Abstract
ATP is known to increase the activity of the type-1 inositol 1,4,5-trisphosphate receptor (InsP3R1). This effect is attributed to the binding of ATP to glycine rich Walker A-type motifs present in the regulatory domain of the receptor. Only two such motifs are present in neuronal S2+ splice variant of InsP3R1 and are designated the ATPA and ATPB sites. The ATPA site is unique to InsP3R1, and the ATPB site is conserved among all three InsP3R isoforms. Despite the fact that both the ATPA and ATPB sites are known to bind ATP, the relative contribution of these two sites to the enhancing effects of ATP on InsP3R1 function is not known. We report here a mutational analysis of the ATPA and ATPB sites and conclude neither of these sites is required for ATP modulation of InsP3R1. ATP augmented InsP3-induced Ca2+ release from permeabilized cells expressing wild type and ATP-binding site-deficient InsP3R1. Similarly, ATP increased the single channel open probability of the mutated InsP3R1 to the same extent as wild type. ATP likely exerts its effects on InsP3R1 channel function via a novel and as yet unidentified mechanism.
Inositol 1,4,5-trisphosphate receptors (InsP3R)3 are a family of large, tetrameric, InsP3-gated cation channels. The three members of this family (InsP3R1, InsP3R2, and InsP3R3) are nearly ubiquitously expressed and are localized primarily to the endoplasmic reticulum (ER) membrane (1–3). Numerous hormones, neurotransmitters, and growth factors bind to receptors that stimulate phospholipase C-induced InsP3 production (4). InsP3 subsequently binds to the InsP3R and induces channel opening. This pathway represents a major mechanism for Ca2+ liberation from ER stores (5). All three InsP3R isoforms are dynamically regulated by cytosolic factors in addition to InsP3 (1). Ca2+ is perhaps the most important determinant of InsP3R activity besides InsP3 itself and is known to regulate InsP3R both positively and negatively (6). ATP, in concert with InsP3 and Ca2+, also regulates InsP3R as do numerous kinases, phosphatases, and protein-binding partners (7–10). This intricate network of regulation allows InsP3R activity to be finely tuned by the local cytosolic environment (9). As a result, InsP3-induced Ca2+ signals can exhibit a wide variety of spatial and temporal patterns, which likely allows Ca2+ to control many diverse cellular processes.
Modulation of InsP3-induced Ca2+ release (IICR) by ATP and other nucleotides provides a direct link between intracellular Ca2+ signaling and the metabolic state of the cell. Metabolic fluctuations could, therefore, impact Ca2+ signaling in many cell types given that InsP3R are expressed in all cells (11, 12). Consistent with this, ATP has been shown to augment IICR in many diverse cell types including primary neurons (13), smooth muscle cells (14), and exocrine acinar cells (15) as well as in immortalized cell lines (16–18). The effects of ATP on InsP3R function do not require hydrolysis because non-hydrolyzable ATP analogues are as effective as ATP (7, 14). ATP is thought to bind to distinct regions in the central, coupling domain of the receptors and to facilitate channel opening (2, 19). ATP is not required for channel gating, but instead, increases InsP3R activity in an allosteric fashion by increasing the open probability of the channel in the presence of activating concentrations of InsP3 and Ca2+ (7, 8, 20).
Despite a wealth of knowledge regarding the functional effects of ATP on InsP3R function, there is relatively little known about the molecular determinants of these actions. ATP is thought to exert effects on channel function by direct binding to glycine-rich regions containing the consensus sequence GXGXXG that are present in the receptors (2). These sequences were first proposed to be ATP-binding domains due to their similarity with Walker A motifs (21). The neuronal S2+ splice variant of InsP3R1 contains two such domains termed ATPA and ATPB. A third site, ATPC, is formed upon removal of the S2 splice site (2, 22). The ATPB site is conserved in InsP3R2 and InsP3R3, while the ATPA and ATPC sites are unique to InsP3R1. Our prior work examining the functional consequences of mutating these ATP-binding sites has yielded unexpected results. For example, mutating the ATPB site in InsP3R2 completely eliminated the enhancing effects of ATP on this isoform while mutating the analogous site in InsP3R3 failed to alter the effects of ATP (23). This indicated the presence of an additional locus for ATP modulation of InsP3R3. In addition, mutation of the ATPC in the S2− splice variant of InsP3R1 did not alter the ability of ATP to modulate Ca2+ release, but instead impaired the ability of protein kinase A to phosphorylate Ser-1755 of this isoform (22).
The ATPA and ATPB sites in InsP3R1 were first identified as putative nucleotide-binding domains after the cloning of the full-length receptor (24). Early binding experiments with 8-azido-[α-32P]ATP established that ATP cross-linked with receptor purified from rat cerebellum at one site per receptor monomer (19). Later, more detailed, binding experiments on trypsinized recombinant rat InsP3R1 showed cross-linking of ATP to two distinct regions of the receptor that corresponded with the ATPA and ATPB sites (17). We and others (16, 22, 23) have also reported the binding of ATP analogues to purified GST fusions of small regions of InsP3R1 surrounding the ATPA and ATPB sites. It is widely accepted, in the context of the sequence similarity to Walker A motifs and biochemical data, that the ATPA and ATPB sites are the loci where ATP exerts its positive functional effects on InsP3R1 function (1–3, 16). Furthermore, the higher affinity of the ATPA site to ATP is thought to confer the higher sensitivity of InsP3R1 to ATP versus InsP3R3, which contains the ATPB site exclusively (25, 26). The purpose of this study, therefore, was to examine the contributions of the ATPA and ATPB sites to ATP modulation of the S2+ splice variant of InsP3R1. We compared the effects of ATP on InsP3R1 and on ATP-binding site mutated InsP3R1 using detailed functional analyses in permeabilized cells and in single channel recordings. Here we report that InsP3R1 is similar to InsP3R3 in that ATP modulates IICR even at maximal InsP3 concentrations and that neither the ATPA nor the ATPB site is required for this effect.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
An expression construct harboring cDNA for rat InsP3R1 was used as templates for mutagenesis. A two-step QuikChange (Stratagene, La Jolla, CA) mutagenesis strategy (27) was used to create the ΔATPA and ΔATPB mutants. The ATPA mutant was generated by introducing G5318C, G5324C, G5330C, G5333C, and G5339C point mutations and the ΔATPBB mutant was made by introducing G6047C, G6053C, and G6062C point mutations into the InsP3R1 cDNA. These mutations code for amino acid substitutions: G1773A, G1775A, G1777, G1778A, and G1780A for ΔATPA and G2016A, G2018, and G2021 for ΔATPB. Correct incorporation of the mutations was confirmed by DNA sequencing (GeneWiz, South Plainfield, NJ).
