Abstract
Numerous post-translational modifications have been identified in histones. Most of these occur within the histone tails, but a few have been identified within the histone core sequences. Histone core post-translational modifications have the potential to directly modulate nucleosome structure and consequently DNA accessibility. Here, we identify threonine 45 of histone H3 (H3T45) as a site of phosphorylation in vivo. We find that phosphorylation of H3T45 (H3T45ph) increases dramatically in apoptotic cells, around the time of DNA nicking. To further explore this connection, we analyzed human neutrophil cells because they are short-lived cells that undergo apoptosis in vivo. Freshly isolated neutrophils contain very little H3T45ph, whereas cells cultured for 20 h possess significant amounts; the kinetics of H3T45ph induction closely parallel those of caspase-3 activation. Cytokine inhibition of neutrophil apoptosis leads to reduced levels of H3T45ph. We identify protein kinase C-δ as the kinase responsible for H3T45ph in vitro and in vivo. Given the nucleosomal position of H3T45, we postulate that H3T45ph induces structural change within the nucleosome to facilitate DNA nicking and/or fragmentation.
In higher organisms, DNA is packaged into a highly ordered and structured complex termed chromatin. The fundamental unit of chromatin is the nucleosome consisting of ∼150 bp of DNA wrapped around a core histone octamer (two each of H2A, H2B, H3, and H4 (1)). It is in this architectural context that DNA processes such as transcription, replication, repair, and recombination must be performed. In general, chromatin is highly repressive to DNA processes as the DNA template tends to be inaccessible to the relevant enzymes and ancillary factors. Thus, chromatin structure needs to be carefully manipulated to facilitate controlled access to the DNA template.
Eukaryotes have developed multiple mechanisms to finely tune chromatin structure. A principle approach involves the post-translational modification (PTM)4 of histones. These modifications include acetylation, methylation, and phosphorylation, all of which are laid down in a dynamic fashion (2–4). In chromatin, histone tails protrude from their own nucleosome, and they can make contact with adjacent nucleosomes (1). It is therefore not too surprising that the histone N-terminal tails are key targets for PTM because the modifications are likely to affect regional protein-DNA and protein-protein interactions (reviewed in Ref. 4). This in turn has the potential to regulate the overall chromatin architecture. For instance, H3K9me is catalyzed by the SUVAR3–9 methyltransferase, and the modified histone then forms a binding platform for the heterochromatin protein HP1, which binds to methylated H3K9 via its chromodomain (5–7). A positive feedback loop is initiated because the H3-bound HP1 recruits further SUVAR3–9, leading to additional methylation of H3K9 residues in nearby nucleosomes and more recruitment of HP1. In this manner, repressive heterochromatin can spread until it is prevented from doing so by regions such as boundary elements (8). This simple example highlights the importance of dynamic histone tail modifications in the regulation of chromatin structure.
Phosphorylation is another dynamic histone PTM that has been associated with chromatin function in processes such as transcription, mitosis, and cellular apoptosis. Importantly, a single phosphorylated site within a histone can be associated with very different chromatin states. For example, phosphorylation of H3S10 (H3S10ph) has been associated with rapidly induced transcription at the immediate early genes, such as Jun and Fos (9, 10). In contrast, the same histone mark is also globally present during mitosis, a stage in the cell cycle where the vast majority of chromatin is tightly condensed (11). It is not clear what governs whether H3S10ph is associated with “active” or “inactive” chromatin. Presumably, the answer is linked, at least in part, to other histone modifications that together with H3S10ph elicit an appropriate biological outcome. Additionally, the presence or absence of factors that recognize and bind H3S10ph may well play a role. Of course, phosphorylation of H3S10 will dramatically affect local electrostatic and ionic potentials, and this will have a direct consequence on nucleosome and chromatin structures. Although H3S10ph is used as an example here, it is likely that similar mechanisms will be operational at other sites of histone phosphorylation.
Historically, the histone tails have been the main focus for investigators attempting to decipher how PTMs affect chromatin structure. Less attention, however, has been given to the potential of PTMs in the histone cores. In this study, we identify an in vivo phosphorylation site in histone H3 at threonine 45 (H3T45ph) as a novel H3 core modification. To further investigate the possible function(s) of this modification, we raised specific polyclonal rabbit antisera against H3T45ph. These antisera enabled us to demonstrate that H3T45ph is associated with apoptosis of HL60 cells and purified human neutrophil cells. Furthermore, we identify protein kinase C-δ (PKCδ) as the kinase responsible for this modification. This is the first link between a histone core PTM and the process of apoptosis.
