Abstract
Diffuse large B-cell lymphoma (DLBCL) with a germinal center B-cell (GCB) phenotype is believed to confer a better prognosis than DLBCL with an activated B-cell (ABC) phenotype. Previous studies have suggested that nuclear factor-κB (NF-κB) activation plays an important role in the ABC subtype of DLBCL, whereas c-REL amplification is associated with the GCB subtype. Using immunohistochemical techniques, we examined 68 newly diagnosed de novo DLBCL cases (median follow-up 44 months, range 1 to 142 months) for the expression of c-REL, BCL-6, CD10, and MUM1/IRF4. Forty-four (65%) cases demonstrated positive c-REL nuclear expression. In this cohort of patients, the GCB phenotype was associated with a better overall survival (OS) than the non-GCB phenotype (Kaplan–Meier survival (KMS) analysis, p = 0.016, Breslow–Gehan–Wilcoxon test). In general, c-REL nuclear expression did not correlate with GCB vs. non-GCB phenotype, International Prognostic Index score, or OS. However, cases with a GCB phenotype and negative nuclear c-REL demonstrated better OS than cases with a GCB phenotype and positive nuclear c-REL (KMS analysis, p = 0.045, Breslow–Gehan–Wilcoxon test), whereas in cases with non-GCB phenotype, the expression of c-REL did not significantly impact the prognosis. These results suggest that c-REL nuclear expression may be a prognostic factor in DLBCL and it may improve patient risk stratification in combination with GCB/non-GCB phenotyping.
Keywords: c-REL, Diffuse large B-cell lymphoma, DLBCL, Prognosis
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common subtype of non-Hodgkin’s lymphoma, and it comprises approximately 30–40% of new lymphoma diagnoses in the USA [1, 2]. Recently, using expression microarray technology, several studies have shown that DLBCL can be divided into three subgroups based on gene expression profiles. These include germinal center B-cell-like (GCB), activated B-cell-like (ABC), and type 3 [3, 4]. Independent of other established prognostic markers such as the International Prognostic Index (IPI) [5], patients with a gene expression profile resembling a GCB type have significantly better overall survival than those with an ABC type; the type 3 group also has a poor prognosis, but it represents a heterogeneous category that is not yet well defined. Immunohistochemistry (IHC), a technology that is more widely available and economically feasible than expression microarrays, has been attempted for classifying DLBCL into GCB and non-GCB subgroups using different combinations of CD10, BCL-6, MUM1, and/or CD138 [6–9]. The majority of these studies have confirmed significantly better survival for GCB group suggesting that IHC is an important tool to validate the microarray findings in pathology practice [6–9].
The nuclear factor-κB (NF-κB) signaling pathway plays an important role in regulating the survival of normal and malignant B-cells by controlling the expression of multiple cell death regulatory pathways [10, 11]. The recent development of NF-κB inhibitors may make this pathway a potential therapeutic target for lymphoma [12]. NF-κB target genes can enhance cell survival by modulating TNFα signaling, inhibiting FAS-mediated apoptosis, and limiting the activity of BCL2 family members [11]. The role of NF-κB pathways in different subgroups of DLBCL has not been well elucidated [12]. Although some studies suggest that constitutive activation of NF-κB contributes to the sustained oncogenicity of the ABC-like DLBCL [12–14], a recent expert review encouraged cautious interpretation of this data [12]. Studies have also suggested that c-REL amplification, a subunit of the REL/NF-κB family, is more commonly observed in GCB-type DLBCL than ABC-type DLBCL [4]; however, c-REL amplification is not frequently associated with NF-κB activation (nuclear c-REL expression) [10, 11, 15].
Activation of the NF-κB pathway in a B-cell lymphoma can be preliminarily assessed by determining the nuclear localization of NF-κB subunits such as c-REL [16–18]. In the inactive status, NF-κB heterodimers (c-REL or REL homolog A and NF-κB1) reside in the cytoplasm bound to an inhibitor of kappa B signaling (I-κB) [11]. During NF-κB activation, I-kappa kinase (I-κK) phosphorylates I-κB resulting in the dissociation of the inhibitor from the cytoplasmic NF-κB heterodimer. Thereafter, the freed (activated) NF-κB heterodimer, including c-REL, translocates to the nucleus where it induces the transcription of NF-κB target genes [11].
