Abstract
Misfolded proteins of the secretory pathway are recognized in the endoplasmic reticulum (ER), retrotranslocated into the cytoplasm, and degraded by the ubiquitin-proteasome system. Right after retrotranslocation and polyubiquitination, they are extracted from the cytosolic side of the ER membrane through a complex consisting of the AAA ATPase Cdc48 (p97 in mammals), Ufd1, and Npl4. This complex delivers misfolded proteins to the proteasome for final degradation. Extraction, delivery, and processing of ERAD (ER-associated degradation) substrates to the proteasome requires additional cofactors of Cdc48. Here we characterize the UBX domain containing protein Ubx4 (Cui1) as a crucial factor for the degradation of polyubiquitinated proteins via ERAD. Ubx4 modulates the Cdc48-Ufd1-Npl4 complex to guarantee its correct function. Mutant variants of Ubx4 lead to defective degradation of misfolded proteins and accumulation of polyubiquitinated proteins bound to Cdc48. We show the requirement of the UBX domain of Ubx4 for its function in ERAD. The observation that Ubx2 and Ubx4 are not found together in one complex with Cdc48 suggests several distinct steps in modulating the activity and localization of Cdc48 in ERAD.
Folding states of secretory proteins undergo proofreading in the endoplasmic reticulum (ER).3 Misfolded proteins are recognized, removed from the secretory process, channeled back into the cytosol, and degraded by the ubiquitin-proteasome system. This process is termed ER quality control and associated degradation (1–6). Delivery of misfolded proteins from the ER to the proteasome depends on Cdc48 (p97 in higher eukaryotes), a member of the AAA ATPase family of molecular chaperones (7–11), which also plays a role in ubiquitin-mediated degradation of cytosolic proteins (12, 13). An array of cofactors regulates the activity of Cdc48 and directs the AAA ATPase to recruitment and processing of substrate proteins (14–16). A common function of Cdc48 is the ATP-coupled disassembly of protein complexes and segregation of proteins from their binding partners (14, 17). An example is the extraction of misfolded polyubiquitinated proteins from the cytosolic side of the ER membrane to deliver them to the proteasome for degradation (ERAD) (2, 3, 6). Other processes requiring Cdc48 are the cell cycle, the activation of transcription factors, and homotypic membrane fusion events (18). Cdc48 can bind polyubiquitinated proteins via its N-terminal domain (12, 19). More usual, ubiquitin binding occurs together with one or more of its many known cofactors. The heterodimeric cofactor Ufd1-Npl4 supports Cdc48 in the recruitment of polyubiquitinated proteins destined for degradation. It is involved in several degradation pathways like the ubiquitin-fusion-degradation pathway (13), the OLE pathway (19), and the ERAD pathway (2–11). Ubiquitin binding is exerted by the zinc finger domain of Npl4 and the N-terminal domain of Ufd1 (20). In yeast, Npl4 does not contain a zinc finger domain, leaving Ufd1 as the ubiquitin-binding protein (21). Npl4 binds Cdc48 via a UBX-related UBD domain, whereas Ufd1 binds Cdc48 via a BS1 domain (21). Interestingly, only one protomer of the homohexameric AAA ATPase Cdc48 is occupied by Ufd1-Npl4 (16). Unoccupied protomers of Cdc48 can bind additional cofactors like Ubx2, an ER membrane protein facing both termini to the cytosol (22, 23). Ubx2 contains an N-terminal UBA domain and a C-terminal UBX domain, which recruits the Cdc48-Ufd1-Npl4 complex to the ER membrane. Thereby it links the Cdc48 complex to E3 ligases and misfolded substrates that appear on the cytosolic side of the ER membrane. The UBX domain is a general motif that binds to the N-terminal domain of Cdc48 (24). Shp1 (Ubx1) is another UBX protein that functions together with Cdc48 in membrane fusion disassembling SNARE complexes (25, 26). Shp1 and Ufd1-Npl4 bind to Cdc48 in a mutually exclusive manner (18, 20). The function of the additional five UBX domain-containing proteins in yeast (Ubx3-Ubx7) is not well characterized. Ubx4, Ubx6, and Ubx7 as well as Shp1 (Ubx1) and Ubx2 are known to be involved in the degradation of the artificial substrate ubiquitin-proline-β-galactosidase (27, 28). Ubx4, Ubx6, and Ubx7 were also found to function in meiosis (27). Here we characterize the function of the protein Ubx4. We show that Ubx4 is required for the continuous degradation of polyubiquitinated proteins of the ERAD pathway. With truncated forms of Ubx4 we identified the UBX domain to be crucial for this degradation event. In addition, in a UBX4-deleted strain, we found increased amounts of polyubiquitinated misfolded proteins bound to the Cdc48 complex in the cytoplasm and at membranes. Our data indicate a function of Ubx4 in the release of polyubiquitinated material from the Cdc48 complex.
EXPERIMENTAL PROCEDURES
Growth Conditions, Yeast Strains, and Plasmids
Genetic and molecular biology techniques were carried out using standard methods (29–31). Cells were grown at 30 °C in synthetic complete media. Saccharomyces cerevisiae strains are based on the genetic background of strain W303 prc1-1 (MATα ade2-1ocre can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 prc1-1). All mutations and tags were integrated into the cell genome. Gene deletion and epitope tagging was generated via homologous recombination (32–34) or strain crossing and subsequent tetrad dissection (29). Yeast strains used in this study are depicted in Table 1. The strains YSA10 and YSA12 were obtained by transformation of W303 prc1-1 with a kanMX deletion module. Strains YSA18, YSA21, YCR1, YCR2, and YCR4 were created by transformation of a his5+ deletion module (32). Crossing of YSA10 and YCR1 and subsequent tetrad dissection generated strain YCR11. Epitope tagging of YSA14, YCR14, YSA27, YSA28, and YSA29 with Myc9 or HA3 tags was done as described (33, 34). YSA31 was prepared by transformation of YSA10 with an HA3 tagging module (34). Deletion of parts of 9xMyc-UBX4 in YSA25 and YSA30 was done using YSA14 as a parent strain and transformation with a LEU2 deletion cassette (32). Tagging of the N terminus and simultaneous loss of the N-terminal amino acids 1–170 of Ubx4 resulted in strain YSA26 (33). Plasmids pRS316 expressing CTL* under the GAL4 promotor and plasmid pSM1911 expressing Ste6*-HA under the PGK1 promotor have been described (35, 36).
