Abstract
Yeast vacuole fusion requires soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), the Rab GTPase Ypt7p, vacuolar lipids, Sec17p and Sec18p, and the homotypic fusion and vacuole protein sorting complex (HOPS). HOPS is a multisubunit protein with direct affinities for SNAREs, vacuolar lipids, and the GTP-bound form of Ypt7p; each of these affinities contributes to HOPS association with the organelle. Using all-purified components, we have reconstituted fusion, but the Rab Ypt7p was not required. We now report that phosphorylation of HOPS by the vacuolar kinase Yck3p blocks HOPS binding to vacuolar lipids, making HOPS membrane association and the ensuing fusion depend on the presence of Ypt7p. In accord with this finding in the reconstituted fusion reaction, the inactivation of Ypt7p by the GTPase-activating protein Gyp1–46p only blocks the fusion of purified vacuoles when Yck3p is present and active. Thus, although Ypt7p may contribute to other fusion functions, its central role is to bind HOPS to the membrane.
Rab proteins are small GTP-binding proteins involved in multiple steps of membrane traffic, including protein sorting, vesicle transport, and SNARE3-dependent membrane fusion (1). Rabs in their GTP-bound state bind proteins that are essential for mediating Rab function, which are therefore termed “effectors.” These effectors are diverse and perform various biochemical functions. For membrane fusion, Rabs and their effectors support tethering, the initial membrane contact that is needed for the subsequent assembly of trans-SNARE complexes between membranes (1, 2). A central question in organelle trafficking, which we now address, is whether Rabs are only required for binding their effectors to the membrane or whether they also activate the bound effector or provide some additional essential function for membrane fusion.
We study membrane fusion using isolated yeast vacuoles (3). Yeast vacuole fusion requires the Rab GTPase Ypt7p, the heterohexameric HOPS complex, four vacuolar SNAREs, the SNARE disassembly chaperones Sec17p and Sec18p, and chemically minor yet functionally essential lipids, termed “regulatory” lipids. The HOPS complex is an effector of Ypt7p (4) and belongs to a group of functionally conserved large multisubunit tethering complexes, many of which are Rab effectors (5). The Vps39p subunit of HOPS is a nucleotide exchange factor for Ypt7p (6). HOPS is also a SNARE chaperone; its Vps33p subunit is a Sec1p/Munc18-1 family (SM) protein, HOPS binds multiple vacuolar SNAREs (7–9), and it proofreads SNARE complex structure (10). HOPS also binds to specific phosphoinositides (8), and these are among the regulatory lipids that are important for fusion (11–13).
We have recently reconstituted membrane fusion using proteoliposomes of pure vacuolar proteins and lipids (13). HOPS and the regulatory lipids are crucial for rapid fusion of proteoliposome pairs bearing the three Q-SNAREs on one proteoliposome and the R-SNARE on the other and are absolutely required when all four SNAREs are present on each proteoliposome and Sec17p and Sec18p are present. Ypt7p is not required, showing that HOPS can stimulate SNARE-dependent fusion in vitro even in the absence of its Rab, although Ypt7p stimulates the fusion of these proteoliposomes.4
Yeast vacuole fusion can be negatively regulated either by GTPase-activating proteins (GAPs) (14, 15) that promote GTP hydrolysis by Ypt7p or by the kinase Yck3p, which phosphorylates the Vps41p subunit of HOPS (16) and the vacuolar SNARE Vam3p (15). Yck3p is a palmitoylated (17), vacuole-localized kinase of the casein kinase I family (18). The complete fragmentation of vacuoles in vivo, indicating a block of fusion, requires both Ypt7p inactivation by a RabGAP and the presence of Yck3p (15). Yck3p is necessary for efficient vacuole inheritance (16) and normal vacuole morphology (19), suggesting that its function is part of the normal mechanism of vacuole segregation during the cell cycle. Although Yck3p clearly regulates vacuole fusion through phosphorylation of HOPS, it remains unclear which activities of HOPS are inhibited by Yck3p phosphorylation and whether Yck3p must also phosphorylate other vacuole fusion proteins such as Vam3p to block fusion.
We now show that phosphorylation of the Vps41p subunit of HOPS by purified Yck3p reduces HOPS binding to membrane lipids, thereby making HOPS association with the membrane and the ensuing fusion of reconstituted proteoliposomes dependent on active Ypt7p. These data with proteoliposomes are supported by assays with purified vacuoles; the RabGAP Gyp1–46p only inhibits the in vitro fusion of yck3Δ vacuoles when purified Yck3p is added. As for Ypt7p and HOPS, the major function of other Rabs may also be to act as membrane receptors for their effectors.
EXPERIMENTAL PROCEDURES
Strain and Plasmid Construction
BJ3505 (MATα ura3-52 trp1-Δ101 his3-Δ200 lys2-801 gal2 (gal3) can1 prb1-Δ1.6R pep4::HIS3) (20) and DKY6281 (MATα ura3-52 leu2-3112 trp1-Δ901 his3-Δ200 lys2-801 suc2-Δ9 pho8::TRP1) (21) have been described previously. CHY53 (BJ3505 yck3Δ::hphMX4) and CHY54 (DKY6281 yck3Δ::hphMX4) were generated by transformation (22) of BJ3505 and DKY6281 to replace YCK3 with a DNA cassette that was formed from a two-round PCR amplification from pAG32 (23) using primer set 1 (Table 1) followed by primer set 2. DNA coding for amino acids 2–516 of Yck3p was PCR-amplified from FY834 (MATα ura3-52 leu2-Δ1 lys2-Δ202 trp1-Δ63 his3-Δ200) (24) genomic DNA using primer set 3 and cloned into the SpeI/XhoI sites of pHIS.Parallel1 (25) to produce pHIS.Parallel-YCK3-(2–516).
TABLE 1.