Creation of Stable InsP3R1-expressing DT40-3KO Cell Lines
Wild type and mutated InsP3R1 were linearized with NruI and introduced into DT40-3KO cells by nucleofection using solution T and program B23 as per the manufacturer's instructions (Amaxa, Cologne, Germany). The cells were incubated in growth medium for 24 h prior to dilution in selection medium containing 2 mg/ml Geneticin (Invitrogen). Cells were then seeded into 96-well tissue culture plates at ∼1000 cells/well and incubated in selection medium for at least 7 days. Wells exhibiting growth after the selection period were picked for expansion.
Permeabilized Cell Ca2+ Measurements
Measurements of ER luminal Ca2+ were performed essentially as described previously (22, 23, 28). InsP3R1-expressing stable DT40-3KO cells were loaded with 20 μm furaptra-AM (Teflabs, Austin, TX) at 39 °C for 30 min in a HEPES-buffered physiological saline solution (HEPES-PSS) containing 5.5 mm glucose, 137 mm NaCl, 0.56 mm MgCl2, 4.7 mm KCl, 1 mm Na2HPO4, 10 mm HEPES (pH 7.4), 1.2 mm CaCl2, and 1% (w/v) bovine serum albumin. Furaptra-loaded cells were permeabilized by superfusion for 1–2 min with 40 μm β-escin in intracellular medium (ICM) containing 125 mm KCl, 19 mm NaCl, 10 mm HEPES, 1 mm EGTA (pH 7.3). Permeabilized cells were then washed in ICM without β-escin for 15 min to facilitate removal of cytosolic dye, and the cells were then superfused in ICM containing 1.4 mm MgCl2, Na2ATP, and 0.650 mm CaCl2 (free [Ca2+] of 200 nm, calculated using Maxchelator) to load the intracellular stores. The free [Ca2+] was subsequently maintained at a constant 200 nm throughout all experimental maneuvers. The free Ca2+ concentration was verified by fluorescent measurement of free Ca2+ in solutions. Prior to application of InsP3, the cells were superfused in ICM without MgCl2 for 1 min to disable SERCA activity. The unidirectional flux of Ca2+ upon InsP3 application was then monitored in the same solution containing various concentrations of InsP3 and ATP by monitoring the emission of the dye above 505 nm following excitation at 340 and 380 nm (exposure for 20 ms, once per second) using a TILL Photonics imaging system. Following removal of InsP3, refilling of the stores to allow repeated stimulations was accomplished by superfusion of ICM containing MgCl2 and ATP. Ca2+ release events were averages of the 30–50 cells in a field of view. Rates of Ca2+ release were estimated from these average responses by fitting the initial 30-s period of decreasing fluorescence to a single exponential function (GraphPad Prism, San Diego, CA).
Single InsP3R1 Channel Measurements in Isolated DT40 Nuclei
Nuclear Preparations
Isolated DT40 nuclei were prepared by homogenization. Homogenization buffer (HB) contained 250 mm sucrose, 150 mm KCl, 3 μm β-mercaptoethanol, 10 mm Tris, 1 mm phenylmethylsulfonyl fluoride, pH 7.5 with a complete protease inhibitor tablet (Roche, Nutley, NJ). Cells were washed and resuspended in HB prior to nuclear isolation using a RZR 2021 homogenizer (Heidolph Instruments) with 25 strokes at 1200 rpm. 3 μl of nuclear suspension were placed in 3 ml of bath solution (BS), which contained 140 mm KCl, 10 mm HEPES, 500 μm BAPTA, and 246 nm free Ca2+, pH 7.1. Nuclei were allowed to adhere to a plastic culture dish for 10 min.
Patch Clamp Experiments
Single InsP3R1 currents were measured in the on-nucleus voltage clamp configuration using PClamp 9 and an Axopatch 200B amplifier (MDS Analytical Technologies, Toronto, Canada). Currents were measured at 20 kHz sampling rate and filtered at 5 kHz with a low-pass 4-pole bessel filter. Pipette solution (PS) contained 10 μm InsP3, 140 mm KCl, 10 mm HEPES, 100 μm BAPTA, 200 nm free Ca2+, and 0–5 mm ATP, pH 7.1. Measurements were made at −100 mV following establishment of a >5 GΩ seal. Data for current-voltage relationships were collected by stepping voltage from 0 mV to potentials between −100 and 100 mV in 10 mV increments. Typically, 1 out of every 5 patches contained reproducible single InsP3R1 channel activity. No activity was evident in patches from DT40-3KO nuclei under standard pipette conditions with 5 mm ATP (40 patches, 407 min of total recording time). Additionally, there was no single channel activity in nuclei from cells expressing InsP3R1 in the absence of InsP3 in the recording pipette (37 patches, 412 min of total recording time).
