Abstract
The MLL-ELL fusion gene results from the translocation t(11;19)(q23;p13.1) that is associated with de novo and therapy-related acute myeloid leukemia. To study its transforming properties, we retrovirally transduced primary murine hematopoietic progenitors and assessed their growth properties both in vitro and in vivo. MLL-ELL increased the proliferation of myeloid colony-forming cells in methylcellulose cultures upon serial replating, whereas overexpression of ELL alone had no effect. We reconstituted lethally irradiated congenic mice with bone marrow progenitors transduced with MLL-ELL or the control MIE vector encoding the enhanced green fluorescent protein. When the peripheral blood of the mice was analyzed 11–13 weeks postreconstitution, we found that the engraftment of the MLL-ELL-transduced cells was superior to that of the MIE controls. At this time point, the contribution of the donor cells was normally distributed among the myeloid and nonmyeloid compartments. Although all of the MIE animals (n = 10) remained healthy for more than a year, all of the MLL-ELL mice (n = 20) succumbed to monoclonal or pauciclonal acute myeloid leukemias within 100–200 days. The leukemic cells were readily transplantable to secondary recipients and could be established as immortalized cell lines in liquid cultures. These studies demonstrate the enhancing effect of MLL-ELL on the proliferative potential of myeloid progenitors as well as its causal role in the genesis of acute myeloid leukemias.
Translocations involving chromosome band 11q23 occur frequently in hematologic malignancies and account for 7–10% of acute lymphoblastic leukemia and 5–6% of acute myeloid leukemia (1). In addition to de novo leukemias, the frequent development of therapy-related leukemias with 11q23 aberrations has been recognized in patients who have previously received chemotherapy drugs that target topoisomerase II (2). The MLL gene has been cloned from the 11q23 chromosomal translocation breakpoint by several groups and is referred to by various names (MLL, HRX, and ALL-1) (3–5). Although more than 20 different recurring cytogenetic aberrations that affect the MLL gene have been described, the functions of most MLL partner genes are not yet known. The critical feature of these chromosomal rearrangements is the generation of a chimeric transcript consisting of 5′ MLL and 3′ sequences of the gene on the partner chromosome. The ELL gene was first identified as a partner gene of MLL in the (11;19)(q23;p13.1) translocation (6). Subsequent studies have revealed that ELL is an RNA polymerase II transcriptional elongation factor (7). It serves to increase the catalytic rate of RNA polymerase II by suppressing transient pausing by the polymerase along the DNA template.
Two different strategies have been used to generate mouse models of 11q23 leukemias. To create an Mll-AF9 “knock-in” mouse, the AF9 cDNA was targeted into the murine Mll genomic locus by homologous recombination in embryonic stem cells, and the majority of the chimeric mice developed acute myeloid leukemia (8). Chimeric mice that expressed only the N-terminal sequences of Mll fused to a MYC tag did not develop leukemia. In addition, retroviral bone marrow (BM) infection has been used to express the MLL-ENL gene in hematopoietic cells (9). In serial replating assays in methylcellulose, the MLL-ENL-transduced cells became immortalized, whereas constructs containing either N-terminal MLL, ENL alone, or vector controls could not maintain proliferative capacity beyond three passages. After transplantation of MLL-ENL transduced BM, recipient mice developed acute myeloid leukemia (9). The normal functions of AF9 and ENL, the two MLL partner genes studied in these mouse models, are not known. However, AF9 and ENL exhibit extensive amino acid similarity. Thus, it is not clear whether similar phenotypes might be observed in mouse models of 11q23 leukemia that express alternative MLL fusions. At this time, mouse models involving other MLL partner genes have not yet been established.
In contrast to the majority of MLL partner proteins, much is now known about the function of ELL. Thus, we sought to develop a mouse model of MLL-ELL leukemia to characterize the pathogenesis of the (11;19)(q23;p13.1) in acute leukemia and to provide further insight into the potential mechanisms of leukemias associated with MLL fusions. We have assembled an MLL-ELL chimeric cDNA that recapitulates the fusion gene expressed in patients with the t(11;19)(q23;p13.1). This MLL-ELL fusion gene contains the AT hooks, methyltransferase domain, and repression domain of 5′ MLL fused to ELL sequences beginning at amino acid 46 that include the elongation domain and lysine-rich motif of ELL. Retroviral transduction of this MLL-ELL fusion gene immortalized primary hematopoietic progenitor cells in vitro. In addition, transplantation of MLL-ELL transduced progenitor cells to congenic mice induced the development of acute myeloid leukemia in all recipient mice. Moreover, enhanced engraftment of the MLL-ELL transduced progenitor cells was demonstrated, suggesting an early growth advantage conferred by expression of the MLL-ELL fusion gene.
Materials and Methods
Retroviral Constructs, Infection of Primitive Hematopoietic Progenitors, and Methylcellulose Colony-Forming Assays.