Primers used for strain and plasmid construction (5′–3′)
| Primer name | Primer |
|---|---|
| 1a | GTGTAAAAGGACGATAAGGAAAGATGCAGCTGAAGCTTCGTACGC |
| 1b | GCGGCATAAGGAAACAGCCAATCAGCATAGGCCACTAGTGGATCTG |
| 2a | AGTGGTATCTCATTCTGAAGAAAAAGTGTAAAAGGACGATAAGGA |
| 2b | TATTGGAAAGTCATAAATTGGGAAAGAGCGGCATAAGGAAACAGC |
| 3a | GCGCACTAGTGTCCCAACGATCTTCACAACAC |
| 3b | GCGCCTCGAGTCATCAATATTTGTATATTTTAGAACAAAATGTATC |
| 4a | GGTCCAGGTGAAAATTTGTATTTTCAAGGTTCTTCTAGAAAAAAAAATATTTTGAAAG |
| 4b | CCCGAATTCCTCTAGCCAATTTATCCTGTCGTTG |
| 5a | CCCAGATCTGTAAAGAGCCCCATTATC |
| 5b | ACCTTGAAAATACAAATTTTCACCTGGACCACGCGGAACCAGATCCGATT |
DNA for the coding sequence and terminator of YPT7 was PCR-amplified from Saccharomyces cerevisiae genomic DNA using primer set 4. DNA encoding the S. cerevisiae GAL1 promoter followed by Schistosoma japonicum GST was amplified from pFA6a-kanMX6-PGAL-GST (26) using primer set 5. The products of these reactions were gel-purified (QIAquick PCR purification kit; Qiagen) and mixed, and an overlapping DNA was PCR-amplified using primers 4b and 5a. This product, encoding the GAL1 promoter, GST, a tobacco etch virus protease cleavage site, the YPT7 coding sequence with the initial methionine replaced by the glycine from the tobacco etch virus protease site, and the YPT7 terminator, was digested with EcoRI and BamHI (New England Biolabs), gel-purified, and ligated (rapid DNA ligation kit; Roche Applied Science) into the same sites in pYC230 (27). The resulting plasmid was transformed (22) into BJ5459 (MATa ura3-52 trp1 lys2-801 leu2-Δ1 his3-Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL) (28) to produce CSY29.
Proteins
His6-Sec17p and His6-Sec18p were purified as described (29). Gyp1–46p (30) was the generous gift of Dr. Vincent Starai (Dartmouth).
To produce recombinant Yck3p, competent Escherichia coli Rosetta2(DE3)pLysS cells (Novagen) were transformed with pHIS.Parallel-YCK3-(2–516), and a colony was used to inoculate 3 liters of LB medium with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol in a 6-liter flask and grown with shaking at 37 °C to A600 ∼1. The culture was transferred to a shaker at 30 °C, and protein expression was induced with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside for 5 h. Cells were harvested by centrifugation (5 min, 4 °C, 5000 rpm, Beckman JLA-10.500 rotor), resuspended in 35 ml of 20 mm HEPES-NaOH, pH 7.4, 250 mm NaCl, 10% glycerol, 20 mm imidazole, 1 mm phenylmethylsulfonyl fluoride, 0.46 μg/ml leupeptin, 3.5 μg/ml pepstatin A, 2.4 μg/ml Pefabloc-SC, and frozen dropwise in liquid N2. Cells were thawed and lysed via French press at 4 °C. The extract was applied to 3 ml of packed nickel-nitrilotriacetic acid agarose (Qiagen) in a 1.5 × 10 cm column (Bio-Rad) equilibrated with 20 mm HEPES-NaOH, pH 7.4, 250 mm NaCl, 10% glycerol, 20 mm imidazole. The resin was washed with 20 ml of 20 mm HEPES-NaOH, pH 7.4, 250 mm NaCl, 10% glycerol, 20 mm imidazole, and protein was eluted with 20 mm HEPES-NaOH, pH 7.4, 250 mm NaCl, 10% glycerol, 250 mm imidazole. The protein pool was dialyzed into 20 mm HEPES-NaOH, pH 7.4, 250 mm NaCl, 10% glycerol, 2 mm dithiothreitol. This protein, His6-Yck3-(2–516)p, lacks the C-terminal eight amino acids of the native protein required for palmitoylation in yeast, which does not occur in E. coli.
To make recombinant Ypt7p protein from yeast, CSY29 was grown at 30 °C in 10 liters of YPD medium containing 200 μg/ml G418 (Mediatech) to an A600 of ∼2. d-Galactose (Difco) was added from a 40% solution to a final concentration of 2%, and the culture was continued at 30 °C for 24 h. Cells were harvested by centrifugation (5 min, 4 °C, 5000 rpm, Beckman JLA-10.500 rotor), washed with 350 ml of RB500+ (20 mm NaHEPES, pH 7.4, 500 mm NaCl, 1 mm MgCl2 10% glycerol), resuspended with a minimal amount of RB500+ containing phenylmethylsulfonyl fluoride at a final concentration of 1 mm, frozen dropwise in liquid N2, broken using a Waring blender in the presence of liquid N2, and stored at −80 °C. After thawing on ice, the lysate was divided into four Type 45 Ti tubes (Beckman), and the tubes were filled with RB500+ with 1 mm phenylmethylsulfonyl fluoride. The lysate was centrifuged for 1 h at 4 °C in a Type 45 Ti rotor (Beckman) at 40,000 rpm, and the pellets were washed twice by resuspension with 50 ml/tube of RB500+ and centrifugation as above. The washed pellets were resuspended with 50 ml/tube of solubilization buffer (RB500+ with 0.5% Thesit) and incubated on ice for 1 h. The suspensions were centrifuged as above, and the supernatant was nutated with 10 ml of glutathione-Sepharose 4B (GE Healthcare) for 1 h. The resin was washed three times with 40 ml of solubilization buffer and three times with 40 ml of cleavage buffer (20 mm Tris-Cl, pH 8.0, 50 mm NaCl, 1 mm MgCl2, 10% glycerol, 0.5% Thesit), “in-batch” in a 50-ml conical tube and brought to a 50% slurry with cleavage buffer. The amount of bound GST-Ypt7p was estimated by SDS-PAGE and Coomassie Brilliant Blue R250 staining, with bovine serum albumin as a standard. Tobacco etch virus protease was added at a 1:5 protease:GST-Ypt7p molar ratio, and the cleavage reaction was nutated overnight at 4 °C. The resin was drained and washed three times with 10 ml of cleavage buffer; washes and cleavage supernatant were pooled and applied at 0.5 ml/min to a MonoQ 5/50 GL column (GE Healthcare) equilibrated with 50 mm NaCl in buffer A (20 mm Tris-Cl, pH 8.0, 10% glycerol, 1% n-octyl-β-d-glucopyranoside, 1 mm MgCl2, 1 mm dithiothreitol). The column was washed with 10 ml of the same buffer then eluted with a 1-ml gradient to 100 mm NaCl in buffer A and a 20-ml gradient to 200 mm NaCl in buffer A. Fractions of 1 ml were collected; fractions containing Ypt7p were identified by SDS-PAGE and Coomassie Brilliant Blue R250 staining and pooled, and the pooled protein was frozen in aliquots in liquid N2.
