Abstract
KatG (catalase-peroxidase) in Mycobacterium tuberculosis is responsible for activation of isoniazid (INH), a pro-drug used to treat tuberculosis infections. Resistance to INH is a global health problem most often associated with mutations in the katG gene. The origin of INH resistance caused by the KatG[S315G] mutant enzyme is examined here. Overexpressed KatG[S315G] was characterized by optical, EPR, and resonance Raman spectroscopy and by studies of the INH activation mechanism in vitro. Catalase activity and peroxidase activity with artificial substrates were moderately reduced (50 and 35%, respectively), whereas the rates of formation of oxyferryl heme:porphyrin π-cation radical and the decay of heme intermediates were ∼2-fold faster in KatG[S315G] compared with WT enzyme. The INH binding affinity for the resting enzyme was unchanged, whereas INH activation, measured by the rate of formation of an acyl-nicotinamide adenine dinucleotide adduct considered to be a bactericidal molecule, was reduced by 30% compared with WT KatG. INH resistance is suggested to arise from a redirection of catalytic intermediates into nonproductive reactions that interfere with oxidation of INH. In the resting mutant enzyme, a rapid evolution of 5-c heme to 6-c species occurred in contrast with the behavior of WT KatG and KatG[S315T] and consistent with greater flexibility at the heme edge in the absence of the hydroxyl of residue 315. Insights into the effects of mutations at residue 315 on enzyme structure, peroxidation kinetics, and specific interactions with INH are presented.
Tuberculosis infection kills nearly 2 million people a year and is the leading cause of death due to infectious diseases in adults and in AIDS patients (1). The infection is usually treatable, and isoniazid (isonicotinic acid hydrazide (INH))4 has been a first line antibiotic against Mycobacterium tuberculosis since 1952 (2). The management of the disease is complicated by the fact that bacterial strains have been steadily acquiring and accumulating mutations that confer resistance to INH and other drugs (3–6). Recently, the appearance of multidrug-resistant tuberculosis, resistant to at least two first line antibiotics, and extensively drug-resistant bacteria (defined as multidrug-resistant tuberculosis plus resistance to at least one fluoroquinolone and at least one of the injectable second line drugs) has made the disease virtually incurable in a growing number of cases (7, 8). Despite the widespread emergence of antibiotic-resistant strains, the molecular mechanisms by which enzyme targets or pro-drug activating enzymes confer resistance are poorly understood.
The pro-drug INH requires activation by M. tuberculosis catalase-peroxidase KatG, a heme enzyme classified in the Class I superfamily of fungal, plant, and bacterial peroxidases (9). KatG is important for the virulence of M. tuberculosis due to its role in oxidative stress management (10). This enzyme exhibits both high catalase activity and a broad spectrum peroxidase activity (9, 11) for which a physiologically relevant substrate has not been identified. In vitro, INH is oxidized by KatG (12–15) to an acylating species, most likely an acyl radical, that forms an adduct (IN-NAD) when it reacts with NAD+ (16). This modified cofactor then acts as a potent inhibitor of the M. tuberculosis enoyl-acyl carrier protein reductase, InhA, and interferes with cell wall biosynthesis (17, 18). The most common INH resistance mutations in M. tuberculosis clinical isolates occur in katG (19), although mutations in other genes, including inhA, and the promoter for this enzyme (mabA-inhA operon) may cause resistance (20–22). Dihydrofolate reductase has also been recently proposed as a target of isoniazid that can be inhibited by an IN-NADP adduct (23, 24). Issues remain to be resolved about INH action as well as resistance in a large set of clinical isolates.
Replacements at residue Ser315 are the most commonly encountered in the mutated katG gene of INH-resistant strains (19, 22, 25–28). Among these, S315T, which confers high level drug resistance (up to a 200-fold increase in minimum inhibitory concentration (MIC) that kills 50% of bacteria (29)) is the most frequent and is found in more than 50% of INH-resistant isolates of M. tuberculosis. In vitro, this mutant enzyme exhibits a very poor rate of peroxidation/activation of the antibiotic, although the enzyme has close to normal catalase activity and peroxidase activity with substrates other than INH (30–32). According to the crystal structure of KatG[S315T] (33), the replacement of serine by threonine leads to a structurally modified substrate access channel. This channel leads from the surface of the enzyme to the heme edge at the propionate of pyrrole IV. Residues Asp137 and Ser315 delimit the narrowest region of the channel, which is reduced in width from 6 to 4.7 Å. The methyl group of threonine effectively restricts accessibility to the heme pocket and apparently interferes with specific interactions required for binding and activation of the drug. Although a binding site for INH in KatG is not specifically defined by x-ray crystallography at this time, a recently reported CCP-INH structure (yeast CCP is a homologous Class I peroxidase) presents what should be an excellent model of drug binding in KatG (34). Hydrogen bonds between the backbone carbonyl of Ser185 (Ser315 in M. tuberculosis KatG), a water molecule, and the pyridine nitrogen of the drug are found in the CCP-INH complex. Thus, it is reasonable that mutations at residue 315 in KatG have an impact on drug binding and activation but little impact on catalase or peroxidase activity with substrates that may not require the same specific interactions as high affinity INH binding.
Beyond these studies, there is a substantial gap in the knowledge of the relationship between INH resistance due to the numerous other mutations in the katG gene and the lost drug activation function of the mutant enzymes. The main goal of the present study was to examine KatG[S315G] in vitro. We report the generation, overexpression, purification, and characterization of this enzyme found in clinical isolates of M. tuberculosis having low level INH resistance with MIC values up to 40-fold higher than WT strains (8 μg/μl versus 0.05 μg/μl) (22, 25). An interesting aspect of the problem is that in KatG[S315T], a steric influence on INH binding strongly interferes with activation, whereas resistance is still present with the glycine replacement of serine 315, which would not be assumed to interfere with substrate access or binding at the same locus.
The application of optical stopped-flow spectrophotometry, isothermal titration calorimetry (ITC), optical titration, EPR spectroscopy, and rapid freeze-quench EPR (RFQ-EPR) allowed us to probe the functional and structural consequences of the mutation on INH activation. Our results strongly suggest that resistance is due to catalytic changes rather than major changes in specific interactions between the enzyme and INH. Importantly, the results demonstrate the validity of an in vitro INH activation approach used here, since we find a correlation between our observations and the in vivo behavior of INH-resistant M. tuberculosis strains for both KatG[S315T] and KatG[S315G].
EXPERIMENTAL PROCEDURES
All standard chemicals and reagents were obtained from Sigma. INH was recrystallized from methanol before use. PAA (32%) was diluted to 10 mm in potassium phosphate buffer and was incubated with 780 units/ml bovine catalase (Roche Applied Science) for 3 h at 37 °C to degrade hydrogen peroxide. After treatment, catalase was removed by ultrafiltration.
