Abstract
A quantitative in vivo method for detecting protein-protein interactions will enhance our understanding of protein interaction networks and facilitate affinity maturation as well as designing new interaction pairs. We have developed a novel platform, dubbed “yeast surface two-hybrid (YS2H),” to enable a quantitative measurement of pairwise protein interactions via the secretory pathway by expressing one protein (bait) anchored to the cell wall and the other (prey) in soluble form. In YS2H, the prey is released either outside of the cells or remains on the cell surface by virtue of its binding to the bait. The strength of their interaction is measured by antibody binding to the epitope tag appended to the prey or direct readout of split green fluorescence protein (GFP) complementation. When two α-helices forming coiled coils were expressed as a pair of prey and bait, the amount of the prey in complex with the bait progressively decreased as the affinity changes from 100 pm to 10 μm. With GFP complementation assay, we were able to discriminate a 6-log difference in binding affinities in the range of 100 pm to 100 μm. The affinity estimated from the level of antibody binding to fusion tags was in good agreement with that measured in solution using a surface plasmon resonance technique. In contrast, the level of GFP complementation linearly increased with the on-rate of coiled coil interactions, likely because of the irreversible nature of GFP reconstitution. Furthermore, we demonstrate the use of YS2H in exploring the nature of antigen recognition by antibodies and activation allostery in integrins and in isolating heavy chain-only antibodies against botulinum neurotoxin.
Protein-protein interactions are essential to virtually every cellular process, and their understanding is of great interest in basic science as well as in the development of effective therapeutics. Existing techniques to detect and screen pairs of interacting proteins in vivo include the yeast two-hybrid system (1) and protein-fragment complementation assay (PCA)2 (2–6), where the association of two interacting proteins either turns on a target gene that is necessary for cell survival or leads to the reconstitution of enzymes or green fluorescence protein (GFP) or its variants. The application of protein-protein interactions that are probed with yeast two-hybrid and PCA has been focused mainly on the interactions occurring in the nucleus or cytosol. To study interactions among secretory proteins and membrane-associated proteins, a variant of yeast two-hybrid has been developed for detecting protein-protein interactions occurring in the secretory pathway (7, 8). However, most existing methods are designed to map connectivity information for pairwise interactions and are not suitable for measuring the affinity between two interacting proteins, comparing interaction strength of different pairs, or ranking multiple binders to the interaction “hub” according to their binding affinity.
Quantitative estimation of protein-protein interactions in vivo will require the amount of the complex to be directly measured or the level of reconstituted reporters to be directly proportional to the strength of the interactions. To achieve quantitative analysis of protein interactions in eukaryotic expression system, we have designed a yeast surface two-hybrid (YS2H) system that can express a pair of proteins, one protein as a fusion to a yeast cell wall protein, agglutinin, and the other in a secretory form. When two proteins interact in this system, they associate in the secretory pathway, and the prey that would otherwise be released into the media is captured on the surface by the bait. We have devised two different schemes to quantitatively estimate the affinity of two interacting molecules: flow cytometric detection of antibody binding to the epitope tags fused to the prey and the bait, and the GFP readout from the complementation of split GFP fragments fused to the prey and the bait. They are induced under a bi-directional promoter to promote a synchronized and comparable level of expression.
Herein we demonstrate the quantitative nature of YS2H in predicting the affinity between two interacting proteins, particularly in the range of 100 pm to 10 μm. This feature allowed us to examine specific interactions between antigen and antibody, to identify hot spots of allosteric activation in integrins, and to isolate camelid heavy chain-only antibodies against botulinum neurotoxin as components of therapeutic agents to treat botulism (9). With the incorporation of PCA technique into the YS2H, our system may be developed into an in vivo tool to measure the kinetics of protein-protein interactions. Potential applications of YS2H include affinity maturation of antibodies, differentiation of weak to high affinity binders to the hub protein in interaction networks, and confirmation of hypothetical interacting pairs of proteins in a high throughput manner.
EXPERIMENTAL PROCEDURES
YS2H Vector Design
Plasmid pCTCON was used as a backbone for constructing the YS2H vector (see Fig. 1a). A PCR fragment containing GAL10 promoter, AGA2, eGFP gene, FLAG tag, and terminator was inserted into the pCTCON by AgeI/KpnI sites. To express prey proteins as secretory forms, AGA2 sequence under the GAL1 promoter was removed by replacing an EcoRI/BamHI fragment with the fragment consisting of a signal sequence, either that of Aga2 or α-1 mating factor, and prey. The cDNA coding for the variable domains of AL-57 was obtained from the expression plasmid (a kind gift from Dr. Shimaoka at Harvard Medical School). The variable domains of TS1/22 were cloned from the hybridoma (ATCC). VH and VL cDNAs were connected with four repeats of a Gly-Gly-Gly-Gly-Ser linker sequence to produce scFv.
FIGURE 1.