Data Analysis
Analyses were performed using the event detection protocol in Clampfit 9. Channel openings were detected by half-threshold crossing criteria. We assumed that the number of channels in any particular patch is represented by the maximum number of discrete stacked events observed during the experiment. Even at low Po, stacking events were evident (data not shown). Only patches with 1 apparent channel were considered for analysis of open probability, and mean open and closed times. The slope conductances were determined from the linear fits of the current-voltage relationships where g = Ik/(V − Vk). Equation parameters were estimated using a non-linear, least squares algorithm.
RESULTS
ATP Enhances InsP3-induced Ca2+ Release in DT40-3KO Cells Expressing Rat InsP3R1
We generated a DT40 triple knock-out (DT40-3KO) cell line stably expressing the rat S2+ InsP3R1 splice isoform (DT40-InsP3R1) containing only the putative ATPA and ATPB sites, to study the effects of ATP on this isoform expressed in isolation. These cells allow the unambiguous examination of isoform-dependent InsP3R function because all three endogenous InsP3R isoforms have been deleted via homologous recombination (29). Furthermore, stable cell lines harboring mutated receptors allow the examination of the effects of removing putative sites for modulation (28).
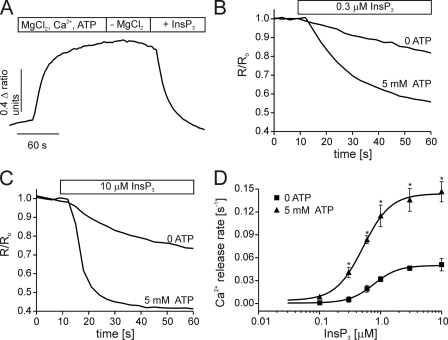
We employed ER calcium measurements in permeabilized DT40-InsP3R1 cells to examine the effect of ATP on IICR in the presence of various InsP3 and ATP concentrations. Similar to other cell lines expressing predominantly InsP3R1 (16) and DT40 cells expressing chicken InsP3R1 in isolation (30), ATP clearly potentiates IICR in DT40-InsP3R1 cells (Fig. 1). Ca2+ release rates were measured over a range of InsP3 concentrations in the presence or absence of 5 mm ATP. As shown in Fig. 1, ATP was required for the maximal rate of IICR in DT40-InsP3R1 cells. This effect of ATP on IICR from InsP3R1 is similar to results obtained from DT40 cells expressing rat InsP3R3 in isolation, but distinctly different from cells expressing InsP3R2 (23). While InsP3R1 and InsP3R3 are regulated by ATP at maximal InsP3 concentrations, InsP3R2 is only modulated by ATP at submaximal InsP3.
FIGURE 1.
ATP enhances IICR from DT40-3KO cells stably expressing rat InsP3R1. ER luminal Ca2+ measurements were obtained from cells stably expressing rat S2+ InsP3R1. Cells were loaded with furaptra and permeabilized with β-escin as described under “Experimental Procedures.” A shows the experimental paradigm. ER stores are loaded with Ca2+ by superfusion with a solution containing Mg2+, Ca2+, and ATP. A solution lacking Mg2+ is then applied to prevent SERCA activity. InsP3 initiates the release of Ca2+ from the ER. B and C show representative traces from cells treated with low (0.1 μm, in B) or high (10 μm, in C) InsP3 in the presence or absence of 5 mm ATP. D shows the concentration-response relationship for InsP3 in the absence (squares) or presence (triangles) of 5 mm ATP. Each point is the mean ± S.E. of at least four separate experiments. The solid lines are the fits of the data. Ca2+ release rates were calculated by fitting the average time course from the first 30 s of InsP3 application from 20–50 cells to a single exponential. (*, p ≤ .05; Student's unpaired t test).
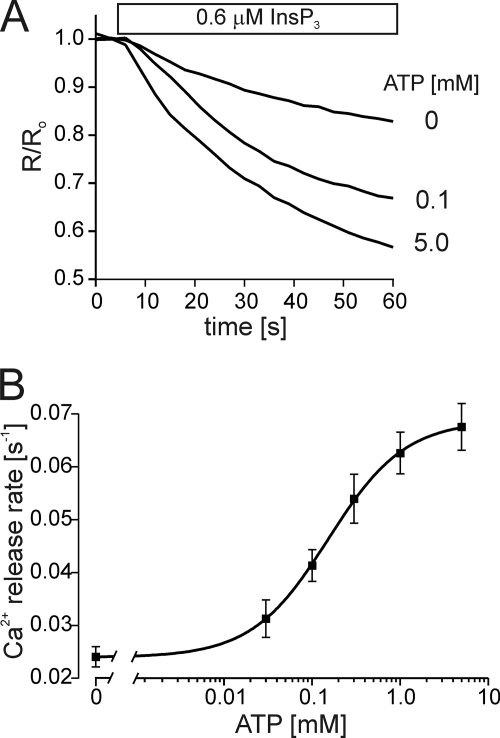
Our prior results also established that InsP3R2 was more sensitive to ATP than InsP3R3 (23). To determine how InsP3R1 compared with the other two isoforms in this regard, we next determined the sensitivity of InsP3R1 to ATP modulation by stimulating IICR at a set InsP3 concentration over a range of ATP concentrations. Under these conditions, ATP increased InsP3R1 activity with an EC50 of ∼150 μm (Fig. 2). The sensitivity of InsP3R1 to ATP is therefore lower than that of InsP3R2, but higher than that of InsP3R3.
FIGURE 2.
The sensitivity of InsP3R1 to ATP. ER luminal Ca2+ measurements were obtained from permeabilized DT40-InsP3R1 cells in the presence of 0.6 μm InsP3 and a range of ATP concentrations. A shows representative Ca2+ release events in the presence of 0, 0.1 mm, and 5 mm ATP as indicated. B shows a concentration-response relationship for ATP. Each point is the mean ± S.E. of at least four separate experiments. The solid line is the fit for the data and yields an EC50 of 150 μm.