The MLL-ELL, MLLdel, ELL cDNAs (6), and MLL-ENL were cloned in the MSCV vector (10). For in vivo expression, MLL-ELL was cloned upstream of the internal ribosomal entry site (IRES) of the MIE [MSCV IRES EGFP (enhanced green fluorescent protein)] vector (11). Production of retroviral supernatants in Bosc23 (12) was performed as described (9). Infection of lineage-depleted (Lin−) BM obtained from BA.1 mice 5 days after 5 fluorouracil treatment and culture of the transduced progenitors in methylcellulose culture has been described (13). Efficiency of transduction by the MIE-based vectors was determined by flow cytometry on the day after the second round of spinoculation. Although the percentage of Lin- cells transduced with the empty MIE vector varied from 40% to 90% between experiments, the intensity of green fluorescence obtained with MLL-ELL- or MLL-ENL-encoding vectors was very low, making it difficult to distinguish the transduced progenitors from the noninfected control by flow cytometry.
Reconstitution Assay and Characterization of Leukemias.
Reconstitution of lethally irradiated B6.SJL mice with transduced progenitors was performed as described (9) with the following modifications. Each mouse was inoculated by tail vein injection with 1–2 × 104 Lin− BM cells transduced with MLL-ELL, together with 105 normal B6.SJL BM cells to ensure radioprotection. In parallel, the transduced Lin− cells were seeded in methylcellulose cultures to determine their content in colony-forming cells (CFCs). The degree of engraftment was assessed by flow cytometric analysis of peripheral blood (PB) stained with anti-Ly5.2 antibodies (PharMingen). MSCVMLL-ELL mice were analyzed by using anti-Ly5.2 conjugated to fluoresceine isothiocyanate as described (9), and MIEMLL-ELL mice were studied with anti-Ly5.2 conjugated to biotin followed by a secondary stain with streptavidin-allophycocyanin (Caltag). PB counts were measured by using a Cell Dyn 3500R (Abbott). Immunophenotypic characterization of the leukemic cells was done by costaining the circulating leukocytes with the anti-Ly5.2 antibodies and phycoerythrin-conjugated antibodies against F4/80 (Caltag), CD11b/Mac-1 Gr-1, CD19, CD3, Thy1 (antibodies against Thy1.1 and Thy1.2 were mixed), or with the IgG2a or IgG2b isotype controls (PharMingen). Stained cells were examined with a FACSCalibur instrument (Becton Dickinson) after exclusion of dead cells by high propidium iodide staining and forward light scatter. Tissues were fixed in formalin and sectioned and stained with hematoxylin and eosin for histological analysis. Blood smears and cytospin preparations of the BM were stained with Wright-Giemsa. The transplantability of tumors was determined by injecting 106 nucleated BM cells from the primary leukemic mice into sublethally irradiated (a single dose of 525 rads) B6.SJL secondary recipients. To establish cell lines, BM was seeded at a density of 5 × 105 cells/ml in RPMI 1640 medium containing 10% FCS and 0.05 mM 2-mercaptoethanol, in the presence or absence of IL-3 (10 ng/ml) and stem cell factor (100 ng/ml). The cells then were maintained up to 4 months.
DNA and Reverse Transcriptase–PCR Analysis.
DNA was prepared from tumor-infiltrated spleens; Southern blot analyses using a MLL probe were performed as described (10). The clonality of the reconstituting cells was assessed in the MIE or MIEMLL-ELL mice by probing the NcoI-digested DNA with the EGFP cDNA. Total RNA was isolated from BM cell lines or spleens by using RNA STAT-60 (Tel-Test, Friendswood, TX) as recommended by the manufacturer. One microgram of RNA was reversed-transcribed into cDNA with enhanced AMV reverse transcriptase (Sigma). PCRs were performed with Taq polymerase by using a forward primer from MLL exon 7 and a reverse primer from ELL exon 2. To exclude a false positive signal from DNA contamination of the RNA, PCRs also were performed on 1-μg aliquots of RNA that had not been incubated with reverse transcriptase. As a positive control, PCRs also were performed by using primers from the actin gene.
Western Blot Analysis.
Nuclear extracts were prepared from BOSC23 cells by the method of Dignam (14). Protein samples were electrophoresed in SDS-7.5% polyacrylamide gels and blotted on nitrocellulose membranes (Bio-Rad) by using a transfer buffer with 25 mM Tris, 192 mM glycine, 0.1% SDS, and 20% methanol, pH 8.3. The membranes were blocked in 5% nonfat dry milk in Tris-buffered saline with 0.05% Tween 20 and incubated with a rabbit polyclonal antiserum to ELL (15). The membranes were washed and incubated with biotin-conjugated goat-anti-rabbit antibody (Santa Cruz Biotechnology), washed, and then incubated with horseradish peroxidase-conjugated streptavidin (Jackson ImmunoResearch). After washes, the protein bands were detected with an enhanced chemiluminescence protocol (ECL) (Amersham Pharmacia).