EXPERIMENTAL PROCEDURES
Cell Culture
HL60 cells were maintained in Iscove's modified Dulbecco's medium (Invitrogen) supplemented with 20% fetal calf serum, penicillin, streptomycin, 2 mm l-glutamine. Cells were passaged to maintain a cell density <1 × 106.
HL60 Cellular Differentiation and Phosphatase Inhibitor Treatment
HL60 cells were seeded at a concentration of 4 × 105/ml in Iscove's modified Dulbecco's medium (Invitrogen) (20% fetal calf serum, penicillin, streptomycin, 2 mm l-glutamine). Cellular differentiation was induced by the addition of 1.3% (v/v) DMSO (Sigma). Inhibition of cellular phosphatases was achieved by treatment with 20 nm calyculin A (Calbiochem 208851). Cells were incubated under standard growing conditions, and samples were removed as required.
Cellular Fractionation of HL60 Cells
Approximately 4 × 106 cells were washed once with PBS and scraped into 10 ml of PBS. Cells were pelleted and washed in 400 μl of buffer A (10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm dithiothreitol, and one CompleteTM protease inhibitor mixture tablet (Roche Applied Science)). The pellet was resuspended in 400 μl of buffer A supplemented with 0.1% (v/v) Nonidet P-40 and then incubated on ice. After 10 min, the sample was vortexed for 10 s and microcentrifuged (13,000 rpm, 1 min, 4 °C). The supernatant contained the cytosolic fraction and was removed. The pellet was then washed with buffer A. After microcentrifugation, the pellet was resuspended in 20–100 μl of buffer B (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 420 mm NaCl, 0.5 mm dithiothreitol, 25% (v/v) glycerol, 0.2 mm EDTA). Samples were vortexed and incubated on ice for 20 min and then microcentrifuged (13,000 rpm, 2 min, 4 °C). The supernatant contained the nucleosolic fraction and was removed. The pellet was washed in buffer B. Chromatin and associated proteins were found within the pellet.
Isolation of Neutrophils
Normal patients were venesected, and 20 ml of whole blood were obtained. Samples were separated on a Ficoll density gradient by centrifugation. The red blood cells and neutrophils were isolated and treated with red cell lysis buffer (150 mm NH4Cl, 10 mm KHCO3, 0.1 mm EDTA, and one CompleteTM protease inhibitor mixture tablet (Roche Applied Science)). Samples were centrifuged, and the pellet containing neutrophils was washed twice in PBS. Cells were analyzed to test purity and incubated in RPMI (Invitrogen) supplemented with 10% fetal calf serum.
Western Blotting
Approximately 1 × 106 cells were lysed by the addition of 0.5 ml of 1× SDS loading buffer. Lysed cells were sonicated at high setting (Bioruptor, Diagenode) for 7 min with 30 s off and 30 s on and boiled for 10 min. 5–30 μl of the whole cell extracts were analyzed by SDS-PAGE and Western blotting.
Proteins were transferred from polyacrylamide gels to nitrocellulose membranes (Whatman) using standard procedures. After the transfer, the nitrocellulose membrane was placed either in milk blocking buffer (Tris-buffered saline with 0.5% (v/v) Tween 20, 5% (w/v) nonfat milk powder) or in BSA blocking buffer (Tris-buffered saline with 0.5% (v/v) Tween 20, 5% BSA) overnight at 4 °C. The antibodies used were anti-H3K4me3 (ab8580, Abcam), anti-H3T6ph (ab14102, Abcam), anti-H3S28ph (ab5169, Abcam), anti-cleaved caspase-3 (Asp175) (CST9664, Cell Signaling Technology), anti-PKCδ (C-20) (sc-937, Santa Cruz Biotechnology), and anti-β-tubulin (T5201, Sigma).
Peptide Synthesis, Antibody Generation, and Characterization
Histone H3 peptides were synthesized as described before (12) that contain 3–4 specific amino acids on each side of the phosphorylated residue (sequences available upon request). Polyclonal anti-H3T45ph was raised in collaboration with Abcam (ab26127) by immunizing a rabbit with a peptide corresponding to H3T45ph and then affinity-purifying the antibody from whole serum against the immunizing peptide (12). The specificity of the anti-H3T45ph antibody was validated by peptide competition using an enzyme-linked immunosorbent pate assay (ELISA) (12) and peptide dot blots.