TABLE 1.
Yeast strains used in this study
| Name | Genotype | Ref. |
|---|---|---|
| W303prc1-1 | MATα ade2-1ocre can1-100 his3-11,15 leu2-3,112 trp1-1ura3-1 prc1-1 | 45 |
| YJB009 | W303prc1-1 der3::HIS3 | 37 |
| YSA10 | W303prc1-1ubx4::kanMX | This study |
| YSA12 | W303prc1-1ubx7::kanMX | This study |
| YSA14 | W303prc1-1 9xMyc-UBX4 | This study |
| YSA18 | W303prc1-1 ubx3::his5+ | This study |
| YSA21 | W303prc1-1 ubx1::his5+ | This study |
| YCR1 | W303prc1-1 ubx2::his5+ | This study |
| YCR2 | W303prc1-1 ubx5::his5+ | This study |
| YCR4 | W303prc1-1 ubx6::his5+ | This study |
| YCR11 | W303prc1-1ubx2::his5+ubx4::kanMX | This study |
| YCR14 | W303prc1-1UBX2-3xHA-His3MX6 | This study |
| YSA25 | W303prc1-1 9xMyc-ubx4-Δ268-416::LEU2 | This study |
| YSA26 | W303prc1-19xMyc-ubx4-Δ1-170 | This study |
| YSA27 | W303prc1-1 9xMyc-UBX4 UBX2-3xHA-His3MX6 | This study |
| YSA28 | W303prc1-1 9xMyc-UBX4 NPL4-3xHA-His3MX6 | This study |
| YSA29 | W303prc1-1 NPL4-3xHA-His3MX6 | This study |
| YSA30 | W303prc1-1 9xMyc-ubx4-Δ171-416::LEU2 | This study |
| YSA31 | W303prc1-1 ubx4::kanMX UBX2-3xHA-His3MX6 | This study |
| YSA33 | W303prc1-1 9xMyc-ubx4-Δ268-416::his5+ | This study |
Growth Tests
Single colonies were inoculated into 2 ml of selection medium (+Leu −Ura) and incubated at 30 °C until stationary phase was reached. Cells were then diluted to 1 OD600 in the same medium, and dilution series of each culture (1:10) was prepared in a 96-well plate. 4 μl of cell culture were then spotted onto plates with a 48-prong replica-plating device. Plates were incubated for 2–4 days at 30 °C or as indicated.
Antibodies
For precipitation of CPY* in pulse-chase analysis, polyclonal anti-CPY antibody (Rockland Immunochemicals, Inc.) was used, and for immunodetection monoclonal CPY antibody (Molecular Probes) was used. c-Myc was detected using specific antibody (clone 9E10, Santa Cruz Biotechnology). Ubiquitin and HA epitopes were detected using specific antibodies (clone P4G7 and clone 16B12, Covance). Antibody against Pgk1 was from Molecular Probes. Der3/Hrd1 was detected using a polyclonal antibody, which was described (37). Sec61 and Cdc48 antibody were gifts from T. Sommer.
Protein Degradation Assays
Pulse-chase experiments of CPY* and Ste6*-HA were carried out essentially as described (36, 38). Diagrams represent data of up to five independent experiments, and the error bars indicate the respective S.E. The procedure to perform cycloheximide chase experiments was described previously (36). SDS-PAGE and immunoblot analysis were performed according to standard protocols (29).
Membrane Extraction Experiment
60 OD cells (OD600 = 1.0) were spheroblasted in sorbitol buffer (1 m sorbitol, 50 mm sodium phosphate, pH 7.4) with 20 μl of Oxalyticase (stock, 5 mg/ml) for 30 min at 30 °C. Spheroblasted cells were then lysed in lysis buffer (0.8 m sorbitol, 10 mm Pipes, 1 mm EDTA, pH 7.2) in a Wheaton homogenizer. After preclearing at 1,500 × g for 5 min, the cell extract was incubated with lysis buffer or lysis buffer containing 0.6 m KOAc, 2 m urea, 1% Triton X-100 or 0.5% SDS, respectively. After incubation for 30 min on ice, samples were centrifuged at 100,000 × g for 1 h to separate them into a pellet (P) and a soluble (S) fraction. Samples were analyzed by SDS-PAGE and immunodetection (29).
Immunoprecipitation Experiments
Immunoprecipitation experiments were essentially performed as described (23, 39) except for the following treatments. For cell lysis 150 OD of cells were taken. The lysates of the digitonin-solubilized membranes of the washed pellet fractions were cleared by centrifugation at 100,000 × g for 30 min prior to immunoprecipitation. Standard methods for SDS-PAGE, Western blotting, and immunodecoration were used (29).