Using c-REL nuclear expression by IHC as a surrogate marker for NF-κB activation, studies have demonstrated that primary mediastinal large B-cell lymphoma cases show homogeneously strong c-REL nuclear staining, whereas the c-REL staining pattern in other DLBCL is more variable [16, 17, 19]. However, these studies have not attempted to correlate c-REL expression with clinical parameters. The main goal of the current study was to evaluate the impact of c-REL expression and GCB status on the clinical outcome in DLBCL patients using IHC.
Materials and methods
Case selection
DLBCL cases from 1992–2001 were retrospectively retrieved from the archives of the Department of Pathology at the Medical College of Wisconsin and The Methodist Hospital. Sixty-eight cases (44 cases from a tissue microarray with each case consisting of three 1.2-mm cores and 24 cases from whole tissue sections) of newly diagnosed de novo DLBCLs with adequate clinical follow-up information were included in the study. This cohort included 15 cases from a previous study [6]. Cases with a preceding diagnosis of low-grade lymphoma or a history of having received immunosuppressive therapy were excluded from the study. This cohort of patients did not include DLBCL cases occurring in the central nervous system (CNS) or mediastinum, because primary DLBCL of the CNS and primary mediastinal large B-cell lymphoma are considered distinct clinicopathologic subgroups of DLBCL by WHO classification [20]. A single case of DLBCL occurring in bone in our file were excluded in this study since we could not determine if this case represented primary lymphoma or secondary involvement of lymphoma based on the clinical information available. The pertinent clinical information for each case was obtained by reviewing the tumor registry records and/or patient medical charts. This study was approved by the Institutional Review Boards of the participating institutions.
Histopathology
All cases were reviewed to confirm that morphologic characteristics fulfilled the criteria of DLBCL by the WHO classification [20]. The H&E-stained sections of all cases show a neoplasm of large B lymphoid cells with nuclear size equal to or exceeding normal macrophage nuclei or more than twice the size of a normal lymphocyte that has a diffuse growth pattern.
Immunohistochemistry
Immunohistochemical staining was performed using antibodies against c-REL and a panel of GC B-cell (CD10 and BCL-6) and activation (MUM1/IRF4) markers as previously published [6]. For c-REL staining, the slides were pretreated with Citrate (DAKO USA, Carpinteria, CA, USA) for 30 min in a steamer. Primary rabbit anti-c-REL(AB-1) antibody (1:25dilution; Calbiochem/EMD Bioscience, Inc, San Diego, CA, USA) was applied in DAKO diluent for 1 h. The slides were washed, and the Dual Envision Kit (DAKO) was then applied for 30 min. After further washing, immunoperoxidase staining was developed using a diaminobenzidine chromogen kit (DAKO). CD10, BCL-6, and MUM1/IRF4 staining were performed as previously described [6].
Two hematopathologists (CVC and CCC) independently reviewed each case and assigned a semiquantitative score for the expression of each antigenic marker without the knowledge of clinical information. Each case was evaluated by visual estimation and recorded in 10% increments. Any discrepancies were resolved by reviewing slides with a multi-headed microscope to achieve consensus. Expression of each marker was considered positive when >20% of large neoplastic lymphocytes stained positively at the appropriate subcellular locations (c-REL, BCL-6, and MUM1/IRF4 with nuclear and/or cytoplasmic staining; CD10 with membranous and/or cytoplamic staining). This cutoff has been commonly used in previous studies evaluating protein expression by IHC [21–23].
Statistical methods
Demographics were summarized using frequencies and percentages for categorical variables and median and range for continuous variables. Survival probabilities were calculated using the Kaplan–Meier analysis. All analyses were done using SAS v 5.0.1.