Mass Spectroscopy
HL60 cells treated with calyculin A or non-treated HeLa cells were lysed using a hypotonic buffer and Dounce homogenization essentially as described (13). Following centrifugation, crude nuclei were lysed by adding two pellet volumes of a high salt (420 mm NaCl) buffer. Following another centrifugation step, the remaining insoluble nuclear pellets were resolubilized in a buffer containing 600 mm NaCl, 50 mm Tris HCl, pH 8.0, 1% Nonidet P-40 supplemented with Benzonase and phosphatase inhibitors. After 20 min of incubation at room temperature in a thermoshaker, solubilized proteins were loaded on a 4–12% bis-tris gradient gel (Invitrogen). Histone bands were excised from the gel and subjected to in-gel trypsin digestion. Phosphopeptides were enriched from the total pool of extracted tryptic peptides using titanium dioxide beads (14). Peptides eluted from these beads were analyzed on an LTQ-Orbitrap instrument connected to an online nanoflow high pressure liquid chromatography (Agilent 1100 system) via a nanoelectrospray ion source (Proxeon Biosystems). The tryptic peptide mixtures were autosampled onto a 15-cm-long 75-μm ID column packed in-house with 3-μm C18-AQUA-Pur Reprosil reversed-phase beads (Dr. Maisch GmbH) and eluted using a 2-h linear gradient from 8 to 40% acetonitrile. The separated peptides were electrosprayed directly into the LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific), which was operated in the data-dependent mode to automatically switch between MS and MS2. Survey spectra were acquired with 60,000 resolution in the Orbitrap while acquiring up to five tandem mass spectra in the LTQ part of the instrument. All full-scan spectra were recalibrated in real-time using the lock-mass option (15). The tandem mass spectra were acquired with the multistage activation enabled for neutral loss of phosphoric acid (32.66, 48.99, and 97.97 atomic mass units) (16). Peak lists were generated from all raw files using the in-house-written raw2msm script, which intensity-weighs all parent masses over their liquid chromatography elution peak and reduces the tandem mass spectra to six fragments per 100 atomic mass units. Fragment spectra were searched using the MASCOT software program (Matrix Science).
Quantitive Real-time PCR
Total RNA was prepared from 1 × 107 cells by using TRIzol (Invitrogen), and subsequent first strand complementary DNA synthesis was achieved with the Superscript III reverse transcriptase kit (Invitrogen) in accordance with the manufacturer's protocols. The cDNA samples were then used as templates for real-time PCR. cDNA purified as described was used to quantify mRNA levels. Quantitative real-time PCR experiments were performed using Taqman® probes (Applied Biosystems) labeled with Fam (reporter) and Tamra (quencher) as a marker for DNA amplification, on an ABI Prism 7000 apparatus (Applied Biosystems). The DNA sequence of the primers used to detect the presence of cd11b mRNA were as follows: forward, 5′-agt tgc cga att gca tcg a-3′, and reverse, 5′-tgg cgt tcc cac cag aga-3′. The Taqman probe sequence was as follows: 5′-Fam-agt tca ggc gca gca caa tgg ggt-Tamra-3′. Standard curves were measured using dilutions of cDNA from control cells.
Immunofluorescence and Confocal Microscopy
Cells were seeded into 20-cm2 dishes containing sterile coverslips. Cells were washed twice with PBS and fixed for 20 min in 4% paraformaldehyde and 2% sucrose for 20 min at room temperature. Cells were washed three times with PBS and then blocked at 37 °C for 15 min with 3% BSA, 0.6% (v/v) Triton in PBS. The primary antibody was diluted accordingly in 3% BSA in PBS and incubated with cells at room temperature for 1 h. Cells were then washed with 3% BSA in PBS and incubated with anti-mouse or anti-rabbit secondary antibody labeled with Alexa Fluor 488 or Alexa Fluor 594 or Texas Red (Molecular Probes/Invitrogen) for 1 h at room temperature in the dark. Cells were washed once with PBS and then incubated with 1 μg/ml of Hoechst 33258 in PBS for 5 min. Cells were finally washed four times with PBS, and coverslips were mounted onto glass slides with the VECTASHIELD mounting medium (Vector Laboratories). Coverslips were fixed with nail varnish. Samples were analyzed by confocal microscopy using a Bio-Rad MRC 1024 or Bio-Rad Radiance 2100 confocal system, on an upright Nikon fluorescence microscope equipped with a ×60 oil lens.