RESULTS
Growth Assays Indicate a Participation of Ubx4 in Protein Degradation
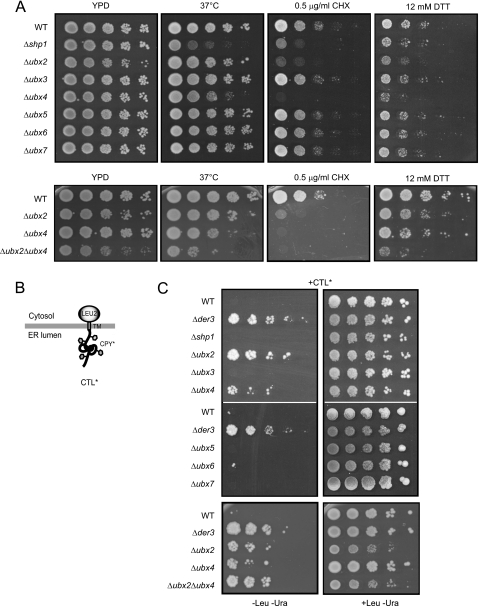
Cdc48 has functions in a variety of different cellular processes like cell cycle regulation, activation of transcription factors, and homotypic membrane fusion. As Cdc48 is also a central component of the ERAD machinery, we were interested whether additional Ubx proteins other than Ubx2 play a role in the ERAD pathway. We constructed yeast strains in the W303 prc1-1 background expressing CPY* and deleted for UBX1 to UBX7, respectively. In these strains we looked for sensitivity toward different stress situations. Shp1 (Ubx1) is required for growth of cells at elevated temperature (28). As shown for Shp1, a strain deleted in UBX4 also exhibits a sensitivity toward elevated temperature (Fig. 1A). Furthermore, we observed hypersensitivity of the Δubx4 strain toward cycloheximide and dithiothreitol (DTT) that was even stronger than for the Δshp1 strain (Fig. 1A). Only when exposed to heat stress at 37 °C was the growth of the Δshp1 strain more disturbed than for the Δubx4 strain. These data indicate a strong link of UBX4 to stress tolerance and proteasomal degradation. In particular, the sensitivity of the Δubx4 strain to the reducing agent DTT points to a disturbed degradation of misfolded proteins of the ER. A multitude of newly synthesized secretory proteins receive disulfide bonds in the ER and are therefore structurally highly sensitive to reducing agents such as DTT. Indeed, the hypersensitivity of strains deleted in UBX2 for DTT (Fig. 1A) underlines the discovered function of Ubx2 in the degradation of misfolded proteins of the ER (ERAD) (22, 23). Thus, the observed DTT sensitivity of Δubx4 mutants also points to a participation of Ubx4 in ERAD (Fig. 1A). A double deletion mutant defective in UBX2 and UBX4 shows a somewhat stronger growth phenotype on all media tested. For analysis of which of the Ubx-cofactor proteins of Cdc48 has a function in the degradation of ERAD substrates, we employed the previously elaborated growth test of cells for detection of a malfunctioning ERAD (36, 38). We expressed the membrane-anchored ERAD substrate CTL* from a plasmid in the yeast strains of the W303 prc1-1 background deleted for UBX1 to UBX7, respectively, containing a leucine auxotrophy (Fig. 1B) (36, 38). CTL* consists of misfolded ER-luminal CPY*, a transmembrane domain and the Leu2 domain located in the cytosol. In wild type cells the misfolded CPY* domain induces complete degradation of CTL*. Because of a defective leucine biosynthetic pathway, these cells cannot grow on medium lacking leucine. In contrast, cells with a defect in a gene essential for ERAD such as DER3/HRD1 are unable to degrade CTL*, therefore allowing growth due to the presence of the Leu2 protein (Fig. 1C). When testing the different UBX deletion strains in this growth assay, we observed growth of the Δubx2 strain and, interestingly, of the Δubx4 strain (Fig. 1C). This indicates that Ubx4 influences degradation of misfolded substrates of the ER. No growth was observed for the Δshp1 (Δubx1), Δubx3, Δubx5, Δubx6, and Δubx7 strains, indicating the presence of a wild type ERAD system in these mutant cells. The Δubx2Δubx4 double deletion strain exhibits a somewhat stronger growth indicating an increased half-life of CTL* (Fig. 1C).
FIGURE 1.
Growth assays indicate a participation of Ubx4 in protein degradation. A, growth assays to analyze Δubx strains for sensitivity toward heat, cycloheximide (CHX), and dithiothreitol (DTT). 10-Fold serial dilutions of the indicated strains were spotted onto plates containing YPD, YPD plus cycloheximide, or YPD plus DTT and grown for 2–4 days at 30 °C or as indicated before being photographed. B, schematic representation of the ERAD substrate CTL*. C, growth test of yeast strains transformed with a plasmid expressing CTL* under the control of the GAL4 promotor. 10-Fold serial dilutions of the indicated strains were spotted onto plates containing selective media without or, as a control, with leucine and grown for 2–3 days at 30 °C before being photographed. The W303 prc1-1 wild type (WT) strain defective in the LEU2 gene fails to grow on medium lacking leucine, whereas strains defective in ERAD are able to grow due to complementation of the leucine deficiency by CTL*. The strain lacking the E3 ligase Der3/Hrd1 serves as a control. Two of the seven screened Δubx deletion strains, Δubx2 and Δubx4, complement the leucine auxotrophy.