HOPS was produced as described previously (10). Phosphorylated-HOPS (P-HOPS) and mock-treated HOPS were produced using this methodology with modifications. After the yeast membrane lysate was passed over the glutathione-Sepharose (Amersham Biosciences) column, the column was washed with 2 bed volumes of lysis buffer. The glutathione-Sepharose was then resuspended in 1 bed volume of lysis buffer, split into two equal fractions in 50-ml conical tubes, and centrifuged (5 min, 4 °C, 1120 × g, clinical centrifuge), and the supernatants were removed. Lysis buffer with 1 mm MgCl2 and 0.5 mm ATP was added to each tube, and then Yck3p was added to the treated sample to a final concentration of 7 μm. After mixing, tubes were incubated at 4 °C for 12 h. The contents of each tube were resuspended, poured into two separate columns, and washed with 2 bed volumes of lysis buffer followed by 3 bed volumes of low Triton X-100 buffer. HOPS and P-HOPS were eluted (10), concentrated (10), and stored at −80 °C in aliquots.
Vacuole Isolation and in Vitro Vacuole Fusion Assay
Yeast vacuoles from YCK3 (BJ3505 and DKY6281) and yck3Δ (CHY53 and CHY54) strains were isolated, and fusion was assayed as described (31). Fusion reactions contained, per 30-μl reaction, 3 μg each of vacuoles lacking either the protease Pep4p (BJ3505 or CHY53) or the phosphatase Pho8p (DKY6281 or CHY54), fusion reaction buffer (20 mm PIPES-KOH, pH 6.8, 125 mm KCl, 5 mm MgCl2, 200 mm sorbitol), ATP-regenerating system (1 mm ATP, 40 mm creatine phosphate, 0.1 mg/ml creatine kinase), 582 nm purified Pbi2p, and 10 μm coenzyme A. For assays with yck3Δ vacuoles, Yck3p was diluted in 20 mm HEPES-NaOH, pH 7.4, 250 mm NaCl, 10% glycerol, 2 mm dithiothreitol, and 1 μl of this buffer or Yck3p at the indicated final concentrations was added.
Liposome Production
Proteoliposomes were prepared as described (13) with modifications. All synthetic phospholipids used were of 16:0–18:1 (PO) acyl chain composition. Where present, yeast Ypt7p was added to the detergent-lipid mixed micellar solutions at either 1 μm or 2 μm/2 mm lipids. 1-Palmitoyl-2-oleoyl phosphatidylcholine/1-palmitoyl-2-oleoyl-phosphatidyl-l-serine liposomes contain 1-palmitoyl-2-oleoyl phosphatidylcholine (82%), 1-palmitoyl-2-oleoyl-phosphatidyl-l-serine (15%), and fluorescent lipids. Protein-free liposomes were made in the same manner but without the addition of SNAREs or Ypt7p.
Lipid Mixing Assays
Lipid mixing assays were performed as described (13). Briefly, reaction mixtures in RB150 were prepared in 384-well plates on ice and were composed of donor (50 μm lipids) and acceptor (400 μm lipids) SNARE proteoliposomes. Proteoliposome lipid composition was vacuole mimic plus regulatory lipids (13) unless otherwise designated.
RESULTS
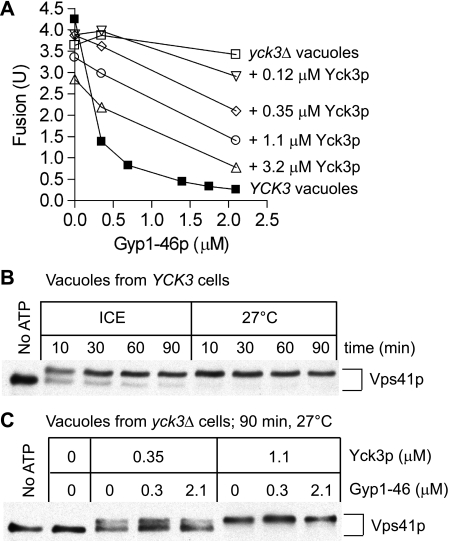
Ypt7p function on isolated vacuoles can be modulated by the addition of a RabGAP (14), triggering hydrolysis of bound GTP to GDP with consequent inactivation of Rab function. Fusion of isolated vacuoles can be blocked by at least two different RabGAPs, Gyp7–47p (14, 30) or Gyp1–46p (30, 32) (Fig. 1A, filled squares). A pioneering study by LaGrassa and Ungermann (16) showed that Gyp7–47p does not inactivate the fusion of vacuoles from yck3Δ strains that lack the vacuolar kinase Yck3p. This result is not GAP-specific as Gyp1–46p also does not inactivate fusion of yck3Δ vacuoles (Fig. 1A, open squares). To test whether this reflects a direct requirement for Yck3p or whether Yck3p acts indirectly (e.g. through regulating the trafficking of a fusion factor to the organelle (33)), we purified recombinant, unpalmitoylated Yck3p from E. coli. The addition of Yck3p to vacuoles from yck3Δ strains had little effect on fusion but restored the capacity of Gyp1–46p to inhibit fusion (Fig. 1A, open symbols). The Vps41p subunit of HOPS undergoes an efficient, rapid, and ATP-dependent shift to higher Mr during in vitro fusion incubation of vacuoles from YCK3 strains (16, 34) (Fig. 1B), but this shift is not seen with vacuoles from yck3Δ strains (16) (Fig. 1C). The addition of recombinant Yck3p restores the Mr shift of Vps41p, unaffected by Gyp1–46p (Fig. 1C).