Identification of Apoptotic Cells
Apoptotic cells were identified using the in situ cell detection kit (Roche Applied Science) according to manufacturer's instructions. 1 × 106 cells were seeded into 20-cm2 dishes containing sterile coverslips. Cells were washed twice with PBS and fixed for 20 min in 4% paraformaldehyde and 2% sucrose for 20 min at room temperature. Cells were washed three times with PBS and then blocked at 37 °C for 15 min with 3% BSA, 0.6% (v/v) Triton in PBS. 100 μl of label and 450 μl of enzyme solution were mixed and dropped onto coverslips. Cells were incubated for 60 min at 37 °C in a humidified atmosphere and then washed twice and incubated with 1 μg/ml Hoechst 33258 in PBS for 5 min. Cells were washed four times with PBS, and coverslips were mounted onto glass slides with VECTASHIELD mounting medium (Vector Laboratories) and fixed with nail varnish. Samples were analyzed by confocal microscopy using a Bio-Rad MRC 1024 or Bio-Rad Radiance 2100 confocal system, on an upright Nikon fluorescence microscope equipped with a 60 × oil lens.
Kinase Assays
PKCδ kinase (7342, Cell Signaling Technology) was used in in vitro kinase assays according to the manufacturer's instructions. Briefly, assays were performed in 50 μl of kinase buffer (5 mm MOPS, pH 7.2, 2.5 mm β-glycerophosphate, 1 mm EGTA, 0.4 mm EDTA, 5 mm MgCl2, 0.1 mm CaCl2, 0.05 mm dithiothreitol, 0.05 mg/ml phosphatidylserine, 0.005 mg/ml diacylglycerol). 370 kBq of [γ-32P]ATP (6000 Ci/mmol, PerkinElmer Life Sciences) was added to the buffer in the radiolabeled kinase assays. Approximately 500 ng of purified recombinant histone H3 (wild type or H3T45A mutant) or 2 mg of total core histone (from calf thymus; Sigma) were used as substrate.
RESULTS
H3T45ph Is a Novel Histone Modification
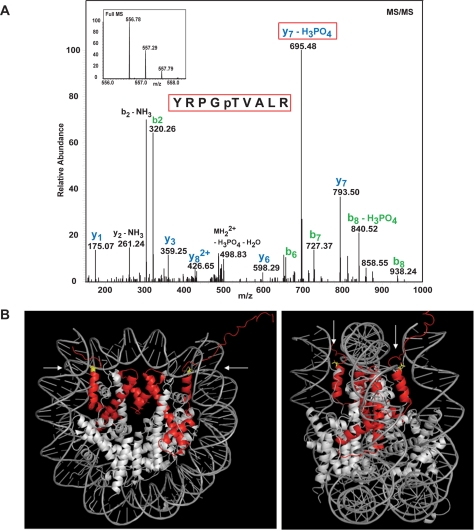
In our attempt to identify novel histone modifications, we decided to focus on phosphorylation. We chose phosphorylation for two practical reasons. Firstly, phosphorylation results in a relatively large increase in mass, which can be unequivocally detected and identified by mass spectroscopy. Secondly, the in vivo equilibrium between kinase and phosphatase action can be very easily biased in favor of kinases via the simple addition of phosphatase inhibitors to cells. We therefore treated human HL60 cells with calyculin A (a broad spectrum phosphatase inhibitor) for a short time (15 min to minimize indirect action), purified histone H3, and subjected the protein to analysis by MS/MS. This approach identified H3T45 as a site that is phosphorylated in vivo (H3T45ph; Fig. 1A). Importantly, a similar analysis also detected H3T45ph at much lower levels in untreated cells (data not shown).
FIGURE 1.
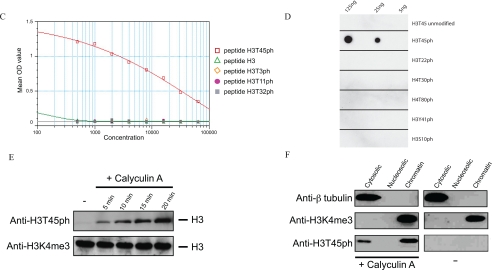
Identification of H3T45ph in chromatin. A, MS/MS identifies H3T45ph in vivo. HL60 cells were treated with calyculin A. Proteins were prepared as described and resolved by SDS-PAGE. Proteins corresponding to the size of H3 were isolated from the acrylamide gel and fragmented by in-gel tryptic digestion. Phosphopeptides were enriched using titanium oxide beads, and after elution, injected into the Orbitrap mass spectrometer. The inset at the top shows the spectrum of the intact peptide of interest encompassing H3T45. This peptide was then subjected to MS/MS shown in the main part of the panel. B, the position of histone H3 (red) within the nucleosome is indicated. The location of H3T45 (yellow) is indicated by the arrows; H3T45 is positioned at the N terminus of the first helix of H3 (the αN1-helix), a position close to where the DNA both enters and exits the nucleosome. Images were constructed using the MacPyMOL software. C, peptide competition analyses confirm the specificity of the anti-H3T45ph antibody. The specificity of the anti-H3T45ph antibody was validated by peptide competition using an ELISA plate assay. The antibody raised to an H3T45ph peptide specifically recognizes only this peptide and fails to recognize the identical but unmodified H3 peptide. Similarly, the anti-H3T45ph antibody does not recognize other H3 phospho-threonine peptides that have previously been shown to be modified on histone H3. OD, optical density. D, the indicated amounts of various phosphorylated histone peptides were spotted to polyvinylidene difluoride membrane and probed with the anti-H3T45ph antibody. The anti-H3T45ph antibody only recognizes the peptide with Thr-45 phosphorylated. E, HL60 cells were incubated with 20 nm calyculin A (Calbiochem 208851) for the times indicated. Whole cell extracts were analyzed by Western blotting using the anti-H3T45ph antibody, and an anti-H3K4me3 (Abcam ab8580) antibody was used as a loading control. F, HL60 cells were treated with (+) or without (−) calyculin A. Protein extracts were prepared by cellular fractionation and analyzed by Western blotting using the anti-H3T45ph, anti-β-tubulin (Sigma), or anti-H3K4me3 (Abcam ab8580) antibodies.