Ubx4 Is Required for Degradation of Misfolded Secretory Proteins
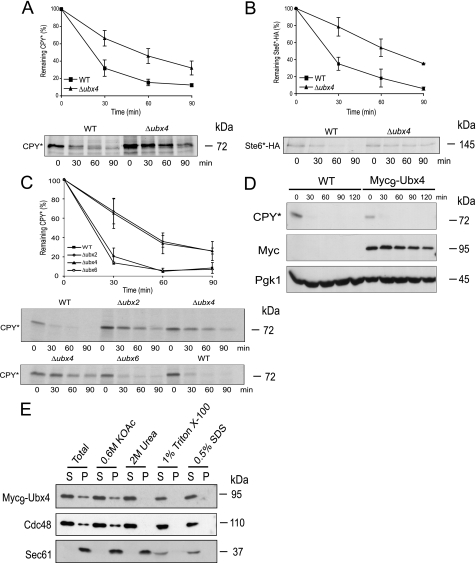
To confirm a possible function of Ubx4 in elimination of substrates of the ERAD pathway, we tested the degradation of two well known ERAD substrates, luminal CPY* (1) and the polytopic transmembrane protein Ste6*-HA (35) by pulse-chase analysis. CPY* is defined as an ERAD-L substrate carrying a misfolded domain localized in the ER lumen, which is polyubiquitinated via the Der3/Hrd1 ligase. Ste6*-HA, on the other hand, is defined as an ERAD-C substrate carrying a C-terminal truncation in the cytoplasm. This misfolded protein is mainly polyubiquitinated via the ubiquitin ligase Doa10 (35, 40). Degradation of the two misfolded ERAD substrates is significantly disturbed in a Δubx4 strain (Fig. 2, A and B). Cells deleted in UBX2 show a similar disturbance of CPY* degradation as known from the literature (Fig. 2C) (22, 23). In contrast, a UBX6-deleted strain does not show a disturbed degradation of CPY* (Fig. 2C) or Ste6*-HA (data not shown). Obviously, the function of Ubx4 is required for the elimination of misfolded proteins of both ERAD pathways, ERAD-L and ERAD-C.
FIGURE 2.
Pulse-chase experiments and membrane extraction assays. A–C, pulse-chase analysis of CPY* and Ste6*-HA degradation. Wild type (WT) and Δubx4 (A and B) and wild type, Δubx2, Δubx4, and Δubx6 mutant cells (C) were grown and radiolabeled. Extracts were prepared, and the substrates were immunoprecipitated with specific antibodies at the indicated time points. Samples were separated via SDS-PAGE, and proteins were detected using a PhosphorImager system. D, cycloheximide chase experiment to analyze if Myc9-Ubx4 is functional regarding degradation of CPY* compared with wild type (WT). Pgk1 served as a loading control. Immunoblotting was done using specific antibodies. E, analysis of Ubx4 distribution in soluble and membrane fractions. Membrane extraction assay was performed in wild type cells expressing Myc9-Ubx4. Spheroblasted cells were homogenized and treated with different agents. Subsequently, the samples were separated by high speed centrifugation into supernatant (S) and pellet (P) fractions and analyzed via immunodetection with Myc, Cdc48, and as a control, Sec61 antibodies.
Ubx4 and Cdc48 Are Membrane-associated
Cdc48 and Ubx4 have been shown to mainly localize to the cytosol and the nucleus, whereas a considerable fraction is also found at membranes of the ER and the nuclear envelope (22, 23, 27, 41). For analysis of Ubx4 via immunological detection, we generated an N-terminal Myc9-tagged version of the protein. This fusion protein is fully functional regarding degradation of CPY* (Fig. 2D). As shown in Fig. 2E, Myc9-Ubx4 and Cdc48 are partly associated with the membrane fraction, most likely the ER membrane. This association can be completely abolished by the treatment with 2 m urea, indicating that both proteins are not membrane-integrated but might associate via a linker molecule in the membrane.
UBX Domain of Ubx4 Is Necessary for Degradation of Misfolded Proteins
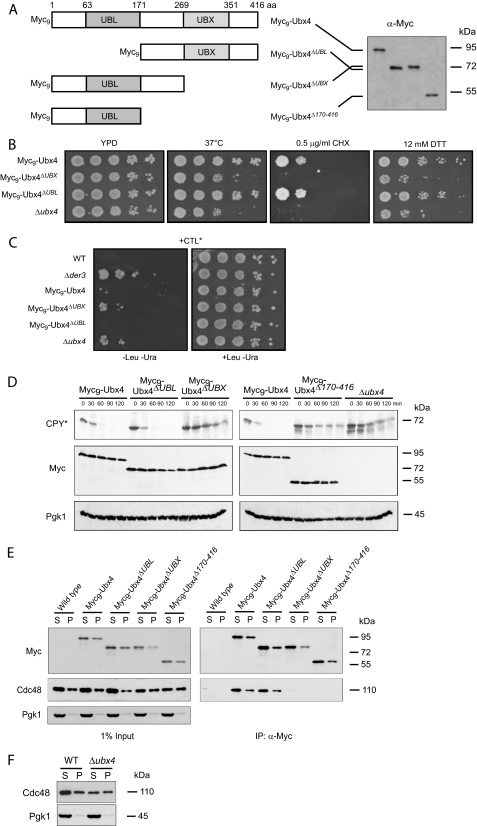
The putative mammalian orthologue of Ubx4 TUG (ASPCR1/UBXD9) contains a UBL domain near the N terminus and a UBX domain located close to the C terminus (15). The UBX domain of Ubx4 in yeast was already published (28). In an alignment we compared the UBL domains of different yeast proteins with the N terminus of Ubx4 (supplemental Fig. 1). In most yeast proteins the UBL domain is usually about 80 amino acids long. One exception is the UBL domain of Usa1, which consists of about 60 amino acids. Interestingly, Ubx4 contains a longer variant of the UBL domain of about 108 amino acids (supplemental Fig. 1). As Ubx4 contains both domains, a UBL and a UBX domain, it is unique among the Ubx proteins in yeast. Both domains share high similarity to ubiquitin (24). We tested if both domains are required for degradation of ERAD substrates. Therefore, N-terminally Myc9-tagged truncations of Ubx4 were created (Fig. 3A). As can be seen from Fig. 3, B and C, deletion of the UBX domain of UBX4 (amino acids 269–416 or 171–416) leads to similar growth phenotypes as found for the Δubx4 deletion strain. The corresponding degradation assays of the UBX domain deleted UBX4 strains resulted in a comparable delay of CPY* degradation as found for the UBX4 deletion (Fig. 3D). Interestingly, a deletion of the UBL domain has no effect on CPY* elimination (Fig. 3D). Immunoprecipitation of soluble Myc9-Ubx4 (S) and membrane solubilized Myc9-Ubx4 (P) showed clearly that the deletion of the UBX domain abrogates the interaction of Ubx4 with Cdc48 (Fig. 3E). This indicates that only the UBX domain is required for Ubx4 cofactor interaction with the AAA ATPase Cdc48. The complex of Cdc48-Ubx4 exists in both fractions. One is found in the soluble, cytoplasmic fraction (S), the other one is membrane-associated (P) (Fig. 3E). The amount of Cdc48 at the ER membrane is identical in wild type and mutant cells, whereas the amount of Cdc48 in the supernatant of Δubx4 mutant cells is always somewhat lower than in the wild type (Fig. 3, E and F). Therefore, we tested if Ubx4 has a role in the degradation of Cdc48. In a cycloheximide chase experiment, no difference in the degradation of Cdc48 between a Δubx4 strain and a wild type strain could be observed (data not shown).