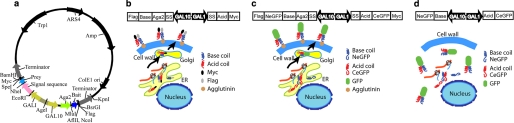
Design of the YS2H system. a, a map of the YS2H vector is drawn with restriction enzyme sites and genes labeled. The bait protein is expressed as a fusion to Aga2 on cell surface, whereas the prey protein is expressed as a secretory form. b and c, schematic diagrams of the expression cassette and protein-protein interactions (acid base coiled coils) via the secretory pathway are depicted. The prey bound to the bait is detected by antibody binding to the Myc tag (b) or by direct GFP readout from split GFP complementation (c). FLAG (DYKDDDDK) and Myc (EQKLISEEDL) epitope tags are fused to the C-terminal of the bait and prey proteins, respectively, and are used to measure the surface expression of the bait and the amount of the prey that is bound to the bait. d, the deletion of signal sequence for the prey and bait proteins leads to their expression in the cytosol.
Yeast Transformation, Magnetic Affinity Cell Sorting, and Library Construction
The plasmid encoding a specific pair of prey and bait proteins was introduced into yeast cells using a commercial reagent (Frozen-EZ Yeast Transformation II Kit, Zymo Research). Transformed yeast cells were grown in a solid medium plate for 48 h. A mutagenesis library of LFA-1 I domain was constructed by electroporation of a mixture of a MluI/NcoI linearized vector and error-prone PCR products of the I domain (Asn-129-Thr-318) into yeast, as described previously (10). After transformation, the yeast libraries were grown in selective dextrose liquid medium at 30 °C with shaking for 24 h and induced in selective galactose media for 24–48 h at room temperature with shaking. To construct the variable domain of heavy chain from heavy chain-only antibody (VHH) yeast library, cDNA encoding VHH library was amplified by PCR using the primers shown in supplemental Table S1, which were designed based on the primers used by Maass et al. (11). VHH cDNA PCR product was first ligated into the YS2H vector using NheI/BamHI sites and then was transformed into XL1-Blue (Stratagene) by electroporation. The plasmids extracted from ∼5 × 106 colonies were transformed into EBY100 by a lithium acetate method (12). A single colony of EBY100 from fresh plate was inoculated into 10 ml of YPDA medium and cultured at 30 °C with shaking at 225 rpm for 16 h. The cells were then inoculated into 100 ml of YPDA at 0.5 A600 and cultured for another 4 h until A600 reaches 2. The cells were washed twice in water and resuspended in 10.8 ml of transformation mix buffer (7.2 ml of 50% polyethylene glycol, 1.1 ml of 1 m LiAc, 1.5 ml of 2 mg/ml single strand carrier DNA, and 150 μg of library plasmid in 1.0 ml water). The mixture was then incubated at 42 °C for 50 min. After incubation, the cells were cultured into 100 ml of selective dextrose liquid medium for 24 h and induced in selective galactose medium for 24–48 h. Library construction by homologous recombination or the lithium acetate method produced a library size of 106–107. The libraries of LFA-1 I domain and VHH were sorted with anti-Myc antibody using magnetic affinity cell sorting as described previously (10).
Immunofluorescence Flow Cytometry
Antibodies used in this study were the anti-c-Myc antibody 9E10 (ATCC), anti-FLAG, and phycoerythrin-labeled goat polyclonal anti-murine antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). To measure the surface expression of specific prey and bait proteins using flow cytometry, one to five colonies from solid medium plate were inoculated together to obtain averaged values. After induction, the cells were harvested, washed in 100 μl of the labeling buffer (phosphate-buffered saline with 0.5% bovine serum albumin), and then incubated with ligands at 10 μg/ml in 50 μl of the labeling buffer for 20 min with shaking at 30 °C. The cells were then washed and incubated with secondary antibodies at 5 μg/ml in 50 μl of the labeling buffer for 20 min at 4 °C. Finally, the cells were washed once in 100 μl and suspended in 100 μl of the labeling buffer for flow cytometry (FACScan, BD Biosciences). For detecting TS1/22 binding (see Fig. 4b), goat polyclonal anti-murine antibody was used as a primary antibody.
FIGURE 4.
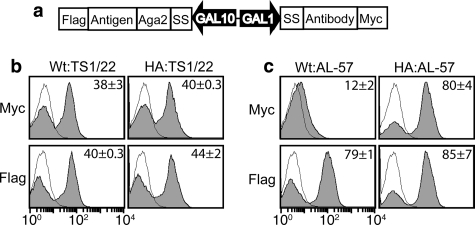
Detection of specific interactions between antibodies and antigens in YS2H. a, schematic diagram of the expression cassette used to study antigen (bait) and antibody (prey) interactions. b and c, shown are the histograms of the interactions of the wild-type and the high affinity (HA) I domains as baits and activation-insensitive antibody, TS1/22 (b) activation-specific antibody, AL-57 (c) as preys. Filled histograms are of antibody binding to Myc and FLAG tags to the induced clones. Thin black lines represent antibody binding to uninduced clones as controls. The numbers in each plot indicate the means ± S.E. of the MFI of the filled histograms from three independent measurements.
Protein Expression
The I domains were expressed in Escherichia coli BL21 DE3 (Invitrogen) as inclusion bodies and refolded and purified by an S75 size exclusion column connected to fast protein liquid chromatography (GE Healthcare) (10). AL-57 as a single-chain format (scFv AL-57) was expressed using the protocol for I domain production, except that 3 mm cystamine and 6 mm cysteamine were added to the refolding buffer. Full-length BoNT/A and BoNT/B-LC encoding DNA (amino acids 1–448 of A-LC and 1–440 of B-LC) were synthesized employing codons optimal for expression in E. coli. A-LC and B-LC containing hexahistidine tags at both termini were produced using a pET14b vector. To express VHHs in soluble forms, we inserted VHH cDNA into the pET20b expression vector (Novagen). Soluble VHH was expressed in E. coli BL21 DE3, extracted by sonication, and purified using a nickel nitrilotriacetic acid column. Eluted VHHs were then injected into an S75 size exclusion column for further purification.