Results
MLL-ELL Induces the Proliferation of Myeloid Progenitors in Vitro.
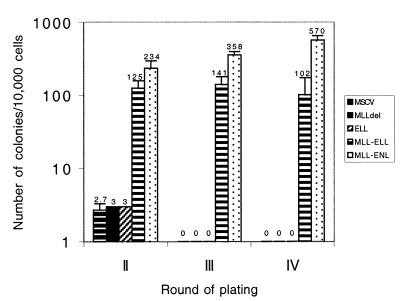
Murine BM enriched in early hematopoietic precursors by depletion of the terminally differentiated cells (this primitive population is referred herein as Lin− cells) was transduced with MLL-ELL by using the MSCVneo retroviral vector. The transforming potential of MLL-ELL was determined by measuring the growth of the transduced cells in a clonogenic assay for myeloid progenitors that was developed to study the proliferative effect of MLL-ENL (9). In this assay, the transduced cells were plated in G418-containing methylcellulose with growth factors that favor the development of myeloid colonies. After 7 days of culture, all of the cells in the culture dish were rinsed from the methylcellulose and replated in new media to test their potential to form myeloid colonies upon secondary plating; this process was repeated for two more passages to determine the frequency of tertiary and quaternary colonies. We compared the growth of cells transduced with the control MSCVneo vector to that of cells transduced with MLL-ENL, MLL-ELL, MLLdel (MLL portion of the MLL fusion products), or ELL (Fig. 1). We first observed that, like MLL-ENL (9), MLL-ELL had a deleterious effect on the retroviral titer and resulted in a 10- to 20-fold reduction in the number of G418-resistant primary colonies compared with the empty MSCVneo vector. We attribute this result to the size of the MLL-ELL transgene as well as to its cellular toxicity, a property observed when attempting to stably express the fusion protein in several cell lines (ref. 16 and M.J.T., unpublished data). Upon serial replating we found, as previously reported, that MSCVneo or MLLdel-transduced cells exhausted virtually all of their proliferative potential after the second passage and generated rare (3 ± 1 colonies/104 cells seeded), small secondary colonies, and the cells transduced with MLL-ENL generated highly proliferative macroscopic colonies that were generated in at least three rounds of replating (570 ± 125 quaternary colonies/104 cells seeded). The cells transduced with MLL-ELL generated serial colonies comparable in size to those obtained with MLL-ENL, but these were less numerous (102 ± 55 quaternary colonies/104 cells seeded). In contrast, the cells transduced with ELL displayed similar growth potential as those transduced with the control MSCVneo construct (Fig. 2). Expression of the MLL-ELL and ELL proteins was confirmed by Western blot analysis of the transfected Bosc23 cells (Fig. 1B). Thus, MLL-ELL confers a proliferative advantage to primary myeloid cells in vitro, and this effect requires the fusion of MLL to ELL.
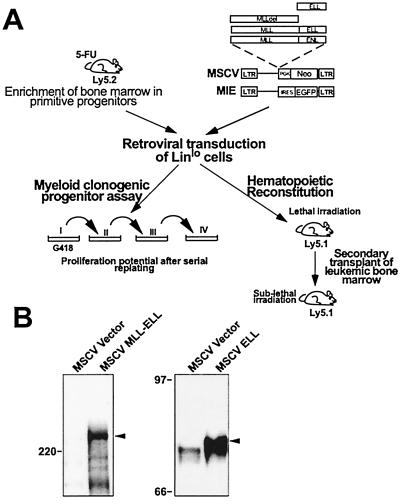
Figure 1.
Schematic representation of the experimental strategy and expression of the constructs. (A) Scheme used to compare the proliferative effect of MLL-ELL to MLL-ENL and to mutants in vitro and to study its leukemic effect in vivo. IRES, internal ribosomal entry site; PGK, phosphoglycerate kinase. (B) Western blot analysis of MLL-ELL and ELL expressed from the MSCV construct in transfected Bosc23 cells by using an anti-ELL antiserum. The arrowheads indicate the specific bands. The MLL-ELL fusion protein migrates at approximately 250 kDa and the ELL protein migrates at approximately 75 kDa. The position of molecular mass markers is shown.
Figure 2.
Proliferative effect of MLL-ELL compared with that of MLLdel, ELL, and MLL-ENL. The bars represent the number of colonies per 104 input cells generated after secondary, tertiary, and quaternary rounds of plating in methylcellulose. Results shown represent the mean ± SD from at least four independent experiments.
Progenitors Retrovirally Infected with MLL-ELL Display an Engraftment Advantage.
