Abstract
Trans-sialidase is an essential enzyme for Trypanosoma cruzi, the causative agent of Chagas’ disease, to escape from the host immune system and to invade the host cells. Therefore, Trypanosoma cruzi trans-sialidase (TcTS) presents a potential and appealing therapeutic target for this lethal disease. Availability of a structurally very similar enzyme with strict hydrolase activity (Trypanosoma rangeli sialidase, TrSA) provides us a unique opportunity to understand the determinants of their structure and catalytic mechanism. In this study, we compare the catalytic cleft plasticity of free (apo) and ligand-bound (holo) forms of the two enzymes using molecular dynamics simulations. We focus on the mouth of the catalytic cleft that is defined by two residues—W312 and Y119 in TcTS, and W312 and S119 in TrSA. Our results indicate that TcTS has a very flexible, widely open catalytic cleft, mostly due to W312 loop motion, in apo form. However, when the catalytic cleft is occupied by a ligand, the flexibility and solvent exposure of TcTS is significantly reduced. On the other hand, TrSA maintains a more open catalytic cleft compared to its crystal structures in both apo and holo forms (and compared to TcTS in holo forms). The reduced solvent exposure of TcTS catalytic cleft, might be partially or fully responsible for TcTS to be a less efficient hydrolase than TrSA.
Keywords: protein flexibility, molecular dynamics, Chagas’ disease, sialidases
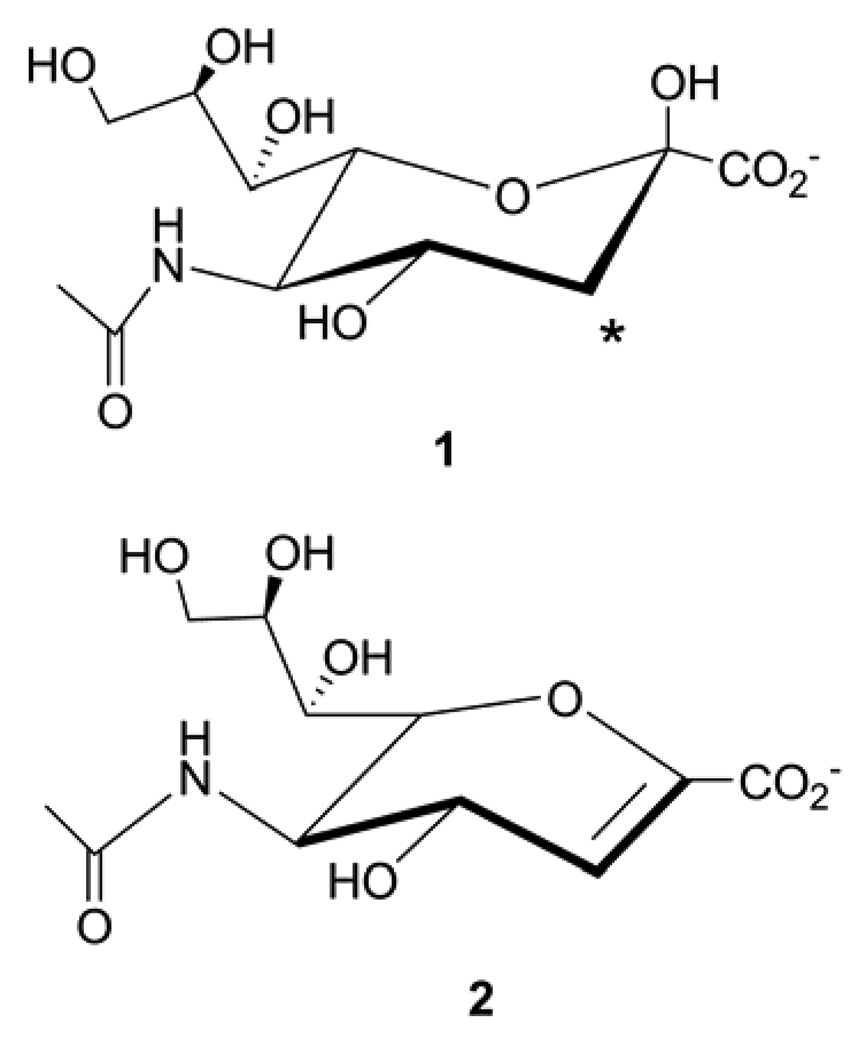
Chagas’ disease is a neglected tropical disease devastating millions of people in South and Central America (1). No effective cure is available for the infected people and the only two drugs identified in early 1970s have severe side effects which make their use for the chronic phase highly controversial (2). Trypanosoma cruzi, a flagellatae protozoan, is identified as the causative agent of this lethal disease (3–5). This parasite is unable to synthesize sialic acids (1, Fig. 1), which are vital for its virulence in human body (6, 7), but expresses an enzyme called trans-sialidase attached to its outer membrane to provide sialic acids. Therefore, the parasite strongly relies on its trans-sialidase enzyme to crop sialic acids from the host body. The lack of trans-sialidase in humans makes TcTS a potential and appealing therapeutic target.
Figure 1.
Chemical structures of sialic acid (1) and 2, 3-dehydro-3-deoxy-Nacetylneuraminic acid (2, abbreviated as DANA). The star shows the location of F atom insertion mentioned in the text.
Sialic acids, which are O- and N-substituted derivatives of a nine-carbon monosaccharide called neuraminic acid, lie at the terminal positions of cell surface glycoproteins and glycolipids, and are used for recognition purposes by the immune system (8, 9). Sialic acid is a general name for the whole family of neuraminic acid derivatives as well as being specificly used for N-acetyl neuraminic acid. We will use the latter throughout our article. Sialidases (a.k.a. neuraminidases) catalyze the hydrolysis of sialic acids (10). Some sialidases—called trans-sialidases—can additionally (and even more efficiently than hydrolysis) transfer sialic acids from donor sialo-glycoconjugates to acceptor glycoconjugates. In the family of sialidases, Trypanosoma cruzi trans-sialidase (TcTS) and Trypanosoma rangeli sialidase (TrSA) have very high structural similarity although they differ in their catalytic function (11–14). TrSA is a strict hydrolase while TcTS catalyzes sialyl-transfer in the presence of acceptor molecules and hydrolysis otherwise (15–19). Their sequence and structural similarity is so high that it was even possible to confer trans-sialidase activity to TrSA with only 5 point mutations (20). Thus, the two enzymes pose a unique opportunity to understand the determinants of their structure and catalytic mechanism.
TcTS is anchored to the cell surface by glycosylphosphatidilinositol (GPI) and consists of a 70 kDa globular core that accommodates the catalytic site and a variable number of highly immunogenic repeats called shed acute phase antigen (SAPA) (21). TrSA lacks SAPA tail but has unusual similarity to the 631-residue-long globular core of TcTS (14). The two enzymes have a 70% sequence identity with only one amino acid insertion in TrSA close to the N-terminus, and the RMSD value is only 0.59 Å when the corresponding Cα atoms of the enzymes are superimposed. Both enzymes consist of two globular domains that are attached by a long α-helical segment (14). Despite the high structural similarity overall and at the catalytic cleft, TcTS differs from TrSA in their inhibitor binding properties as well. 2,3-dehydro-3-deoxy-N-acetylneuraminic acid (DANA) (2, Fig. 1), which is a structural analog of sialic acid oxocarbenium ion and an efficient inhibitor for TrSA and sialidases in general, does not inhibit TcTS (Ki: 12.29 mM for TcTS, 0.0015 mM for TrSA) (20, 22). The same trend is also seen for inhibitors like zanamivir and oseltamivir (23). This fact points to either structural or dynamical differences between TcTS and TrSA that are not revealed by crystal structures.