FIGURE 1.
RabGAP sensitivity and Vps41p phosphorylation are restored to yck3Δ vacuoles treated with Yck3p. A, vacuoles isolated from BJ3505 and DKY6281 or BJ3505 yck3Δ (CHY53) and DKY6281 yck3Δ (CHY54) were assayed for fusion in the presence of the indicated concentrations of Gyp1–46p and Yck3p. B, vacuoles were isolated from BJ3505 and DKY6281 and then either incubated without ATP on ice or incubated under fusion reaction conditions (see “Experimental Procedures”) (except that vacuole concentrations were 6 μg of each per 30-μl reaction) for the indicated times on ice or at 27 °C. Reactions were incubated with 0.4% SDS at 90 °C for 5 min followed by immunoblot for Vps41p. C, vacuoles were isolated from yck3Δ strains CHY53 and CHY54 and then either incubated without ATP on ice or incubated under fusion reaction conditions (see “Experimental Procedures”). Reactions were incubated with 0.4% SDS at 90 °C for 5 min followed by immunoblot for Vps41p. These samples were from some of the reactions of panel A.
These data suggest that HOPS might be able to support fusion without Ypt7p function, whereas phosphorylated HOPS is strictly Ypt7p-dependent. Although this has been established by further experiments (below), these data (Fig. 1) were also consistent with other models. For example, without phosphorylation, HOPS might have blocked Gyp1–46p from acting on Ypt7p. Alternatively, Yck3p phosphorylation of Vam3p (15), or other vacuole proteins, might have contributed to the Yck3p inhibition of fusion. Other, more complex models can be envisioned. To distinguish among these possibilities, we turned to the chemically defined reconstitution of proteoliposome fusion, using purified vacuole components4 (13).
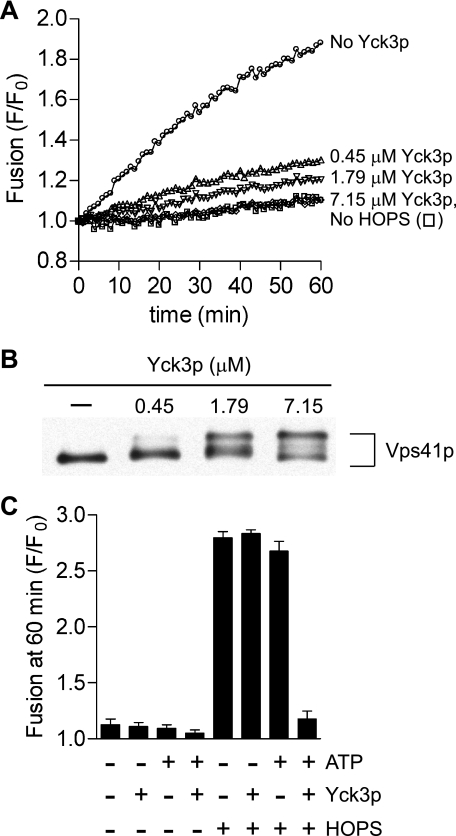
We first assayed whether Yck3p would act in this reconstituted fusion reaction. Proteoliposomes of vacuolar lipid composition, including phosphoinositides, which bear the two integral vacuolar Q-SNAREs (Vam3p, Vti1p) on one set of liposomes and the vacuolar R-SNARE (Nyv1p) on the others, were prepared by the standard dialysis method (13). The soluble Q-SNARE Vam7p was added to initiate fusion reactions. The HOPS-dependent fusion of these proteoliposomes was progressively inhibited by increasing amounts of added Yck3p (Fig. 2A), corresponding to modification of an increasing portion of Vps41p (Fig. 2B). Inhibition required both Yck3p and ATP (Fig. 2C), and thus, likely results from Yck3p activity as a protein kinase.
FIGURE 2.
Recombinant Yck3p inhibits Ypt7p-independent reconstituted proteoliposome fusion and directly phosphorylates the Vps41p of pure HOPS. A, lipid mixing between liposomes bearing Qab-SNAREs (acceptor) and R-SNARE (donor). All reactions contained 1 mm MgCl2 and 0.5 mm ATP. HOPS (66.3 nm) and Yck3p (0.45–7.15 μm) were added as indicated. Vam7p (300 nm) was added to all wells at time 0, which followed a 10-min preincubation of the plate at 27 °C. B, selected reactions from panel A were incubated with 0.4% SDS at 90 °C for 5 min followed by SDS-PAGE and immunoblot for Vps41p. C, lipid mixing between liposomes bearing Qab-SNAREs (acceptor) and R-SNARE (donor). Reactions without ATP contained 0.5 mm MgCl2, and reactions with ATP contained 1 mm MgCl2 and 0.5 mm ATP. Yck3p (7.15 μm) and HOPS (66.3 nm) were added as indicated. Vam7p (300 nm) was added to all tubes at time 0, which followed a 10-min preincubation of the plate at 27 °C. Results are the mean of three independent experiments ± S.D.
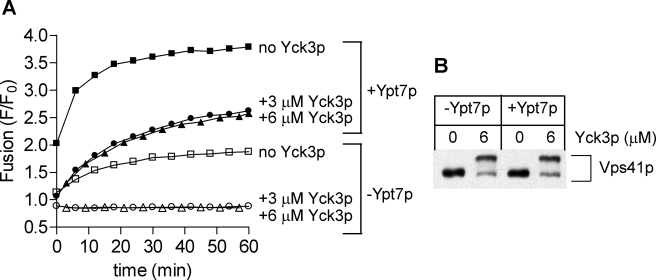
Yck3p only blocks vacuole fusion when Ypt7p is inactivated (16) (Fig. 1A). We therefore asked whether the inclusion of purified prenylated Ypt7p during proteoliposome preparation would allow fusion in the presence of Yck3p. Ypt7p stimulated fusion (Fig. 3A, squares), and concentrations of Yck3p that fully inhibit Ypt7p-independent fusion (Fig. 3A, open symbols) only partially inhibit the fusion of proteoliposomes bearing Ypt7p (Fig. 3A, filled symbols). Recent studies of intact vacuoles by Merz and colleagues (15) suggested that Ypt7p might influence the phosphorylation of Vps41p by Yck3p. In the reconstituted reactions shown in Fig. 3A, the Mr shift of Vps41p caused by Yck3p was equivalent in the presence or absence of Ypt7p (Fig. 3B). Thus, under our conditions, Ypt7p promotes the activity of phosphorylated HOPS rather than inhibiting the phosphorylation of Vps41p.