To determine the effect of MLL-ELL on repopulating stem cells in vivo, we reconstituted lethally irradiated mice with congenic cells transduced with MLL-ELL. Lin− BM cells from Ly5.2 donor mice were transduced and injected into lethally irradiated Ly5.1 recipients along with 105 normal Ly5.1 BM cells to ensure radioprotection. We first studied a cohort of 10 mice that each received 104 Lin− BM cells transduced with MSCVMLL-ELL (experiment 1). This inoculum contained 2.5 × 103 CFCs (Table 1), among which approximately 230 were resistant to G418, hence transduced. When these mice were bled 13 weeks posttransplantation, a significant proportion of the circulating leukocytes was found to be of donor origin (59 ± 6% SEM, Ly5.2 cells). This level of engraftment was superior (approximately 2-fold higher) to that observed in similar competitive repopulation experiments conducted with other genes in our laboratory (data not shown). To further document a possible engraftment advantage of MLL-ELL-transduced stem cells, we performed two additional transplantation experiments (experiments 2 and 3, Table 1) using the MIE bicistronic virus derived from MSCV that encodes EGFP and thus facilitates the detection of transduced hematopoietic cells in the recipient mice. As with the MSCVneo vector, the MLL-ELL-encoding MIE vector gave much lower titers than the empty MIE vector. This resulted in a great reduction in both the percentage of Lin− cells expressing EGFP and the mean intensity of EGFP expression per cell. PB from the mice was analyzed 10–11 weeks postinoculation for engraftment and vector expression. Although EGFP expression was readily detectable in the MIE-reconstituted mice (ranging from 30% to 70% of the donor leukocytes), its expression was below detectable levels in the MIEMLL-ELL-reconstituted mice. However, we found that the MIEMLL-ELL animals consistently displayed a higher level of engraftment than the control mice inoculated with the empty MIE vector-transduced BM. Although this engraftment advantage was marginal in experiment 2 (42% versus 31% Ly5.2, P = 0.17), it was significant in experiment 3 (79% versus 65%, P = 0.002). This difference between experiments is probably attributable to variations in transduction efficacy of the progenitor cells. Importantly, the animals did not display any significant elevation in the percentage of circulating myeloid cells of donor origin at the time those engraftment levels were measured (Table 1). Although we cannot rule out that the higher transduction efficiency achieved with the MIE vector might adversely affect engraftment, our results suggest that MLL-ELL confers a growth advantage to the repopulating cells.
Table 1.
Numbers of cells transplanted per mouse and engraftment
| Experiment | Total cells injected | CFC injected | Construct (n of mice) | Engraftment* | Myeloid donor cells† |
|---|---|---|---|---|---|
| 1 | 10 × 103 | 2.5 × 103 | MSCVMLL-ELL (10) | 59 ± 19% | 16 ± 13% |
| 2 | 20 × 103 | 4.2 × 103 | MIE (10) | 31 ± 21% | 12 ± 4% |
| MIEMLL-ELL (10) | 42 ± 13% | 17 ± 9% | |||
| 3 | 15 × 103 | 5.4 × 103 | MIE (10) | 65 ± 7% | 8 ± 3% |
| MIEMLL-ELL (9) | 79 ± 9% | 9 ± 2% |
Engraftment was determined by the mean percentage of Ly5.2 cells (±SD) in the PB measured at 13 weeks (experiment 1), 11 weeks (experiment 2), or 10 weeks (experiment 3) postreconstitution.
The mean percent of Mac1- and/or Gr-1-positive cells (±SEM) among the donor (Ly5.2) PB leukocytes, measured simultaneously with engraftment.
MLL-ELL-Reconstituted Mice Develop Acute Myeloid Leukemias.
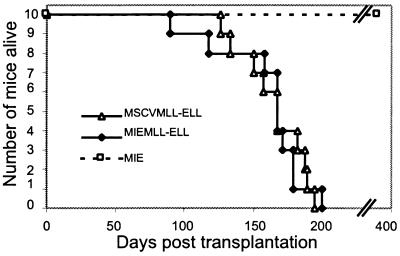
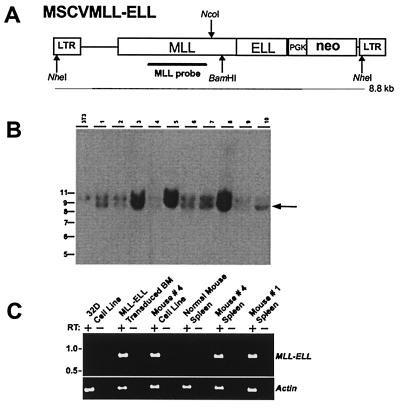
We monitored the mice reconstituted in experiments 1 and 2 and found that all recipients of MLL-ELL transduced cells died of myeloid leukemias within 100–200 days posttransplant (Fig. 3), whereas the 10 mice reconstituted with the MIE empty vector remained healthy for more than 1 year. To determine the presence and integrity of the MLL-ELL-encoding provirus in the leukemic mice we performed Southern blot analyses of splenic DNA digested with the NheI restriction enzyme that cuts in each long terminal repeat (LTR) (Fig. 4A). The expected fragment of 8.8 kb hybridizing to an MLL probe was detected in all of the animals (MSCVMLL-ELL mice shown in Fig. 4B and MIEMLL-ELL mice not shown). The presence of this band confirmed that no gross deletion or rearrangement had occurred during the retroviral production and expansion of the leukemic cells. To verify that the MLL-ELL gene was expressed in the MSCVMLL-ELL mice, we used oligonucleotide primers flanking the MLL-ELL fusion site to perform reverse transcription–PCR analysis on RNA isolated from the spleens and from cell lines established from the BM of these animals (Fig. 4C). The presence of EGFP was readily detectable by flow cytometry in the PB, BM and spleen of the MIEMLL-ELL mice after the development of leukemias (see Fig. 5). This finding is in contrast to the analysis 10–13 weeks posttransplantation when EGFP expression could not be detected. The expression of EGFP and the presence of unrearranged proviral copies indicate that the bicistronic transcript encoding MLL-ELL was being correctly expressed in the MIEMLL-ELL leukemias.