The ATPA and ATPB Sites Are Not Required for ATP Modulation of InsP3R1
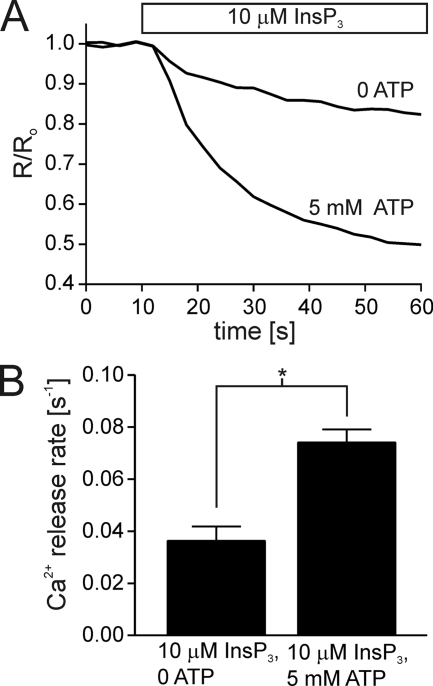
The ATPB site is the only ATP-binding site conserved in all three mammalian InsP3R isoforms (2). Even though this site is conserved in InsP3R2 and InsP3R3, the effects of mutating the sites in these isoforms were dramatically different. Abrogation of the ATPB site in InsP3R2 eliminated the enhancing effects of ATP, but ATP was able to modulate InsP3R3 even after the ATPB site was disrupted (23). We next generated a stable cell line expressing ATPB-mutated InsP3R1 (DT40-InsP3R1-ΔATPB) to test whether this site was required for ATP modulation of InsP3R1. In our earlier studies, mutation of the central glycine to alanine was sufficient to completely abrogate specific TNP-ATP binding to a GST fragment containing the ATPB site (22). In the present studies, all three glycine residues present in the ATPB site (Gly-2016, Gly-2018, Gly-2021) were mutated to alanine residues to ensure that ATP binding would be eliminated unquestionably. Cells expressing this mutated receptor were tested for ATP modulation in the permeabilized DT40 cells under identical experimental conditions as described for the wild type receptor. Similar to experiments with wild type InsP3R1, ATP (5 mm) increased the rate of IICR in DT40-InsP3R1-ΔATPB stimulated with 10 μm InsP3 (Fig. 3).
FIGURE 3.
Ablation of the ATPB site fails to eliminate the enhancing effects of ATP on InsP3R1. A stable cell line was generated expressing InsP3R1-ΔATPB (G2016A, G2018A, and G2021A). A shows representative ER Ca2+ measurements from permeabilized DT40-InsP3R1-ΔATPB cells treated with 10 μm InsP3 in the absence or presence of 10 μm InsP3. B shows the mean ± S.E. for each condition from five separate experiments (*, p ≤ .05; Student's unpaired t test).
The results presented in Fig. 3 suggest that the ATPB site is not required for ATP modulation of InsP3R1. One possible explanation for this observation is that the presence of the ATPA site, still present in the mutated receptor, might mask any possible effects of the ATPB site mutation. This would be expected based on results from bilayer measurements of the opisthotonos mutant InsP3R1 (InsP3R1-opt) (25). This spontaneously mutated receptor is devoid of a stretch of amino acids (Gly-1732 to Gln-1839), which contains the ATPA (Gly-1773 to Gly-1780) as well as the PKA phosphorylation site (Ser-1755) (31). When this mutated receptor was incorporated into bilayers it exhibited a diminished sensitivity to ATP compared with the wild type receptor (25). This result led the authors to conclude that the ATPA site conferred the high sensitivity of InsP3R1 to ATP whereas the ATPB site conferred a lower sensitivity to ATP in the absence of the ATPA site.
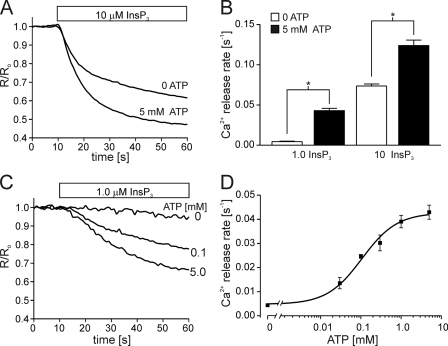
We tested whether the ATPA site is indeed determining the functional effects of ATP in the DT40-InsP3R1-ΔATPB cells, using a stable cell line expressing InsP3R1 mutated at both the ATPA and ATPB sites (DT40-InsP3R1-ΔATPA/B). Again, mutation of 2 glycine residues to alanine in a GST fragment harboring the ATPA was earlier shown to completely abrogate TNP-ATP binding to the ATPA site (22). In the present study, all glycine residues (5 in total) in the motif were mutated to alanine. This mutant receptor is, therefore, devoid of any putative ATP-binding sites. As shown in Fig. 4, however, ATP was able to increase the rate of IICR from permeabilized DT40-InsP3R1-ΔATPA/B. This effect was evident over a range of InsP3 concentrations similar to the effects of ATP on IICR from DT40-InsP3R1 and DT40-InsP3R1-ΔATPB cells. ATP also increased IICR in DT40-InsP3R1-ΔATPA/B with an EC50 of 100 μm, which was similar to the sensitivity of wild type InsP3R1 (EC50 of 150 μm). These results indicate that the known ATP-binding sites in InsP3R1 are not required for ATP modulation of this isoform. This is in contrast to the results described above for InsP3R1-opt and could reflect the different methodologies to obtain the data (lipid bilayers measurements versus ER Ca2+ measurements). Alternatively, the results may suggest a role for the other residues removed in the InsP3R1-opt in conferring ATP sensitivity of InsP3R1. It is important to note, however, that the opt mutation had numerous effects on the single channel properties of InsP3R1. For example, changes in the single channel conductance, Ca2+ regulation, and the sensitivity to ATP were apparently induced by the opt deletion (25).