FIGURE 3.
Ypt7p diminishes the inhibition of proteoliposome fusion by Yck3p but does not inhibit the Yck3p phosphorylation of Vps41p. A, lipid mixing between liposomes bearing Qab-SNAREs (acceptor) and R-SNARE (donor) ± Ypt7p (200 nm for acceptor proteoliposomes and 25 nm for donor proteoliposomes). All reactions contained 6 mm MgCl2 and 1 mm ATP. HOPS (75 nm) and Yck3p (3 or 6 μm) was added as indicated. Vam7p (600 nm), Sec17p (1.2 μm), and Sec18p (1.4 μm) were added to all tubes at time 0, which followed a 10-min preincubation of the plate at 27 °C. B, selected reactions from panel A were incubated with 0.4% SDS at 90 °C for 5 min followed by SDS-PAGE and immunoblot for Vps41p.
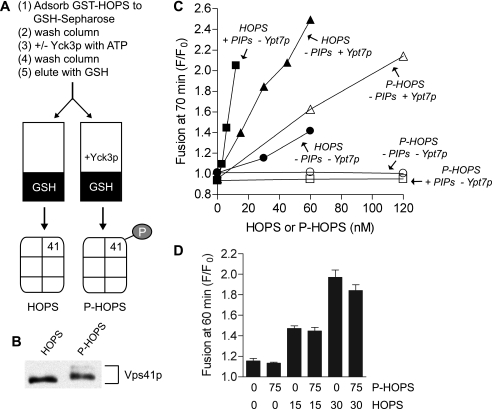
Because Yck3p can phosphorylate both the Vps41p subunit of HOPS (16) and the Vam3p SNARE (15), which are both essential for fusion (35, 36), we phosphorylated HOPS in isolation to study its role. To produce P-HOPS, GST-tagged HOPS was bound to glutathione-Sepharose, treated with Yck3p, and then washed to remove the Yck3p before eluting the P-HOPS (Fig. 4A). Mock-treated HOPS, which was bound, incubated without kinase in the same buffer, and eluted, was used as the control for all experiments employing P-HOPS. The Vps41p of P-HOPS had slower electrophoretic mobility than that of HOPS (Fig. 4B). Although the majority of the Vps41p of P-HOPS only shifted to an intermediate species, P-HOPS has activities (below) consistent with Yck3p addition to proteoliposome reactions. Fig. 2B clearly shows that Vps41p phosphorylation by Yck3p results in multiple species, indicating multiple sites of phosphorylation. This is consistent with the finding that multiple serine residues need to be mutated to abolish Vps41p electrophoretic mobility shift after in vitro incubation of membranes with ATP (37). However, it remains unclear which of these residues are actually phosphorylated by Yck3p and whether phosphorylation of only these serines is sufficient to cause the observed maximal shift in Vps41p mobility. In this report, we will focus on the effect of Vps41p phosphorylation on HOPS activity and how this relates to the need for Ypt7p.
FIGURE 4.
Phosphorylated-HOPS (P-HOPS) only stimulates SNARE-dependent proteoliposome fusion when proteoliposomes bear Ypt7p. A, scheme for the preparation of HOPS and P-HOPS. B, immunoblot for Vps41p of HOPS and P-HOPS preparations. C, lipid mixing between liposomes bearing Qab-SNAREs (acceptor) and R-SNARE (donor). Proteoliposomes had no phosphoinositides or Ypt7p (−PIPs − Ypt7p), phosphoinositides and no Ypt7p (+ PIPs − Ypt7p), or Ypt7p (400 nm for acceptor proteoliposomes and 50 nm for donor proteoliposomes) and no phosphoinositides (− PIPs + Ypt7p), as indicated. All reactions contained 1.5 mm MgCl2 and 1 mm ATP. HOPS and P-HOPS were added at the indicated concentrations. Vam7p (350 nm), Sec17p (1.2 μm), and Sec18p (1.4 μm) were added to all tubes at time 0, which followed a 10-min preincubation of the plate at 27 °C. D, lipid mixing between liposomes bearing Qab-SNAREs (acceptor) and R-SNARE (donor). All reactions contained 0.5 mm MgCl2. P-HOPS was added to a mixture of donor and acceptor liposomes 10 min before HOPS, all on ice. Vam7 (300 nm) was added at time 0, which followed a 10-min preincubation of the plate at 27 °C. Results are the mean of three independent experiments ± S.D.
HOPS and P-HOPS were added to fusion reactions with liposomes either bearing bulk vacuolar lipids or supplemented with phosphoinositides or Ypt7p. As reported, HOPS supports proteoliposome fusion (13) (Fig. 4C, filled circles), and either phosphoinositides (filled squares) or Ypt7p (filled triangles), which support HOPS association with the membrane (8) (Fig. 5), stimulate the rate of fusion. In contrast, P-HOPS strictly requires Ypt7p to support any fusion at all (Fig. 4C, open symbols). The simplest explanation for these findings would be that P-HOPS has lost its affinity for vacuolar lipids while retaining its affinity for Ypt7p. In accord with this idea, excess P-HOPS does not interfere with the capacity of HOPS to support the fusion of SNARE proteoliposomes that lack Ypt7p (Fig. 4D).