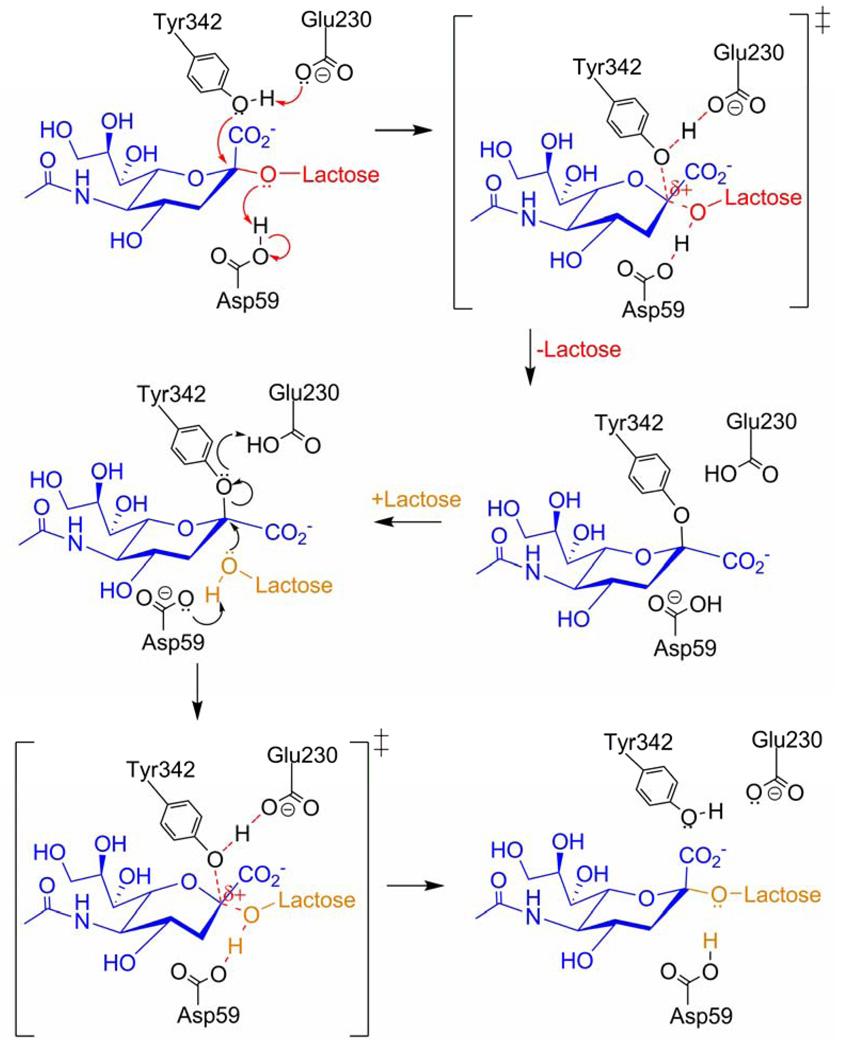
Experimental evidence suggests that a relatively long-lived covalent intermediate forms in the sialyl-transfer mechanism of TcTS (Fig. 2) (24–27). In this scenario, sialic acid is scavenged from the host’s glycoconjugates (donor) and remains covalently bound to the enzyme until the parasite’s glycoconjugate (acceptor) binds at the active site. TcTS sialyl-transfer and TrSA hydrolysis share the same mechanism up to the covalent intermediate formation step (Fig. 2) (14, 28, 29). Since TcTS can also catalyze hydrolysis (in the absence of an acceptor ligand) just like TrSA, it is not clear how the TcTS covalent intermediate is protected from hydrolysis until the acceptor ligand arrives. Differential solvent exposure of TcTS and TrSA catalytic clefts could be a possible explanation for the enzymes’ catalytic difference, but no sign of this is seen in the crystal structures except that TcTS has two more hydrophobic residues at the active site than TrSA as stated in Buschiazzo et al. (30).
Figure 2.
The proposed mechanism for trans-sialidase catalysis reaction of TcTS. The donor and acceptor sugar moieties are colored in red and orange, respectively. In sialidase catalysis reaction of TcTS, a water molecule takes the role of the acceptor lactose.
In this study, we focus on the behavior of the catalytic clefts of TcTS and TrSA—alone and in complex with different ligands. Structural details of sialyl-lactose and DANA binding to TcTS and TrSA will be presented in a future publication. In our analysis, we find that the catalytic cleft of TcTS has high plasticity—especially in the apo form—and visits more open forms as well as the closed form found in the crystal structures. The mobility of the W312 loop mainly provides this opening/closure motion of TcTS catalytic cleft. On the other hand, MD simulations of TrSA—in all the apo and holo forms (except in the case of sialyl-lactose-bound TrSA, as will be described in the Results and Discussion section)—show that TrSA catalytic cleft always opens and maintains a more open form than the crystal structures. Ligand-bound forms of TrSA maintain a more open catalytic cleft also with respect to corresponding TcTS forms in the simulations. The reduced solvent exposure of the catalytic cleft of TcTS compared to that of TrSA can be partially or entirely responsible for the differential catalytic function; sialyl-transfer or hydrolysis.
This study also demonstrates that some structural information—like the high flexibility of W312 loop or the differential solvent exposure of TcTS and TrSA catalytic clefts—can be veiled by the crystal structures especially at the flexible loop regions of proteins. Construction of the unit cell of TrSA reveals close crystal contacts exactly at the catalytic cleft which are very likely to bias the crystal structure towards the closed form. Islam and Weaver (31) claims that approximately 25–30% of a typical protein surface is in contact with the molecules in their crystallographic environment and thus, is affected by crystal packing forces. Based on our data, TcTS and TrSA are no exceptions to this and their catalytic clefts should be handled cautiously for docking and inhibitor design studies by taking into account the crystallographic biases and flexibilities.
Methods
The initial structures of different forms of TcTS and TrSA for MD simulations are obtained from the Protein Databank and all these 7 structures correspond to different points in the catalytic process (Table 1). Only the catalytic domain—the first 371 residues for TcTS, the first 375 residues for free and DANA-bound TrSA, the first 373 residues for TrSA covalent intermediate—and the ligand, if any, are kept and the rest of the proteins and any solvent molecules are excluded while preparing the systems for simulation. In addition to the above 7 structures available in the Protein Databank, it would be interesting to simulate sialyl-lactose-bound TrSA. Since there was no such crystal structure available, we modelled sialyl-lactose-bound TrSA in silico by superimposing crystal structures of DANA-bound TrSA (1N1T.pdb) and the sialyllactose-bound TcTS (1S0I.pdb) in VMD program (32) and imitating the sialyl-lactose pose seen in TcTS.
Table 1.
The crystal structures used in MD simulations.
| Protein Data Bank Code | Description |
|---|---|
| 1MS3 | Empty TcTS |
| 2AH2 | TcTS covalent intermediate |
| 1MS1 | DANA-bound TcTS |
| 1S0I | Sialyl-lactose-bound TcTS |
| 1N1S | Empty TrSA |
| 2A75 | TrSA covalent intermediate |
| 1N1T | DANA-bound TrSA |
While preparing the input files for MD simulations several changes are performed on the original crystal structure files to imitate the natural catalysis event better. In order to obtain a crystal structure of sialyl-lactose-bound TcTS, the base catalyst D59 was mutated to an alanine protecting the ligand from hydrolysis (30). Thus, we mutated A59 in the crystal structure back to an aspartic acid for our simulations. If the crystal structure was obtained as a homodimer, we only preserved chain A and deleted chain B. Withers group pioneered a method of using modified (fluorinated) substrates to trap the covalent intermediates of β-glucosidases (33). This method, which is successfully used to trap covalent intermediates of TcTS and TrSA (24, 29), caused the relevant crystal structures to contain a F atom bound to the C atom adjacent to the anomeric C atom (Fig. 1). We altered this F atom in the crystal structures to a H atom.
The resulting file is then fed into the Leap module of Amber9 (34) molecular dynamics package where the necessary H atoms and any other missing atoms are added as well as being solvated with ≈7,600 TIP3P (35) explicit water molecules and in truncated octahedral box.
Since D59 is responsible for acid/base catalysis, its protonation state is changed to the aspartic acid form instead of the aspartate form which is found normally under physiological conditions. The protonation states of D59, E230 and Y342 change over the course of the reaction and are adjusted in the covalent intermediates accordingly: D59 is in the aspartate form while E230 is in the glutamic acid form instead of the glutamate form due to the H transfer from Y342 to E230 in the reaction. In all other systems—free enzymes and Michaelis complexes—D59 and Y342 are protonated while E230 is unprotonated.
Amber.FF99SB (36, 37) and Glycam04 (38–40) force fields are used to construct the topology files. Since the inhibitor DANA is not a standard residue in Glycam04, the parameters for this molecule had to be constructed. First, the geometry for DANA is obtained modifying the standard terminal sialic acid geometry from Glycam04 using Hyperchem (41) program and minimizing it to get planarity around its anomeric C atom. HF/6-31G* in Gaussian03 (42) package is used to produce RESP charges for DANA and these charges are then fed into the Antechamber module of Amber9 package to produce the parameter and topology files using GAFF (43) force field.
The PMEMD module of Amber9 package is used first to relax the systems under periodic boundary conditions with a procedure explained in detail in the Supporting Information.
Following the relaxation, 50 ns (130 ns for unligated TcTS) of production run is performed following the same procedure as in the last step of the relaxation. Replicate molecular dynamics simulations of approximately 80 ns are also performed after assigning different initial velocities to each atom of the relaxed systems by using a different seed for the random number generator. The stability of enzymatic systems throughout MD simulations is demonstrated by root mean square deviation plots of Fig. S1.