Results
The cohort contained 36 females (53%) and 32 males (47%), ranging in age from 16 to 88 years (mean 63 years). The median follow-up was 43 months (range 0–142 months) and the median follow-up among survivors was 66 months (range 14–142 months). All patients received Cytoxan, Doxorubicin, Vincristine, and Prednisone- (CHOP-) based chemotherapy without Rituximab. We were able to abstract IPI scores in 34 cases. Twenty-three had low IPI scores (≤2, low risk and low-intermediate risk groups) and 11 had high IPI scores (>3, high-intermediate risk and high risk groups).
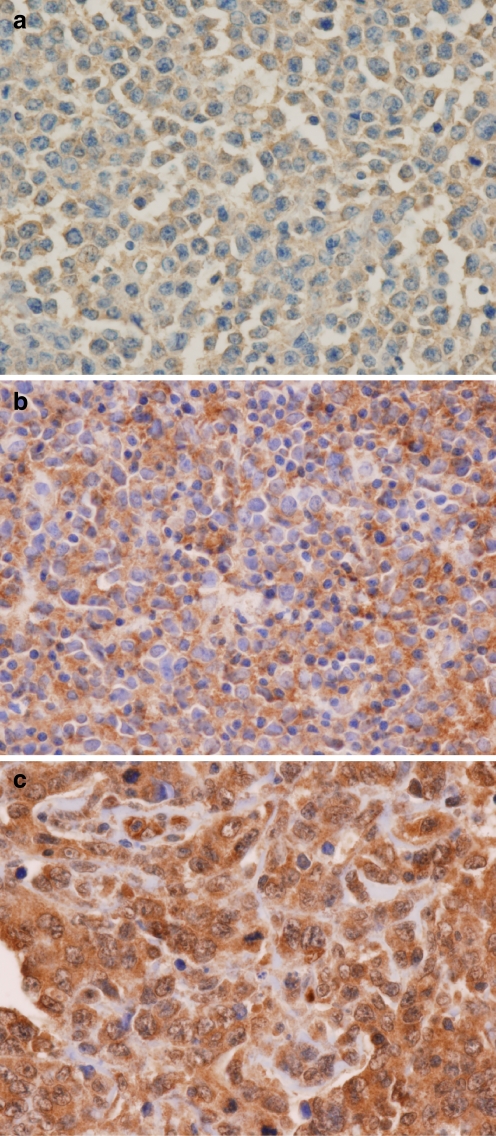
Forty-four cases (65%) were positive for c-REL nuclear expression. They showed moderate to strong nuclear and cytoplasmic staining of 20% to 100% of the large neoplastic lymphocytes (Fig. 1). The remaining 24 cases (35%) were negative for c-REL expression. The majority of the c-REL negative cases showed weak to moderate cytoplasmic staining in large neoplastic lymphocytes with rare scattered nuclei showing weak staining (Fig. 1). The frequencies of expression for CD10, BCL-6, and MUM1 in the cases studied were as follows: 29 of 68 (43%), 50 of 68 (74%), and 44 of 68 (65%; Table 1). The frequencies of antigen expression were quite comparable between the cases using tissue microarray and the cases using whole tissue section, suggesting that there was no sampling bias using tissue microarray. Based on these results, 24 cases (35%) were GCB phenotype (CD10+ and/or BCL-6+, MUM1-), and 44 cases (65%) were non-GCB phenotype (MUM1+, regardless of CD10 and BCL-6 expression) using the classification scheme by Chang et al. [6]. Using Hans’s classification methods [7], 34 cases (50%) were GCB phenotype (CD10+ or CD10-/BCL6+/MUM1-) and 34 cases (50%) were non-GCB phenotype (CD10-/BCL6- or CD10-/BCL6+/MUM1+).
Fig. 1.
Representative photomicroghs of c-REL expression in DLBCL (original magnification, ×400; immunohistochemical c-REL stain). a, b Negative c-REL expression cases: Negative nuclear expression with weak (a) and moderate (b) cytoplasmic expression, respectively. c Case scored as positive c-REL showing both nuclear and cytoplasmic staining; note small lymphocytes without nuclear or cytoplasmic staining
Table 1.