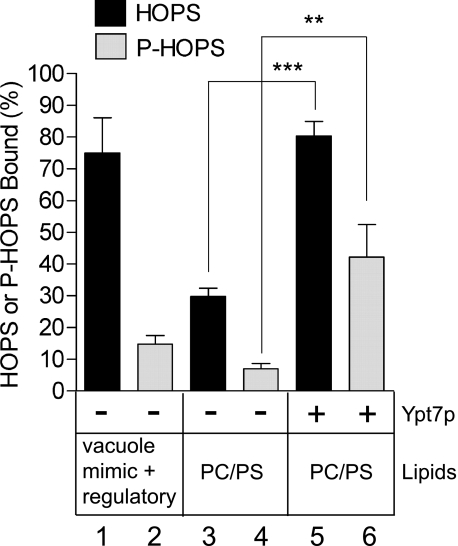
FIGURE 5.
P-HOPS is deficient in binding vacuole membrane lipids but retains the ability to bind Ypt7p. SNARE-free liposomes (450 μm lipids final) of the indicated compositions (Ypt7p was 450 nm where present) with NBD-PE (N-(7-nitro-2,1,3-benzoxadiazole-4-yl)-phosphatidylethanolamine) and Rh-PE (N-(lissamine rhodamine B sulfonyl) phosphatidylethanolamine) were mixed on ice in RB150 with 0.5 mm MgCl2, 2 mg/ml defatted bovine serum albumin, and HOPS (75 nm final) or P-HOPS (75 nm final) in a total volume of 20 μl in Microfuge tubes. The tubes were incubated for 10 min at 27 °C and then on ice for 5 min. Histodenz was added to 40% from a 54% stock in RB150, the tubes were centrifuged (Eppendorf 5415 C Microfuge, 1500 rpm, 2 min, 4 °C), and the mixture (80 μl) was transferred to a 7 × 20-mm tube (Beckman). This mixture was overlaid with 80 μl of 35% Histodenz in RB150, 80 μl of 30% Histodenz in RB150, and 50 μl of RB150 and then centrifuged (50,000 rpm, 1.5 h, 4 °C, TLS-55 rotor with adaptors for 7 × 20-mm tubes). Liposomes (80 μl) were recovered from the top of the gradient and solubilized in 0.1% Thesit to determine lipid recovery or incubated with 0.4% SDS at 90 °C for 5 min followed by SDS-PAGE and immunoblotting for Vps11p. Liposome-bound HOPS or P-HOPS was quantified using the UN-SCAN-IT software (Silk Scientific) with a standard curve of P-HOPS. Lipid recovery was measured by fluorescence (λex/λem 460/538 nm) of the Thesit-solubilized liposomes and used to adjust the percentage of HOPS or P-HOPS bound. Results are the mean of three independent experiments ± S.D. ***, p < 0.0001, **, p < 0.01 using unpaired t test (GraphPad Prism). PC/PS, phosphatidylcholine/phosphatidylserine.
In direct binding assays, P-HOPS had greatly diminished capacity for stable association with liposomes of vacuolar lipids, including phosphoinositides (Fig. 5, lanes 1 and 2). In contrast to the very stable association of HOPS observed with vacuole lipid liposomes, much less HOPS bound to phosphatidylcholine/phosphatidylserine (PC/PS) liposomes (lane 3). However, the inclusion of Ypt7p in phosphatidylcholine/phosphatidylserine liposomes enhanced P-HOPS binding (lane 6 versus lane 4) as much as HOPS binding (lane 5 versus lane 3). Thus, binding of HOPS to liposomes can be mediated by lipids, Ypt7p, or both, whereas P-HOPS requires Ypt7p for stable binding. These differences in binding explain why P-HOPS requires Ypt7p for reconstituted fusion, whereas HOPS does not.
DISCUSSION
Rabs have been proposed to function in membrane fusion through multiple mechanisms. Many Rabs have a multisubunit tethering complex as an effector (1, 5). It has been unclear whether tethering complexes require Rabs for functions beyond supporting their membrane localization, e.g. for their allosteric activation. Also unclear is whether Rabs function downstream of tethering, in catalysis of trans-SNARE complex assembly or bilayer destabilization, for example. Current systems to study Rabs have particular shortcomings. With in vivo studies, the absence of a specific Rab may be compensated by a non-cognate Rab (38) and confounded by aberrant Rab-dependent biosynthetic trafficking (1). Organelles used for in vitro fusion reactions, such as the yeast vacuole, are of complex composition, hindering the identification of the minimal components required for fusion. Using all-purified components, whether vacuolar4 or endosomal,5 reconstituted fusion systems that are affected by Rabs and their effectors are attractive for dissecting the molecular events performed by these conserved factors. Here we used such a reconstitution system to show that the major function of the Rab Ypt7p is to bind its effector, HOPS, to the membrane, a function that HOPS can fulfill on its own through direct lipid binding when the Vps41p subunit of HOPS is not phosphorylated by the vacuolar kinase Yck3p.
The involvement of the Rab Ypt7p in vacuole fusion has been well established both in vivo (39) and in vitro (40). Purified vacuoles bear Ypt7p (40). The active Ypt7p is in the GTP-loaded state and can be inactivated by RabGAPs (14). Ypt7p is needed for the formation and maintenance of the “vertex ring” domain, a specialized membrane microdomain enriched in the proteins and lipids crucial for fusion (12, 41, 42). HOPS, also enriched at the vertex ring domain, binds to Ypt7p, SNAREs, and regulatory lipids, and each of these interactions likely contribute to maintenance of this domain (12). Thus, although HOPS is the only known effector of Ypt7p, Ypt7p indirectly localizes the SNAREs and regulatory lipids through its localization of HOPS. Despite the essential role of Ypt7p in vacuole fusion under physiological conditions, its fusion function can be bypassed by elevated levels of each of the vacuolar SNARE proteins (43). However, vacuoles with these high SNARE densities undergo lysis as well as fusion, although lysis is not seen when fusion is driven by the Rab and normal levels of the SNAREs (43). Thus, one function of the Rab is to localize the proper levels of SNAREs to sites of fusion, allowing the cell to maintain overall low levels of SNAREs, which would destabilize membranes at high densities. This model of Rab function in fusion agrees with the more general paradigm that Rabs are involved in the maintenance of membrane identity and specific function (44).