Results and Discussion
Catalytic Clefts of TcTS and TrSA
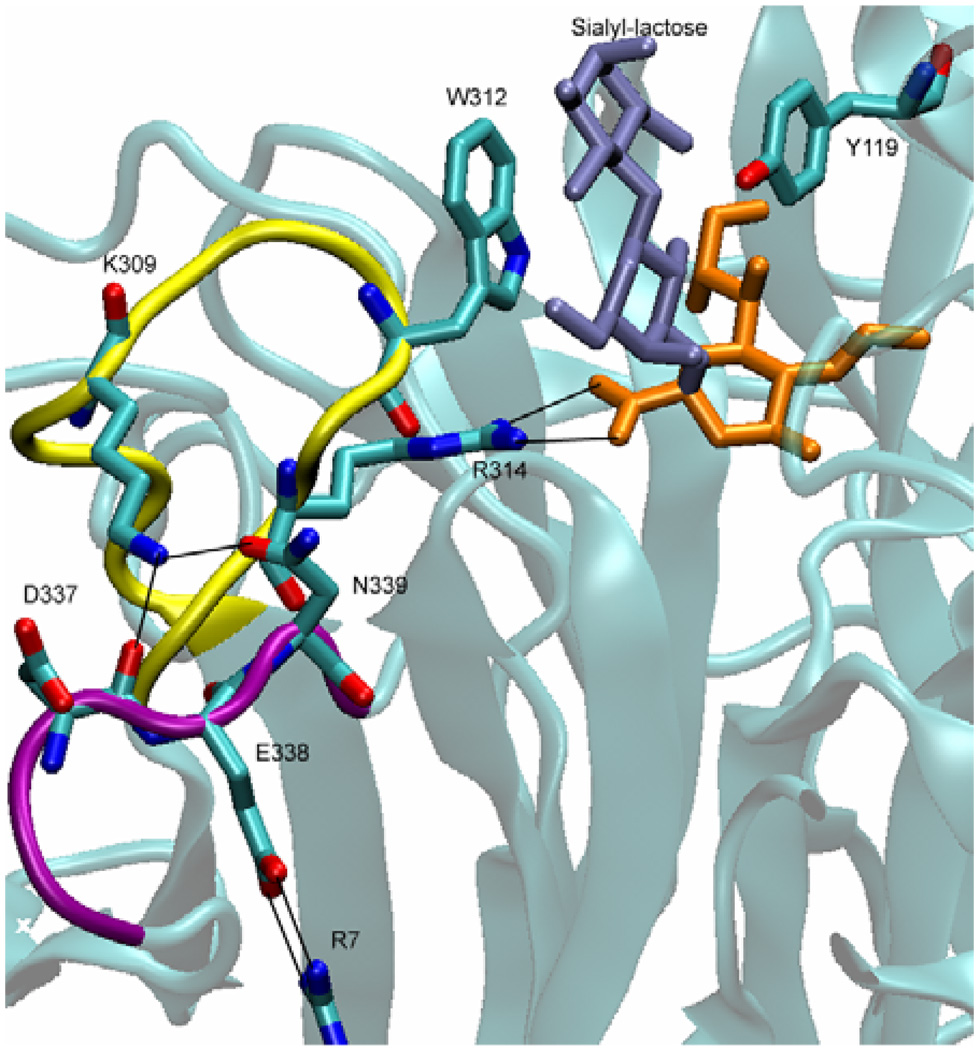
The catalytic cleft of TcTS consists of two regions: The region that embraces the lactose (which is used in crystal structures in place of long glycoconjugates) part of the ligand, and the region that embraces the sialic acid part of the ligand. The latter region is buried deep in the enzyme while the lactose-binding region is defined by two residues, namely W312 and Y119, which lie on flexible loops at the periphery of the enzyme (Fig. 3). The residue numbering in TcTS will be referred for equivalent positions of both enzymes throughout this work. As observed in the crystal structure of sialyl-lactose-bound TcTS, the lactose part of the ligand is tightly sandwiched between the parallel-lying aromatic side chains of W312 and Y119. TrSA’s catalytic cleft is very similar to that of TcTS; when crystal structures are superimposed, their backbones exactly overlap and the residues and their conformations are identical with only a few exceptions. The most relevant ones for this study are that the conformation of W312—which can be described with a particular pair of χ1 and χ2 dihedral angles—differs in the crystal structures of TcTS and TrSA and that TrSA has a serine residue instead of Y119 in TcTS.
Figure 3.
The interactions that clamp W312 loop (in yellow) to R7 shown for TcTS bound to sialyl-lactose (sialic acid is colored orange and lactose is colored ice blue). K309 and E338 connect the two flexible loops (in yellow and purple) to R7 that lies on a β-sheet structure. R314 serves to fix W312 loop by interacting with the carboxylate group of sialic acid in the presence of a ligand.
Flexibility of W312 Loop
Molecular dynamics simulations show that especially when TcTS is in apo form, the loop of Y119 remains mostly intact while the loop of W312 is very mobile (indicated by the RMSF plots included in the Supporting Information as Figure S2). This loop extending from residue 306 to residue 316 in the lactose-binding region rearranges to accommodate the catalytic cleft opening and closure which significantly changes the solvent-exposure (Fig. 4). When the catalytic cleft opens, W312 loop is highly mobile and W312 is always further away from Y119 compared to the crystal structure.
Figure 4.
Closed and open forms of W312 loop. The initial closed form of the loop (red) rearranges and creates a more open catalytic cleft (blue) in apo form of TcTS. W312 visits all possible different conformations once W312 loop leaves the closed form.
A set of strong ionic bridges involving K309 side chain and residues D337, E338, N339, R7 keep the W312 loop fixed except for the middle part of the loop extending from residue 310 to residue 314 (Fig. 3). This mobility of residues 310-314 is important since W312 which resides on this flexible region is required to be in a certain position and conformation—corresponding to the closed form of the catalytic cleft seen in the crystal structures—in order to provide a binding site for the lactose part of the ligand (our unpublished results). The lactose binding site of the catalytic cleft is especially important for the acceptor sugar which depends solely on the interactions with the lactose binding site since it lacks the interactions of a terminal sialic acid that are effective in sialyl-lactose binding.
Monitoring the Extent of W312 Loop Flexibility in Apo and Holo Forms of TcTS
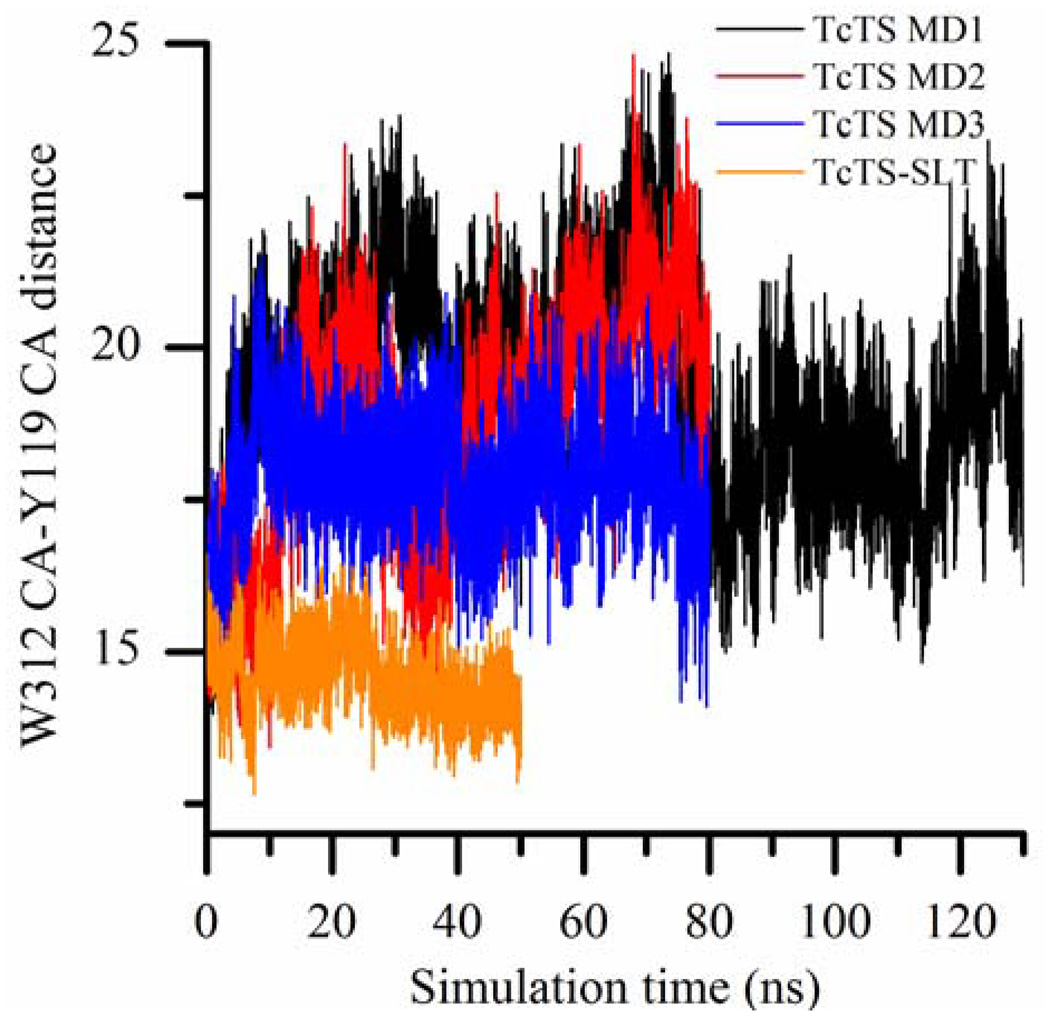
The extent of the catalytic cleft opening can be measured by monitoring the distance between the Cα atoms of W312 and Y119. To provide a reference, the W312-Y119 distances of TcTS crystal structures 1MS3, 1MS1 and 1S0I are 14.7 Å, 14.8 Å and 14.7 Å, respectively, and the catalytic clefts with similar distances will be referred to be in the “closed” form from now on. In the sialyl-lactose-bound TcTS MD simulation, a distance of 14.6 Å (± 0.6 Å) is preserved between these two Cα atoms—corresponding to a closed catalytic cleft—throughout 50 ns (Fig. 5).
Figure 5.
W312 Cα-Y119 Cα distance (in Å) in the molecular dynamics simulations of apo and sialyl-lactose bound forms of TcTS.
However, in all 3 independent MD simulations of apo form of TcTS, W312 loop which is initially in the closed form shuttles between the closed form and open forms whose W312-Y119 distance can become as large as 25 Å (Fig. 5). Due to the lack of interaction with a ligand, R314 side chain is highly mobile and allows extra mobility for the W312 loop in the MD simulations of apo form of TcTS. R314 side chain is so mobile in these simulations that it even interacts with the carboxyl group of Q282—causing W312 loop to move further away from Y119.