Correlation of nuclear c-REL expression with other immunohistochemical markers in DLBCL
| Immunohistochemical markers | Parameter (n) | Nuclear c-REL expression | P value | |
|---|---|---|---|---|
| Positive (n = 44) | Negative (n = 24) | |||
| CD10 | ||||
| Positive | 29 | 20 | 9 | 0.5262a |
| Negative | 39 | 24 | 15 | |
| BCL6 | ||||
| Positive | 50 | 30 | 20 | 0.1759a |
| Negative | 18 | 14 | 4 | |
| MUM1/IRF4 | ||||
| Positive | 44 | 29 | 15 | 0.7786a |
| Negative | 24 | 15 | 9 | |
| GCB vs non-GCB phenotype | ||||
| Non-GCB | 44 | 29 | 15 | 0.7786a |
| GCB | 24 | 15 | 9 | |
aChi Square
Of note, there was no statistically significant difference in the frequency of c-REL expression between the GCB and non-GCB phenotype and other immunohistochemical markers (CD10, BCL-6, or MUM1; Table 1). We further evaluated multiple clinical parameters in relation to the presence or absence of nuclear c-REL staining. No statistically significant correlation was observed between c-REL nuclear expression and age, sex, extranodal sites, or IPI (Table 2).
Table 2.
Comparison of clinical features in patients with or without positive nuclear c-REL Expression in DLBCL
| Clinical parameters | Parameter (n) | Nuclear c-REL expression | P value | |
|---|---|---|---|---|
| Positive (n = 44) | Negative (n = 24) | |||
| Age | ||||
| <60 | 23 | 15 | 8 | 0.9497a |
| ≥60 | 45 | 29 | 16 | |
| Sex | ||||
| Male | 32 | 19 | 13 | 0.3858a |
| Female | 36 | 25 | 11 | |
| Site | ||||
| Nodal | 37 | 21 | 16 | 0.1340a |
| Extranodal | 31 | 23 | 8 | |
| IPI | ||||
| Low (0–2) | 23 | 16 | 7 | 0.8499a |
| High (3–5) | 11 | 8 | 3 | |
aChi Square
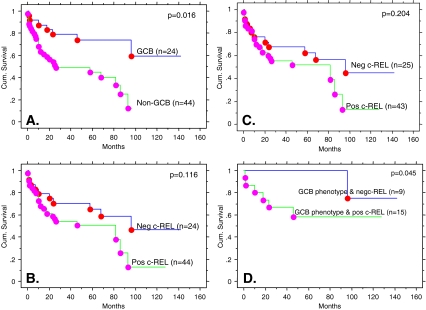
The GCB subgroup showed better overall survival (OS) than the non-GCB subgroup (50% cumulative survival not reach for GCB compared to 25–26 months for non-GCB, Kaplan–Meier survival analysis, p = 0.016, Breslow–Gehan–Wilcoxon test, Fig. 2a), using Chang’s classification scheme. The results were similar using Han’s scheme for defining GC and non-GC status (data not shown). Using only c-REL expression, no correlation with OS was observed (Fig. 2b). The finding of no correlation with OS remained the same when the data was analyzed using the 50% cutoff as defined by Rodig et al. [16] for c-REL expression (Fig. 2c). However, when combining c-REL nuclear expression with GCB vs. non-GCB phenotype as defined by Chang et al., cases with a GCB phenotype and negative nuclear c-REL had a better OS (only one out of nine patients died at 96 months with a median follow-up of 72 months) than cases with a GCB phenotype and positive nuclear c-REL (six out of 16 patients died at a median of 18 months, range 1 to 46 months, p = 0.045, Breslow–Gehan–Wilcoxon test (Fig. 2d)). When the GCB or non-GCB were defined using Han’s classification, the similar trend of having a better OS toward the group of patients with GCB phenotype and negative nuclear c-REL was observed, but the difference was not statistically significant (data not shown). Of noted, there was no statistically difference of IPI among c-REL positive or c-REL negative in the GCB phenotype subgroups (data not shown). Among the patients with non-GCB phenotype, c-REL expression did not impact the OS (data not shown).
