrose L, Lauder I. Detection of immunoglobulin heavy chain gene rearrangements in Hodgkin's disease using PCR. J Clin Pathol. 1993;46(10):940–2. doi: 10.1136/jcp.46.10.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angel CA, Pringle JH, Naylor J, West KP, Lauder I. Analysis of antigen receptor genes in Hodgkin's disease. J Clin Pathol. 1993;46(4):337–40. doi: 10.1136/jcp.46.4.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al Saati T, Galoin S, Gravel S, Lamant L, Roda D, Chittal SM, Delsol G. IgH and TcR-gamma gene rearrangements identified in Hodgkin disease by PCR demonstrate lack of correlation between genotype, phenotype, and Epstein–Barr virus status. J Pathol. 1997;181(4):387–93. doi: 10.1002/(SICI)1096-9896(199704)181:4<387::AID-PATH781>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 9.Manzanal AI, Santon A, Acevedo A, Aguilera B, Oliva H, Bellas C. Molecular analysis of the IgH gene in 212 cases of Hodgkin's disease: correlation of IgH clonality with the histologic and the immunocytochemical features. Mod Pathol. 1997;10(7):679–85. [PubMed] [Google Scholar]
- 10.Kanzler H, Kuppers R, Hansmann ML, Rajewsky K. Hodgkin and Reed–Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med. 1996;184:1495–1505. doi: 10.1084/jem.184.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marafioti T, Hummel M, Foss HD, Laumen H, Korbjuhn P, Anagnostopoulos I, Lammert H, Demel G, Theil J, Wirth T, Stein H. Hodgkin and Reed–Sternberg cells represent an expansion of a single clone originating from a germinal center B-cell with functional immunoglobulin gene rearrangements but defective immunoglobulin transcription. Blood. 2000;95(4):1443–50. [PubMed] [Google Scholar]
- 12.Halldórsdóttir AM, Zehnbauer BA, Burack WR. Application of BIOMED-2 clonality assays to formalin-fixed paraffin embedded follicular lymphoma specimens: superior performance of the IGK assays compared to IGH for suboptimal specimens. Leuk Lymphoma. 2007;48(7):1338–43. doi: 10.1080/10428190701377022. [DOI] [PubMed] [Google Scholar]