FIGURE 3.
Function of the UBL and UBX domain of Ubx4 in ERAD. A, schematic representation of the Ubx4 domain structure and the deletions generated as well as their molecular masses determined by SDS-PAGE, subsequent Western blotting, and immunodetection using Myc antibodies. B, cells expressing Myc9-tagged Ubx4, Myc9-tagged fragments of Ubx4 deleted in the UBX or the UBL domain, as well as cells deleted in UBX4 were grown on different media as described in legend to Fig. 1A. C, growth test of cells expressing Myc9-tagged Ubx4, Myc9-tagged fragments of Ubx4 deleted in the UBX or the UBL domain, as well as cells deleted in UBX4, and for control, wild type and DER3 deleted cells were transformed with a plasmid expressing CTL* under control of the GAL4 promotor. The test was done as described in legend to Fig. 1C. D, cycloheximide chase experiments of CPY* degradation in different UBX4 deletion mutants. Detection of Myc signals served as control for expression and stability of Ubx4 and its fragments. Pgk1 (3-phosphoglycerate kinase) protein was used as a loading control. Immunoblotting was done using the respective specific antibodies. E, interaction of N-terminal Myc9-tagged Ubx4 and its truncated variants carrying deletions of the UBX or the UBL domain with Cdc48. Cells expressing Myc9-Ubx4, its Myc9-tagged truncated variants, and as a control wild type Ubx4 were lysed and separated into soluble (S) and membrane (P) proteins. Membrane proteins (P) were solubilized by digitonin treatment. Interaction of the Myc9-Ubx4 and mutant proteins with Cdc48 was followed by immunoprecipitation (IP) using Myc-specific antibodies. After SDS-PAGE and Western blotting, proteins were detected using specific antibodies. Pgk1 was used as a loading control. F, amount of Cdc48 in wild type (WT) cells compared with Δubx4 cells after fractionation of cell lysates by high speed centrifugation. Loading control was done using Pgk1 antibody.
Ubx4 Is in Complex with the Substrate Recruiting Cofactor Ufd1- Npl4
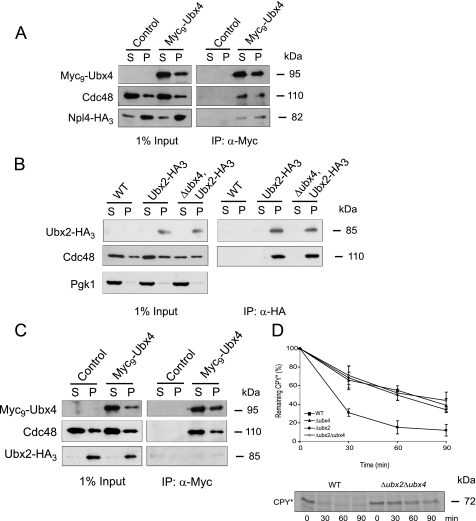
We wanted to know whether the Ubx4-containing Cdc48 complex also binds the heterodimeric cofactor Ufd1-Npl4. The binding of Ufd1 to Cdc48 is necessary and a prerequisite for the interaction of Cdc48 with Npl4 (21). We tagged Npl4 at the C terminus with an HA3 epitope and immunoprecipitated cytosolic Myc9-Ubx4 (S) and membrane-associated Myc9-Ubx4 (P) via their Myc tags. Myc9-Ubx4 coimmunoprecipitates with Cdc48 and Npl4-HA3 of both, the cytosolic and the membrane bound complex, indicating that Ubx4 is indeed a component of the Cdc48-Ufd1-Npl4 complex (Fig. 4A).
FIGURE 4.