Fig. 2.
Overall survival (OS) curves by Kaplan–Meier survival analysis. a OS curves using GCB versus non-GCB phenotypes. b Negative c-REL versus positive c-REL expression by our criteria. c Negative c-REL versus positive c-REL expression by Rodig’s criteria. d Only cases classified as GCB phenotype: negative c-REL versus positive c-REL expression
Discussion
Using c-REL nuclear expression by immunohistochemistry as a surrogate marker for NF-κB activation, our results suggest that DLBCL with a GCB phenotype and negative c-REL expression may have a better clinical outcome. Furthermore, c-REL expression does not correlate with either GCB or non-GCB phenotype or IPI score. Taken together, c-REL expression in combination with GCB or non-GCB phenotype evaluation may improve patient risk stratification in DLBCL. The detailed mechanisms leading to a better prognosis in the GCB subtype cases without c-REL expression remain to be elucidated. One possible mechanism would be increased susceptibility to chemotherapy imparted by a lack of NF-κB activation and its resultant reduction in anti-apoptotic signals in the neoplastic lymphocytes. Additionally, the observed decreased OS among cases of GCB phenotypes with c-REL nuclear expression suggests that the lymphoma cells, even with a good prognostic GCB phenotype, may become resistant to chemotherapeutic agents once the NF-κB pathway is activated. Therefore, these cases may benefit from the addition of an NF-κB inhibitor to the CHOP regimen. However, in cases with the non-GCB phenotype, the c-REL nuclear expression (NF-κB activation) does not correlate with OS suggesting that other pathways leading to the more aggressive clinical course may also be involved. It should be noted that our findings are somewhat limited by the nature of the retrospective study design since the treatment and clinical evaluation may be relatively heterogeneous.
Our c-REL immunohistochemical staining results largely correlate with those of previous studies. Others have also reported that c-REL expression in DLBCL is heterogeneous [16, 17, 19]. Furthermore, in agreement with previous studies, we do not observe any correlation between clinical parameters and c-REL nuclear expression [15]. However, the frequency of c-REL nuclear expression appeared higher in our study (65% vs. 18% to 44%, respectively) [16]. This observation may be attributed to the use of different c-REL antibody dilution, different staining methodologies, different scoring criteria, and/or case selection bias. For example, Rodig et al. used more stringent nuclear scoring criteria with a cutoff of 50% of cells staining positive to be considered positive [16]. These scoring criteria may have underestimated the number of cases with activated NF-κB signaling. When we used the same strict criteria to analyze our data, the results of the statistical analysis remained unchanged (Fig. 2c). Furthermore, we cannot compare our OS data with these studies since the correlation between c-REL nuclear expression and OS has not been attempted in these studies.
Our results suggest that there is no correlation between c-REL nuclear expression and GCB or non-GCB phenotypes by immunohistochemical staining. This is in contrast to previous experiments which reported that c-REL nuclear expression by immunofluorescence staining is more commonly observed in the ABC phenotype determined by expression microarray analysis [15]. However, the difference described by Houldsworth et al. was not statistically significant (c-REL nuclear expression, 6/18 in GCB vs. 7/11 in non-GCB, p = 0.143, Fisher’s Exact). These differences may be caused by the different c-REL detection methodologies (immunofluorescence vs. IHC) or the different GCB/non-GCB phenotype classification methodologies (expression microarray vs. IHC). Further studies using IHC to study the expression of NF-κB targeted gene activation are ongoing in our laboratory. These experiments may provide valuable insight into the role of NF-κB activation in the molecular pathogenesis of the different subtypes of DLBCL.