The plasmid pKAT II was used as an overexpression vector for KatG (35) as well as a mutagenesis template. Escherichia coli strain UM262 (36) was used for overexpression. UM262 and pKAT II were both gifts from Stewart Cole (Ecoles Polytechniques Fédérales de Lausanne). Mutagenesis was performed using the QuikChange II site-directed mutagenesis kit from Stratagene (La Jolla, CA). The pairs of complementary primers (synthesized and purified by Operon Biotechnologies, Inc.) were designed to introduce the S315G mutation. The oligonucleotide pairs (mutated codons are in boldface type) were 5′-925GGTAAGGACGCGATCACCGGCGGCATCGAGGTCG958-3′ and 5′-958CGACCTCGATGCCGCCGGTGATCGCGTCCTTACC925-3′. Mutagenesis was performed according to the manufacturer's protocol, and the reaction products were transformed into the E. coli XL1-Blue strain for selection purposes. The presence of the mutated 315 codon in the katG gene was confirmed by DNA sequencing (Gene Wiz, Inc.), and the mutated plasmid was electroporated into E. coli strain UM262 for protein overexpression. KatG[S315G] was purified as previously described for WT KatG (37) in potassium phosphate buffer, pH 7.2. The pure enzyme had an optical purity ratio (A405/A280) of ∼0.45. The proportion of heme-deficient enzyme, which is a catalytically active dimer containing only one heme, represented more than half of the total overexpressed enzyme isolated in these experiments. In the case of WT KatG, only a small percentage of total enzyme is found as heme-deficient dimer (37). The overexpression conditions are such that polypeptide synthesis outpaces the availability of heme despite supplementation of the medium with the heme precursor δ-ALA. The unknown conditions in M. tuberculosis carrying KatG[S315G] precludes commenting on the physiological significance of these observations. Here, we have focused only on the properties of the holoenzymes.
Protein concentration was determined using a heme extinction coefficient of ∼85 mm−1 cm−1 at 405 nm for the mutant enzyme and 100 mm−1 for WT KatG catalase, and peroxidase activities were determined at 25 °C according to published procedures (31, 38) in 20 mm potassium phosphate buffer, pH 7.2, and 50 mm sodium acetate buffer, pH 5.5, respectively.
Spectrophotometric measurements were obtained using an NT14 UV-visible spectrophotometer interfaced to a personal computer (Aviv Associates). A rapid scanning diode array stopped-flow apparatus (HiTech Scientific model SF-61DX2) was used for kinetics measurements. Data acquisition and analyses were performed using the Kinet-Asyst software package (HiTech Scientific). All reactions were carried out at 25 °C in potassium phosphate buffer at pH 7.2. For calculation of second order rates for conversion of resting enzyme into the catalytic intermediate oxyferryl heme:porphyrin π-cation radical (Fe(IV)=OPor+·), the change in absorbance in the Soret region upon reaction with varying concentrations of PAA was followed for 2 s, and data were fit to a single exponential decay function. Observed rates (kobs) were used to calculate second order rate constants from the plot of kobs versus PAA concentration. Double mixing stopped-flow experiments were performed as previously reported (30, 37) to follow the reaction of Fe(IV) = OPor+· after it was preformed with a small excess of PAA, with INH, or with additional PAA as reducing substrates added in the second mixing step.
The interaction of INH with ferric KatG was analyzed using optical difference spectroscopy in the Soret region, similar to published procedures (32, 39). The change in the difference in absorbance evaluated between 378 and 411 nm was plotted against free isoniazid concentration for titrations conducted at 25 °C in 20 mm phosphate buffer, pH 7.2. Data were fit to a single site saturation curve using Sigma Plot version 8.0 software.
Isothermal titration calorimetry was performed using a MicroCal VP ITC calorimeter. Data analysis was performed using ORIGIN software supplied with the instrument. KatG[S315G] and titrant (INH) solutions were prepared in the same 20 mm potassium phosphate buffer, pH 7.2, to ensure minimal background from buffer mismatch. Reliable titration data were obtained using 20 μm enzyme in the sample cell (1.4 ml) and an INH concentration in the ligand delivery syringe that gave an INH/heme ratio of ∼0.15:1 per injection. A total of 16 injections (10 μl/injection) were made at 10-min intervals because of a very slow equilibration period, and the heat of reaction per injection (microcalories/s) was determined by integration of the peak areas. The software provided the best fit values for ΔH, ΔS, the stoichiometry of binding (n)/mol of heme, and the dissociation constant (Kd) from plots of heat evolved/mol of substrate injected versus the drug/heme molar ratio. (ITC titrations of KatG with INH and other hydrazide ligands may overestimate the affinity based on heat released because of background reactions (39)).
IN-NAD Formation
The rate of IN-NAD adduct generation catalyzed by KatG[S315G] was examined in a spectrophotometric assay as a function of INH concentration according to a previously published method (33). In this method, KatG (0.5 μm), NAD+ (50 μm), glucose oxidase (10 milliunits/ml), and glucose (5 mm) were incubated in the presence of varying amounts of INH (10, 20, 30, 50, 100, 250, 500, 1000, and 2000 μm), and the increase in absorbance at 326 nm due to IN-NAD adduct (ϵ326 nm = 6900 m−1 cm−1 (40)) was recorded for 20 min. The reference cuvette contained all components except NAD+ to correct for background.
EPR Spectroscopy
X-band EPR spectra were recorded using a Bruker E500 ElexSys EPR spectrometer with data acquisition and manipulation performed using XeprView and WinEPR software (Bruker). Low temperature spectra were recorded using an Oxford Spectrostat continuous flow cryostat and ITC503 temperature controller. The spectra of KatG[S315G] (100 μm) were recorded at 4 K in 20 mm potassium phosphate buffer, pH 7.2. Experimental parameters are given in the figure legends.
RFQ-EPR
The RFQ-EPR samples (aerobic) were prepared using an Update Instrument, Inc. model 1000 chemical-freeze-quench apparatus, as described previously (41). Solutions of enzyme (typically 100 μm heme) and peroxyacetic acid (300 μm) in 20 mm potassium phosphate buffer, pH 7.2, were mixed in a 1:1 ratio, and the mixture was incubated for the indicated time periods followed by freeze-quenching in isopentane at ∼−130 °C. PAA solutions were freshly prepared before each experiment from H2O2-free stock solutions stored at −80 °C. EPR samples were maintained in liquid nitrogen after removal of isopentane, and spectra were recorded at 77 K using a finger Dewar flask held in the EPR cavity. Samples were also prepared anaerobically by thoroughly degassing enzyme and peroxide solutions and filling syringes for the apparatus under continuous argon purging.