Figure 3.
Survival curves of mice reconstituted with stem cells transduced by MSCV-MLL-ELL, MIEMLL-ELL, or MIE.
Figure 4.
Proviral structure, integration, and expression in MSCVMLL-ELL mice. (A) Structure of the MSCVMLL-ELL retrovirus. Indicated are the diagnostic NheI, BamHI, and NcoI restriction endonuclease recognition sites that permit assessment of provirus integrity and copy number. PGK, phosphoglycerate kinase. (B) Southern blot analysis of NheI-digested genomic DNA with an MLL probe showing that leukemic spleens from MSCVMLL-ELL mice harbor nonrearranged (8.8-kb NheI band indicated by arrow) proviruses. The 10-kb fragment is representative of the endogenous MLL gene. At left is sizes (in kb) of the molecular weight standards. (C) Expression by reverse transcription–PCR of retroviral-derived MLL-ELL. Expression was studied in a negative control cell line (32D), MLL-ELL-transduced BM, a cell line derived from the leukemic BM of mouse #4 as well as in spleen from leukemic mice (spleen from mice #1 and #4 are shown), or from a control normal mouse.
Figure 5.
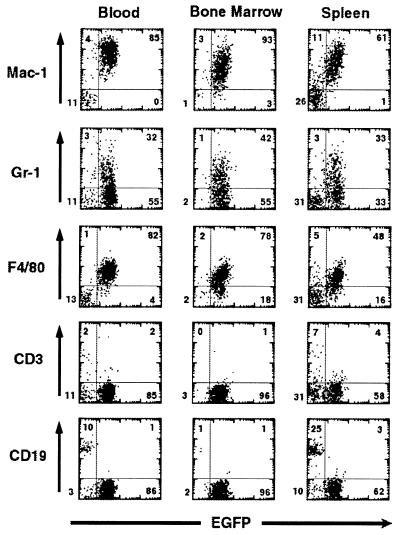
Immunophenotyping of MLL-ELL-associated leukemias by flow cytometry. Analysis of EGFP expression versus expression of lineage-specific markers (Mac-1 and F4/80 for macrophages, Gr-1 for granulocytes, CD3 for T cells, or CD19 for B cells) from the PB, BM or spleen of a representative MIEMLL-ELL mouse. The numbers indicate the percentage of cells that are present in each quadrant.
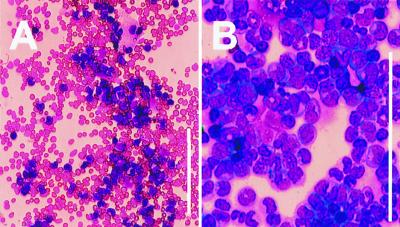
Hematological analyses and pathological examination were performed on the diseased animals (see Table 2). In the majority of the cases, we observed an elevation in the number of white blood cells (ranging from 15 to 210 103/μl in 13 of 20 mice) although some animals were instead leukopenic. Almost all of the mice (19 of 20) showed abnormal differential counts with a sharp increase in the proportion of monocytes (ranging from 20% to 72%, compared with 5 ± 1% for the MIE control mice). The mice were also generally anemic and thrombocytopenic (Table 2). The circulating leukemic cells were always of donor origin (Ly5.2), and their myeloid nature was ascertained by immunofluocytometry; 100% expressed Mac-1, with the majority positive for F4/80 (60–90%) and/or Gr-1 (40–80%). However, none were recognized by antibodies reacting with T or B lymphocytes such as Thy1, CD3, or CD19 (Fig. 5) or with the erythroid marker Ter119 (data not shown). Morphologically, these cells were characterized by a predominance of partially differentiated myeloid band forms and bilobed forms with immature chromatin (Fig. 6). In the majority of the mice, the BM was essentially effaced with leukemic cells with moderate to high nuclear to cytoplasmic ratios, open chromatin, and generally indented nuclei. At necropsy, the mice displayed pronounced splenomegaly (1.5- to 8-fold increase in spleen weight) and frequent enlargement of lymph nodes. Histological examination revealed that the leukemic infiltrate was present in the kidneys and liver where it invaded both the periportal regions and the parenchyma, and it largely replaced the normal architecture of the spleen, thymus, and lymph nodes.