When a ligand binds to the active site of TcTS, R314 guanidinium group forms salt bridges with the carboxylate group of the ligand’s sialic acid and this strong interaction decreases the mobility of W312 loop by fixing the position of R314. This is seen in MD simulations of DANA-bound TcTS and TcTS covalent intermediate for which the W312-Y119 distance is mostly ~16 Å and can hardly become 20 Å at most (Fig. 6 and 7). The catalytic cleft is in a semi-open form in these simulations.
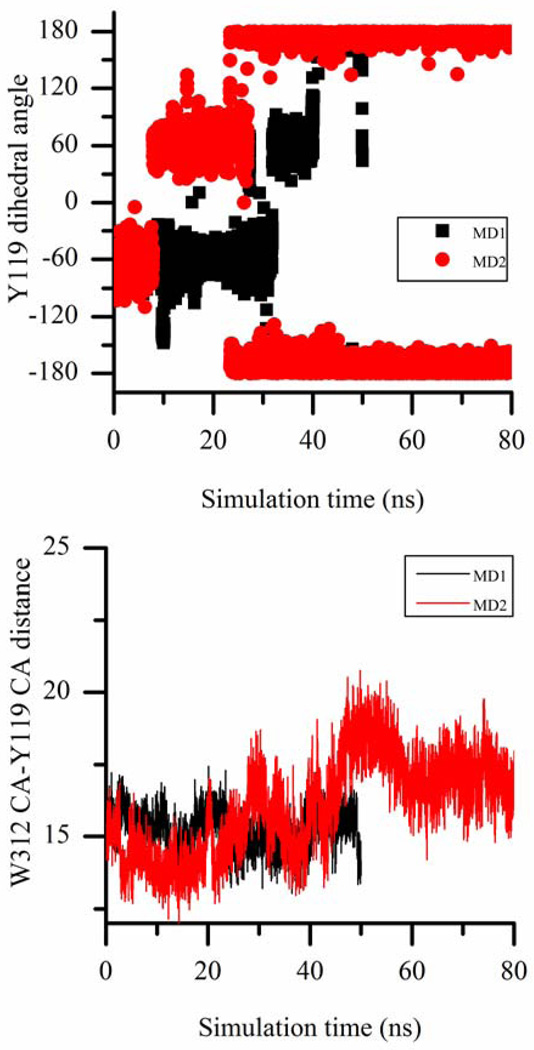
Figure 6.
The correlation of Y119 χ1 dihedral angle change (in the top panel) with the opening of W312 loop (in the bottom panel) monitored by the distance between Cα atoms of W312 and Y119 in two separate MD simulations of DANA-bound TcTS. W312 loop opening does not occur unless Y119 side chain swings into the catalytic cleft (for which χ1=180°). The dihedral angles are in degrees and distances are in Angstroms.
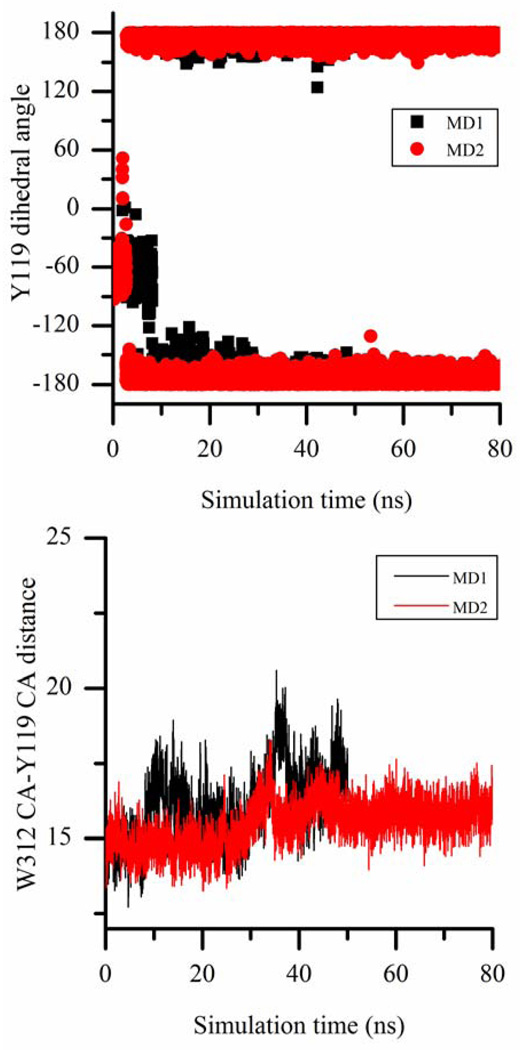
Figure 7.
The correlation of Y119 χ1 dihedral angle change (in the top panel) with the opening of W312 loop (in the bottom panel) monitored by the distance between Cα atoms of W312 and Y119 in two separate MD simulations of TcTS covalent intermediate. The dihedral angles are in degrees and distances are in Angstroms.
So, in our simulations, the catalytic cleft of TcTS very mobile and widely open in the apo form, but the mobility and the extent of cleft opening are gradually reduced as we go from the apo form of TcTS to partially-occupied catalytic clefts (DANA-bound TcTS and TcTS covalent intermediate) and lastly to fully-occupied catalytic cleft (sialyllactose-bound TcTS) (Table 2).
Table 2.
The extent of catalytic cleft opening and mobility in TcTS and TrSA MD simulations.
| Description | Catalytic cleft | Mobility |
|---|---|---|
| Empty TcTS | Open | +++ |
| TcTS covalent intermediate | Semi-open | + |
| DANA-bound TcTS | Semi-open | + |
| Sialyl-lactose-bound TcTS | Closed | − |
| Empty TrSA | Open | + |
| TrSA covalent intermediate | Open | + |
| DANA-bound TrSA | Open | + |
| Sialyl-lactose-bound TrSA | Closed | − |
Decrease in W312 Loop Flexibility after Sialic Acid Binding Enhances Lactose Affinity
Our finding about differential mobility of the W312 loop can help us explain a puzzling experimental result. Our data show that occupation of the sialic acid binding region of the catalytic cleft by a ligand decreases the mobility of W312 loop since the carboxylate group of the ligand strongly interacts with R314 side chain fixing its position. This decrease in W312 loop mobility can account for the surface plasmon resonance experiments by Buschiazzo and coworkers which show that TcTS can bind lactose only after it is preequilibrated with sialic acid or sialyl-lactose (30). These experimental results were hard to explain since lactose is a natural acceptor in TcTS catalysis reaction and must bind at some point in the catalytic cycle. However, based on our findings, W312 loop is very mobile when the sialic acid binding site is empty and thus, no stable lactose binding site is available. And once the sialic acid binding site is occupied by sialic acid, R314 interaction with sialic acid (or its analog) decreases the mobility of W312 loop and approaches the loop to a conformation more appropriate for binding. One could argue that presence of sialic acid and lactose together in the catalytic active site should cause a steric clash because 3-OH group of lactose and the hydroxyl group bound to the sialic acid’s anomeric C want to occupy the same space. However, several different experimental studies (including the one that crystallized TcTS which was preequilibrated by sialic acid and soaked with lactose) showed that TcTS can produce DANA as a by-product when it reacts with sialic acid or sialyl-lactose (30, 44). DANA differs from sialic acid only by lacking the hydroxyl group—which causes the mentioned steric clash—at the anomeric C and having a double bond there instead (Fig. 1). Thus, DANA should be the species that is produced and stored in the catalytic site after preequilibration of TcTS with sialic acid. Preequilibration with sialyl-lactose could also produce either the covalent intermediate or DANA. In summary, preequilibration produces a sialic acid analog that provides the carboxylate group to bind R314 reducing W312 loop mobility and making TcTS-lactose-binding easier. Buschiazzo et al. (30) suggested Y119 conformational motion to be a link between the sialic acid binding and the lactose binding affinity but noted that “further structural modifications of the protein may contribute to modulate lactose affinity”. We present here that the change in W312 loop mobility due to sialic acid binding can modulate the lactose binding affinity.
Correlation of W312 Loop Motion with Y119 Conformation in TcTS
Y119 side chain has 3 stable conformations that differ in χ1 dihedral angle; two of these are solvent-exposed and project the phenol side chain to the outer milieu of the enzyme (“outward” conformations which can be specified with χ1=60° or χ1=−60°, Fig. 8) while the third one projects the phenol side chain into the catalytic cleft (the “inward” conformation which can be specified with χ1=180°, Fig. 8). In the crystal structures of apo form of TcTS, DANA-bound TcTS and TcTS covalent intermediate, double conformations of Y119 (one inward, one outward conformation) are seen.