SPR Analysis
A protein-coupled or a control mock-coupled CM5 sensor chip was prepared using an amine coupling kit (BIAcore, Piscataway, NJ), as described previously (10). SPR was measured using a Biacore (BIA2000). I domains were injected over the chip in 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 10 mm MgCl2 at a flow rate of 10 μl/min at room temperature. VHHs were injected over the chip in 20 mm Tris-HCl, pH 8.0, 150 mm NaCl at a flow rate of 10 μl/min at room temperature. The chip surface was regenerated by flowing 20 μl of 10 mm Tris-glycine, pH 1.5 buffer.
RESULTS
The Design of the YS2H
YS2H is built on a yeast display system (13, 14), which expresses, under the control of the GAL1 promoter, a protein of interest as a fusion to Aga2. Aga2 connects to the β-glucan linked Aga1 to form a cell wall protein called agglutinin. To extend this methodology to the expression of a pair of proteins, we have inserted into the yeast display vector, pCTCON (13), an additional expression cassette under the GAL10 promoter (15). We observed comparable expression of eGFP by GAL1 and GAL10 promoters using two different plasmids that were constructed to express eGFP under either GAL1 or GAL10 promoters (data not shown). The final YS2H vector (Fig. 1a) was designed to express the bait protein under the GAL10 promoter as a fusion to Aga2 and the prey protein under the GAL1 promoter without Aga2 fusion. The signal sequence used is either that of Aga2 (for the data in Figs. 2 and 3) or the α-1 mating factor (for the data in Figs. 4–6) (16). The expression level of prey proteins with α-1 mating factor was comparable with those containing Aga2 signal sequence (data not shown). FLAG and Myc tags are fused to the C-terminal of the bait and prey proteins, respectively, and are used to examine the surface expression of the bait and the amount of the prey that is bound to the bait (Fig. 1b). To incorporate the PCA technique into the YS2H system, we inserted the sequence encoding enhanced eGFP fragments (3) downstream of the bait (NeGFP contains residues Val-2 to Ala-155) and the prey (CeGFP with Asp-156 to Lys-239) to monitor their interaction by GFP readout (Fig. 1c). The deletion of the secretory signal sequence of the prey and bait proteins causes this pair to express in the cytosol (Fig. 1d), which can be used to compare protein-protein interactions occurring in the secretory pathway versus cytosol.
FIGURE 2.
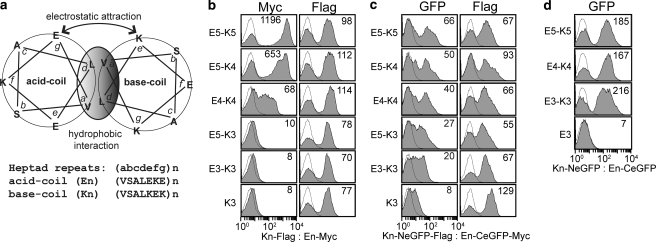
Detection of coiled coil interactions by epitope expression and GFP complementation. a, a schematic diagram (adapted from the Fig. 1 by De Crescenzo et al. (17)) of the acid (En)-base (Kn) coiled coils with n indicating the number of heptad repeats. b and c, the detection of coiled coil interactions by antibody binding to Myc tag (b) or direct GFP readout (c) using flow cytometry. Antibody binding to the FLAG tag measures the level of the base coil expression on cell surface. d, shown are the plots of GFP complementation caused by the coiled coil interactions occurring inside the cells. The numbers in each plot (b–d) indicate the MFI of an entire population shown in filled histogram. The thin lines represent the histograms of uninduced clones. The pairs of bait and prey are denoted for each column as bait:prey. The labels K3 and E3 indicate that the other coil is deleted from the expression vector.
FIGURE 3.
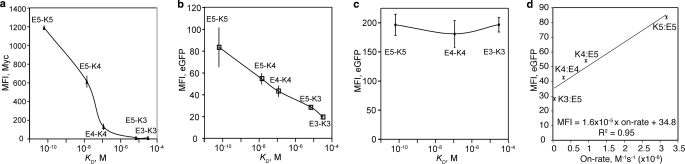
The correlation of the affinity measured by SPR (17) with the detection by epitope tag or direct readout of GFP complementation caused by coiled coil interactions occurring in the secretory pathway (a, b, and d) or in the cytosol (c). The data are from three independent experiments involving different clones (means ± S.E.). The smooth solid lines are drawn by connecting data points. d, the MFI of eGFP complementation from the coiled coil interactions is plotted as a function of their on-rate, measured by SPR (17). The solid line represents a least square fit to the data points.
FIGURE 5.