RESULTS
Characteristics of the Resting Mutant Enzyme
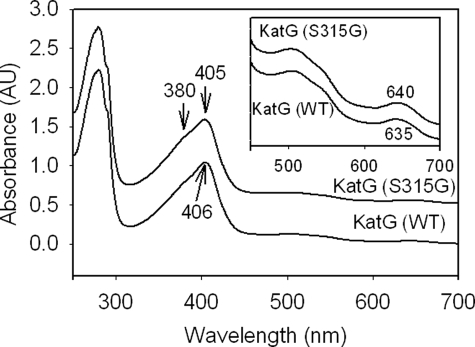
Purified mutant enzyme from multiple preparations consistently showed a reduced optical purity ratio (A405/A280 = 0.40–0.45) compared with that of WT KatG (0.50–0.55) but similar to that of KatG[S315T] (30). Although a significantly higher amount of heme-deficient KatG[S315G] enzyme was consistently found during purification, this form of the enzyme resolves completely from the holoenzyme upon hydrophobic chromatography; therefore, contamination of the holoenzyme with heme-deficient enzyme is not the origin of the low ratio. Another factor that would reduce the optical purity ratio is the presence of a high proportion of 5-c heme versus 6-c heme. The blue-shifted Soret peak and red-shifted CT1 band (>640 nm) in the optical spectrum of the freshly isolated mutant enzyme compared with these bands in WT KatG confirm the high proportion of 5-c heme (Fig. 1). The accumulation of six-coordinate KatG during storage of purified enzymes leads to a Soret extinction coefficient ∼30% higher than that of the freshly isolated enzyme and a blue-shifted CT1 band (42). Interestingly, although KatG[S315G] samples contained a greater proportion of 5-c heme than was typically found for WT KatG immediately after purification, the conversion to 6-c heme was notably faster than in the WT enzyme. For example, after 1 week of storage at 4 °C, the 6-c heme content in the mutant protein doubled, according to resonance Raman spectroscopy (not shown); for WT KatG, similar changes require more than 3 weeks, and the proportion of 6-c heme in WT KatG has not been found to reach the levels in the mutant even after long term storage. This so-called “aging” of M. tuberculosis KatG has been previously described, although specific structural differences between the 5-c and 6-c enzyme forms have not been defined beyond the change in heme iron coordination number (42).
FIGURE 1.
UV-visible spectrum of resting (ferric) WT KatG and KatG[S315G]. Optical absorption spectra of 10 μm wild-type KatG and KatG[S315G] in 20 mm potassium phosphate buffer, pH 7.2, at 25 °C are shown. Spectra are offset for presentation purposes. Differences in the absorbance maxima marked by arrows are relevant to the greater abundanceof 5-c heme in the mutant. AU, absorbance units. Inset, expanded visible region of spectra.
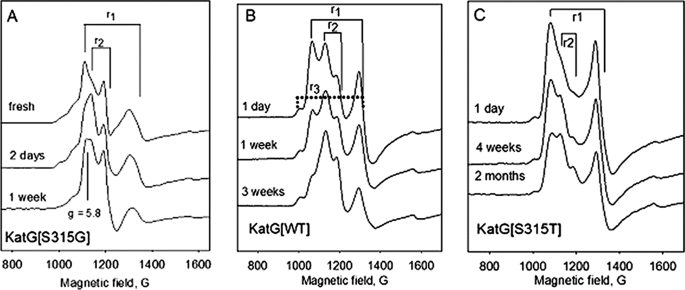
Low temperature EPR spectra provided additional evidence for the rapid evolution of 6-c heme in KatG[S315G]. The EPR spectrum of KatG[S315G] frozen immediately after purification has two rhombic signals that account for nearly all of the intensity; signal r1 (g1 = 6.3, g2 = 5.1, g3 ∼ 2.00) was previously assigned to a 5-c heme species in freshly isolated WT KatG and in KatG[S315T], whereas signal r2 (g1 = 6.04, g2 = 5.54, g3 ∼ 2.00) assigned to 6-c heme becomes predominant after short term storage; in KatG[S315T], the latter signal only develops after very long term storage (Fig. 2) (30, 43). An axial signal (g⊥ = 5.83), probably from another 6-c species, also becomes evident in spectra of the S315G mutant. Little change was observed in the intensity of a second 5-c heme signal (r3; g1 = 6.60, g2 = 5.01, g3 ∼ 2.00 shown in Fig. 2B but present in all three examples) (42). These observations are consistent with conversion of 5-c heme species into 6-c heme more rapidly in KatG[S315G] than in either WT KatG or KatG[S315T].
FIGURE 2.
Low temperature EPR spectra as a function of storage time after purification. A, KatG[S315G]; B, wild-type KatG; C, KatG[S315T] (from Ref. 43). Experimental conditions were as follows: temperature, 4 K; microwave power, 1 milliwatt; modulation amplitude, 4 G; frequency, 9.38 GHz. g-value assignments are as follows: for r2, g1 = 6.04, g2 = 5.54, and g3 ∼ 2.00; for r1, g1 = 6.30, g2 = 5.1, and g3 ∼ 2; for r3, g1 = 6.68, g2 = 5.1, and g3 ∼ 2 was present in each case.
The low temperature EPR spectra in general show a greater abundance of 6-c heme relative to 5-c heme than the estimated amounts of these species from room temperature resonance Raman spectra, but this type of quantification is difficult, and its temperature and pH dependence are not yet understood. The coordination number has been previously addressed based on EPR, optical, and resonance Raman spectra and on the presence of 6-c heme in the x-ray crystal structure of WT KatG (44), and the assumption here is that 6-c species contain a water molecule associated with heme iron in the sixth coordination position.
As a first step to assess the possible altered function of this mutant responsible for low level isoniazid resistance, we proceeded to study catalytic mechanisms.
Catalytic Functions
The catalase- and peroxidase-specific activities of KatG[S315G] were lower by ∼46% (2039 ± 461 IU/mg) and ∼33% (0.61 ± 0.15 IU/mg), compared with WT KatG (Table 1). Similar reductions in the activities of other KatG mutants, including enzymes from INH-resistant isolates, have been reported based on assays with artificial substrates (30, 31, 45, 46). However, new insights into INH resistance mechanisms must be specifically gained in each case. Since we suggest that there is a high affinity binding site for INH in KatG that could be disrupted by mutations, this binding site may not be common to other peroxidase substrates used in assays. For strains with the KatG[S315G] mutation, the MIC is up to 40-fold higher compared with WT (22), whereas the peroxidase activity measured in vitro with o-dianisidine is only reduced about 2-fold. Further exploration of the properties of the mutant enzyme reveals structural and catalytic features relevant to understanding the INH resistance mechanism in detail.
TABLE 1.