H3T45 lies in a structurally important region in the nucleosome, being positioned at the extreme N terminus of the first helix of H3 (αN1-helix: Fig. 1B). This region of H3 forms the platform from which the H3 N-terminal tail exits the nucleosome and, as it does so, the tail makes contacts with the nearby DNA. It is highly likely therefore that this region of H3 will play an important role in nucleosome structure/stability. Given the potential importance of its location, we decided to further study the phosphorylation of H3T45. To this end, we raised rabbit polyclonal antisera to H3 peptides encompassing a phosphorylated Thr-45 residue. The resulting immune serum was affinity-purified on a column bearing H3 peptides containing H3T45ph. An ELISA indicated that the resulting IgG population specifically binds an H3T45ph peptide but not the non-phosphorylated version of the peptide (Fig. 1C). Furthermore, peptide competition analyses using ELISA and peptide dot blots indicate that the antibody preparation does not cross-react to other sites of histone phosphorylation (Fig. 1, C and D; see also Fig. 4C for additional antibody specificity data). We refer to this purified antibody preparation as anti-H3T45ph antibody.
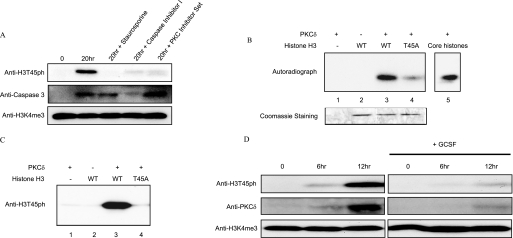
FIGURE 4.
H3T45 is phosphorylated by PKCδ. A, neutrophils were isolated and cultured for the indicated times in the presence or absence of the following protein kinase inhibitors: 200 nm InSolutionTM staurosporine (Calbiochem, 569396), 25 μm InSolutionTM caspase-3 inhibitor I (Calbiochem, 235427), and protein kinase C inhibitor set (Calbiochem, 539573). Whole cell extracts were analyzed by Western blot using anti-H3T45ph, anti-cleaved caspase-3, or anti-H3K4me3. B, radioactive in vitro kinase assay. A mixture of core histones (2 mg), recombinant WT H3 (500 ng), and recombinant H3T45A (500 ng) was used as substrate in the in vitro kinase assay using [γ-32P]ATP and recombinant PKCδ (7342, Cell Signaling Technology). Equal loading of histones is demonstrated by the Coomassie Blue-stained gel. WT, wild type. C, in vitro kinase assay. Recombinant WT H3 (500 ng) and H3T45A (500 ng) were used as substrates in the in vitro kinase assay using non-radioactive ATP. The reaction products were resolved by SDS-PAGE, and phosphorylation was then analyzed by Western blotting with the anti-H3T45ph antibody. D, human neutrophils were isolated and cultured for the indicated times in the presence or absence of 100 ng/ml granulocyte colony-stimulating factor (GCSF) (G0407 Sigma). Whole cell extracts were then analyzed by Western blotting using anti-H3T45ph, anti-PKCδ, or anti-H3K4me3 (as a loading control) antibodies.