Binding of cofactors to the Cdc48 complex. A, interaction of the Cdc48-Ubx4 complex with Npl4-HA3. Cells expressing Myc9-Ubx4 and Npl4-HA3 and, as a control, cells only expressing Npl4-HA3 were lysed and separated into soluble (S) and membrane (P) proteins. Membrane proteins (P) were solubilized by digitonin treatment. Immunoprecipitation (IP) of Myc9-Ubx4 (right panel) was done using monoclonal Myc antibody. Npl4-HA3 was detected using monoclonal HA antibody. 1% yeast extract (left panel) was immunoblotted for control. B, Cdc48-Ubx2 interaction is not affected in a Δubx4 strain. Ubx2-HA3 was expressed in wild type and Δubx4 cells and coprecipitated Cdc48. In the control strain Ubx2-HA3 was not expressed. Cell lysis and preparation of soluble and membrane proteins were done as described in A. Pgk1 was used as loading control. C, binding of Ubx4 excludes Ubx2 from the Cdc48 complex. Cells expressing Myc9-Ubx4 and Ubx2-HA3 and, as a control, cells only expressing Ubx2-HA3 were lysed and separated into soluble (S) and membrane (P) proteins. Cell lysis and preparation of soluble and membrane proteins were done as in A. Coimmunoprecipitation experiments were done using monoclonal Myc antibodies. Proteins were separated on SDS-PAGE and detected on immunoblots using Myc, Cdc48, or HA antibodies. D, pulse-chase analysis of CPY* degradation in wild type (WT), Δubx2, Δubx4, and Δubx2Δubx4 mutants. Cells were grown and radiolabeled; extracts were prepared, and the substrate was immunoprecipitated with specific CPY antibodies at the indicated time points. Samples were separated via SDS-PAGE, and the protein was detected using a PhosphorImager system.
Cdc48-Ubx2 Complex Formation Does Not Depend on Ubx4
To get more insight into the function of Ubx4, we tested whether the absence of Ubx4 has any influence on the binding of Cdc48 to the ER membrane via the known recruiting cofactor Ubx2. When immunoprecipitating membrane-solubilized Ubx2-HA3 isolated from wild type cells and the Δubx4 deletion strain, we observed equal amounts of Cdc48 coprecipitated with Ubx2-HA3 (Fig. 4B (P)). As expected, no Cdc48-Ubx2-HA3 complex was found in the fractions (S) containing soluble proteins because Ubx2 is a membrane protein. This shows that the absence of Ubx4 has no influence on the Cdc48-Ubx2 interaction.
Ubx4 Is Not in Complex with Ubx2
A part of Ubx4 is localized together with the Cdc48 complex at membranes (Fig. 4A). Therefore, we analyzed whether this Cdc48-Ubx4 complex is also linked via Ubx2 to the ER membrane. For this purpose we precipitated soluble and membrane-solubilized Myc9-Ubx4. Astonishingly, no Ubx2-HA3 protein was found in the membrane-associated Cdc48-Ubx4 complex (Fig. 4C). This indicates that binding of Ubx4 to the Cdc48-Ufd1-Npl4 complex excludes concomitant binding of Ubx2. Pulse-chase experiments in the deletion mutants of UBX2, UBX4, or both showed a delayed degradation of CPY* compared with wild type cells. The degradation kinetics are similar in all three mutant strains (Fig. 4D) and may suggest that Ubx2 and Ubx4 act in the same pathway.
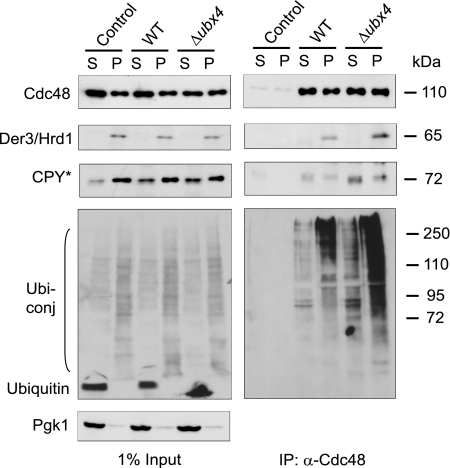
Increased Amounts of CPY* and Polyubiquitinated Proteins Coprecipitate with Cdc48 in a Δubx4 Strain
It has been shown that the function of the Cdc48 complex in the ERAD pathway resides in its capacity to remove polyubiquitinated misfolded proteins from the ER and deliver them to the proteasome (7–11). Considering this, the delayed degradation of ERAD substrates in a Δubx4 strain may be due to the inability of the Cdc48-Ufd1-Npl4 complex to deliver polyubiquitinated substrates properly to proteasomal proteolysis. To address this question, we analyzed the amount of the misfolded substrate CPY* bound to the Cdc48 complex (Fig. 5). Immunoprecipitation with Cdc48 antibodies resulted in a considerable increased amount of CPY* bound to the Cdc48 complex in a Δubx4 strain compared with wild type in both the soluble (S) and membrane-solubilized (P) fractions (Fig. 5). In addition, the total amount of coimmunoprecipitated polyubiquitinated proteins bound to the Cdc48 complex is remarkably increased in a Δubx4 deletion strain as compared with wild type. This difference was especially striking in the precipitates isolated from membrane-solubilized proteins of Δubx4 cells (Fig. 5). The amount of the ubiquitin ligase Der3/Hrd1 coprecipitated with the Cdc48 complex is enhanced in the Δubx4 deletion strain (Fig. 5). Obviously, in the absence of Ubx4 the Cdc48 complex binds stronger or longer to membrane components of the ER and accumulates misfolded CPY* and polyubiquitinated material. This indicates a disturbance in the release of misfolded proteins from Cdc48 when Ubx4 is missing.
FIGURE 5.
Δubx4 mutants exhibit an increased load of CPY * and polyubiquitinated proteins on the Cdc48 complex. Wild type and Δubx4 cells were lysed and separated into soluble (S) and membrane (P) proteins. Membrane proteins (P) were solubilized by digitonin treatment. Coimmunoprecipitation (IP) of CPY*, Der3/Hrd1, and polyubiquitinated proteins with Cdc48 was done using Cdc48-specific antibodies. After SDS-PAGE and blotting onto nitrocellulose, protein detection was done using specific antibodies against CPY, Der3/Hrd1, and ubiquitin following ECL detection. Pgk1 was used as loading control.