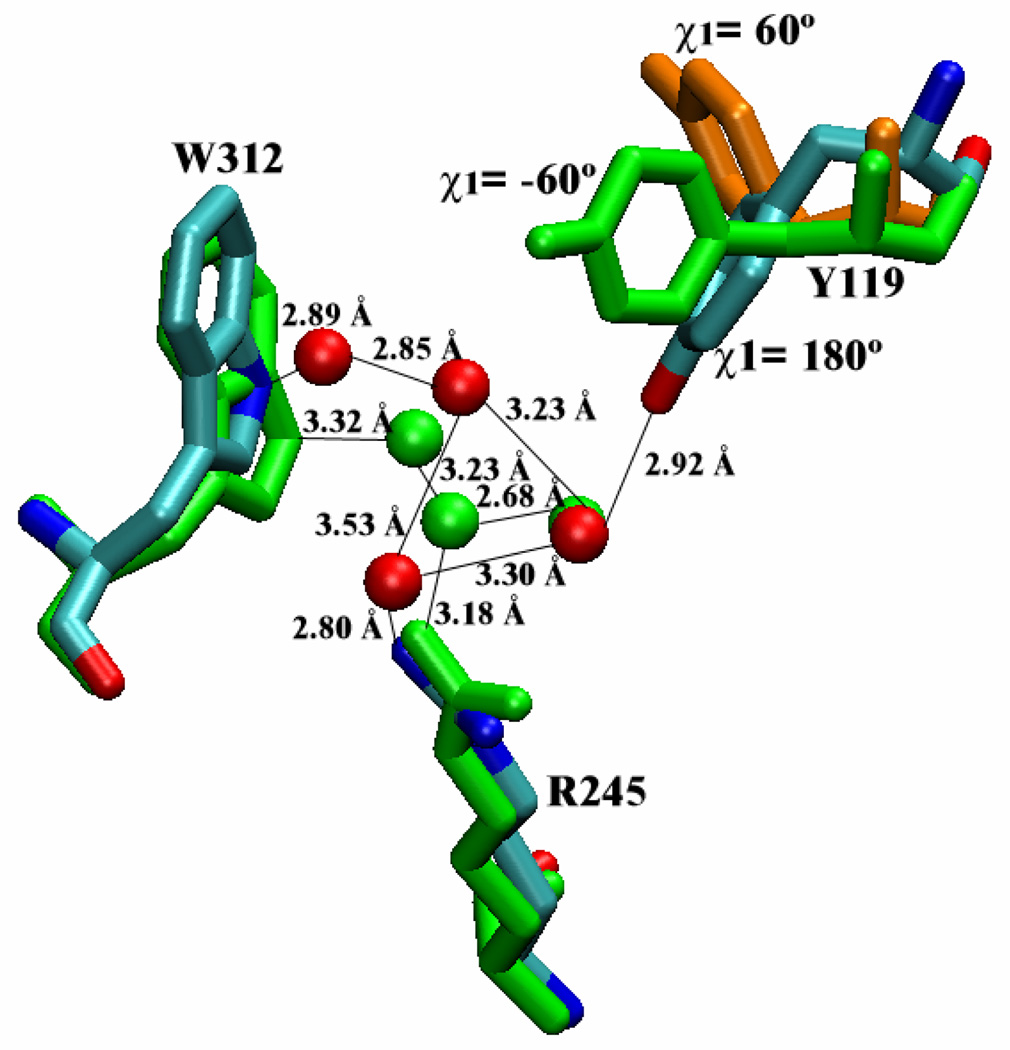
Figure 8.
Water network around R245 that connects Y119 conformational motion to W312 loop motion. The water network at the beginning of simulation (shown in green) is distorted (colored according to atom type) when Y119 moves from χ1=−60° to χ1=180°. The third possible Y119 conformation is also shown (in orange).
Interestingly, we observe that Y119 conformational change triggers W312 loop motion in TcTS simulations. Unless the lactose binding site is occupied, the inward conformation of Y119 is found to be the dominant one among the three. In the 2 MD simulations of DANA-bound TcTS and the 2 MD simulations of TcTS covalent intermediate, Y119 is initially in one of the outward conformations and the catalytic cleft maintains its closed form until Y119 moves to inward conformation, and only after that, W312 loop begins to rearrange forming a more open catalytic cleft (Fig. 6 and 7). In the MD simulation of sialyl-lactose-bound TcTS, Y119 is always in one of the outward conformations and the catalytic cleft is always in the closed form. Additionally, in all three MD simulations of apo form of TcTS, Y119 is initially in the inward conformation (which is maintained for the entire simulation) and thus, W312 loop starts opening the catalytic cleft at the beginning of the production run as shown in Fig. 5. Based on all these data, after the sialic acid binding region is occupied (by either sialic acid or DANA) fixing R314 position, the closed form of the catalytic cleft is stable as long as Y119 is in one of the outward conformations. And once Y119 side chain swings into the catalytic cleft, it disrupts this stability and causes free motion of W312 loop and catalytic cleft opening. But this opening is to a smaller extent compared to that of apo form of TcTS and forms a semi-open catalytic cleft.
The best candidate to link Y119 conformation and W312 loop motion is found to be the water network around R245 guanidinium group (Fig. 8). In the closed form of the catalytic site, the side chain N atom of W312 interacts with the water site right above R245. When Y119 side chain swings from one of the outward conformations into the inward conformation, its hydroxyl group replaces one water molecule and the motion of Y119 side chain into the catalytic cleft distorts the water network and weakens the water-mediated hydrogen bonding between W312 and R245 side chains, consequently triggering W312 loop motion from the closed form to more open forms.