We next sought to confirm the mass spectroscopy results by analyzing cell extracts via Western blotting with the anti-H3T45ph antibody. HL60 cell extracts were prepared from cultures that were either untreated or treated with calyculin A phosphatase inhibitor for various times as indicated (Fig. 1E, upper panel). The results from these Western blots clearly show that the anti-H3T45ph antibody detects a band corresponding to the mobility of H3 that is induced upon phosphatase inhibition. It should also be noted that a band of the same mobility of H3 is also detected in the untreated extract following a much longer exposure of the membrane to film (data not shown). To ensure equal loading, the same samples were also stained with Ponceau S after blotting to nitrocellulose (data not shown) and then probed with an H3K4me3 antibody. The results clearly indicate that all samples contained equivalent levels of H3 (Fig. 1E, lower panel). Cellular fractionation was then performed to demonstrate that the Thr-45-phosphorylated H3 was associated with chromatin. HL60 cells were fractionated into cytosolic, nucleosolic, and chromatin preparations, and the resulting extracts were resolved by SDS-PAGE and Western blotted with the anti-H3T45ph antibody (Fig. 1F). The results indicate that the majority of H3T45ph is indeed associated with chromatin-bound H3, but there is also a small amount detectable in the cytosol. The integrity of the fractionated extracts was confirmed by probing the same blots with an anti-β-tubulin antibody (cytosolic marker) and an anti-H3K4me3 antibody (chromatin marker). Taken together, these data clearly indicate that H3T45 is phosphorylated in a highly dynamic manner in the chromatin of cultured human cells.
H3T45ph Is an Abundant Marker of Apoptotic Cell
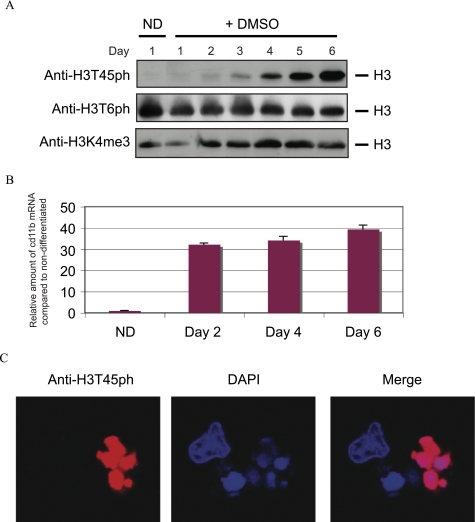
Our next goal was to link H3T45ph to a physiological event. In this regard, we took advantage of the ability of HL60 cells in culture to differentiate. It is well documented that the addition of retinoic acid or DMSO to these cells induces them to differentiate toward a granulocytic lineage (17). We therefore induced HL60 cells to differentiate by treating them with DMSO for a 6-day period. We harvested cells daily for the duration of the time course. Total cellular protein extracts were prepared from each time point, and the extracts were Western blotted using the anti-H3T45ph antibody. Fig. 2A (upper panel) shows that the levels of H3T45ph steadily increase over the 6-day time course, reaching their highest level at day 6. As a loading control, the extracts were also probed with an anti-H3K4me3 antibody (Fig. 2A, lower panel). In contrast, an antibody against a different phosphorylated site within histones, H3T6ph, does not show the same DMSO-induced trend as observed with the anti-H3T45ph antibody (Fig. 2A, middle panel). Thus, H3T45ph is induced in HL60 cells following the addition of DMSO to the cells. A DMSO-induced increase in cd11b mRNA, a granulocytic marker (18), confirms that the HL60 cells were differentiating (Fig. 2B). Immunofluorescence was then performed with the anti-H3T45ph antibody and 6-day DMSO-exposed HL60 cells (Fig. 2C). The antibody strongly stains cells with lobulated nuclei, but it does not stain cells with a morphology more typical of undifferentiated HL60 cells. These data indicate that H3T45ph appears in DMSO-treated HL60 cells, particularly in cells possessing an unusual lobulated nuclear morphology.
FIGURE 2.
H3T45ph is induced as HL60 cells differentiate. A, HL60 cells were treated with 1.3% (v/v) DMSO for 6 days to induce granulocytic differentiation, and samples were taken daily. ND represents the non-differentiated control cells. Whole cell extracts were analyzed by Western blotting using antibodies against H3T45ph, H3T6ph (Abcam ab14102), or anti-H3K4me3 (Abcam ab8580). B, DMSO-induced increase in cd11b mRNA confirms HL60 cell differentiation. mRNA levels from HL60 cells described in panel A were measured using primers specific to the granulocytic marker, cd11b, and to β-microglobulin (for normalization). Error bars represent the S.D. C, confocal microscopy. Cells were fixed with 4% paraformaldehyde and stained with anti-H3T45ph and anti-rabbit labeled with Alexa 594 (Molecular Probes A11012) antibodies. 4′,6-Diamidino-2-phenylindole (DAPI) was included to identify DNA.