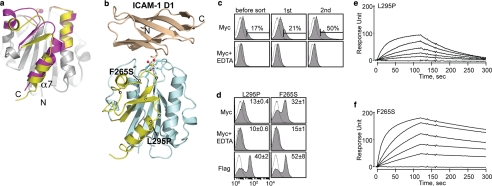
Discovery of allosteric activation in the I domain. a, cartoon diagrams of low (inactive) and high affinity (active) conformations of the LFA-1 I domains. The regions that are structurally conserved between two states are colored gray. The regions that differ structurally are colored in magenta and yellow for the inactive and the active conformations, respectively. The metal ions in the metal ion-dependent adhesion site are shown as spheres. The N and C termini and α7-helix are labeled. b, the structure of the I domain is shown in complex with the first domain of ICAM-1 (D1). Gray spheres with a white center display the positions for the hot spots for allosteric activation found in our previous study (10). The metal ion and three oxygen atoms of water molecules are depicted as spheres. The residues that coordinate to the metal ion are shown in stick models. The structures of the I domains and the complex of I domain with the ICAM-1 were modeled based on the crystal structures, as described previously (31). c, Myc expression of the I domain library before sort and after first and second sort are shown. The numbers indicate the percentage of the clones within the gated region. Antibody binding was measured with 10 mm MgCl2 or no metal ions with 10 mm EDTA. d, two activating mutations from the second sort were of F265S and L295P. The numbers in each plot indicate means ± S.E. of the MFI of the filled histograms from three independent measurements. e and f, SPR measurements of L295P (e) and F265S (f) binding to scFv AL-57. I domains were injected over the scFv AL-57-coated chip as a series of 2-fold dilutions beginning at 500 nm.
FIGURE 6.
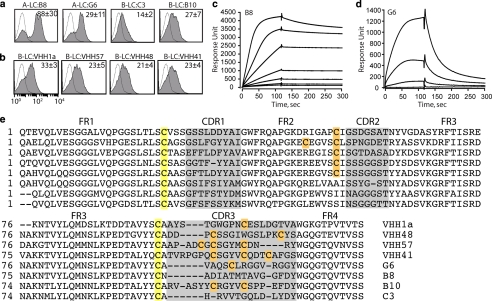
Detection of VHH binding to BoNT LC protease. a, specific binding of the VHHs against A-LC and B-LC was confirmed in YS2H by Myc expression. b, New VHHs against B-LC protease were isolated by YS2H. The numbers in each plot indicate means ± S.E. of the MFI of the filled histograms from three independent measurements. c and d, SPR measurements of B8 (c) and G6 (d) binding to BoNT/A-LC. A-LC was injected at a series of 2-fold dilutions beginning at 160 nm to the B8-coated and 400 nm to the G6-coated chip. e, cysteines are highlighted in yellow for the pair that forms a conserved disulfide bond or in orange that forms extra disulfide bonds. The framework region (FR) and complementarity determining regions (CDR) are noted.
The Validation of the Yeast Surface Two-hybrid System Using Coiled Coil Interaction
To validate that antibody binding to the Myc tag or GFP readout correlates with the strength of molecular interactions in YS2H, we expressed five pairs of acid (En) and base (Kn) α-helices of varying heptad repeats (n) that associate into coiled coils (Fig. 2). These coiled coils have been designed de novo to have affinities (KD) in the range of 100 pm (E5/K5) to 100 μm (E3/K3) with higher affinity for longer helices through hydrophobic interactions at the interface and electrostatic attraction between the oppositely charged residues from each helix (17). Myc expression (mean fluorescence intensity (MFI), measured by antibody binding to Myc tag) exhibited a strong correlation with the interaction affinity within the range of 100 pm to 10 μm KD for E5-K5 to E5-K3 (Figs. 2b and 3a). With GFP complementation, this correlation extended beyond 10 μm KD, and the difference between E5-K3 and E3-K3, corresponding to the affinity range of 10 μm to 100 μm, was clearly discernible (Figs. 2c and 3b). The Myc expression and GFP complementation were close to the level of background when the acid coil (K3 in Fig. 2, b and c) was deleted, indicating a lack of spontaneous complementation of the two split GFP. The level of surface expression of the bait protein measured by antibody binding to the FLAG tag was relatively invariant (Fig. 2, b and c), supporting the idea that the difference in the amount of the prey protein is solely due to the difference in its affinity to the bait. In contrast to a quantitative correlation between the strength of protein-protein interactions and GFP complementation, the acid and base coil interactions occurring in the cytosol (expression of the coils without secretory signal sequence) led to the complementation of split GFP that lacks correlation with the strength of coiled coil interactions (Figs. 2d and 3c). However, GFP complementation for these pairs was still due to specific interaction between acid and base coils, evidenced by the absence of fluorescence when the base coil was deleted from NeGFP (Fig. 2d).
YS2H Detects Specific Interactions of Antibodies and Antigens
To investigate a potential use of YS2H for antibody discovery, we first examined whether YS2H can detect specific interactions of known pairs of antigen and antibody. As a model system, we chose a ligand-binding domain of the integrin LFA-1, known as the Inserted or I domain, and monoclonal antibodies specific to LFA-1 I domain (Fig. 4). The I domain exists in two distinct conformations that correspond to low and high affinity states to its ligand, intercellular adhesion molecule-1 (ICAM-1) (Fig. 5, a and b). Although the I domain in isolation is predominantly in an inactive, low affinity conformation, the mutations that would favor the active conformation were found to induce high affinity binding of the I domain to the ICAM-1. For example, the mutations of K287C and K294C (high affinity or HA I domain) designed to stabilize by disulfide bond the position of the α7-helix into active conformation led to an increase in the affinity to ICAM-1 by 10,000-fold over the wild-type I domain (18).