Table 2.
Characteristics of the diseases in recipients of MLL-ELL-transduced BM cells
| Mouse | Latency, days | WBC, 103/μl | Percent monocyte‡ | RBC, 106/μl | PLT, 103/μl | Spleen weight, g | Ly5.2 WBC expressing Mac1, % |
|---|---|---|---|---|---|---|---|
| MSCVMLL-ELL# 3 | 126 | 73 | 44 | 4.4 | 162 | 0.37 | 88 |
| MSCVMLL-ELL# 2 | 133 | 4 | 2 | 8.9 | 137 | 0.32 | 80 |
| MSCVMLL-ELL# 5 | 150 | 17 | 55 | 9.4 | 225 | 0.35 | 76 |
| MSCVMLL-ELL# 7 | 157 | 12 | 20 | 2.6 | 34 | 0.35 | 88 |
| MSCVMLL-ELL# 8 | 167 | 33 | 48 | 6.2 | 142 | 0.65 | 86 |
| MSCVMLL-ELL# 9 | 167 | 80 | 43 | 6.7 | 514 | 0.46 | 93 |
| MSCVMLL-ELL# 1 | 182 | 36 | 30 | 1.35 | 25 | 0.39 | 94 |
| MSCVMLL-ELL# 4 | 187 | 3.9 | 23 | 9.6 | 50 | 0.13 | 39 |
| MSCVMLL-ELL# 10 | 189 | 185 | 60 | 1.7 | 129 | 0.52 | 94 |
| MSCVMLL-ELL# 6 | 194 | 210 | 52 | 4.6 | 242 | 0.5 | 78 |
| MSCVMLL-ELL mice* | 165 ± 23 | 65 ± 744 | 38 ± 18 | 5.5 ± 3 | 166 ± 143 | 0.40 ± 0.14 | 82 ± 16 |
| MIEMLL-ELL mice* | 160 ± 32 | 25 ± 25 | 39 ± 26 | 4.0 ± 2 | 342 ± 280 | 0.46 ± 0.16 | 77 ± 27 |
| MIE mice* | NA | 8 ± 2.6 | 4.8 ± 2.8 | 9.4 ± 0.5 | 945 ± 124 | 0.090 ± 0.01† | 16 ± 6 |
RBC, red blood cells; PLT, platelets; NA, not applicable; WBC, white blood cells.
Values shown are the mean (±SD) from the 10 MSCVMLL-ELL mice, nine MIEMLL-ELL mice at the time of death, or the 10 control MIE mice bled 140 days after reconstitution.
Spleen weight from age-matched mice.
These differential counts are an approximation as they were measured by the Cell-Dyn counter.
Figure 6.
Morphology of MLL-ELL leukemic cells. Wright-Giemsa stain of (A) PB smear showing marked leukocytosis consisting predominantly of mature and immature myeloid cells and (B) BM cytospin revealing preponderance of myeloblasts (mouse #1). (Bar = 133 μm.)
Transplantation of MLL-ELL Leukemias to Secondary Recipients and Establishment of Continuous Cultures.
To further establish the malignant nature of the MLL-ELL disease, BM cells from five moribund MSCVMLL-ELL primary recipient mice were transplanted into sublethally irradiated secondary recipients (4–5 recipients per primary donor). All of the secondary recipients developed leukemias with similar characteristics as those observed in the primary animals after an average latency of 43 days.
To further characterize the properties of the leukemic cells, BM from primary leukemic mice or secondary recipients were seeded in culture with or without growth factors (IL-3 and stem cell factor) in an attempt to establish cell lines and to determine their growth factor dependence. The establishment of continuous cultures in media supplemented with growth factors was successful in all cases except for one (mouse MSCVMLL-ELL #10). Growth factor-independent cell expansion was observed in three of 11 primary mice tested and three of five secondary mice. These variations in growth potential among the leukemic cells originating from different animals indicate that although the leukemias appear quite homogeneous in their clinical and phenotypic characteristics, they possess distinct biological potentials.
Clonality of the MLL-ELL Leukemias.
To determine the number of malignant clones present in the MLL-ELL tumors, we performed Southern blot analyses of genomic DNA from the spleens of the MSCVMLL-ELL leukemic mice. The DNA was digested with BamHI or NcoI, which cleave the integrated provirus between the LTRs and release fragments unique to the integration sites (Fig. 4). Based on the number of bands and their unequal autoradiographic intensities we estimated the number of clones in the infiltrating leukemic cells and found that the neoplasias were always monoclonal or pauciclonal (data not shown).
Discussion
Immortalizing Properties of MLL-ELL.