FIGURE 4.
IICR is enhanced by ATP in cells expressing Walker A site-deficient InsP3R1. A stable cell line was generated expressing InsP3R1-ΔATPA/B (G1773A, G1775A, G1777A, G1778A, G1780A, G2016A, G2018A, G2021A). A shows a representative ER Ca2+ measurement from permeabilized DT40-InsP3R1-ΔATPA/B treated with 10 μm InsP3 in the presence or absence of 5 mm ATP as indicated. B shows the mean ± S.E. of four experiments measuring the effects of 5 mm ATP on Ca2+ release rates induced by 1 μm InsP3 or 10 μm InsP3. C shows the time course of Ca2+ release induced by 1 μm InsP3 in the presence of 0, 0.1, or 5 mm ATP. D shows the concentration-response relationship for ATP. Each point is the mean ± S.E. of at least four experiments, and the solid line is the fit of the data and yields an EC50 for ATP of 100 μm (*, p ≤ .05; Student's unpaired t test).
ATP Enhances the Single Channel Open Probability of InsP3R1-ΔATPA/B
Mutating the ATPA and ATPB sites in InsP3R1 did not alter the ability of ATP to regulate IICR from permeabilized cells expressing this mutant. Macroscopic measurements from pools of permeabilized cells, as described above, may not provide the resolution to detect subtle changes in ATP modulation of InsP3R1-ΔATPB. Because ATP is thought to exert effects on InsP3R activity by altering the channel open probability (7, 8), we next compared the effects of ATP on the single channel properties of wild type and InsP3R1-ΔATPA/B.
We utilized the nuclear membrane patch-clamp methodology developed by Foskett and co-workers (1, 32, 33) to analyze the effects of ATP on wild type and ΔATPA/B-mutated InsP3R1. In this configuration, high resistance seals are made between the pipette and the outer nuclear membrane. Nuclei were isolated from DT40 cell lines stably expressing InsP3R1 as described under “Experimental Procedures.” InsP3R single channels were readily detected in these preparations in the presence of 10 μm InsP3, ∼200 nm free Ca2+ and 5 mm ATP. No channel activity was evident if InsP3 was excluded from the pipette solution. Similarly, there was no detectable activity in nuclei prepared from DT40-3KO cells, indicating that the single channels were, indeed, recorded as a consequence of InsP3R activity.
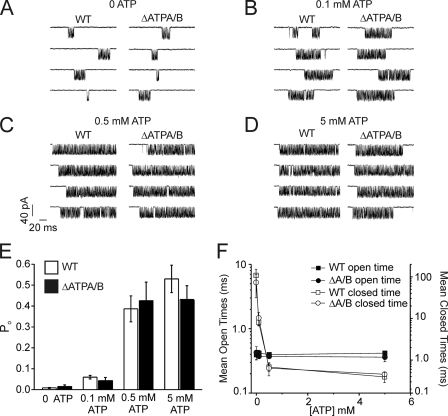
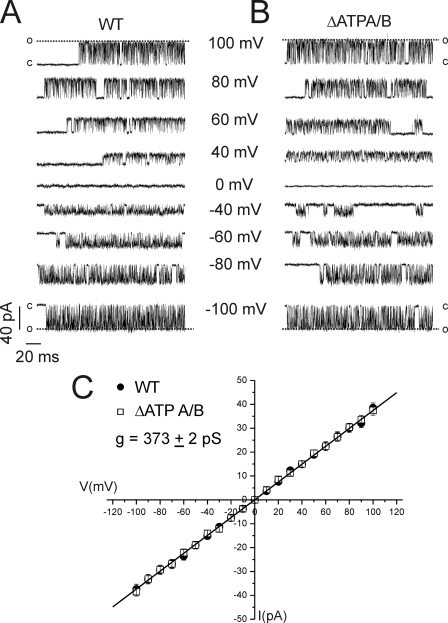
Fig. 5 shows representative 200-ms sweeps from patch recordings of nuclei prepared from DT40-InsP3R1 and DT40-InsP3R1-ΔATPA/B cells in the presence of 10 μm InsP3 and at various concentrations of ATP. Similar to prior results (7, 8), no channel activity was stimulated by ATP in the absence of InsP3 in 40 separate patches (>400 min of recording time). This indicates that ATP alone is not sufficient to stimulate channel activity. Channel open probability (Po) induced by 10 μm InsP3 was, however, increased by more than 5-fold with the inclusion of 5 mm ATP in the pipette solution (Fig. 5). Similar to the effects of ATP on IICR in the permeabilized cell experiments above, 5 mm ATP increased the Po of InsP3R1-ΔATPA/B channels to the same extent as wild type (Fig. 5). Wild type and mutated InsP3R1 Po was also increased by ATP in a concentration-dependent manner in both preparations. The major effect of increasing [ATP] on both the WT and ΔATPA/B receptor was to decrease the mean closed times (Fig. 5F). Increasing [ATP] had no effect on the mean open times of either receptor (Fig. 5F). Disruption of these sites had no additional observable effects on channel function as evidenced by similar single channel conductances of the two receptors (Fig. 6). These results clearly demonstrate enhancing effects of ATP on InsP3R1-ΔATPA/B single channel function and further strengthen our contention that the Walker A-type ATP-binding motifs in InsP3R1 are dispensable for channel modulation by ATP.
FIGURE 5.