The antibodies that were expressed as scFv formats include the activation-insensitive antibody (TS1/22) (19), binding to both inactive and active I domains, and the activation-dependent antibody (AL-57) (10, 20), which binds only to the active I domain. The interaction between antigen and antibody was measured by the detection of Myc tag fused to the antibody at the C terminus (Fig. 4). A tag-based assay was chosen instead of GFP complementation because it was found that the I domain fused to NeGFP did not express (no antibody binding to FLAG tag), presumably because of the quality control machinery in protein secretion (21) that prohibits misfolded proteins to be secreted (data not shown). This is in contrast to the expression of split GFP with a fusion of short coils, e.g. K3-NeGFP in Fig. 2c. Therefore, it appears that when NeGFP is fused to the I domain that by itself requires proper folding for secretion, the I domain fusion to NeGFP becomes completely misfolded and does not pass the quality control for secretion.
Myc tag expression in YS2H was in agreement with the specificities of monoclonal antibody AL-57 and TS1/22 against the LFA I domain; although the clones expressing TS1/22 displayed Myc expression either with the wild-type or with the HA I domains as antigens (Fig. 4b), the AL-57 clones exhibited Myc expression only with the HA I domain (Fig. 4c).
Discovery of Activating Mutations in the LFA-1 I Domain
Next, we examined the ability of our system in isolating activating mutations in antigens that exhibit two different activation states. With the expression of AL-57 scFv and the error-prone PCR products of the wild-type I domain in YS2H, yeast library was constructed and sorted with anti-Myc antibody using a magnetic affinity cell sorter. With successive sorting, there was a gradual increase in the percentage of the population of cells that showed Myc expression above the background level (Fig. 5c). After two rounds of sorting, the cells were plated to yield individual clones, from which four clones were sequenced and tested for Myc expression. Of the four, three contained a mutation of F265S, and one contained L295P (Fig. 5d). These two mutations belonged to a long list of activation hot spots that were identified in our previous study (10), where a large number of yeast cells were sorted and analyzed for their binding to exogenous AL-57 or ICAM-1-Fcγ.
The mutations of L295P and F265S were previously found to contribute to an increase in the binding of the I domain to ICAM-1 at 6 and 152%, respectively, of the HA I domain binding to ICAM-1 (10). To directly measure the affinity of scFv AL-57 to I domain variants, we used a SPR technique (Fig. 5, e and f). A first order Langmuir adsorption equation was fitted to the sensograms to obtain the kinetic and equilibrium binding constants. The equilibrium dissociation constants (KD) of L295P and F265S to scFv AL-57 were 243 and 15.7 nm, respectively, in agreement with higher Myc expression with F265S in our system. AL-57 binding to the LFA-1 I domain depends on the presence of metal ions at the top of the I domain, known as the metal ion-dependent adhesion site (Fig. 5b) (22). This was also confirmed by the decrease in the Myc tag expression when EDTA was added at 10 mm to the cells during labeling (Fig. 5, c and d).
Antibody Discovery: VHH against Botulinum Neurotoxin Protease
Approximately half of the IgGs in camelid sera are heavy chain-only antibodies devoid of light chains (11, 23). Because of the lack of light chains, antigenic specificity of the heavy chain-only antibodies is limited to a variable domain of the heavy chain. We are seeking VHH agents that bind and inhibit the LC protease domains of Botulinum neurotoxins (BoNTs) as components in therapeutic agents for the treatment of botulism. In prior work (Maass et al. (11),3 we immunized alpacas with BoNT LCs of serotype A (A-LC) followed by serotype B (B-LC). We then used phage display techniques to identify VHHs from these alpacas with affinity for the BoNT LC proteases. We selected two A-LC binding VHHs (B8 and G6) and two B-LC binding VHHs (B10 and C3) for testing with the YS2H system (Fig. 6a). Myc tag expression was found to be highest in the clone expressing VHH-B8 and BoNT/A-LC, whereas the binding level of the other VHHs was lower with MFI ranges from 14 to 29.
To confirm that the level of Myc expression correlates with the solution affinity of VHH to LC, we used a SPR technique (Fig. 6, c and d). A series of 2-fold dilutions of A-LC was injected into a chip coated with B8 and G6. The KD values of B8 and G6 to BoNT/A-LC were estimated to be 2.3 and 230 nm, respectively. The 100-fold difference in the affinity was mainly due to the 34-fold difference in the dissociation rate (koff = 5.18 × 10−4 s−1 for B8 versus 1.82 × 10−2 s−1 for G6), with the association rate differing by only 3-fold (kon = 2.26 × 105 m−1 s−1 for B8 versus 7.61 × 104 m−1 s−1 for G6).
To validate the use of YS2H for antibody discovery, a yeast antibody library was constructed by transforming cells with the alpaca immune cDNA library as prey and the BoNT/B-LC gene as bait. Yeast cells enriched after two rounds of magnetic affinity cell sorting with anti-Myc antibody began to show an increase in Myc tag expression (data not shown). Of 30 clones that were tested individually, 14 clones displayed positive expression of Myc tag. Eleven of these 14 clones were found to be unique clones, including B10 and C3, which were originally isolated by phage display. Anti-Myc antibody binding of the newly isolated nine clones was in the range of 21–33 MFI units, which is lower than that of B8 binding to A-LC (Myc expression of four selected clones are shown in Fig. 5b). Overall, anti-B-LC VHHs isolated by both the phage display and YS2H were low affinity binders (KD = 100 nm to 1 μm), suggesting a lack of the high affinity binders to B-LC protease in alpaca immune library.