What is the basis for in vitro fusion of yck3Δ vacuoles in the presence of Ypt7p inhibitors (16) (Fig. 1A) or proteoliposomes lacking Ypt7p in the absence of Yck3p activity (13) (Fig. 2A)? The binding of HOPS to vacuolar lipids (Fig. 5) provides sufficient affinity for HOPS association with the membrane. Most studies of in vitro vacuole fusion have used vacuoles bearing active Yck3p, which rapidly and efficiently phosphorylates Vps41p when the vacuoles are incubated with ATP (Fig. 1B). HOPS binding to lipids is therefore reduced under these conditions, making the interaction with the Rab essential for fusion. These data are consistent with studies showing that more HOPS is released from vacuoles bearing Yck3p than those lacking Yck3p when treated with Gyp7–47p or Gdi1p (16). This model also explains why Gyp1–46p-treated vacuoles assayed for fusion without ATP, in the presence of recombinant Vam7p, show less inhibition of fusion than Gyp1–46p-treated vacuoles assayed for fusion with ATP (32).
The fraction of HOPS that is phosphorylated in vivo, where ATP is ample and phosphatases are present, remains unclear. The fact that Yck3p is required for proper vacuole homeostasis (16, 19) suggests that HOPS spends significant time in the phosphorylated state. Understanding HOPS phosphorylation will also require the identification of a phosphatase dedicated to removing the inhibitory phosphate(s) from Vps41p. It will be interesting to determine whether Ypt7p always acts in concert with lipid binding to maintain HOPS on the organelle or whether Ypt7p is largely binding phosphorylated HOPS.
It has been suggested that HOPS phosphorylation might weaken the interaction of HOPS with Ypt7p (5, 15, 16). However, we find that this interaction is maintained and that the binding of HOPS to lipids is altered by phosphorylation (Fig. 5). One model incorporating Yck3p activity has suggested that active Ypt7p opposes HOPS phosphorylation by Yck3p (15). In our reconstituted system, Yck3p phosphorylation was equivalent in the presence and absence of active Ypt7p (Fig. 3B). Despite active Yck3p, proteoliposomes with Ypt7p retain significant fusion activity (Fig. 3A), consistent with our finding that Ypt7p can bind phosphorylated HOPS to membranes (Fig. 5). Vacuoles might bear factors not present in our reconstitution that could influence this system. One possibility (15) is that a vacuolar phosphatase might be localized and/or activated by active Ypt7p. However, Gyp1–46p did not affect the ability of added Yck3p to phosphorylate Vps41p from yck3Δ vacuoles (Fig. 1C). It should be noted that the recombinant Yck3p used in our work is lacking the palmitoylation modification found on the native protein, which may be important for the regulation of Yck3p on vacuoles. However, Ypt7p need not oppose HOPS phosphorylation to stimulate fusion in the presence of Yck3p (Fig. 3A). Our model is in agreement with the finding that Ypt7p overproduction can rescue the vacuole fragmentation phenotype of yeast expressing a mutant Vps41p that mimics Vps41p that has been phosphorylated by Yck3p (37).
Other Rab effector tethering complexes are also regulated by phosphorylation. The Rab5 effectors EEA1 and rabenosyn-5 are phosphorylated by the p38 mitogen-activated protein (MAP) kinase during stimulated exocytosis of μ-opoid receptors (45). EEA1 is phosphorylated within its FYVE domain, which is responsible for phosphatidylinositol-3-phosphate binding, and this phosphorylation seems to enhance the membrane localization and fusion activity of EEA1. The activity of the coiled-coil protein p115, the mammalian homologue of yeast Uso1p, is also enhanced when phosphorylated (46). In this case, the stimulation by phosphorylation of Golgi reassembly after mitotic fragmentation appears to be through enhanced protein-protein interaction. Recently, phosphorylation of the exocyst during insulin stimulation has been reported, yet the functional effect, if any, of this phosphorylation is unknown (47). Thus, although phosphorylation of tethering complexes is a common theme, the molecular interactions affected and the direction of the effect vary considerably.
In this report, we have shown that the major role of Ypt7p in fusion is to bind HOPS to the vacuole membrane. HOPS does not require Ypt7p to stimulate fusion when it can associate with vacuoles or proteoliposomes via binding vacuolar lipids, suggesting either that Ypt7p does not allosterically activate HOPS or that an allosteric activation of HOPS by Ypt7p is not required for fusion. Our work with purified vacuoles shows that Ypt7p is also important for the formation and maintenance of the vertex ring membrane microdomain where fusion occurs (12, 41, 42). It is likely that the proteoliposomes used in our current investigations have properties more like the vertex ring domain than the overall vacuole (e.g. higher SNARE densities, higher concentrations of regulatory lipids, and perhaps greater membrane curvature due to their smaller diameters), reducing the need for membrane microdomain formation.
Supplementary Material
Acknowledgments
We thank Drs. Joji Mima (Dartmouth) and Vincent Starai (Dartmouth) for reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1GM23377 through the NIGMS (to W. W) and by National Institutes of Health Training Grants T32GM008704 and T32AR007576 (to C. M. H.).
This article was selected as a Paper of the Week.
C. Stroupe, C. M. Hickey, J. Mima, A. S. Burfeind, and W. Wickner, manuscript in preparation.
M. Zerial, personal communication.
- SNARE
- soluble NSF attachment protein receptor
- Q-SNARE
- SNARE with glutamine
- R-SNARE
- SNARE with arginine
- NSF
- N-ethylmaleimide-sensitive factor
- HOPS
- homotypic fusion and vacuole protein sorting complex
- P-HOPS
- phosphorylated HOPS
- GAP
- GTPase-activating protein
- GST
- glutathione S-transferase
- PIPES
- 1,4-piperazinediethanesulfonic acid.