We showed that MLL-ELL increases the proliferative potential of primary myeloid progenitors in methylcellulose culture. This property requires the fusion of ELL with MLL because neither the expression of the truncated MLLdel mutant nor the overexpression of ELL had any proliferative effect on myeloid CFCs. In contrast, ELL has been reported to induce anchorage-independent growth of Rat1 fibroblasts and reduce their requirement for growth factors (17). Such discrepancy between results obtained from different transformation assays has been described for other leukemic genes and highlights the influence of the cellular context in which these studies are performed and the importance of using primary hematopoietic cells (18).
The proliferative effect of MLL-ELL on myeloid CFCs is, however, moderate compared with that of MLL-ENL. Indeed the numbers of third- and fourth-passage MLL-ELL colonies was significantly lower than that observed with MLL-ENL (9). In addition, the establishment of immortalized cell lines from pooled tertiary colonies transformed by MLL-ELL was rarely successful. This lower transforming activity in vitro seemed paradoxical in view of our previous transplantation studies with MLL-ENL where although the onset of disease was quicker (latency of 73–119 days) only approximately 50% of the mice developed leukemias (9). However, the technique used in this previous work differed in several aspects; in particular, the retroviral transduction was targeted to purified stem cells and a low number of these cells were inoculated per recipient mice. To compare more directly the leukemogenic activity of MLL-ENL with MLL-ELL, we have performed a transplantation study of these two fusion genes side by side by using the MIE vector and method described in this report (unpublished data). Because the level of EGFP expression was too low to accurately measure the percentage of cells transduced by MLL-ELL or MLL-ENL, we do not know whether a similar number of cells transduced with either construct was infused per mouse. However, we found that all of the mice (n = 10) reconstituted with MLL-ENL died of myeloid leukemias within 60–90 days, suggesting that MLL-ENL's malignant potential is indeed stronger than that of MLL-ELL and that the methylcellulose transformation assay is a valid model to compare genes involved in myeloid leukemias.
Possible Effect of MLL Fusion Genes on Hematopoietic Stem Cells.
Before the appearance of leukemic symptoms, the MLL-ELL-reconstituted mice displayed a higher degree of engraftment compared with mice reconstituted with the empty vector. This finding raises the possibility that independently from its leukemic effect, MLL-ELL increases the repopulating potential of the transplanted cells. This finding is concordant with studies of MLL-AF9 knock-in chimeric mice (8), which showed that this fusion gene induced the preferential expansion of embryonic stem cell-derived hematopoietic cells. Furthermore we have shown that hematopoietic stem cells transduced by MLL-ENL and expanded as clonal populations for more than 3 months in culture could maintain their ability to contribute to both lymphoid and myeloid lineages upon transplantation into congenic mice (C.L. and C.D., unpublished data). Taken together, these findings indicate that MLL fusion proteins favor the proliferation of stem cells with in vivo repopulating ability.
Is the Transforming Effect of MLL-ELL Limited to the Myeloid Lineage?
The in vitro transforming assay we have used in this study is designed to favor the growth and maturation of myeloid progenitors. Because of the lack of adequate growth factors, this in vitro assay would preclude the emergence of transformed lymphoid precursors. On the other hand, the in vivo reconstitution assay can read out cellular transformation affecting any hematopoietic lineage. The presence of EGFP in all of the cell types of the PB of the control mice reconstituted with the MIE vector demonstrates that retroviral expression, regulated by the MSCV promoter at the transcription level, and by the internal ribosomal entry site at the translation level, is not limited to the myeloid lineage in vivo. The fact that all 20 mice reconstituted with MLL-ELL developed acute myeloid leukemias indicates that the fusion protein is predominantly (or exclusively) oncogenic in myeloid committed cells. This also reflects the leukemia phenotype of humans with the t(11;19)(q23;p13.1) translocation (19).
Do Additional Mutations Participate in the Progression of MLL-ELL Leukemias?
This study demonstrates that MLL-ELL causes myeloid malignancies in mice. However, several features of the murine disease suggest that additional genetic alterations are probably necessary. First, the leukemias occur after a relatively long latency period (100–200 days), suggesting that they are the result of a selective process. Second, the finding that all of the tumors are either monoclonal or pauciclonal suggests that they have acquired additional rare mutations. Although this must be tempered by the fact that reconstitution experiments with retrovirally tagged stem cells have shown that only one or a few marked clones participate to the hematopoietic system of the host (our observation and ref. 20). In addition, the low titers obtained with the MLL-ELL-encoding viruses and the cellular toxicity of the fusion protein further limit the number of engrafting cells that have been transduced. Third, although it was difficult to establish immortalized cell lines from pooled methylcellulose colonies transformed by MLL-ELL in vitro, malignant clones of MLL-ELL leukemic mice showed a high propensity to form continuous cell lines upon transfer into liquid culture, suggesting that additional mutations occurred in vivo to contribute to this phenotype. Fourth, the leukemic mice displayed some heterogeneity with regard to the disease latency and the white blood cell counts (Table 2) and the growth factor-dependence of the leukemic cells in culture was variable as well. In the case of the MIEMLL-ELL leukemias, we could not see any correlation between these parameters and the intensity of the EGFP marker (data not shown), making it unlikely that varying levels of MLL-ELL expression caused by differences in site of viral integration could account for this heterogeneity.