Catalase- and peroxidase-specific activities of KatG
| WT KatG | KatG[S315G] | |
|---|---|---|
| Catalase activity (IU/mg) | 3800 ± 344 | 2039 ± 461 |
| Peroxidase activity (IU/mg) | 0.9 ± 0.1 | 0.6 ± 0.15 |
| CmpdI formation (PAA) (m−1s−1) | 3 × 104 | 7.4 × 104 |
| CmpdI decay (PAA) (s) | ∼80 s | ∼40 s |
| INH affinity (optical) (μm) | 1.0 | 0.7 |
| INH affinity (ITC) (μm) | 1.6 | 1.4 |
| Decay of 5-c heme (aging) (weeks) | ∼3 | ∼1 |
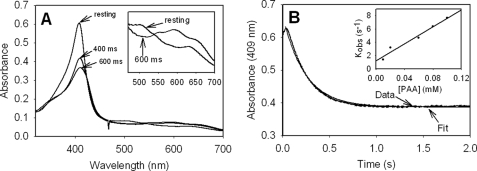
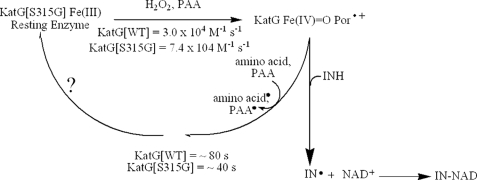
Evaluation of the rate of formation of Fe(IV)=OPor+·, which is a heme species catalytically competent for INH oxidation, is among our approaches to characterize catalytic function of KatG enzymes. The optical spectrum of this species formed upon turnover of ferric KatG[S315G] with small excesses of alkyl peroxides (PAA, chloroperoxybenzoic acid, tert-butyl hydroperoxide) was essentially the same as that reported for WT KatG and showed the characteristic 40–50% hypochromicity and red shift to 411 nm in the Soret region and peaks at 550 and 590 nm, together with a shoulder at 665 nm (Fig. 3), similar to other reports (37, 47). In the case of PAA, a second order rate constant was determined from the linear plot of the kobs values as a function of the concentration of peroxide. The rate was ∼2.5 times faster than that for WT KatG (7.4 × 104 versus 3.0 × 104 m−1 s−1) under similar conditions. This small increase does not, however, enhance INH activation (see below) or the peroxidase-specific activity as usually assayed.
FIGURE 3.
Formation of Fe(IV)=OPor+· in KatG[S315G]. A, absorption spectra recorded for resting enzyme and after the addition of 25 μm PAA to 5 μm (final concentrations) resting enzyme in 20 mm potassium phosphate buffer, pH 7.2, at 25 °C in a stopped-flow experiment; B, absorbance versus time recorded at 409 nm. The time course at 409 nm was fitted with a first order exponential decay function. Inset, linear dependence of the observed rates (kobs) on PAA concentration, giving a second order rate constant equal to 7.4 × 104 m−1 s−1.
Interestingly, the S315G mutant was more readily damaged during turnover of PAA than the WT enzyme, evidenced by the losses in absorbance intensity of enzyme samples after incubation with a 10-fold excess of PAA (not shown). For WT KatG, similar damage was only noted using twice the level of PAA. The origin of this higher susceptibility to damage by peroxide in KatG[S315G] is addressed below.
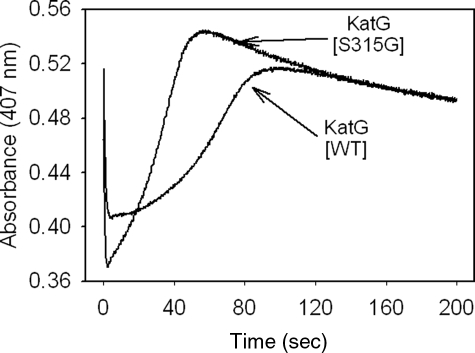
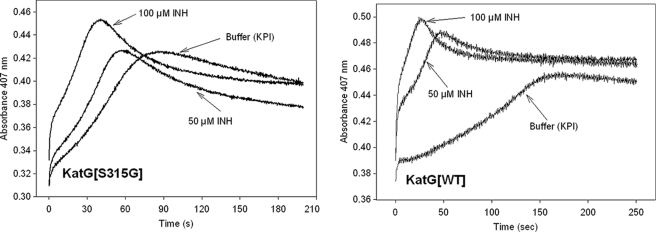
The spontaneous return to the resting enzyme after formation of the Fe(IV)=OPor+· intermediate is also of interest, since this process is known to involve endogenous electron transfers producing radicals on amino acids (40, 41, 48) and could have an impact on INH activation. Interestingly, the time course for this return was ∼2 times faster in the mutant than in WT KatG (Fig. 4); this was estimated from the slope of the increasing absorbance versus time at 407 nm after turnover of resting enzyme with a small excess of PAA. Since KatG[S315G] and the WT enzyme contain the same redox-active amino acids, this faster return implicates reactions other than electron transfers from these residues. To test this hypothesis, a double mixing optical stopped flow experiment was used to follow the rate of reaction of Fe(IV)=OPor+· with additional PAA. For a given amount of PAA added to the preformed intermediate in KatG[S315G], the loss of the intermediate was accelerated by around 2.5-fold relative to that in WT KatG (Fig. S1). This observation demonstrates an enhanced reactivity of the mutant enzyme in reaction mixtures containing excess peroxide.
FIGURE 4.
Formation and decay of Fe(IV) = OPor+· in WT KatG and KatG[S315G].5 Extended time traces (at 407 nm) recorded after mixing 5 μm resting WT KatG or KatG[S315G] with 25 μm PAA (final concentrations) in 20 mm potassium phosphate buffer, pH 7.2, at 25 °C. The initial rapid decrease is followed by the slow return to the starting absorbance value around 0.5.
RFQ-EPR provided further evidence that PAA could be the species responsible for initiating such decay, since evidence for the peroxyl radical, which would be formed from single electron oxidation of PAA, was consistently found in RFQ-EPR samples (Fig. S2). Furthermore, the RFQ-EPR results did not show an increase in the yield of protein-based radicals, such as the tyrosyl and tryptophanyl radicals assigned in WT KatG (40, 41), which are formed under similar conditions in the presence of PAA. One additional finding was that anaerobic preparation of RFQ-EPR samples of enzyme treated with PAA did not alter the identity of the signals observed to any great extent (not shown) compared with aerobic samples, arguing against a peroxyl radical produced from reaction of dioxygen with an amino acid radical. This is a known source of peroxyl radical in other proteins treated with peroxides (49, 50). Although these observations are considered evidence for more facile turnover of catalytic intermediates in KatG[S315G] relative to WT enzyme, such turnover does not extend to an enhancement of INH activation, since the mutant enzyme is responsible for resistance to the antibiotic. Therefore, the results indicate that there may be competition between peroxide and other substrates for the intermediates that would otherwise be available to oxidize the drug and lead to IN-NAD formation.
Is INH a Poor Substrate of KatG[S315G]?
In order to directly examine the catalytic competence for INH activation by Fe(IV) = OPor+· in KatG[S315G], double mixing stopped-flow experiments were performed according to previously published methods (30). The heme intermediate, preformed by the reaction of 10 μ m WT KatG or KatG [S315G] with 50 μm PAA, was mixed after a 2-s delay with increasing concentrations of INH (20, 50, 100, 200, and 500 μm), after which the absorbance changes at 407 nm were followed. As expected, INH accelerated the return to the ferric enzyme in both cases, compared with the rate after the second mixing with buffer alone (Fig. 5). For the WT protein, the rate of return after the second mixing step was accelerated 2-fold using 50 μm INH and 5-fold using 100 μm INH, relative to the rate after the addition of buffer alone. For KatG[S315G], the rate of acceleration was only 1.3-fold (50 μm INH) and 2-fold (100 μm INH), directly demonstrating the deficient turnover of the drug by the mutant enzyme (Scheme 1). Also, as can be seen in Fig. 5, the increased absorbance (“overshoot”) observed after the consumption of PAA in the single mixing experiment is also present but is more pronounced in the presence of INH, because excess drug added in the second mixing step binds to the 6-c heme enzyme product and converts the heme to the 5-c form (30).5 An approach to examine INH activation using biomimetic conditions provides evidence consistent with the observations above and with the low level resistance reported from in vivo treatment of M. tuberculosis with INH (33).