Sequence analysis of the VHHs identified by phage or YS2H revealed that VHHs contain two, four, or six cysteines, which would result in up to two extra disulfide bonds in addition to the one that is conserved in all immunoglobulin fold domains (Fig. 6e). We found that of the nine VHHs newly isolated by our YS2H system, three VHHs contain six cysteines, and the other six VHHs contain four cysteines. In contrast, either two or four cysteines were dominant in VHHs that were isolated by phage display. The VHHs identified by a phage display system may be limited to those that fold properly in a bacterial expression system. VHHs containing extra disulfides may fold improperly in bacteria, whereas the formation of correct disulfide bonds is much less problematic in yeast.
DISCUSSION
Here we demonstrate that our novel system, YS2H, is highly efficient in the detection and discovery of protein-protein interactions. The quantitative nature of YS2H is from the fact that protein-protein interactions occur via a secretory pathway, and the amount of the prey protein in complex with the bait is determined by the equilibrium affinity between the two. With the use of a PCA technique, our system may be designed to discriminate different pairs of protein-protein interactions according to their kinetics of binding.
The utility of in vivo methods for quantitative estimation of binding affinity extends to the cases where one aims to increase the affinity of weak interactions between antigen and antibody and to engineer high affinity ligands and receptors that can potentially serve as agonists or antagonists. As an example, a prokaryotic system capable of co-expression of antigens anchored on the inner membrane of bacteria and single chain variable fragments (scFv) as soluble form was efficient in affinity screening and maturation (24). Therefore, co-expression of antigen and antibody through eukaryotic secretory system will further enable screening of antibody libraries against the proteins that require eukaryotic folding machinery or that undergo post-translational modifications.
The amount of the prey bound to the bait in our system follows the Langmuir binding isotherm model. With the expression system used in this study, the concentration of prey proteins released into the media is far larger than that of the bait proteins, which is fused to Aga2. Under the mating conditions, the number of agglutinin goes up to 10,000 copies/cell (25), which approximates the concentration of the bait proteins to be 1.7 nm at 108 cells/1 ml of culture medium. The Langmuir equation is then given by [bait:prey]/[bait] = [prey]/([prey] + KD) where [bait:prey], [bait], and [prey] denote the concentrations of the bait in complex with the prey, the bait, and the prey, respectively. By replacing [bait:prey] and [bait] with antibody binding to Myc (MFI_Myc) and FLAG tag (MFI_FLAG), respectively, and taking into consideration of the MFI ratio (α) of anti-Myc to anti-FLAG antibody binding to equal copies of Myc and FLAG tags, the Langmuir equation rearranges into 1/MFI_Myc = α−1 (1+ KD/[prey])/MFI_FLAG. From this equation (with measured values of α = 15 and [prey] = 10 nm), the KD values predicted for coiled coil interactions (E5-K5, E5-K4, E4-K4, and E5-K3) closely approximated the KD values measured by SPR (17) (Table 1). The quantitative nature of YS2H in measuring protein-protein interactions extends to antigen and antibody interactions. The binding affinity of scFv TS1/22 to the wild-type and HA I domain, scFv AL-57 to the HA I domain, and the F265S ranged between 148 and 237 nm KD, whereas it was 440 nm for the binding of L295P to scFv AL-57. SPR measurement of the binding of I domain variants to scFv AL-57 estimated that although F265S and HA I domains have comparable affinity to scFv AL-57, L295P showed much lower affinity (Fig. 5, e and f, and Table 1). The predicted affinity for VHH-B8 and VHH-G6 binding to A-LC is 38 and 143 nm, compared with the measured KD of 2.3 and 230 nm, respectively. Overall, the affinity predicted by the level of antibody binding to Myc and FLAG tag in our system agreed well with the measured affinity in the range of 1 nm to 1 μm KD (Table 1).
TABLE 1.
Comparison of equilibrium dissociation constants (KD) predicted from YS2H versus directly measured using surface plasmon resonance
| Interaction pairs | Predicted from YS2H(means ± S.E.) | SPR measurements |
|---|---|---|
| nm | nm | |
| E3-K3 | 1269.5 ± 53 | 32000 ± 3000a |
| E5-K3 | 1292.8 ± 37 | 7000 ± 800a |
| E4-K4 | 150.7 ± 48 | 116 ± 8a |
| E5-K4 | 18.3 ± 3 | 14 ± 1a |
| E5-K5 | 0.49 ± 0.2 | 0.063 ± 0.005a |
| HA-AL57 | 150.9 ± 8 | 32.6 ± 0.28b |
| F265S-AL57 | 237.4 ± 11.6 | 15.7 ± 0.03b |
| L295P-AL57 | 439.7 ± 13.6 | 243 ± 3.9b |
| VHH-B8-A-LC | 38.2 ± 13.3 | 2.3 ± 0.08b |
| VHH-G6-A-LC | 143.0 ± 41.9 | 230 ± 1.3b |
a The values are from the paper by De Crescenzoet al.(17). Shown are the means ± 95% confidence interval.
b The values are measured from this study. Shown are the means ± S.E., estimated from BIAevaulation software from Biacore.