ATP increases the Po of InsP3R1-ΔATPA/B. Single InsP3R channel measurements on isolated DT40 nuclei were performed as described under “Experimental Procedures.” A–D show representative 200 ms sweeps from nuclei isolated from DT40-InsP3R1 (WT) or DT40-InsP3R1-ΔATPA/B (ΔATPA/B) cells. A shows recordings from 4 nuclei from each cell line in the absence of ATP. B–D shows results from nuclei (4 different cells for each condition) where 0.1 mm, 0.5 mm, or 5 mm ATP was included in the recording pipette as indicated. The holding potential was −100 mV for all experiments. E shows the mean ± S.E. Po recorded under the indicated conditions from at least 4 cells for each ATP concentration. F shows the mean ± S.E. open (black) and closed (white) times for InsP3R1-WT (square) and InsP3R1-ΔATPA/B single channels (circles).
FIGURE 6.
Elimination of ATPA and ATPB is without effect on the single channel conductance of InsP3R1. A and B shows representative single channel recordings at various holding potentials in nuclei prepared from WT (A) or ΔATPA/B (B) InsP3R1-expressing cells. C shows the current-voltage relationships obtained from WT (closed circles) or ΔATPA/B (open circles) channels. Each point is the mean ± S.E. of current amplitudes from at least four separate nuclei at each holding potential.
Unlike the opt mutant, therefore, the targeted deletion of the ATPA site had no effect on the single channel properties of InsP3R1. One explanation for the lack of an effect in the ΔATPA/B receptor is that the mutations did not completely eliminate ATP binding. This is not likely given our prior results examining TNP-ATP binding to GST fusion proteins of InsP3R1 fragments containing the ATPA or ATB sites (22). Fusion proteins containing wild type ATPA or ATPB sequences readily bound TNP-ATP, while purified fragments harboring mutated Walker-A motifs completely eliminated TNP-ATP binding. Another possibility is that some ATP binding is contained in the region deleted in the opt mutant. Again, this is improbable since the binding experiments described above were performed on purified InsP3R1 fragments that spanned the opt deletion region. This indicates that ATP binding to this region of InsP3R1 is dependent on the presence of the ATPA site. Despite clear, demonstrable binding, the present data lead us to the conclusion that this binding is unlikely to mediate any functional effect.
DISCUSSION
InsP3R are regulated by multiple cytosolic factors including ATP and related nucleotides (34). The combined contributions of InsP3, Ca2+, ATP, and other interacting regulatory mechanisms likely facilitates a multitude of possible activation states of the receptor. InsP3R activity and the resulting Ca2+ signals can, therefore, be tightly controlled and tuned to the specific needs of a given cell. A complete understanding of the molecular underpinnings of these regulatory mechanisms will permit insight into the manner in which cells control intracellular Ca2+ signaling via modulation of InsP3R. This study, combined with the results from our prior work (22, 23), allows the first direct comparison of the effects of ATP on mammalian wild type versus ATP-binding site-mutated InsP3R family members expressed in isolation. The three isoforms exhibit some similarities in their respective responses to ATP, but there are also striking differences with regard to the manner in which ATP modulates InsP3R function. For example, while Ca2+ release activity is increased by ATP in all three isoforms, the major defining feature is that ATP regulates IICR from InsP3R1 and InsP3R3 at all InsP3 concentrations, but augments IICR from InsP3R2 exclusively at submaximal [InsP3].
Another major isoform-dependent difference is the range of ATP sensitivities displayed by the three isoforms. InsP3R2 exhibits the highest sensitivity with an EC50 for ATP of ∼40 μm (23), followed by InsP3R1 (EC50 ∼ 150 μm, Fig. 2), and InsP3R3 (EC50 ∼ 400 μm (23)). The differences in the relative sensitivities of the three InsP3R isoforms to ATP would be important in cell types that express a significant majority of one isoform over the others. In fact, our prior work has demonstrated that the high sensitivity of InsP3R2 to ATP dominates in cell types that predominantly express this isoform including isolated mouse pancreatic acinar cells and cultured AR42J cells (15). Cells that express mostly InsP3R3, such as pancreatic acinar cells from InsP3R2 knock-out mice and RinM5F cells, exhibit a 10-fold higher EC50 for ATP, indicating sensitivities more in line with that of InsP3R3 (15).
All mammalian cells express some amount of InsP3R1 (11). Neurons are unique in expressing this isoform in a large majority over InsP3R2 and InsP3R3 with InsP3R1 being better than 95% of total neuronal InsP3R (11). There are a range of reported ATP sensitivities for InsP3R1. Purified cerebellar InsP3R, which is almost exclusively InsP3R1, is reported to display a high sensitivity to ATP (half-maximal value of 40 μm) (7). More recent experiments under similar conditions with recombinant InsP3R1 expressed in Sf9 cell nuclei displayed sensitivity to ATP more in line with the present data (half-maximal value of 150 μm in (26) and a half-maximal value of 240 μm in Ref. 25). Our experimental design is similar to these bilayers studies in that ATP sensitivity was determined at set InsP3 and Ca2+ concentrations. An additional report established a dissociation coefficient of 270 μm for ATP activation of Xenopus InsP3R1 using nuclear patch clamp recordings (8). The sensitivity to ATP in this study was established by analyzing the effects of ATP on the Ca2+-dependent activation of InsP3R single channel activity.
The reported sensitivities of InsP3R1 to ATP are well below the presumed total ATP concentration in healthy neurons, which ranges from 3–10 mm (35). The majority of cytosolic ATP is expected to be in complex with Mg2+. As has been noted previously, however, the reported sensitivities of InsP3R1 to ATP are more in line with the predicted levels of free ATP in cells (8). This distinction is important considering that, while MgATP was as effective as free ATP in enhancing InsP3R activity in some studies (7, 16), it was ineffective in others (8, 20). Even if the sensitivity of InsP3R1 allows it to be maximally stimulated by physiological concentrations of ATP, cellular ATP levels have been shown to fall below 100 μm in cerebral cortex tissue in response to acute hypoxic treatments (36). InsP3R1 activity would therefore be expected to be impaired under these conditions.