FIGURE 5.
Reaction of WT KatG (right) and KatG[S315G] Fe(IV) = OPor+· (left) with INH.5 Absorbance versus time traces (at 407 nm) for double mixing stopped-flow experiments are shown. Resting enzymes were prereacted with PAA to form Fe(IV) = OPor+·, followed by the addition of increasing concentrations of INH. The final concentrations of enzyme and PAA were 5 and 25 μm, respectively; the final concentrations of INH are as labeled in the figure for each time trace.
Scheme. 1.
Reaction pathways relevant to INH activation by KatG. Rates of Fe(IV) = OPor+· formation using PAA (7.4 × 104 m−1 s−1 for KatG[S315G]; 3.0 × 104 m−1 s−1 for WT KatG) and decay of heme intermediates back to resting enzyme (40 s, KatG[S315G]; 80 s, WT KatG) are shown. Results suggest that increased rate or efficiency of single electron reactions with peroxide(s) leads to a decreased activation (oxidation) of INH and therefore to decreased IN-NAD adduct formation, ultimately producing drug resistance in strains harboring the S315G mutation.
IN-NAD Formation
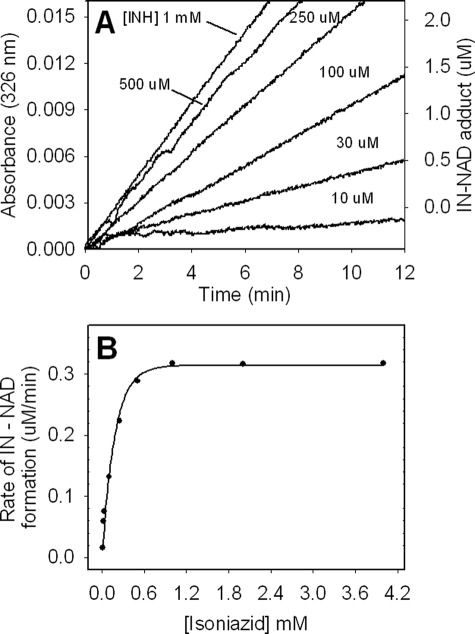
According to a consensus model for INH action in mycobacteria, an acyl radical formed upon oxidation of INH by KatG reacts with NAD+ at the C-4 position of the nicotinamide ring to form an adduct (IN-NAD) (12, 16, 17, 33, 51–53). NAD+ does not bind to KatG, nor is it a substrate of peroxidation, suggesting that the rate of formation of IN-NAD depends exclusively on the rate of formation of an INH-derived radical that can react nonenzymatically with NAD+. The ability of WT KatG to catalyze formation of IN-NAD in a physiologically relevant biomimetic reaction system and the very poor function of KatG[S315T] in this reaction were described in a previous study (33). Here, IN-NAD adduct formation is evaluated using the same biomimetic approach, in which dilute H2O2 produced enzymatically initiates peroxidase catalysis in the presence of excess NAD+ and INH. In this protocol using WT KatG, the rate of appearance of IN-NAD depended on the concentration of INH up to a saturating amount (0.5 mm INH) under conditions in which H2O2 was generated at 2 μm/min (33). In the case of KatG[S315G], 1 mm INH was required to produce a maximum rate (Fig. 6) (both cases in the presence of 50 μm NAD+, previously shown not to be rate-limiting (33)). Also, a decrease in the maximum rate of IN-NAD formation by ∼30% was found (0.3 μm/min versus 0.4 μm IN-NAD/min, saturating INH). These observations are consistent with the double mixing stopped-flow kinetics results above, in which it was shown that INH was an apparently poorer substrate of the mutant enzyme compared with WT KatG. Overall, the results are consistent with only small losses in INH activation by the mutant enzyme, consistent with low level resistance in vivo.
FIGURE 6.
Effect of INH concentration on the rate of production of the IN-NAD adduct. A, IN-NAD adduct formation was followed at 326 nm. KatG[S315G] (0.5 μm), NAD+ (50 μm), and H2O2 (2 μm/min generated enzymatically using Glu/glucose oxidase were incubated with varying concentrations of INH. B, rates of IN-NAD adduct formation as a function of INH concentration.
Optical and ITC Titrations
The impact on high affinity drug binding of mutations in KatG is important to consider in explaining INH resistance. This binding can be monitored in the resting enzyme by changes in optical spectral features, as first reported by Wengenack et al. (32), and more directly by ITC. Titration of KatG with INH produces an increase in the absorbance at 380 nm and a decrease at 411 nm consistent with conversion of 6-c heme to 5-c heme species (39). Dissociation constants can be determined from these optical changes. The ITC approach directly measures the heat evolved upon binding and provides the number of binding sites and the enthalpy and entropy of binding.
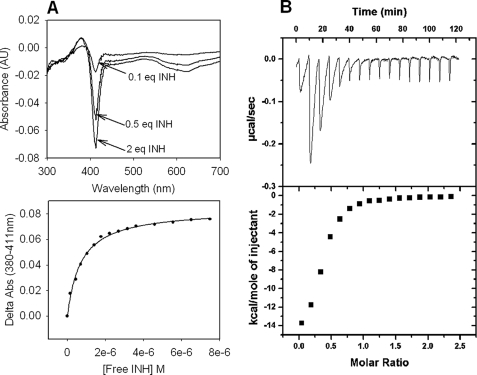
Both optical titrations and ITC results show that less than 1 eq of INH was sufficient to achieve apparent saturation of KatG (Fig. 7). This is reasonable if only a fraction of the total available enzyme provides an appropriate binding site for the drug, as reported previously for WT KatG (34). For this reason, the n value obtained from ITC experiments (∼0.3–0.4) was used to estimate the concentration of available binding sites to allow calculation of dissociation constants from the optical titration results. Apparent Kd values obtained from the optical and ITC titrations for KatG[S315G] were 0.7 and 1.4 μm, respectively, very similar to those reported for the WT enzyme (1.0 and 1.6 μm) (39). These results do not reveal any large change in the affinity of the drug for the enzyme and lead to greater confidence that the mechanistic changes identified above should be considered central to the origin of INH resistance.
FIGURE 7.
Binding of INH to KatG[S315G]. A, optical titrations with INH. Optical titrations were performed using 5 μm enzyme and increasing concentrations of INH (top panel). Difference spectra were obtained by subtracting the spectrum of free KatG from that of the INH-bound enzyme for each increment of addition. Binding curves were generated by plotting the absorbance difference for the peak minus the trough absorbance values at 380 and 411 nm, respectively, versus the concentration of free INH (bottom panel). B, isothermal titration of KatG[S315G] with INH. ITC experiments were carried out at 25 °C in phosphate buffer, pH 7.2, using 10 μm KatG. The top panel shows the isothermal traces measured from a series of injections of INH into enzyme. Heat (integrated values in μcal/s/injection; lower panel) were fitted to a single binding site model. The Kd value calculated from the optical titration was 0.7 μm, and the value from the ITC titration was 1.4 μm.