In the current study, all the data were analyzed using two published algorithms (one by our group and the other by Han et al.) for classifying GCB vs. non-GCB phenotype of DLBCL using IHC [6, 7]. Not surprisingly, two algorithms in general agree with each other relatively well since the only difference is that we consider any cases with MUM1 and CD10 expression as non-GCB while Han et al. consider CD10 expression regardless of MUM1 status as GCB. Nevertheless, the statistical significance of c-REL expression in GCB was observed using our classification algorithm but not by Han et al. although the latter algorithm also showed the similar trend. It would be of interest for the future similar studies to continue to use both algorithms to analyze the data to maximize the potential discoveries.
The patients in the current study were treated with CHOP without Rituximab rather than the current standard therapy of CHOP with Rituximab; the implication of our results in the latter setting will require further validation. Rituximab has an impact on the use of other prognostic markers such as BCL-2 or BCL-6 [24, 25]. Nevertheless, the prognostic significance of GCB vs. ABC subtype of DLBCL remains valid in a recent study by LLMPP consortium using microarray data [26], although the prognostic significance of GCB vs. non-GCB using IHC remains not well defined [27]. Our work should prompt the interest of the investigators to study c-REL expression in their patient cohort to evaluate the significance of our findings in the Rituximab era.
In summary, our data suggest that the evaluation of c-REL nuclear expression combined with GCB and non-GCB phenotyping by IHC may improve patient risk stratification in DLBCL. Further studies are needed to substantiate the clinical significance of our findings.
Acknowledgements
The study was supported in part by a grant support from NIH/NCI P50 CA126752 to CC. The authors appreciate Brandi Smith-Irving for editing this paper.
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.No authors listed (1997) A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin’s lymphoma. The Non-Hodgkin’s Lymphoma Classification Project. Blood 89(11):3909–3918. Jun 1 [PubMed]
- 2.Coiffier B. Diffuse large cell lymphoma. Curr Opin Oncol. 2001;13(5):325–334. doi: 10.1097/00001622-200109000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 5.No authors listed (1993) A predictive model for aggressive non-Hodgkin’s lymphoma. The International Non-Hodgkin’s Lymphoma Prognostic Factors Project. N Engl J Med 329(14):987–994. Sep 30 [DOI] [PubMed]
- 6.Chang CC, McClintock S, Cleveland RP, et al. Immunohistochemical expression patterns of germinal center and activation B-cell markers correlate with prognosis in diffuse large B-cell lymphoma. Am J Surg Pathol. 2004;28(4):464–470. doi: 10.1097/00000478-200404000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 8.Colomo L, Lopez-Guillermo A, Perales M, et al. Clinical impact of the differentiation profile assessed by immunophenotyping in patients with diffuse large B-cell lymphoma. Blood. 2003;101(1):78–84. doi: 10.1182/blood-2002-04-1286. [DOI] [PubMed] [Google Scholar]
- 9.Oh YH, Park CK. Prognostic evaluation of nodal diffuse large B cell lymphoma by immunohistochemical profiles with emphasis on CD138 expression as a poor prognostic factor. J Korean Med Sci. 2006;21(3):397–405. doi: 10.3346/jkms.2006.21.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2(4):301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 11.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat Immunol. 2002;3(3):221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 12.Gilmore TD, Kalaitzidis D, Liang MC, Starczynowski DT. The c-Rel transcription factor and B-cell proliferation: a deal with the devil. Oncogene. 2004;23(13):2275–2286. doi: 10.1038/sj.onc.1207410. [DOI] [PubMed] [Google Scholar]