DISCUSSION
We identified Ubx4, a UBX domain containing cofactor of Cdc48, as a novel member influencing the ERAD pathway in yeast. First hints for an influence of Ubx4 on protein degradation came from a study by Decottignies et al. (27). The authors showed that a Δubx4Δubx6Δubx7 triple knock-out strain stabilizes Ub-P-βGal, suggesting a redundant role of the proteins Ubx4, Ubx6, and Ubx7 on the degradation of cytosolic substrates. Additional hints link Ubx4 to proteasomal degradation and showed that the deletion of UBX4 and RPN4, a transcription factor for proteasomal genes, leads to synthetic lethality (42). A link to the ERAD pathway came from the study of Travers et al. (43). The authors found that the expression of Ubx4 is up-regulated during the unfolded protein response (43). This up-regulation is comparable with the ones found for the known ERAD components Der1, Ubx2, and Hrd3. The hypersensitivity toward DTT and the disturbed degradation of the ERAD substrate CTL* show for the first time that the ERAD pathway is affected in a Δubx4 knock-out strain (Fig. 1, A and C). Furthermore, our pulse-chase experiments with mutants deleted in UBX4 show a defective degradation of a luminal (ERAD-L) and a polytopic membrane (ERAD-C) substrate, CPY* and Ste6*-HA, respectively (Fig. 2). Only one of the two domains of Ubx4, the UBX domain, is required for binding to Cdc48 and subsequently for ERAD (Fig. 3). Deletion of the UBL domain of Ubx4 does not affect binding to Cdc48 nor degradation of CPY*. Up to now the function of the UBL domain of Ubx4 remains unknown.
We show that Ubx4 binds to the Cdc48-Ufd1-Npl4 complex (Fig. 4A), which is the molecular unit pulling polyubiquitinated proteins away from the ER membrane (9, 19). Binding of these two cofactors, Ubx4 and Ufd1-Npl4, to Cdc48 at the same time is not mutually exclusive, in contrast to the well established exclusive binding of either Ufd1-Npl4 or Shp1 to Cdc48 (44). For proper ERAD function the Cdc48-Ufd1-Npl4 complex is bound to the ER membrane via the UBX domain containing protein Ubx2 (22, 23). This interaction is not disturbed in a Δubx4 strain (Fig. 4B). Interestingly, the membrane linker Ubx2 seems to be excluded from the membrane-associated Cdc48-Ubx4 complex (Fig. 4C). These data indicate the existence of a Ubx2-independent Cdc48-Ubx4 complex and of further unknown factors for Cdc48-Ufd1-Npl4 recruitment to the ER membrane. Such an unknown membrane-recruiting factor has also been suggested by Neuber et al. (23). The authors reported that cells lacking Ubx2 exhibit no major reduction in the amount of membrane-bound Cdc48 (23). Our data show that the Cdc48-Ufd1-Npl4 complex exists in different physiological states. The Cdc48-Ufd1-Npl4 complex bound to Ubx2 at the ER membrane defines one state, the Cdc48-Ufd1-Npl4 complex bound to Ubx4 at the membrane and in the cytosol define novel additional states. Our data indicate that these different states of Cdc48 complexes exert a function affecting the ER-associated degradation pathway.
In the absence of Ubx4, the membrane-bound Cdc48 complex as well as the soluble Cdc48 complex carry more misfolded CPY* and polyubiquitinated proteins, indicating that they are unable to release and deliver them properly to the downstream components (Fig. 5). We suggest that both fractions of this complex, one bound to the membrane and the other located in the cytosol, are involved in the delivery of the polyubiquitinated misfolded proteins to the downstream components and to the proteasome. Absence of Ubx4 might also affect other proteolytic pathways that depend on the pulling function of the Cdc48 complex. The altered growth of a UBX4-deleted strain under stress conditions of higher temperature or of toxic agents like cycloheximide and dithiothreitol might underline this conclusion. Increased levels of misfolded polyubiquitinated proteins at Cdc48 in the absence of Ubx4 could also affect different cellular processes that require a functional Cdc48 system.
Supplementary Material
Acknowledgments
We thank Susan Michaelis for providing plasmid pSM1911 and Thomas Sommer for providing Sec61 and Cdc48 antibodies.
This work was supported by the European Network of Excellence RUBICON and the Deutsche Forsch ungs ge mein schaft, Bonn.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- DTT
- dithiothreitol
- HA
- hemagglutinin
- Pipes
- 1,4-piperazinediethanesulfonic acid.