Our studies extend the available mouse models of 11q23 leukemia to include partner genes outside of the AF9/ENL gene family. We have found that expression of the chimeric MLL-ELL fusion gene in murine hematopoietic progenitor cells followed by transplantation into congenic mice produces a myeloid leukemia that closely recapitulates the leukemias observed in AML cases that exhibit the t(11;19)(q23;p13.1). Although secondary genetic events are likely necessary for the development of acute leukemia in these mice, expression of the MLL-ELL fusion protein appears to be sufficient as an initiating event to transform primary hematopoietic cells. As ELL has well-characterized functional domains, this model provides the basis for future structure-function studies of ELL.
Acknowledgments
We are very grateful to Scott Kogan for histological analyses and Ivan Plavec for critically reading this manuscript. This work was supported by grants to M.J.T. from the National Institutes of Health (CA78431), the Burroughs Wellcome Fund, the Leukemia Research Foundation, the American Society of Hematology, and the family of Robert A. Chapski.
Abbreviations
- BM
bone marrow
- CFC
colony-forming cell
- Lin−
lineage-depleted
- PB
peripheral blood
- EGFP
enhanced green fluorescent protein
- LTR
long terminal repeat
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190167297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190167297
References
- 1.Thirman M J, Gill H J, Burnett R C, Mbangkollo D, McCabe N R, Kobayashi H, Ziemin-van der Poel S, Kaneko Y, Morgan R, Sandberg A A, et al. N Engl J Med. 1993;329:909–914. doi: 10.1056/NEJM199309233291302. [DOI] [PubMed] [Google Scholar]
- 2.Thirman M J, Larson R A. Hematol Oncol Clin North Am. 1996;10:293–320. doi: 10.1016/s0889-8588(05)70340-3. [DOI] [PubMed] [Google Scholar]
- 3.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C M, Canaani E. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 4.Tkachuk D C, Kohler S, Cleary M L. Cell. 1992;71:691–700. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 5.Ziemin van-der Poel S, McCabe N R, Gill H J, Espinosa R I, Patel Y, Harden A, Rubinelli P, Smith S D, LeBeau M M, Rowley J D, Diaz M O. Proc Natl Acad Sci USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thirman M J, Levitan D A, Kobayashi H, Simon M C, Rowley J D. Proc Natl Acad Sci USA. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shilatifard A, Lane W S, Jackson K W, Conaway R C, Conaway J W. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 8.Corral J, Lavenir I, Impey H, Warren A J, Forster A, Larson T A, Bell S, McKenzie A N J, King G, Rabbitts T H. Cell. 1996;85:853–861. doi: 10.1016/s0092-8674(00)81269-6. [DOI] [PubMed] [Google Scholar]
- 9.Lavau C, Szilvassy S, Slany R, Cleary M L. EMBO J. 1997;16:4226–4237. doi: 10.1093/emboj/16.14.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawley R G, Lieu F H L, Fong A Z C, Hawley T S. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 11.Du C, Redner R L, Cooke M P, Lavau C. Blood. 1999;94:793–802. [PubMed] [Google Scholar]
- 12.Pear W, Nolan G, Scott M, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slany R K, Lavau C, Cleary M L. Mol Cell Biol. 1998;18:122–129. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thirman M J, Diskin E B, Bin S S, Ip H S, Miller J M, Simon M C. Proc Natl Acad Sci USA. 1997;94:1408–1413. doi: 10.1073/pnas.94.4.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joh T, Kagami Y, Yamamoto K, Segawa T, Takizawa J, Takahashi T, Ueda R, Seto M. Oncogene. 1996;13:1945–1953. [PubMed] [Google Scholar]
- 17.Kanda Y, Mitani K, Kurokawa M, Yamagata T, Yasaki Y, Hirai H. J Biol Chem. 1998;273:5248–5252. doi: 10.1074/jbc.273.9.5248. [DOI] [PubMed] [Google Scholar]
- 18.Ghaffari S, Daley G Q, Lodish H F. Leukemia. 1999;13:1200–1206. doi: 10.1038/sj.leu.2401467. [DOI] [PubMed] [Google Scholar]
- 19.Huret J L, Brizard A, Slater R, Charrin C, Bertheas M F, Guilhot F, Hahlen K, Kroes W, Leeuwen E V, Schoot E V D, et al. Leukemia. 1993;7:152–160. [PubMed] [Google Scholar]
- 20.Szilvassy S J, Cory S. Blood. 1994;84:74–83. [PubMed] [Google Scholar]