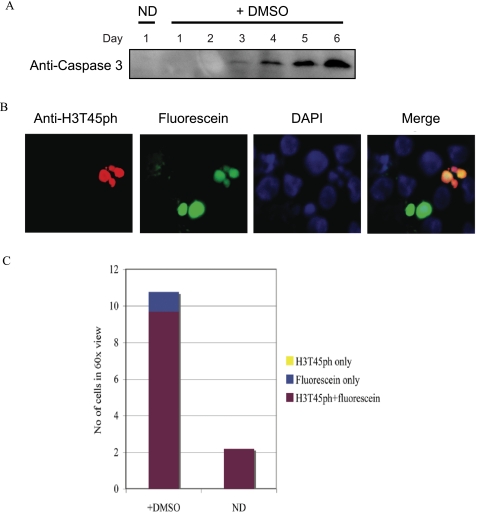
We reasoned that the H3T45ph-positive cells with lobulated nuclei may be neutrophils, a particular type of granulocyte that exhibits multilobed nuclei, the appearance of which have previously been described during HL60 granulopoiesis (Ref. 19 and references therein). On the other hand, we also noted that the H3T45ph-positive cells possess morphology similar to that of apoptotic cells. To distinguish between these possibilities, we decided to directly test whether H3T45ph was associated with apoptotic cells. We first asked whether DMSO induces the appearance of apoptotic markers in HL60 cells. We tested for the appearance of cleaved caspase-3, a well characterized marker of cells undergoing apoptosis (Ref. 20 and references therein). We find that cleaved caspase-3 is induced by DMSO in HL60 cells (Fig. 3A) with kinetics very similar to the induction of H3T45ph (compare Fig. 3A with Fig. 2A). Next, we performed a terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling (TUNEL) assay linked to immunofluorescence on 6-day DMSO-treated HL60 cells to directly visualize cells that were undergoing apoptosis (Fig. 3B). In this assay, TUNEL-positive cells are labeled with fluorescein and appear green (Fig. 3B, second panel). Significantly, H3T45ph-positive multilobed cells are shown to be apoptotic using this approach (Fig. 3B). The results were quantified by analyzing random fields of vision and scoring for H3T45ph and/or fluorescein staining (Fig. 3C). The quantitation confirms that a significant number of DMSO-treated HL60 cells undergo apoptosis. Strikingly, all H3T45ph-positive cells co-stain with fluorescein, indicating that all cells containing H3T45ph are apoptotic. However, a few apoptotic cells do not contain H3T45ph. Considered together, these data indicate a link between H3T45ph and apoptosis, with the histone phosphorylation occurring concomitant with, or downstream of, DNA nicking, a relatively late event in the process of apoptotic cell death.
FIGURE 3.
H3T45ph is within apoptotic cells. A, HL60 differentiation activates caspase-3. Cleaved caspase-3, an apoptotic marker, was detected in HL60 cells treated with 1.3% (v/v) DMSO over 6 days. ND represents the non-differentiated control cells. The same whole cell extracts as used in Fig. 2A were analyzed by Western blotting using anti-cleaved caspase-3 (CST 9664) antibodies. B, apoptotic cells were identified using the in situ cell death detection kit (Roche Applied Science). Cells were co-stained with anti-H3T45ph and anti-rabbit labeled with Texas Red 594 (Invitrogen T2767) antibodies and identified by confocal microscopy. C, cells were scored for fluorescein, H3T45ph, and both fluorescein and H3T45ph staining. 4′,6-Diamidino-2-phenylindole (DAPI) was included to identify DNA. A number of random fields of view were scored, and the results shown represent the mean average.
Our results clearly demonstrate an induction of H3T45ph in apoptotic cells following the administration of DMSO to HL60 cells. We questioned whether this effect may be mimicking a similar in vivo process. We chose to analyze human neutrophil cells because they are short-lived cells that undergo apoptosis in vivo (21). Moreover, neutrophils can be harvested from blood, maintained in a suitable medium, and readily monitored. We therefore isolated human neutrophils and looked for the presence of H3T45ph (Fig. 4A). Notably, freshly isolated neutrophils possessed only very low levels of the histone modification. This is completely consistent with the results from our analysis of HL60 cells where we observe little, if any, H3T45ph in healthy (TUNEL-negative) undifferentiated cells. However, isolated neutrophils that are maintained in culture for 20 h, and which therefore will have entered the apoptotic process (21) (indicated by activation of caspase-3, Fig. 4A), possess a considerable amount of H3T45ph (Fig. 4A, top panel). In contrast, no change in the levels of H3T6ph or H3S28ph was observed (data not shown). These data strongly associate H3T45ph with cellular apoptosis. Indeed, our observations with human primary cells reinforce our conclusions derived from studies with the HL60 cell line.