The major finding presented here, is that the known ATP-binding domains present in InsP3R1 are not the major determinants of ATP modulation of this isoform. This result is in contrast to the case of InsP3R2 in which the ATPB site is required for ATP modulation (23). The identity of the functionally relevant ATP-binding domains in InsP3R1 and InsP3R3, therefore, remain to be determined. The most complete analysis of ATP binding to InsP3R1 demonstrated that 8-azido-[α32-P]ATP cross-linked with InsP3R1 in at least two distinct regions of trypsin-digested InsP3R1 (17). Similarly, the same study reported that ATP cross-linked with at least one region of InsP3R3. The identified regions corresponded to the areas containing the ATPA and ATPB sites in InsP3R1 and the ATPB site in InsP3R3 leading the authors to conclude that these were the only ATP-binding sites present in the receptors (17). The results were not, however, confirmed by performing the ATP cross-linking assays on mutated receptors. These types of experiments will need to be attempted to determine if additional ATP-binding sites are present on ATPA- and ATPB-deficient InsP3R1 and InsP3R3. Of note, recent work has demonstrated that incubation of InsP3R1 with Ca2+ to promote receptor activation causes large pockets of the receptor to become exposed (37). These structural changes in response to activation may lead to the exposure of additional ATP-binding sites. Cross-linking 8-azido-[α32-P]ATP to InsP3R1 and InsP3R3 under these activating conditions may help uncover additional ATP-binding sites not apparent in the original binding studies. Because the ATPB mutated InsP3R2 is insensitive to ATP modulation, a functional comparison of chimerical InsP3R may also yield insights into the regions required for ATP modulation. Regions of InsP3R1 could be transposed onto the ATPB-mutated InsP3R2 backbone to identify areas of the receptor that rescue ATP sensitivity. These approaches may also help determine if the functionally relevant ATP-binding domain is present in the InsP3R1 sequence or contained in one of the many protein-binding partners of the receptor.
The ultimate identification of the functionally important ATP-binding site(s) in InsP3R1 would likely not be a trivial undertaking, but could yield insight into novel mechanisms of ATP modulation of ion channels besides InsP3R. Most notably, members of the related ryanodine receptor (RyR) family are known to be modulated by ATP (38). Similar to the InsP3R, ATP analogues can cross-link with distinct regions of RyR1 (39). Furthermore, electron-spin resonance (ESR) spectroscopy on skeletal muscle RyR also demonstrated ATP binding (40). In this study, the authors determined that a maximum of 8 spin-labeled ATP analogues bound to the tetrameric receptor, thus predicting two ATP-binding sites per RyR monomer. Sequence analysis of the RyR predicts at least 8 putative ATP-binding domains (41). If there are indeed two ATP-binding sites per monomer as suggested by the ESR studies, then a subset of these motifs or alternatively, equally possible, none of the motifs at all may confer ATP sensitivity. Similar to our current study, mutations in putative ATP-binding sites in RyR1 did not alter perturb the effects of ATP on channel function (41). Given the functional and structural homology of InsP3R with RyR, the identification of novel ATP-binding domains in InsP3R may shed light on the molecular determinants of ATP regulation of RyR.
In summary, this study represents the completion of mutagenic analyses of the known ATP-binding sites in the InsP3R family. To date, only two of the known ATP-binding sites have been assigned a function. ATP binding to the ATPB site of InsP3R2 confers ATP sensitivity to this isoform (23) and ATP binding to the ATPC site of the S2− isoform of InsP3R1 facilitates PKA phosphorylation of Ser-1755 of this splice variant (22). The mechanistic details and functional consequences of ATP modulation of InsP3R are isoform-dependent. Any role of ATP in regulating InsP3-induced Ca2+ signaling in cells will, therefore, be dependent on which isoforms are expressed. The emphasis of further work will be to identify the ATP-binding sites responsible for ATP modulation of InsP3R1 and InsP3R3.
Acknowledgments
We thank Dr. T. Kurosaki for providing the DT40-3KO cells, Dr. S. Joseph for providing the rat InsP3R1 expression construct, and Lyndee Knowlton for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-DK054568 and R01-DE016999 (to D. I. Y.).
- InsP3R
- InsP3 receptor
- InsP3
- inositol 1,4,5-trisphosphate
- IICR
- InsP3-induced Ca2+ release
- Po
- open probability
- RyR
- ryanodine receptor
- GST
- glutathione S-transferase
- WT
- wild type
- ER
- endoplasmic reticulum
- TNP-ATP
- 2′(3′)-O-(2,4,6-trinitrophenyl)adenosine 5′-triphosphate.