W312 Loop Flexibility in Apo and Holo Forms of TrSA
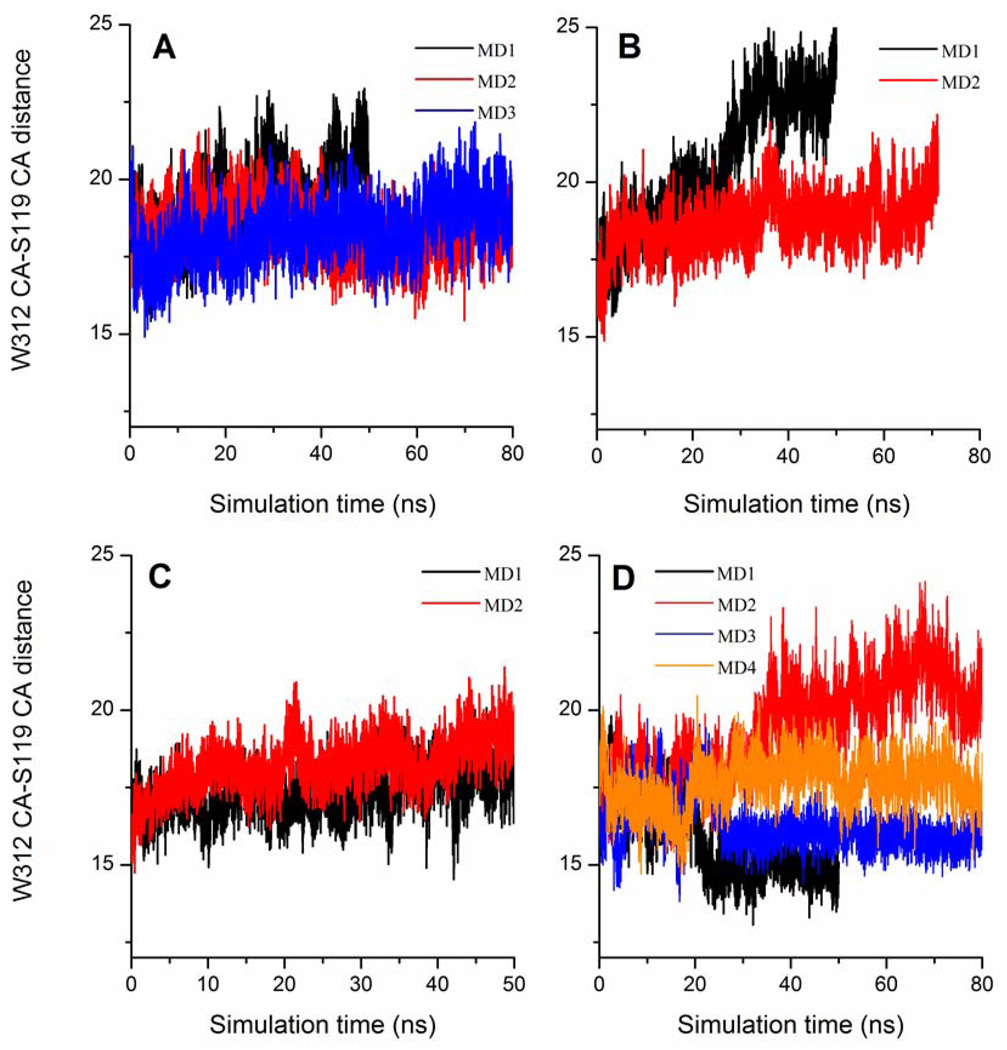
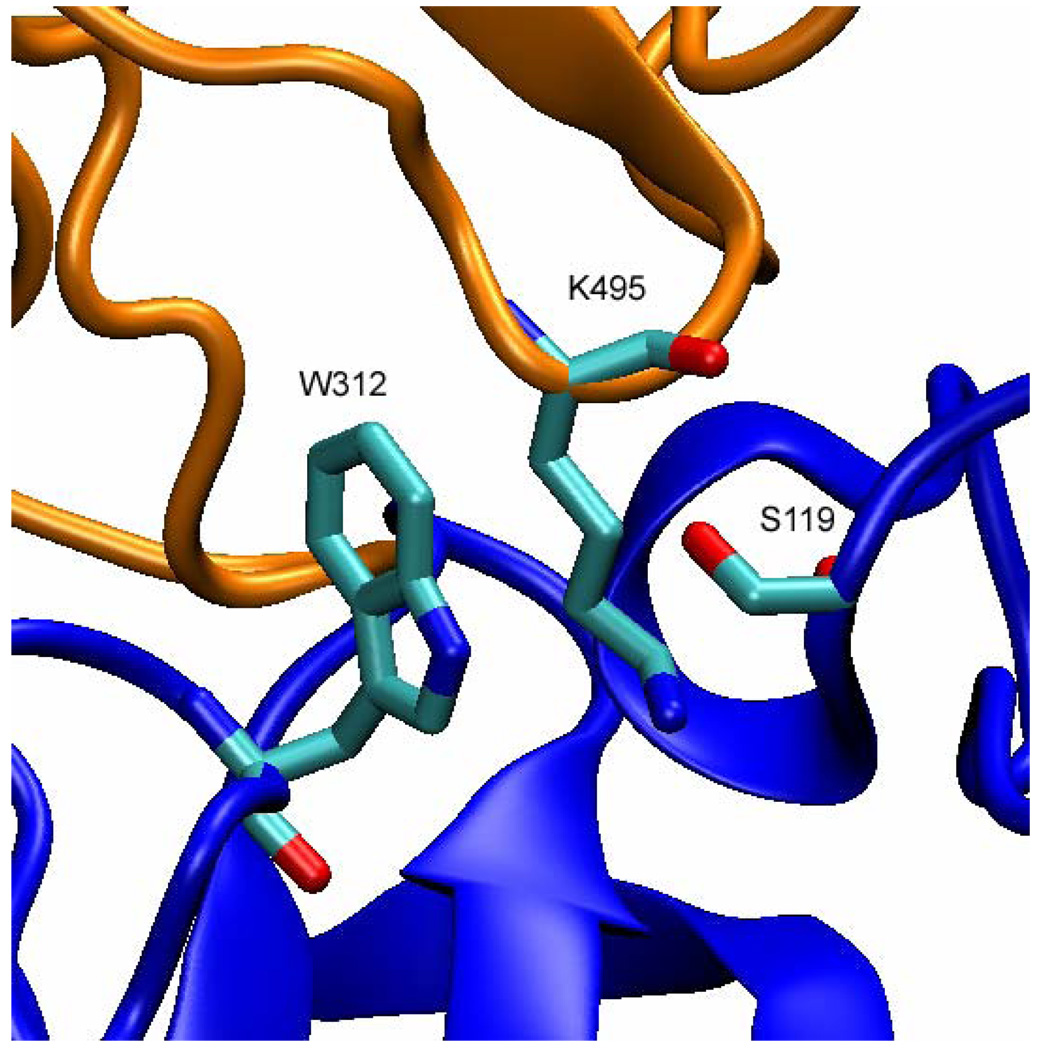
Similar analysis of W312 loop motion in TrSA MD simulations shows that the catalytic cleft is never in the closed form unless the catalytic cleft is fully occupied by the ligand (e.g. sialyl-lactose) (Fig. 9). For example, W312-S119 distances in 50-ns MD simulations are 19.4 Å (± 1.2 Å) for apo form of TrSA (Fig. 9, panel A), 20.6 Å (± 2.1 Å) for TrSA covalent intermediate (Fig. 9, panel B), 17.3 Å (± 0.7 Å) for DANA-bound TrSA (Fig. 9, panel C) and 15.7 Å (± 1.2 Å) for sialyl-lactose-bound TrSA (Fig. 9, panel D). However, in the crystal structures, the catalytic cleft of TrSA is always found in the closed form just like in the case of TcTS. The W312-S119 distances of TrSA crystal structures 1N1S, 1N1T and 2A75 are 15.8 Å, 15.6 Å and 15.6 Å, respectively. The open and closed forms of the TrSA catalytic cleft are depicted in Fig. 10. Construction of the crystallographic environment of the enzyme in the unit cell using appropriate symmetry operations reveals that there are crystal contacts of TrSA with a neighboring enzyme exactly at the catalytic cleft. K495 side chain from the neighboring enzyme extends into the catalytic cleft of TrSA occupying the space between W312 and S119 (Fig. 11). These crystal contacts can explain the difference in the catalytic clefts between MD simulations and crystal structures of TrSA.
Figure 9.
Catalytic cleft opening in TrSA MD simulations monitored by the distance between W312 CA and S119 CA (in Å). A) TrSA B) TrSA covalent intermediate C) DANA-bound TrSA D) Sialyl-lactose-bound TrSA
Figure 10.
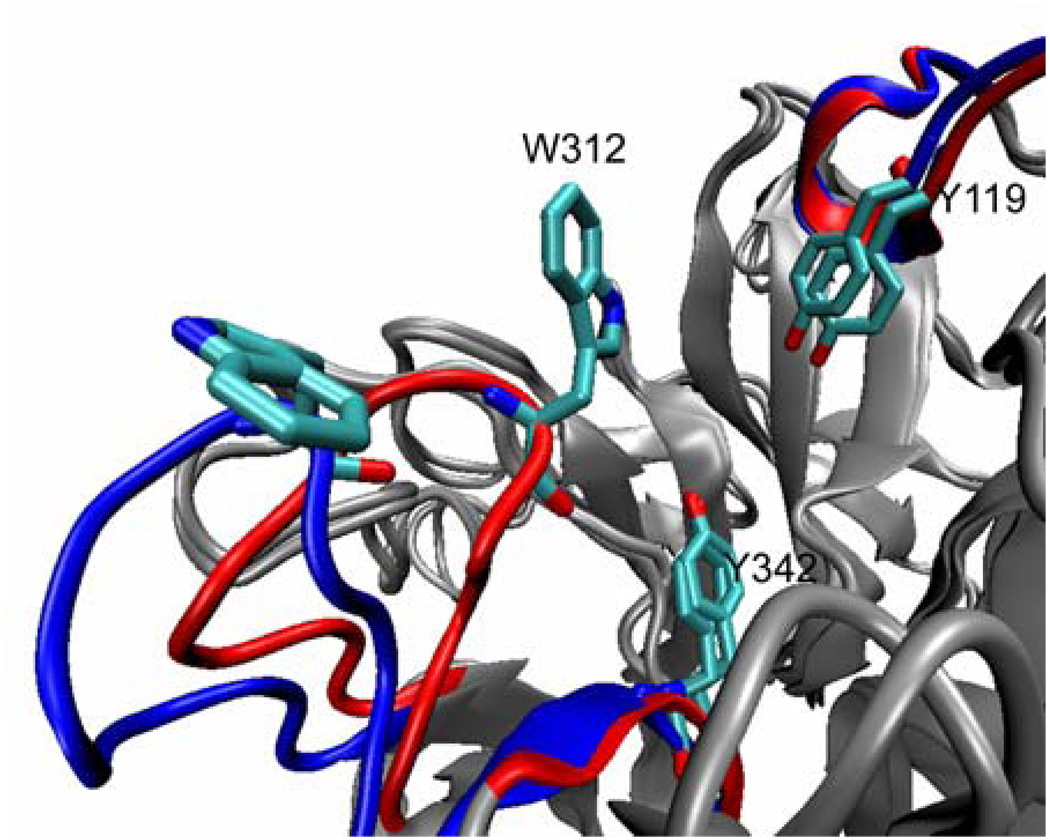
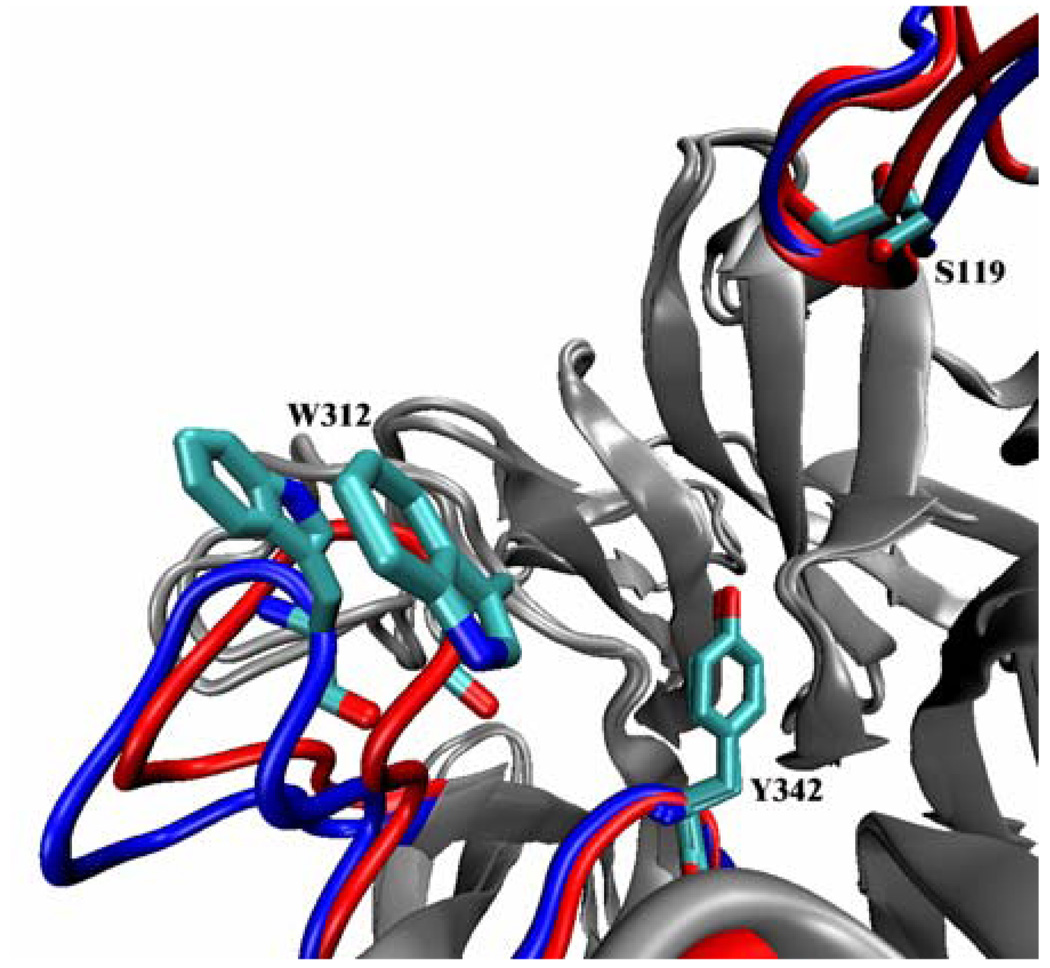
Closed and open forms of TrSA catalytic cleft. The initial closed form and the more open average structure of the apo form of TrSA (in the 50-ns simulation) are superimposed and the important regions of the catalytic cleft are depicted in red and blue, respectively.