- 13.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194(12):1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenwald A, Staudt LM. Gene expression profiling of diffuse large B-cell lymphoma. Leuk Lymphoma 44 Suppl. 2003;3:S41–S47. doi: 10.1080/10428190310001623775. [DOI] [PubMed] [Google Scholar]
- 15.Houldsworth J, Olshen AB, Cattoretti G, et al. Relationship between REL amplification, REL function, and clinical and biologic features in diffuse large B-cell lymphomas. Blood. 2004;103(5):1862–1868. doi: 10.1182/blood-2003-04-1359. [DOI] [PubMed] [Google Scholar]
- 16.Rodig SJ, Savage KJ, LaCasce AS, et al. Expression of TRAF1 and nuclear c-Rel distinguishes primary mediastinal large cell lymphoma from other types of diffuse large B-cell lymphoma. Am J Surg Pathol. 2007;31(1):106–112. doi: 10.1097/01.pas.0000213334.40358.0e. [DOI] [PubMed] [Google Scholar]
- 17.Rodig SJ, Savage KJ, Nguyen V, et al. TRAF1 expression and c-Rel activation are useful adjuncts in distinguishing classical Hodgkin lymphoma from a subset of morphologically or immunophenotypically similar lymphomas. Am J Surg Pathol. 2005;29(2):196–203. doi: 10.1097/01.pas.0000149689.75462.ff. [DOI] [PubMed] [Google Scholar]
- 18.Barth TF, Martin-Subero JI, Joos S, et al. Gains of 2p involving the REL locus correlate with nuclear c-Rel protein accumulation in neoplastic cells of classical Hodgkin lymphoma. Blood. 2003;101(9):3681–3686. doi: 10.1182/blood-2002-08-2577. [DOI] [PubMed] [Google Scholar]
- 19.Feuerhake F, Kutok JL, Monti S, et al. NFkappaB activity, function, and target-gene signatures in primary mediastinal large B-cell lymphoma and diffuse large B-cell lymphoma subtypes. Blood. 2005;106(4):1392–1399. doi: 10.1182/blood-2004-12-4901. [DOI] [PubMed] [Google Scholar]
- 20.Gatter KC, Warnke RA. Diffuse large B-cell Lymphoma. In: Jaffe ES, Harris NL, Stein H, Vardiman JW, editors. Pathology and genetics of tumours of haematopoietic and lymphoid tissues. Lyon: IARC; 2001. pp. 171–174. [Google Scholar]
- 21.Natkunam Y, Warnke RA, Montgomery K, Falini B, Rijn M. Analysis of MUM1/IRF4 protein expression using tissue microarrays and immunohistochemistry. Mod Pathol. 2001;14(7):686–694. doi: 10.1038/modpathol.3880373. [DOI] [PubMed] [Google Scholar]
- 22.Hedvat CV, Hegde A, Chaganti RS, et al. Application of tissue microarray technology to the study of non-Hodgkin’s and Hodgkin’s lymphoma. Hum Pathol. 2002;33(10):968–974. doi: 10.1053/hupa.2002.127438. [DOI] [PubMed] [Google Scholar]
- 23.Milanes-Yearsley M, Hammond ME, Pajak TF, et al. Tissue micro-array: a cost and time-effective method for correlative studies by regional and national cancer study groups. Mod Pathol. 2002;15(12):1366–1373. doi: 10.1097/01.MP.0000036345.18944.22. [DOI] [PubMed] [Google Scholar]
- 24.Mounier N, Briere J, Gisselbrecht C, et al. Rituximab plus CHOP (R-CHOP) overcomes bcl-2–associated resistance to chemotherapy in elderly patients with diffuse large B-cell lymphoma (DLBCL) Blood. 2003;101(11):4279–4284. doi: 10.1182/blood-2002-11-3442. [DOI] [PubMed] [Google Scholar]
- 25.Winter JN, Weller EA, Horning SJ, et al. Prognostic significance of Bcl-6 protein expression in DLBCL treated with CHOP or R-CHOP: a prospective correlative study. Blood. 2006;107(11):4207–4213. doi: 10.1182/blood-2005-10-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campo E (2008) Insights from expression arrays and genomic profiling of aggressive B-cell lymphoma. Society for hematopathology-companion meeting at United States and Canadian Academy of Pathology annual meeting. Denver, USA, pp 2–8
- 27.Nyman H, Adde M, Karjalainen-Lindsberg ML, et al. Prognostic impact of immunohistochemically defined germinal center phenotype in diffuse large B-cell lymphoma patients treated with immunochemotherapy. Blood. 2007;109(11):4930–4935. doi: 10.1182/blood-2006-09-047068. [DOI] [PubMed] [Google Scholar]