REFERENCES
- 1.Foskett J. K., White C., Cheung K. H., Mak D. O. ( 2007) Physiol. Rev. 87, 593– 658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel S., Joseph S. K., Thomas A. P. ( 1999) Cell Calcium 25, 247– 264 [DOI] [PubMed] [Google Scholar]
- 3.Bezprozvanny I. ( 2005) Cell Calcium 38, 261– 272 [DOI] [PubMed] [Google Scholar]
- 4.Berridge M. J., Irvine R. F. ( 1984) Nature 312, 315– 321 [DOI] [PubMed] [Google Scholar]
- 5.Berridge M. J., Bootman M. D., Roderick H. L. ( 2003) Nat. Rev. Mol. Cell Biol. 4, 517– 529 [DOI] [PubMed] [Google Scholar]
- 6.Bezprozvanny I., Watras J., Ehrlich B. E. ( 1991) Nature 351, 751– 754 [DOI] [PubMed] [Google Scholar]
- 7.Bezprozvanny I., Ehrlich B. E. ( 1993) Neuron 10, 1175– 1184 [DOI] [PubMed] [Google Scholar]
- 8.Mak D. O., McBride S., Foskett J. K. ( 1999) J. Biol. Chem. 274, 22231– 22237 [DOI] [PubMed] [Google Scholar]
- 9.Patterson R. L., Boehning D., Snyder S. H. ( 2004) Annu. Rev. Biochem. 73, 437– 465 [DOI] [PubMed] [Google Scholar]
- 10.Mikoshiba K. ( 2007) J. Neurochem. 102, 1426– 1446 [DOI] [PubMed] [Google Scholar]
- 11.Wojcikiewicz R. J. ( 1995) J. Biol. Chem. 270, 11678– 11683 [DOI] [PubMed] [Google Scholar]
- 12.Taylor C. W., Genazzani A. A., Morris S. A. ( 1999) Cell Calcium 26, 237– 251 [DOI] [PubMed] [Google Scholar]
- 13.Kaplin A. I., Snyder S. H., Linden D. J. ( 1996) J. Neurosci. 16, 2002– 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iino M. ( 1991) J. Gen. Physiol. 98, 681– 698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park H. S., Betzenhauser M. J., Won J. H., Chen J., Yule D. I. ( 2008) J. Biol. Chem. 283, 26081– 26088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maes K., Missiaen L., De Smet P., Vanlingen S., Callewaert G., Parys J. B., De Smedt H. ( 2000) Cell Calcium 27, 257– 267 [DOI] [PubMed] [Google Scholar]
- 17.Maes K., Missiaen L., Parys J. B., De Smet P., Sienaert I., Waelkens E., Callewaert G., De Smedt H. ( 2001) J. Biol. Chem. 276, 3492– 3497 [DOI] [PubMed] [Google Scholar]
- 18.Deleted in proof
- 19.Maeda N., Kawasaki T., Nakade S., Yokota N., Taguchi T., Kasai M., Mikoshiba K. ( 1991) J. Biol. Chem. 266, 1109– 1116 [PubMed] [Google Scholar]
- 20.Mak D. O., McBride S., Foskett J. K. ( 2001) J. Gen. Physiol. 117, 447– 456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker J. E., Saraste M., Runswick M. J., Gay N. J. ( 1982) EMBO J. 1, 945– 951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner L. E., 2nd, Betzenhauser M. J., Yule D. I. ( 2006) J. Biol. Chem. 281, 17410– 17419 [DOI] [PubMed] [Google Scholar]
- 23.Betzenhauser M. J., Wagner L. E., 2nd, Iwai M., Michikawa T., Mikoshiba K., Yule D. I. ( 2008) J. Biol. Chem. 283, 21579– 21587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furuichi T., Yoshikawa S., Miyawaki A., Wada K., Maeda N., Mikoshiba K. ( 1989) Nature 342, 32– 38 [DOI] [PubMed] [Google Scholar]
- 25.Tu H., Miyakawa T., Wang Z., Glouchankova L., Iino M., Bezprozvanny I. ( 2002) Biophys. J. 82, 1995– 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tu H., Wang Z., Nosyreva E., De Smedt H., Bezprozvanny I. ( 2005) Biophys. J. 88, 1046– 1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W., Malcolm B. A. ( 2002) Methods Mol. Biol. 182, 37– 43 [DOI] [PubMed] [Google Scholar]
- 28.Betzenhauser M. J., Wagner L. E., 2nd, Won J. H., Yule D. I. ( 2008) Methods 46, 177– 182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugawara H., Kurosaki M., Takata M., Kurosaki T. ( 1997) EMBO J. 16, 3078– 3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyakawa T., Maeda A., Yamazawa T., Hirose K., Kurosaki T., Iino M. ( 1999) EMBO J. 18, 1303– 1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Street V. A., Bosma M. M., Demas V. P., Regan M. R., Lin D. D., Robinson L. C., Agnew W. S., Tempel B. L. ( 1997) J. Neurosci. 17, 635– 645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boehning D., Joseph S. K., Mak D. O., Foskett J. K. ( 2001) Biophys. J. 81, 117– 124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mak D. O., Foskett J. K. ( 1994) J. Biol. Chem. 269, 29375– 29378 [PubMed] [Google Scholar]
- 34.Foskett J. K., Mak D. O. ( 2004) Biol. Res. 37, 513– 519 [DOI] [PubMed] [Google Scholar]
- 35.Erecińska M., Silver I. A. ( 1994) Prog. Neurobiol. 43, 37– 71 [DOI] [PubMed] [Google Scholar]
- 36.Abe K., Kogure K., Yamamoto H., Imazawa M., Miyamoto K. ( 1987) J. Neurochem. 48, 503– 509 [DOI] [PubMed] [Google Scholar]
- 37.Anyatonwu G., Joseph S. K. ( 2009) J. Biol. Chem. 284, 8093– 8102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meissner G. ( 1994) Annu. Rev. Physiol. 56, 485– 508 [DOI] [PubMed] [Google Scholar]
- 39.Zarka A., Shoshan-Barmatz V. ( 1993) Eur. J. Biochem. 213, 147– 154 [DOI] [PubMed] [Google Scholar]
- 40.Dias J. M., Szegedi C., Jóna I., Vogel P. D. ( 2006) Biochemistry 45, 9408– 9415 [DOI] [PubMed] [Google Scholar]
- 41.Du G. G., Oyamada H., Khanna V. K., MacLennan D. H. ( 2001) Biochem. J. 360, 97– 105 [DOI] [PMC free article] [PubMed] [Google Scholar]