DISCUSSION
To date, KatG[S315T] was the only example for which details have been reported of how structural changes impact upon INH activation and lead to resistance to the drug (30, 33). Mutations at the 315-position are very common in clinical isolates, suggesting some unique features of the KatG mutants bearing replacements at this position. Centrally important may be that such replacements do not seriously interfere with the physiological roles of the enzyme in M. tuberculosis, the most important of which is believed to be its catalase activity; no obligatory peroxidase function of the enzyme has yet been identified. The Ser315 residue, located at the heme edge, is not a direct participant in catalytic or redox processes but has effects on heme structure, such as the stability of 5-c versus 6-c heme iron, and on interactions with INH.
One remarkable feature of the KatG[S315G] mutant is the very rapid rate at which 6-c heme evolves from the 5-c form initially found in the freshly isolated enzyme. Prior analyses showed this “aging” of WT KatG to be extremely slow in the KatG[S315T] mutant (39, 42, 43). The process, which was extensive within the first day or two of storage of KatG[S315G], was evident in markers found in optical, resonance Raman, and low temperature EPR spectra (54). The behavior of the mutant is most likely dependent upon changes in the access to the distal cavity through a substrate access channel described in crystal structures of M. tuberculosis KatG enzymes (33, 44) as well as changes in the constraints on the architecture of the active site that must accompany interconversion between 5-c and 6-c heme iron. These constraints probably involve a hydrogen bond between the side chain hydroxyl of residue 315 and the carboxyl of the propionate of pyrrole IV evident in the crystal structure (30, 33). The shift of the heme from 5-c to 6-c (39, 42, 43) will induce changes in geometry that require structural adjustments at the heme edge; for example, recent resonance Raman results demonstrate that the heme is more planar in 6-c KatG than in 5-c enzyme (55) and therefore will have different dimensions. The observations on the glycine mutant here are consistent with release of constraints in the absence of the hydroxyl group of residue 315. Further evidence for this conclusion comes from the behavior of the KatG[S315C] mutant reported elsewhere (43), which also exhibits facile conversion to 6-c heme. In this mutant, the thiol group will form weaker hydrogen bonds than the hydroxyl of serine and may therefore impose weaker constraints on heme geometry. In contrast to this, in KatG[S315T], the presence of the hydroxyl group and the added methyl group creates more constraints than those in WT KatG and thus greatly inhibits the formation of 6-c heme (30, 43).
Our previous reports on the origins of INH resistance in KatG[S315T] ascribed it to a decrease of the dimensions of the substrate access channel that interfered with high affinity INH binding (30, 33). The recent publication of a CCP-INH crystal structure (Protein Data Bank code 2V2E) illustrates the importance of a key set of hydrogen bonds between the pyridine nitrogen of INH, a water molecule, and the backbone carbonyl of Ser315 that are probably critical for high affinity binding and efficient activation of the drug. In the case of the S315G mutant, our results did not demonstrate seriously altered binding parameters. However, the stopped-flow and biomimetic activation results both suggest that the drug is a poorer substrate of the mutant enzyme, meaning that less activated drug and less IN-NAD adduct would be produced during catalytic turnover. In order to reconcile these sets of observations (i.e. the apparently normal binding of INH to the resting enzyme but still poor activation), we consider that catalytically important intermediates of the peroxidase cycle are less efficient due to a change in their structure and or reactivity in the mutant and also that the drug may have altered affinity for these intermediates. The optical spectrum of Fe(IV) = OPor+· in KatG[S315G] is nearly identical to that in WT KatG, arguing against changes in the structure of this species in the mutant. A change in interaction between this intermediate and INH is supported by the kinetics results, although only small changes were evident. The possibility that the function of Fe(IV) = OPor+· is in fact altered is supported by the finding that it is more reactive in KatG[S315G] compared with the behavior of this intermediate in the WT enzyme. Recall that the results above showed that its lifetime in the mutant is about half of that in WT enzyme and also that it reacts more rapidly with PAA. Thus, for a given concentration of peroxide, less enzyme intermediate would be available for activation of INH because of competition by peroxide. Evidence confirming the validity of this suggestion is the finding that in the biomimetic protocol for IN-NAD formation, the rate cannot be driven to the rate catalyzed by WT KatG even in the presence of saturating amounts of INH.
In the absence of reducing substrates, the reaction of WT KatG with excess alkyl peroxides generates tyrosyl and tryptophanyl radicals (40, 41), which contributes to the quenching of heme intermediates. However, no direct evidence for an increased yield of amino acid-based radicals was found in RFQ-EPR experiments with KatG[S315G]; the maximum total yield of radical in the case of the mutant was in fact lower by 60% during the first 200 ms of the reaction, compared with the yield in WT KatG. Furthermore, an axial EPR signal (g‖ = 2.035 and g⊥ = 2.006) characteristic of peroxyl radicals was the major species detected (Fig. S2) rather than the doublet and singlet signals characteristic of tyrosyl and tryptophanyl radicals found in WT KatG under similar conditions. Peroxyl radicals can be formed from amino acid radicals initially produced by electron transfers to peroxidase intermediates, followed by reaction of the radicals with dioxygen (49, 56), or, in our experiments with PAA, can be evidence for alkyl peroxyl radical formed on PAA acting as a single electron substrate of KatG. The latter idea is supported by the double mixing stopped-flow experiments. Furthermore, the available amino acids that form radicals that could go on to the peroxyl radical are the same in the mutant and WT KatG, in which the evidence for the peroxyl radical is scarce and highly variable. These observations argue against the importance of increased endogenous electron transfers in the turnover processes in KatG[S315G]. Overall, the results suggest that under the artificial conditions with PAA and, by extension, in the biomimetic system, turnover of peroxide competes with INH activation in the mutant enzyme. Excess drug would be expected to overcome a simple loss in affinity for binding to catalytic intermediates, which was not observed. Therefore, a decreased availability of such intermediates for INH oxidation is reasonably consistent with all of the observations we have reported. Greater flexibility of the substrate access channel near the heme edge, among other potential structural changes, would give rise to such a change in catalytic behavior.
One provocative hypothesis growing out of the results here is that the observed decrease in protein-based radicals in KatG[S315G], which have potential catalytic function, contributes to reduced activation of INH. This may be ruled out, since KatG[S315T] is responsible for high level resistance yet exhibits WT behavior in RFQ-EPR experiments designed to follow the formation of protein-based radicals (57).
The M. tuberculosis strain in which the KatG[S315G] mutation was first reported also bore a second mutation associated with low level INH resistance. This was found in the promoter region of the inhA structural gene. InhA is the target enzyme of INH action, which is inhibited by the IN-NAD adduct. The promoter mutations cause overexpression of InhA and confer low level resistance to INH due to a “titration” effect (22) in which IN-NAD is sequestered by its tight binding to InhA but achieves incomplete inhibition of the pool of this enzyme target. Thus, strains of M. tuberculosis overexpressing InhA from a multicopy plasmid show low levels of INH resistance (for example, MIC values of 4 μg/ml or less have been reported (21). MIC values of 8 μg/ml reported for the strain bearing KatG[S315G] have not been reported for strains having only inhA promoter mutations (20, 22). Furthermore, KatG[S315G] has been found in M. tuberculosis strains infecting patients in the Georgian Republic as the only mutation correlated with INH resistance (26).6 These observations strongly suggest that the INH resistance observed in clinical isolates harboring KatG[S315G] can be caused exclusively or in part by the faulty catalysis of INH activation by the mutant KatG enzyme.