REFERENCES
- 1.Grosshans B. L., Ortiz D., Novick P. ( 2006) Proc. Natl. Acad. Sci. U. S. A. 103, 11821– 11827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao X., Ballew N., Barlowe C. ( 1998) EMBO J. 17, 2156– 2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostrowicz C. W., Meiringer C. T., Ungermann C. ( 2008) Autophagy 4, 5– 19 [DOI] [PubMed] [Google Scholar]
- 4.Seals D. F., Eitzen G., Margolis N., Wickner W. T., Price A. ( 2000) Proc. Natl. Acad. Sci. U. S. A. 97, 9402– 9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markgraf D. F., Peplowska K., Ungermann C. ( 2007) FEBS Lett. 581, 2125– 2130 [DOI] [PubMed] [Google Scholar]
- 6.Wurmser A. E., Sato T. K., Emr S. D. ( 2000) J. Cell Biol. 151, 551– 562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins K. M., Thorngren N. L., Fratti R. A., Wickner W. T. ( 2005) EMBO J. 24, 1775– 1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stroupe C., Collins K. M., Fratti R. A., Wickner W. ( 2006) EMBO J. 25, 1579– 1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dulubova I., Yamaguchi T., Wang Y., Südhof T. C., Rizo J. ( 2001) Nat. Struct. Biol. 8, 258– 264 [DOI] [PubMed] [Google Scholar]
- 10.Starai V. J., Hickey C. M., Wickner W. ( 2008) Mol. Biol. Cell 19, 2500– 2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer A., Scheglmann D., Dove S., Glatz A., Wickner W., Haas A. ( 2000) Mol. Biol. Cell 11, 807– 817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fratti R. A., Jun Y., Merz A. J., Margolis N., Wickner W. ( 2004) J. Cell Biol. 167, 1087– 1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mima J., Hickey C. M., Xu H., Jun Y., Wickner W. ( 2008) EMBO J. 27, 2031– 2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eitzen G., Will E., Gallwitz D., Haas A., Wickner W. ( 2000) EMBO J. 19, 6713– 6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brett C. L., Plemel R. L., Lobinger B. T., Vignali M., Fields S., Merz A. J. ( 2008) J. Cell Biol. 182, 1141– 1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaGrassa T. J., Ungermann C. ( 2005) J. Cell Biol. 168, 401– 414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meiringer C. T., Ungermann C. ( 2006) Methods 40, 171– 176 [DOI] [PubMed] [Google Scholar]
- 18.Sun B., Chen L., Cao W., Roth A. F., Davis N. G. ( 2004) Mol. Biol. Cell 15, 1397– 1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seeley E. S., Kato M., Margolis N., Wickner W., Eitzen G. ( 2002) Mol. Biol. Cell 13, 782– 794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones E. W. ( 2002) Methods Enzymol. 351, 127– 150 [DOI] [PubMed] [Google Scholar]
- 21.Haas A., Conradt B., Wickner W. ( 1994) J. Cell Biol. 126, 87– 97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gietz R. D., Schiestl R. H. ( 1995) Methods Mol. Cell Biol. 5, 255– 269 [Google Scholar]
- 23.Goldstein A. L., McCusker J. H. ( 1999) Yeast 15, 1541– 1553 [DOI] [PubMed] [Google Scholar]
- 24.Winston F., Dollard C., Ricupero-Hovasse S. L. ( 1995) Yeast 11, 53– 55 [DOI] [PubMed] [Google Scholar]
- 25.Sheffield P., Garrard S., Derewenda Z. ( 1999) Protein Expr Purif. 15, 34– 39 [DOI] [PubMed] [Google Scholar]
- 26.Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. ( 1998) Yeast 14, 953– 961 [DOI] [PubMed] [Google Scholar]
- 27.Olesen K., Franke Johannesen P., Hoffmann L., Bech Sorensen S., Gjermansen C., Hansen J. ( 2000) Yeast 16, 1035– 1043 [DOI] [PubMed] [Google Scholar]
- 28.Jones E. W. ( 1991) Methods Enzymol. 194, 428– 453 [DOI] [PubMed] [Google Scholar]
- 29.Haas A., Wickner W. ( 1996) EMBO J. 15, 3296– 3305 [PMC free article] [PubMed] [Google Scholar]
- 30.Albert S., Will E., Gallwitz D. ( 1999) EMBO J. 18, 5216– 5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haas A. ( 1995) Methods Cell Sci. 17, 283– 294 [Google Scholar]
- 32.Thorngren N., Collins K. M., Fratti R. A., Wickner W., Merz A. J. ( 2004) EMBO J. 23, 2765– 2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anand V. C., Daboussi L., Lorenz T. C., Payne G. S. ( 2009) Mol. Biol. Cell 20, 1592– 1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price A., Seals D., Wickner W., Ungermann C. ( 2000) J. Cell Biol. 148, 1231– 1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price A., Wickner W., Ungermann C. ( 2000) J. Cell Biol. 148, 1223– 1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nichols B. J., Ungermann C., Pelham H. R., Wickner W. T., Haas A. ( 1997) Nature 387, 199– 202 [DOI] [PubMed] [Google Scholar]
- 37.Cabrera M., Ostrowicz C. W., Mari M., LaGrassa T. J., Reggiori F., Ungermann C. ( 2009) Mol. Biol. Cell 20, 1937– 1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ballew N., Liu Y., Barlowe C. ( 2005) Mol. Biol. Cell 16, 1839– 1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wada Y., Ohsumi Y., Anraku Y. ( 1992) J. Biol. Chem. 267, 18665– 18670 [PubMed] [Google Scholar]
- 40.Haas A., Scheglmann D., Lazar T., Gallwitz D., Wickner W. ( 1995) EMBO J. 14, 5258– 5270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L., Seeley E. S., Wickner W., Merz A. J. ( 2002) Cell 108, 357– 369 [DOI] [PubMed] [Google Scholar]
- 42.Wang L., Merz A. J., Collins K. M., Wickner W. ( 2003) J. Cell Biol. 160, 365– 374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starai V. J., Jun Y., Wickner W. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 13551– 13558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zerial M., McBride H. ( 2001) Nat. Rev. Mol. Cell Biol. 2, 107– 117 [DOI] [PubMed] [Google Scholar]
- 45.Macé G., Miaczynska M., Zerial M., Nebreda A. R. ( 2005) EMBO J. 24, 3235– 3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dirac-Svejstrup A. B., Shorter J., Waters M. G., Warren G. ( 2000) J. Cell Biol. 150, 475– 488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyons P. D., Peck G. R., Kettenbach A. N., Gerber S. A., Roudaia L., Lienhard G. E. ( 2009) Biosci. Rep., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.