Although antibody binding to the Myc tag for protein-protein interactions higher than 10 μm KD reduced to the level of background (Fig. 3a), the detection by GFP complementation spanned a larger range of affinities, exhibiting a linear decrease in the fluorescence with an increase in KD in log scale (Fig. 3b). This is attributed to the fact that reconstituted GFP does not dissociate (or complementation is irreversible), such that the complemented GFP is functional whether or not the prey and the bait exist as a complex on the cell surface. Therefore, the dominant factor that determines GFP reconstitution will be the rate at which two coils associate (on-rate) to initiate split GFP assembly. Notably, when the MFI of reconstituted GFP was plotted against the measured on-rates (17) for E5-K5, E5-K4, E4-K4, and E5-K3 (on-rate is unavailable for E3-K3), a liner trend was obtained with a R2 value of 0.95 (Fig. 3d). However, the use of GFP complementation was limited to the study of coiled coil interactions, because the I domain fused to the split GFP did not express on the surface. Therefore, to apply a PCA technique to detect diverse protein-protein interactions through the secretory pathway, split GFP needs to be optimized not to interfere with the folding of the bait and prey proteins. The additional parameter to be optimized is the length of the linker connecting split GFP to the proteins to enable GFP complementation for a wide range of size variation in proteins and topological variation between the binding interface and the GFP fusion site (6).
The fact that GFP complementation occurs by protein-protein interaction through the secretion process explains its quantitative correlation with the strength of protein-protein interactions. This is in contrast to a previous finding (4) that GFP complementation from protein interactions occurring in the cytosol only indicates the presence of the interaction, and fluorescence intensity is relatively invariant with the affinity of two proteins. Indeed, when the two coils were expressed in the cytosol with the deletion of the secretory signal sequence, we found that overall fluorescent intensity was higher, and the complementation of split GFP lacked correlation with the strength of coiled coil association (Figs. 2d and 3c). Because of the irreversible complementation of split GFP, after 24–48 h of induction, it is the concentration of two interacting proteins in the cytosol that determines the GFP complementation rather than their interaction strength.
Systems such as ribosomal (26), phage (27), and yeast (14) displays provide efficient means to couple genotype and phenotype and to screen library for protein engineering and antibody discovery. A typical screening process of antibody libraries requires exogenous antigens in their soluble form. Co-expression of two proteins within the same display system, e.g. the fusion of antigen and antibody into split phage coat protein (28) and the expression of antigen and antibody in bacterial periplasm as bait and prey proteins (24), can be particularly useful if target antigens are hard to express or unstable in solution. Our new system will offer a method to select antibodies against antigens that need to be expressed in eukaryotes. Other applications of YS2H may include expression of heterodimeric proteins, as demonstrated by similar platforms for expression of heterodimeric mammalian proteins such as major histocompatibility complex II α and β subunits (29) and antibodies in Fab format (30). The use of our system to quantify and discover protein-protein interactions is not necessarily limited to the study of secretory proteins, because many proteins in nonsecretory cellular compartments or cytosol will maintain native conformations and interactions.
An in vivo tool to map protein interactions has generated a large set of protein interactions, particularly among yeast proteins (6). The readouts from the assays such as yeast two-hybrid and PCA are of cell growth caused by the expression of auxotrophic markers or reconstitution of enzymes and fluorescent proteins and are suitable for determining the presence or absence of protein interactions. In the case of protein network “hubs” in the binary protein interactome, i.e. the proteins interacting with a large number protein partners, the information on the strength of pairwise interactions may provide an important insight into the flow of biological signals orchestrated by the protein hubs. Our newly developed YS2H system is well poised to implement such tasks. For example, in YS2H the hub proteins and known interacting partners are expressed as a pair of the bait and the prey, respectively, and the strength of pairwise interactions can be quantitatively estimated by antibody binding to fusion tags. Additionally, one can discover unknown interaction partners by expressing a library of hypothetical interacting partners in YS2H.
Supplementary Material
Acknowledgments
We acknowledge Róisín Owens for helpful comments on this manuscript and Xiaoling Gu for helping us to produce VHHs.
This work was supported, in whole or in part, by National Institutes of Health Grant NO1-AI30050 (to C. B. S.). This work was also supported by an American Heart Association Scientist Development grant (to M. M. J.) and a Northeast Biodefense Center grant (to M. M. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
X. Hu, C. B. Shoemaker, and M. M. Jin, manuscript in preparation.
- PCA
- protein-fragment complementation assay
- YS2H
- yeast surface two-hybrid
- GFP
- green fluorescence protein
- SPR
- surface plasmon resonance
- scFv
- single chain variable fragments
- eGFP
- enhanced GFP
- MFI
- mean fluorescence intensity
- LFA-1
- lymphocyte function-associated antigen-1
- ICAM-1
- intercellular adhesion molecule-1
- HA
- high affinity
- VHH
- variable domain of heavy chain from heavy chain-only antibody
- LC
- light chain
- BoNT
- Botulinum neurotoxin.