We emphasize here that a moderate deficiency in INH activation in vitro by a mutant KatG enzyme, such as S315G, would be expected to correlate with a moderately increased MIC in vivo. For KatG[S315T], a much more dramatic loss in interactions with INH occurs, consistent with a greater increase in the MIC (30). Without adjustment of standard dosing in clinical settings, the reduction in activation is sufficient to lead to failure of INH therapy for patients carrying either of the S315 mutant M. tuberculosis strains. These results are consistent with the fact that the mutated KatG enzymes, although they are stable and functional in INH activation, cannot produce sufficient IN-NAD for bactericidal effects under normal treatment regimens.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AI060014 (to R. S. M.) from NIAID.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
A notable feature of the observed time course is that the absorbance intensity at 407 nm rises to a value greater than the initial absorbance before it finally levels off at the starting absorbance value. A similar trend is also observed for WT KatG and KatG[S315T], but the effect is not as pronounced (30). This “overshoot” in the Soret absorbance is evidence for a species having a greater extinction coefficient than the species found after Fe(IV) = OPor+· is fully decayed. This species is probably 6-c heme containing a water ligand produced during reduction of oxoferryl heme intermediates; the water molecule dissociates and regenerates the 5-c state of the starting enzyme after the redox reactions terminate. For the mutant, the more obvious effect relative to WT is due to the fact that the starting enzyme contained a greater abundance of 5-c than 6-c heme, and the proportions of these two species are reestablished after complete decay of intermediates.
Y. Wei Tang, personal communication.
- INH
- isonicotinic acid hydrazide (isoniazid)
- 5-c
- five-coordinate
- 6-c
- six coordinate
- PAA
- peroxyacetic acid
- ITC
- isothermal titration calorimetry
- RFQ-EPR
- rapid freeze-quench EPR
- WT
- wild type.
REFERENCES
- 1.Kochi A. ( 1991) Tubercle 72, 1– 6 [DOI] [PubMed] [Google Scholar]
- 2.Robitzek E. H., Selikoff I. J. ( 1952) Am. Rev. Tuberc. 65, 402– 428 [DOI] [PubMed] [Google Scholar]
- 3.Sepkowitz K. A. ( 2001) Emerg. Infect. Dis. 7, 259– 262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sepkowitz K. A. ( 1995) Clin. Infect. Dis. 20, 232– 242 [DOI] [PubMed] [Google Scholar]
- 5.Hutchison D. C., Drobniewski F. A., Milburn H. J. ( 2003) Respir. Med. 97, 65– 70 [DOI] [PubMed] [Google Scholar]
- 6.Phyu S., Ti T., Jureen R., Hmun T., Myint H., Htun A., Grewal H. M., Bjorvatn B. ( 2003) Emerg. Infect. Dis. 9, 274– 276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. ( 2007) MMWR Morb. Mortal Wkly. Rep. 56, 250– 253 [PubMed] [Google Scholar]
- 8.Gandhi N. R., Moll A., Sturm A. W., Pawinski R., Govender T., Lalloo U., Zeller K., Andrews J., Friedland G. ( 2006) Lancet 368, 1575– 1580 [DOI] [PubMed] [Google Scholar]
- 9.Welinder K. G. ( 1991) Biochim. Biophys. Acta 1080, 215– 220 [DOI] [PubMed] [Google Scholar]
- 10.Ng V. H., Cox J. S., Sousa A. O., MacMicking J. D., McKinney J. D. ( 2004) Mol. Microbiol. 52, 1291– 1302 [DOI] [PubMed] [Google Scholar]
- 11.Nagy J. M., Cass A. E., Brown K. A. ( 1997) J. Biol. Chem. 272, 31265– 31271 [DOI] [PubMed] [Google Scholar]
- 12.Wengenack N. L., Rusnak F. ( 2001) Biochemistry 40, 8990– 8996 [DOI] [PubMed] [Google Scholar]
- 13.Timmins G. S., Deretic V. ( 2006) Mol. Microbiol. 62, 1220– 1227 [DOI] [PubMed] [Google Scholar]
- 14.Sinha B. K. ( 1983) J. Biol. Chem. 258, 796– 801 [PubMed] [Google Scholar]
- 15.Engleder M., Regelsberger G., Jakopitsch C., Furtmüller P. G., Rüker F., Peschek G. A., Obinger C. ( 2000) Biochimie 82, 211– 219 [DOI] [PubMed] [Google Scholar]
- 16.Johnsson K., King D. S., Schultz P. G. ( 1995) J. Am. Chem. Soc. 117, 5009– 5010 [Google Scholar]
- 17.Rozwarski D. A., Grant G. A., Barton D. H., Jacobs W. R., Jr., Sacchettini J. C. ( 1998) Science 279, 98– 102 [DOI] [PubMed] [Google Scholar]
- 18.Quémard A., Sacchettini J. C., Dessen A., Vilcheze C., Bittman R., Jacobs W. R., Jr., Blanchard J. S. ( 1995) Biochemistry 34, 8235– 8241 [DOI] [PubMed] [Google Scholar]
- 19.Hazbón M. H., Brimacombe M., Bobadilla del Valle M., Cavatore M., Guerrero M. I., Varma-Basil M., Billman-Jacobe H., Lavender C., Fyfe J., García-García L., León C. I., Bose M., Chaves F., Murray M., Eisenach K. D., Sifuentes-Osornio J., Cave M. D., Ponce de León A., Alland D. ( 2006) Antimicrob. Agents Chemother. 50, 2640– 2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morlock G. P., Metchock B., Sikes D., Crawford J. T., Cooksey R. C. ( 2003) Antimicrob. Agents Chemother. 47, 3799– 3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen M. H., Vilchèze C., Kremer L., Besra G. S., Parsons L., Salfinger M., Heifets L., Hazbon M. H., Alland D., Sacchettini J. C., Jacobs W. R., Jr. ( 2002) Mol. Microbiol. 46, 453– 466 [DOI] [PubMed] [Google Scholar]
- 22.Cardoso R. F., Cooksey R. C., Morlock G. P., Barco P., Cecon L., Forestiero F., Leite C. Q., Sato D. N., Shikama Mde L., Mamizuka E. M., Hirata R. D., Hirata M. H. ( 2004) Antimicrob. Agents Chemother. 48, 3373– 3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Argyrou A., Vetting M. W., Aladegbami B., Blanchard J. S. ( 2006) Nat. Struct. Mol. Biol. 13, 408– 413 [DOI] [PubMed] [Google Scholar]