REFERENCES
- 1.Hiller M. M., Finger A., Schweiger M., Wolf D. H. ( 1996) Science 273, 1725– 1728 [DOI] [PubMed] [Google Scholar]
- 2.Kostova Z., Wolf D. H. ( 2003) EMBO J. 22, 2309– 2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahner A., Brodsky J. L. ( 2004) Trends Cell Biol. 14, 474– 478 [DOI] [PubMed] [Google Scholar]
- 4.Schäfer A., Kostova Z., Wolf D. H. ( 2008) in The Ubiquitin-Proteasome System and Disease, ( Mayer R. J., Ciechanover A., Rechsteiner M. eds) pp. 123– 143, Wiley-VCH, Weinheim, Germany [Google Scholar]
- 5.Raasi S., Wolf D. H. ( 2007) Semin. Cell Dev. Biol. 18, 780– 791 [DOI] [PubMed] [Google Scholar]
- 6.Meusser B., Hirsch C., Jarosch E., Sommer T. ( 2005) Nat. Cell Biol. 7, 766– 772 [DOI] [PubMed] [Google Scholar]
- 7.Ye Y., Meyer H. H., Rapoport T. A. ( 2001) Nature 414, 652– 656 [DOI] [PubMed] [Google Scholar]
- 8.Rabinovich E., Kerem A., Fröhlich K. U., Diamant N., Bar-Nun S. ( 2002) Mol. Cell. Biol. 22, 626– 634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarosch E., Taxis C., Volkwein C., Bordallo J., Finley D., Wolf D. H., Sommer T. ( 2002) Nat. Cell Biol. 4, 134– 139 [DOI] [PubMed] [Google Scholar]
- 10.Braun S., Matuschewski K., Rape M., Thoms S., Jentsch S. ( 2002) EMBO J. 21, 615– 621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bays N. W., Wilhovsky S. K., Goradia A., Hodgkiss-Harlow K., Hampton R. Y. ( 2001) Mol. Biol. Cell 12, 4114– 4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai R. M., Li C. C. ( 2001) Nat. Cell Biol. 3, 740– 744 [DOI] [PubMed] [Google Scholar]
- 13.Ghislain M., Dohmen R. J., Levy F., Varshavsky A. ( 1996) EMBO J. 15, 4884– 4899 [PMC free article] [PubMed] [Google Scholar]
- 14.Jentsch S., Rumpf S. ( 2007) Trends Biochem. Sci. 32, 6– 11 [DOI] [PubMed] [Google Scholar]
- 15.Schuberth C., Buchberger A. ( 2008) Cell. Mol. Life Sci. 65, 2360– 2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pye V. E., Beuron F., Keetch C. A., McKeown C., Robinson C. V., Meyer H. H., Zhang X., Freemont P. S. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 467– 472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beuron F., Dreveny I., Yuan X., Pye V. E., McKeown C., Briggs L. C., Cliff M. J., Kaneko Y., Wallis R., Isaacson R. L., Ladbury J. E., Matthews S. J., Kondo H., Zhang X., Freemont P. S. ( 2006) EMBO J. 25, 1967– 1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye Y. ( 2006) J. Struct. Biol. 156, 29– 40 [DOI] [PubMed] [Google Scholar]
- 19.Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S. ( 2001) Cell 107, 667– 677 [DOI] [PubMed] [Google Scholar]
- 20.Meyer H. H., Wang Y., Warren G. ( 2002) EMBO J. 21, 5645– 5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruderer R. M., Brasseur C., Meyer H. H. ( 2004) J. Biol. Chem. 279, 49609– 49616 [DOI] [PubMed] [Google Scholar]
- 22.Schuberth C., Buchberger A. ( 2005) Nat. Cell Biol. 7, 999– 1006 [DOI] [PubMed] [Google Scholar]
- 23.Neuber O., Jarosch E., Volkwein C., Walter J., Sommer T. ( 2005) Nat. Cell Biol. 7, 993– 998 [DOI] [PubMed] [Google Scholar]
- 24.Buchberger A., Howard M. J., Proctor M., Bycroft M. ( 2001) J. Mol. Biol. 307, 17– 24 [DOI] [PubMed] [Google Scholar]
- 25.Latterich M., Fröhlich K. U., Schekman R. ( 1995) Cell 82, 885– 893 [DOI] [PubMed] [Google Scholar]
- 26.Rabouille C., Kondo H., Newman R., Hui N., Freemont P., Warren G. ( 1998) Cell 92, 603– 610 [DOI] [PubMed] [Google Scholar]
- 27.Decottignies A., Evain A., Ghislain M. ( 2004) Yeast 21, 127– 139 [DOI] [PubMed] [Google Scholar]
- 28.Schuberth C., Richly H., Rumpf S., Buchberger A. ( 2004) EMBO Rep. 5, 818– 824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. ( 1994) Current Protocols in Molecular Biology, John Wiley & Sons, Inc., New York [Google Scholar]
- 30.Sambrook J., Frisch E., Maniatis T. ( 1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31.Guthrie C., Fink G. R. ( 1991) Methods in Enzymology, Academic Press, San Diego [Google Scholar]
- 32.Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. ( 2002) Nucleic Acids Res. 30, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gauss R., Trautwein M., Sommer T., Spang A. ( 2005) Yeast 22, 1– 12 [DOI] [PubMed] [Google Scholar]
- 34.Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. ( 1999) Yeast 15, 963– 972 [DOI] [PubMed] [Google Scholar]
- 35.Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. ( 2004) J. Biol. Chem. 279, 38369– 38378 [DOI] [PubMed] [Google Scholar]
- 36.Medicherla B., Kostova Z., Schaefer A., Wolf D. H. ( 2004) EMBO Rep. 5, 692– 697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bordallo J., Plemper R. K., Finger A., Wolf D. H. ( 1998) Mol. Biol. Cell 9, 209– 222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buschhorn B. A., Kostova Z., Medicherla B., Wolf D. H. ( 2004) FEBS Lett. 577, 422– 426 [DOI] [PubMed] [Google Scholar]
- 39.Biederer T., Volkwein C., Sommer T. ( 1996) EMBO J. 15, 2069– 2076 [PMC free article] [PubMed] [Google Scholar]
- 40.Vashist S., Ng D. T. ( 2004) J. Cell Biol. 165, 41– 52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuberth C. ( 2006) UBX Domain Proteins as a New Family of Cofactors of the AAA ATPase Cdc48. Ph.D. thesis, Ludwig-Maximilians-Universität, Munich [Google Scholar]
- 42.Pan X., Ye P., Yuan D. S., Wang X., Bader J. S., Boeke J. D. ( 2006) Cell 124, 1069– 1081 [DOI] [PubMed] [Google Scholar]
- 43.Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. ( 2000) Cell 101, 249– 258 [DOI] [PubMed] [Google Scholar]
- 44.Meyer H. H., Shorter J. G., Seemann J., Pappin D., Warren G. ( 2000) EMBO J. 19, 2181– 2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knop M., Finger A., Braun T., Hellmuth K., Wolf D. H. ( 1996) EMBO J. 15, 753– 763 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.