REFERENCES
- 1.Fields S., Song O. ( 1989) Nature 340, 245– 246 [DOI] [PubMed] [Google Scholar]
- 2.Ghosh I., Hamilton A. D., Regan L. ( 2000) J. Am. Chem. Soc. 122, 5658– 5659 [Google Scholar]
- 3.Hu C. D., Chinenov Y., Kerppola T. K. ( 2002) Mol. Cell 9, 789– 798 [DOI] [PubMed] [Google Scholar]
- 4.Magliery T. J., Wilson C. G., Pan W., Mishler D., Ghosh I., Hamilton A. D., Regan L. ( 2005) J. Am. Chem. Soc. 127, 146– 157 [DOI] [PubMed] [Google Scholar]
- 5.Remy I., Michnick S. W. ( 2004) Methods Mol. Biol. 261, 411– 426 [DOI] [PubMed] [Google Scholar]
- 6.Tarassov K., Messier V., Landry C. R., Radinovic S., Molina M. M. S., Shames I., Malitskaya Y., Vogel J., Bussey H., Michnick S. W. ( 2008) Science 320, 1465– 1470 [DOI] [PubMed] [Google Scholar]
- 7.Stagljar I., Fields S. ( 2002) Trends Biochem. Sci. 27, 559– 563 [DOI] [PubMed] [Google Scholar]
- 8.Nyfeler B., Michnick S. W., Hauri H. P. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 6350– 6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnon S. S., Schechter R., Inglesby T. V., Henderson D. A., Bartlett J. G., Ascher M. S., Eitzen E., Fine A. D., Hauer J., Layton M., Lillibridge S., Osterholm M. T., O'Toole T., Parker G., Perl T. M., Russell P. K., Swerdlow D. L., Tonat K. ( 2001) J. Am. Med. Assoc. 285, 1059– 1070 [DOI] [PubMed] [Google Scholar]
- 10.Jin M., Song G., Carman C. V., Kim Y. S., Astrof N. S., Shimaoka M., Wittrup D. K., Springer T. A. ( 2006) Proc. Natl. Acad. Sci. U. S. A. 103, 5758– 5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maass D. R., Sepulveda J., Pernthaner A., Shoemaker C. B. ( 2007) J. Immunol. Methods 324, 13– 25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gietz R. D., Schiestl R. H. ( 2007) Nat. Protoc. 2, 38– 41 [DOI] [PubMed] [Google Scholar]
- 13.Colby D. W., Kellogg B. A., Graff C. P., Yeung Y. A., Swers J. S., Wittrup K. D. ( 2004) Methods Enzymol. 388, 348– 358 [DOI] [PubMed] [Google Scholar]
- 14.Boder E. T., Wittrup K. D. ( 1997) Nat. Biotechnol. 15, 553– 557 [DOI] [PubMed] [Google Scholar]
- 15.Johnston M., Davis R. W. ( 1984) Mol. Cell Biol. 4, 1440– 1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egel-Mitani M., Hansen M. T., Norris K., Snel L., Fiil N. P. ( 1988) Gene 73, 113– 120 [DOI] [PubMed] [Google Scholar]
- 17.De Crescenzo G., Litowski J. R., Hodges R. S., O'Connor-McCourt M. D. ( 2003) Biochemistry 42, 1754– 1763 [DOI] [PubMed] [Google Scholar]
- 18.Shimaoka M., Xiao T., Liu J. H., Yang Y., Dong Y., Jun C. D., McCormack A., Zhang R., Joachimiak A., Takagi J., Wang J. H., Springer T. A. ( 2003) Cell 112, 99– 111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez-Madrid F., Krensky A. M., Ware C. F., Robbins E., Strominger J. L., Burakoff S. J., Springer T. A. ( 1982) Proc. Natl. Acad. Sci. U. S. A. 79, 7489– 7493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang L., Shimaoka M., Rondon I. J., Roy I., Chang Q., Po M., Dransfield D. T., Ladner R. C., Edge A. S., Salas A., Wood C. R., Springer T. A., Cohen E. H. ( 2006) J. Leukocyte Biol. 80, 905– 914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagihara Y., Kim P. S. ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 6619– 6624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimaoka M., Kim M., Cohen E. H., Yang W., Astrof N., Peer D., Salas A., Ferrand A., Springer T. A. ( 2006) Proc. Natl. Acad. Sci. U. S. A. 103, 13991– 13996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E. B., Bendahman N., Hamers R. ( 1993) Nature 363, 446– 448 [DOI] [PubMed] [Google Scholar]
- 24.Jeong K. J., Seo M. J., Iverson B. L., Georgiou G. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 8247– 8252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dranginis A. M., Rauceo J. M., Coronado J. E., Lipke P. N. ( 2007) Microbiol. Mol. Biol. Rev. 71, 282– 294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanes J., Plückthun A. ( 1997) Proc. Natl. Acad. Sci. U. S. A. 94, 4937– 4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith G. P. ( 1985) Science 228, 1315– 1317 [DOI] [PubMed] [Google Scholar]
- 28.Krebber C., Spada S., Desplancq D., Krebber A., Ge L., Pluckthun A. ( 1997) J. Mol. Biol. 268, 607– 618 [DOI] [PubMed] [Google Scholar]
- 29.Boder E. T., Bill J. R., Nields A. W., Marrack P. C., Kappler J. W. ( 2005) Biotechnol. Bioeng. 92, 485– 491 [DOI] [PubMed] [Google Scholar]
- 30.van den Beucken T., Pieters H., Steukers M., van der Vaart M., Ladner R. C., Hoogenboom H. R., Hufton S. E. ( 2003) FEBS Lett. 546, 288– 294 [DOI] [PubMed] [Google Scholar]
- 31.Jin M., Andricioaei I., Springer T. A. ( 2004) Structure 12, 2137– 2147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.