- 24.Argyrou A., Jin L., Siconilfi-Baez L., Angeletti R. H., Blanchard J. S. ( 2006) Biochemistry 45, 13947– 13953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parsons L. M., Salfinger M., Clobridge A., Dormandy J., Mirabello L., Polletta V. L., Sanic A., Sinyavskiy O., Larsen S. C., Driscoll J., Zickas G., Taber H. W. ( 2005) Antimicrob. Agents Chemother. 49, 2218– 2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gegia M., Mdivani N., Mendes R. E., Li H., Akhalaia M., Han J., Khechinashvili G., Tang Y. W. ( 2008) Antimicrob. Agents Chemother. 52, 725– 729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marttila H. J., Mäkinen J., Marjamäki M., Ruutu P., Soini H. ( 2008) Int. J. Tuberc. Lung Dis. 12, 338– 343 [PubMed] [Google Scholar]
- 28.Dalla Costa E. R., Ribeiro M. O., Silva M. S., Arnold L. S., Rostirolla D. C., Cafrune P. I., Espinoza R. C., Palaci M., Telles M. A., Ritacco V., Suffys P. N., Lopes M. L., Campelo C. L., Miranda S. S., Kremer K., da Silva P. E., Fonseca Lde S., Ho J. L., Kritski A. L., Rossetti M. L. ( 2009) BMC Microbiol. 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouse D. A., DeVito J. A., Li Z., Byer H., Morris S. L. ( 1996) Mol. Microbiol. 22, 583– 592 [DOI] [PubMed] [Google Scholar]
- 30.Yu S., Girotto S., Lee C., Magliozzo R. S. ( 2003) J. Biol. Chem. 278, 14769– 14775 [DOI] [PubMed] [Google Scholar]
- 31.Saint-Joanis B., Souchon H., Wilming M., Johnsson K., Alzari P. M., Cole S. T. ( 1999) Biochem. J. 338, 753– 760 [PMC free article] [PubMed] [Google Scholar]
- 32.Wengenack N. L., Todorovic S., Yu L., Rusnak F. ( 1998) Biochemistry 37, 15825– 15834 [DOI] [PubMed] [Google Scholar]
- 33.Zhao X., Yu H., Yu S., Wang F., Sacchettini J. C., Magliozzo R. S. ( 2006) Biochemistry 45, 4131– 4140 [DOI] [PubMed] [Google Scholar]
- 34.Metcalfe C., Macdonald I. K., Murphy E. J., Brown K. A., Raven E. L., Moody P. C. ( 2008) J. Biol. Chem. 283, 6193– 6200 [DOI] [PubMed] [Google Scholar]
- 35.Johnsson K., Froland W. A., Schultz P. G. ( 1997) J. Biol. Chem. 272, 2834– 2840 [DOI] [PubMed] [Google Scholar]
- 36.Loewen P. C., Stauffer G. V. ( 1990) Mol. Gen. Genet. 224, 147– 151 [DOI] [PubMed] [Google Scholar]
- 37.Chouchane S., Lippai I., Magliozzo R. S. ( 2000) Biochemistry 39, 9975– 9983 [DOI] [PubMed] [Google Scholar]
- 38.Marcinkeviciene J. A., Magliozzo R. S., Blanchard J. S. ( 1995) J. Biol. Chem. 270, 22290– 22295 [DOI] [PubMed] [Google Scholar]
- 39.Zhao X., Yu S., Magliozzo R. S. ( 2007) Biochemistry 46, 3161– 3170 [DOI] [PubMed] [Google Scholar]
- 40.Ranguelova K., Girotto S., Gerfen G. J., Yu S., Suarez J., Metlitsky L., Magliozzo R. S. ( 2007) J. Biol. Chem. 282, 6255– 6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chouchane S., Girotto S., Yu S., Magliozzo R. S. ( 2002) J. Biol. Chem. 277, 42633– 42638 [DOI] [PubMed] [Google Scholar]
- 42.Chouchane S., Girotto S., Kapetanaki S., Schelvis J. P., Yu S., Magliozzo R. S. ( 2003) J. Biol. Chem. 278, 8154– 8162 [DOI] [PubMed] [Google Scholar]
- 43.Ranguelova K., Suarez J., Metlitsky L., Yu S., Brejt S. Z., Brejt S. Z., Zhao L., Schelvis J. P., Magliozzo R. S. ( 2008) Biochemistry 47, 12583– 12592 [DOI] [PubMed] [Google Scholar]
- 44.Bertrand T., Eady N. A., Jones J. N., Jesmin Nagy J. M., Jamart-Grégoire B., Raven E. L., Brown K. A. ( 2004) J. Biol. Chem. 279, 38991– 38999 [DOI] [PubMed] [Google Scholar]
- 45.Yu S., Chouchane S., Magliozzo R. S. ( 2002) Protein Sci. 11, 58– 64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wengenack N. L., Uhl J. R., St Amand A. L., Tomlinson A. J., Benson L. M., Naylor S., Kline B. C., Cockerill F. R., 3rd, Rusnak F. ( 1997) J. Infect. Dis. 176, 722– 727 [DOI] [PubMed] [Google Scholar]
- 47.Jakopitsch C., Vlasits J., Wiseman B., Loewen P. C., Obinger C. ( 2007) Biochemistry 46, 1183– 1193 [DOI] [PubMed] [Google Scholar]
- 48.Ivancich A., Jakopitsch C., Auer M., Un S., Obinger C. ( 2003) J. Am. Chem. Soc. 125, 14093– 14102 [DOI] [PubMed] [Google Scholar]
- 49.Svistunenko D. A. ( 2001) Biochim. Biophys. Acta 1546, 365– 378 [DOI] [PubMed] [Google Scholar]
- 50.Kelman D. J., Mason R. P. ( 1992) Free Radic. Res. Commun. 16, 27– 33 [DOI] [PubMed] [Google Scholar]
- 51.Lei B., Wei C. J., Tu S. C. ( 2000) J. Biol. Chem. 275, 2520– 2526 [DOI] [PubMed] [Google Scholar]
- 52.Broussy S., Bernardes-Génisson V., Coppel Y., Quémard A., Bernadou J., Meunier B. ( 2005) Org. Biomol. Chem. 3, 670– 673 [DOI] [PubMed] [Google Scholar]
- 53.Ghiladi R. A., Medzihradszky K. F., Rusnak F. M., Ortiz de Montellano P. R. ( 2005) J. Am. Chem. Soc. 127, 13428– 13442 [DOI] [PubMed] [Google Scholar]
- 54.Kapetanaki S., Chouchane S., Girotto S., Yu S., Magliozzo R. S., Schelvis J. P. ( 2003) Biochemistry 42, 3835– 3845 [DOI] [PubMed] [Google Scholar]
- 55.Kapetanaki S. M., Zhao X., Yu S., Magliozzo R. S., Schelvis J. P. ( 2007) J. Inorg. Biochem. 101, 422– 433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelman D. J., DeGray J. A., Mason R. P. ( 1994) J. Biol. Chem. 269, 7458– 7463 [PubMed] [Google Scholar]
- 57.Girotto S. ( 2004) Application of EPR Spectroscopy to Study the Resting State Structure and the Mechanism of Mycobacterium tuberculosis Catalase-Peroxidase (KatG). Ph.D. thesis, The Graduate Center of the City University of New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.