Figure 11.
Close crystal contacts in the unit cell of apo form of TrSA. Orange and blue ribbons show the two neighboring enzymes. K495 side chain lies inside the active site of TrSA hindering W312 to move freely. S119 is also shown to clarify the relative orientation.
Comparison of Catalytic Clefts of TcTS and TrSA and Its Consequences
A more open catalytic cleft observed in TrSA covalent intermediate compared to TcTS covalent intermediate in MD simulations is also important since it can directly affect the catalytic activity of an enzyme. For a hydrolysis reaction—in which TrSA is very efficient while TcTS is not—, solvent exposure of the catalytic cleft should be important. The covalent intermediate is in fact the most sensitive phase in the catalytic process for solvent exposure. Thus, higher solvent exposure of TrSA covalent intermediate compared to TcTS covalent intermediate might be the reason for TrSA to be more efficient in hydrolysis. Mutagenesis studies showing that a single S119Y mutation decreases the sialidase activity of TrSA by 50% (Table 3 in Paris et al. (18)) can also support this hypothesis since serine can decrease solvent-exposure of the catalytic cleft for two reasons: it is smaller in size and lacks the large aromatic side chain of a tyrosine residue.
Conclusion
Analysis of MD simulations of TcTS clearly demonstrates the plasticity of its catalytic cleft especially in the apo form. This plasticity, which is mainly provided by W312 loop (residues 306-316) opening/closure motion, must be the reason why none of the proposed inhibitors so far are effective (23) since the inhibitors are designed against a static crystal structure. Similar flexibility was observed also in the case of avian influenza neuraminidase simulations (45).
Additionally, TrSA catalytic cleft is always found to adopt a more open form in MD simulations compared to the crystal structures unless the catalytic cleft is fully occupied by the ligand (e.g. sialyl-lactose). The catalytic cleft of TrSA covalent intermediate prefers a more open form compared to TcTS covalent intermediate. This effect is proposed here as the reason for the differential catalytic functions of the two enzymes since a hydrolysis reaction would be directly affected by water accessibility of the catalytic cleft.
None of the above information has been revealed by the crystal structures available. Although the bias in TrSA can easily be accounted for by the crystal contacts found at the catalytic cleft, no such crystal contacts are found at the TcTS catalytic cleft. However, a recent study showed that crystal packing effects can modify atomic fluctuations even at the regions that do not have direct crystal contacts (46). Lately, the flexibility of enzymes has acquired significant attention and resulted in its inclusion in the drug design process (47–50). For example, McCammon and coworkers (51) found inhibitors for Trypanosoma brucei RNA-editing ligase by explicitly targeting dynamics and flexibility of the enzyme and Carlson and coworkers (52) designed HIV-protease inhibitors targeting a binding site that is only revealed during protein motion. Thus, it is now widely accepted that although the crystal structures of enzymes are an asset, they should not be considered as a rigid scaffold and should be complemented by computational simulations to modify biased conformations and to locate flexible regions which can be targeted for inhibitor design. Here, we demonstrate that TcTS and TrSA are cases where one should be very cautious about the structure and flexibility of loops at the catalytic sites especially for purposes of docking and inhibitor design studies.
Our data additionally offers a possible explanation for the experimental results that indicate differential lactose affinity of TcTS depending on whether the enzyme is preequilibrated with sialic acid/ sialyl-lactose or not. Binding of a sialic acid analog (e.g. DANA) decreases the flexibility of the W312 loop and provides a more preassembled active site to facilitate lactose binding to the TcTS catalytic cleft.
Lastly, we report a correlation between Y119 conformation and W312 loop motion. Once the sialic acid binding region is occupied by sialic acid or DANA, W312 loop mobility is decreased but is still affected by the conformation of Y119. W312 loop keeps a conformation maintaining the closed form of the catalytic cleft as long as Y119 side chain is in one of the “outward” conformations. However, when Y119 side chain swings into the catalytic cleft, W312 loop tends to open the catalytic cleft forming a semi-open catalytic cleft. Most likely, the motion of Y119 side chain into the catalytic cleft distorts the water network and weakens the water-mediated hydrogen bonding between W312 and R245 side chains, consequently triggering W312 loop motion from the closed form to more open forms.
Supplementary Material
Details of the molecular dynamics relaxation procedure and root mean square deviations (RMSD) of Cα atoms of the systems during MD simulations. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGEMENTS
Supercomputer time is granted by Large Allocations Resource Committee (TGMCA05T010). We thank Prof. Pedro Alzari for sharing unpublished X-ray data with us. We are very grateful to Prof. Nicole Horenstein for introducing us to TcTS and for very useful discussions.
Abbreviations
- DANA
2,3-dehydro-3-deoxy-N-acetylneuraminic acid
- MD
Molecular dynamics
- RMSD
root mean square deviation
- RMSF
root mean square fluctuation
- SAPA
Shed acute phase antigen
- TcTS
Trypanosoma cruzi trans-sialidase
- TrSA
Trypanosoma rangeli sialidase
Footnotes
This work was funded in part by NIH grant 1R01AI073674-01 and TG-MCA05T010
REFERENCES
- 1. http://www.who.int/TDR.
- 2.Castro JA, Montalto de Mecca M, Bartel LC. Toxic side effects of drugs used to treat Chagas' disease (American trypanosomiasis) Hum. Exp. Toxicol. 2006;25:471–479. doi: 10.1191/0960327106het653oa. [DOI] [PubMed] [Google Scholar]
- 3.Chagas C. New human trypanosomiasis. Studies about the morphology and evolutive cycle of Schizotripanum cruzi, etiological agent of a new morbid entity of man. Mem. Inst. Oswaldo Cruz. 1909;1:159–218. [Google Scholar]
- 4.Kirchhoff LV. Chagas-Disease - American Trypanosomiasis. Infect. Dis. Clin. N. Am. 1993;7:487–502. [PubMed] [Google Scholar]
- 5.Zeledon R, Rabinovich JE. Chagas-Disease - an Ecological Appraisal with Special Emphasis on Its Insect Vectors. Annu. Rev. Entomol. 1981;26:101–133. doi: 10.1146/annurev.en.26.010181.000533. [DOI] [PubMed] [Google Scholar]
- 6.Previato J, Andrade AFB, Pessolani MCV, Mendonca-Previato L. Incorporation of sialic acid into Trypanosoma cruzi macromolecules. A proposal for a new metabolic route. Mol. Biochem. Parasitol. 1985;16:85–96. doi: 10.1016/0166-6851(85)90051-9. [DOI] [PubMed] [Google Scholar]
- 7.Schauer R, Reuter G, Muehlpfordt H, Andrade AFB, Pereira MEA. The occurrence of N-acetyl- and N-glycoloyineuraminic acid in Trypanosoma cruzi. H.-S. Z. Physiol. Chem. 1983;364:1053–1057. doi: 10.1515/bchm2.1983.364.2.1053. [DOI] [PubMed] [Google Scholar]
- 8.Schauer R. Sialic acids: fascinating sugars in higher animals and man. Zoology. 2004;107:49–64. doi: 10.1016/j.zool.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Varki A. Sialic acids as ligands in recognition phenomena. Faseb J. 1997;11:248–255. doi: 10.1096/fasebj.11.4.9068613. [DOI] [PubMed] [Google Scholar]
- 10.Taylor G. Sialidases: Structures, biological significance and therapeutic potential. Curr. Opin. Struct. Biol. 1996;6:830–837. doi: 10.1016/s0959-440x(96)80014-5. [DOI] [PubMed] [Google Scholar]
- 11.Schenkman S, Eichinger D, Pereira MEA, Nussenzweig V. Structural and Functional Properties of Trypanosoma Trans-Sialidase. Annu. Rev. Microbiol. 1994;48:499–523. doi: 10.1146/annurev.mi.48.100194.002435. [DOI] [PubMed] [Google Scholar]
- 12.Buschiazzo A, Tavares GA, Campetella O, Spinelli S, Cremona ML, Paris G, Amaya MF, Frasch ACC, Alzari PM. Structural basis of sialyltransferase activity in trypanosomal sialidases. EMBO J. 2000;19:16–24. doi: 10.1093/emboj/19.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buschiazzo A, Cremona ML, Campetella O, Frasch ACC, Sanchez DO. Sequence of a Trypanosoma rangeli gene closely related to Trypanosoma cruzi trans-sialidase. Mol. Biochem. Parasitol. 1993;62:115–116. doi: 10.1016/0166-6851(93)90183-x. [DOI] [PubMed] [Google Scholar]
- 14.Amaya MF, Buschiazzo A, Nguyen T, Alzari PM. The High Resolution Structures of Free and Inhibitor-bound Trypanosoma rangeli Sialidase and its Comparison with T. cruzi Trans-sialidase. J. Mol. Biol. 2003;325:773–784. doi: 10.1016/s0022-2836(02)01306-2. [DOI] [PubMed] [Google Scholar]
- 15.Ferrero-Garcia MA, Trombetta SE, Sanchez DO, Reglero A, Frasch ACC, Parodi AJ. The action of Trypanosoma cruzi trans-sialidase on glycolipids and glycoproteins. Eur. J. Biochem. 1993;213:765–771. doi: 10.1111/j.1432-1033.1993.tb17818.x. [DOI] [PubMed] [Google Scholar]
- 16.Scudder P, Doom J, Chuenkova M, Manger I, Pereira M. Enzymatic characterization of beta-D-galactoside alpha 2,3-trans- sialidase from Trypanosoma cruzi. J. Biol. Chem. 1993;268:9886–9891. [PubMed] [Google Scholar]
- 17.Buschiazzo A, Campetella O, Frasch ACC. Trypanosoma rangeli sialidase: cloning, expression and similarity to T.cruzi trans-sialidase. Glycobiology. 1997;7:1167–1173. doi: 10.1093/glycob/7.8.1167. [DOI] [PubMed] [Google Scholar]
- 18.Paris G, Cremona ML, Amaya MF, Buschiazzo A, Giambiagi S, Frasch ACC, Alzari PM. Probing molecular function of trypanosomal sialidases: single point mutations can change substrate specificity and increase hydrolytic activity. Glycobiology. 2001;11:305–311. doi: 10.1093/glycob/11.4.305. [DOI] [PubMed] [Google Scholar]
- 19.Pontes de Carvalho LC, Tomlinson S, Nussenzweig V. Trypanosoma-Rangeli Sialidase Lacks Trans-Sialidase Activity. Mol. Biochem. Parasitol. 1993;62:19–25. doi: 10.1016/0166-6851(93)90173-u. [DOI] [PubMed] [Google Scholar]
- 20.Paris G, Ratier L, Amaya MF, Nguyen T, Alzari PM, Frasch ACC. A Sialidase Mutant Displaying trans-Sialidase Activity. J. Mol. Biol. 2005;345:923–934. doi: 10.1016/j.jmb.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 21.Parodi AJ, Pollevick GD, Mautner M, Buschiazzo A, Sanchez DO, Frasch ACC. Identification of the gene(s) coding for the trans-sialidase of Trypanosoma cruzi. EMBO J. 1992;11:1705–1710. doi: 10.1002/j.1460-2075.1992.tb05221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandekerckhove F, Schenkman S, Pontes de Carvalho L, Tomlinson S, Kiso M, Yoshida M, Hasegawa A, Nussenzweig V. Substrate specificity of the Trypanosoma cruzi trans-sialidase. Glycobiology. 1992;2:541–548. doi: 10.1093/glycob/2.6.541. [DOI] [PubMed] [Google Scholar]
- 23.Neres J, Bryce RA, Douglas KT. Rational drug design in parasitology: trans-sialidase as a case study for Chagas disease. Drug Discov. Today. 2008;13:110–117. doi: 10.1016/j.drudis.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Amaya MF, Watts AG, Damager I, Wehenkel A, Nguyen T, Buschiazzo A, Paris G, Frasch AC, Withers SG, Alzari PM. Structural Insights into the Catalytic Mechanism of Trypanosoma cruzi trans-Sialidase. Structure. 2004;12:775–784. doi: 10.1016/j.str.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 25.Horenstein BA, Yang JS, Bruner M. Mechanistic variation in the glycosyltransfer of N-acetylneuraminic acid. Nukleonika. 2002;47:S25–S28. [Google Scholar]
- 26.Watts AG, Damager I, Amaya ML, Buschiazzo A, Alzari P, Frasch AC, Withers SG. Trypanosoma cruzi Trans-sialidase Operates through a Covalent Sialyl-Enzyme Intermediate: Tyrosine Is the Catalytic Nucleophile. J. Am. Chem. Soc. 2003;125:7532–7533. doi: 10.1021/ja0344967. [DOI] [PubMed] [Google Scholar]
- 27.Yang J. Department of Chemistry. Gainesville, FL: University of Florida; 2001. Transition state and mechanistic study of Trypanosoma cruzi trans-sialidase; p. 181. [Google Scholar]
- 28.Watts AG, Oppezzo P, Withers SG, Alzari PM, Buschiazzo A. Structural and kinetic analysis of two covalent sialosyl-enzyme intermediates on Trypanosoma rangeli sialidase. J. Biol. Chem. 2006;281:4149–4155. doi: 10.1074/jbc.M510677200. [DOI] [PubMed] [Google Scholar]
- 29.Watts AG, Withers SG. The synthesis of some mechanistic probes for sialic acid processing enzymes and the labeling of a sialidase from Trypanosoma rangeli. Can. J. Chem. 2004;82:1581–1588. [Google Scholar]
- 30.Buschiazzo A, Amaya MF, Cremona ML, Frasch AC, Alzari PM. The Crystal Structure and Mode of Action of Trans-Sialidase, a Key Enzyme in Trypanosoma cruzi Pathogenesis. Mol. Cell. 2002;10:757–768. doi: 10.1016/s1097-2765(02)00680-9. [DOI] [PubMed] [Google Scholar]
- 31.Islam SAW. Molecular interactions in protein crystals: Solvent accessible surface and stability. Proteins. 1990;8:1–5. doi: 10.1002/prot.340080103. [DOI] [PubMed] [Google Scholar]
- 32.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 33.Wicki J, Rose DR, Withers SG. Enzyme Kinetics and Mechanism, Pt F: Detection and Characterization of Enzyme Reaction Intermediates. San Diego: Academic Press Inc; 2002. Trapping covalent intermediates on beta-glycosidases; pp. 84–105. [DOI] [PubMed] [Google Scholar]
- 34.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollmann PA. Amber9. San Fransisco, CA: University of California; 2006. [Google Scholar]
- 35.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 36.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A 2nd Generation Force-Field for the Simulation of Proteins, Nucleic-Acids, and Organic-Molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 37.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basma M, Sundara S, Calgan D, Vernali T, Woods RJ. Solvated ensemble averaging in the calculation of partial atomic charges. J. Comput. Chem. 2001;22:1125–1137. doi: 10.1002/jcc.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirschner KN, Woods RJ. Solvent interactions determine carbohydrate conformation. P. Natl. Acad. Sci. USA. 2001;98:10541–10545. doi: 10.1073/pnas.191362798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirschner KN, Woods RJ. Quantum mechanical study of the nonbonded forces in water-methanol complexes. J. Phys. Chem. A. 2001;105:4150–4155. doi: 10.1021/jp004413y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hypercube I. Hyperchem, Professional 7.51 ed. Gainesville, FL: [Google Scholar]
- 42.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian03, RevisionC.02 ed. Wallingford, CT: Gaussian Inc.; 2004. [Google Scholar]
- 43.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 44.Todeschini AR, Mendonca-Previato L, Previato JO, Varki A, Halbeek Hv. Trans-sialidase from Trypanosoma cruzi catalyzes sialoside hydrolysis with retention of configuration. Glycobiology. 2000;10:213–221. doi: 10.1093/glycob/10.2.213. [DOI] [PubMed] [Google Scholar]
- 45.Amaro RE, Minh DDL, Cheng LS, Lindstrom WM, Olson AJ, Lin JH, Li WW, McCammon JA. Remarkable loop flexibility in avian influenza N1 and its implications for antiviral drug design. J. Am. Chem. Soc. 2007;129:7764–7765. doi: 10.1021/ja0723535. [DOI] [PubMed] [Google Scholar]
- 46.Hinsen K. Structural flexibility in proteins: impact of the crystal environment. Bioinformatics. 2008;24:521–528. doi: 10.1093/bioinformatics/btm625. [DOI] [PubMed] [Google Scholar]
- 47.Carlson HA. Protein flexibility and drug design: how to hit a moving target. Curr. Opin. in Chem. Biol. 2002;6:447–452. doi: 10.1016/s1367-5931(02)00341-1. [DOI] [PubMed] [Google Scholar]
- 48.Carlson HA, McCammon JA. Accommodating Protein Flexibility in Computational Drug Design. Mol. Pharmacol. 2000;57:213–218. [PubMed] [Google Scholar]
- 49.Meagher KL, Carlson HA. Incorporating Protein Flexibility in Structure-Based Drug Discovery: Using HIV-1 Protease as a Test Case. J. Am. Chem. Soc. 2004;126:13276–13281. doi: 10.1021/ja0469378. [DOI] [PubMed] [Google Scholar]
- 50.Hornak V, Simmerling C. Targeting structural flexibility in HIV-1 protease inhibitor binding. Drug Discov. Today. 2007;12:132–138. doi: 10.1016/j.drudis.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amaro RE, Schnaufer A, Interthal H, Hol W, Stuart KD, McCammon JA. Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. P. Natl. Acad. Sci. USA. 2008;105:17278–17283. doi: 10.1073/pnas.0805820105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Damm KL, Ung PMU, Quintero JJ, Gestwicki JE, Carlson HA. A poke in the eye: Inhibiting HIV-1 protease through its flap-recognition pocket. Biopolymers. 2008;89:643–652. doi: 10.1002/bip.20993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of the molecular dynamics relaxation procedure and root mean square deviations (RMSD) of Cα atoms of the systems during MD simulations. This material is available free of charge via the Internet at http://pubs.acs.org.