PKCδ Phosphorylates H3T45
Inhibition of purified neutrophil apoptosis by caspase inhibitor blocks the activation of caspase-3 and significantly reduces the levels of H3T45ph (Fig. 4A). The addition of staurosporine to the neutrophils severely abrogates the induction of H3T45ph but does not prevent the activation of caspase-3 (Fig. 4A). This suggests that staurosporine inhibits a kinase downstream of caspase-3 activation, a relatively late event in the apoptotic process. Importantly, the use of a PKC inhibitor set mimicked the results with staurosporine, strongly implicating a PKC kinase family member in the pathway leading to H3T45ph (Fig. 4A). It should be noted that the use of many other kinase inhibitors (including KN-62 and STO-609 (calcium/calmodulin protein kinase II), Purvalanol (cyclin-dependent kinases (CDKs)), 2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT) (casein kinase II), Y27632 (Rho-associated protein kinase II), GSK-3 inhibitor XV (GSK-3), PD98059 (MEK1), SB203580 (p38-mitogen-activated protein kinase (p38-MAPK)), JNK inhibitor II (JNK), and H-89 (cAMP-dependent protein kinase)) had no effect on H3T45ph levels (data not shown). Together, these data suggest that the H3T45ph kinase is a PKC family member that is activated downstream of caspase-3 cleavage. We therefore considered the possibility that the kinase responsible for H3T45ph was PKCδ because it is required for neutrophil apoptosis (22, 23), and its cleavage by nuclear caspase-3 generates a constitutively active form of the kinase whose nuclear accumulation rapidly stimulates apoptosis (24). We tested whether purified PKCδ could in vitro phosphorylate H3 in a kinase assay utilizing [γ-32P]ATP. Fig. 4B clearly demonstrates that purified PKCδ efficiently phosphorylates H3. Importantly, the ability to phosphorylate H3 is severely abrogated when H3T45 is converted to alanine (Fig. 4B, compare lane 3 with lane 4). Thus, H3T45 is the main site within H3 for phosphorylation by PKCδ in vitro. We also find that H3 is the main histone substrate for PKCδ when core histones (an equimolar mixture of H3, H2A, H2B, and H4) are used as substrate (Fig. 4B, lane 5).
We performed similar in vitro kinase assays to those described above but with non-radioactive ATP. In this case, the reaction products were Western blotted and probed with the anti-H3T45ph antibody (Fig. 4C). The antibody strongly detects phosphorylation of wild type H3 but not of H3 mutated at Thr-45. These results demonstrate that PKCδ phosphorylates H3T45 in vitro, and they testify to the specificity of the H3T45ph antibody.
Next, we attempted to link the activation of PKCδ to H3T45ph in apoptotic neutrophils. First, we tested whether we could detect the caspase-3-cleaved PKCδ using an antibody that specifically detects the C terminus of the kinase. We find that the cleaved form of PKCδ appears in cultured neutrophils with very similar kinetics to the appearance of H3T45ph (Fig. 4D). Second, it is well documented that the addition of granulocyte colony-stimulating factor to neutrophil culture medium inhibits the apoptotic process in these cells (25). We find that it also inhibits H3T45ph and the appearance of cleaved PKCδ (Fig. 4D). Together, our data provide evidence that PKCδ phosphorylates H3T45 in neutrophils, an event linked to the latter stages of apoptosis.
DISCUSSION
In this work, we have identified in human cells a new histone H3 modification, namely H3T45ph, and we have raised antisera capable of specifically detecting this histone mark. A combination of mass spectroscopy data and results with the anti-H3T45ph antibody indicate that the mark exists in “normal” cycling cells in culture. Firstly, mass spectroscopy of histones from untreated cells detects phosphorylation at H3T45 (data not shown). Secondly, the phosphorylation is very rapidly detected (in less than 15 min) by the anti-H3T45ph antibody following the addition of various phosphatase inhibitors to the cells. This strongly suggests that the phosphate group on H3T45 is rapidly turning over in untreated cells and that the addition of phosphatase inhibitors to the cells quickly pushes the reaction equilibrium toward the phosphorylated state. In addition to its presence in untreated cells, our results also indicate that H3T45ph is induced in the latter phases of apoptosis. These observations raise the interesting question of whether there is a functional link between the turnover of this mark in untreated cells and the robust appearance of it in late apoptosis, but the answer will require significant further work and is beyond the scope of the present study. Furthermore, we have immunologically detected H3T45ph in a number of cell lines, including HeLa and 293T cells following stress stimuli, but whether the appearance of this modification in these cells is linked to apoptosis remains to be determined.
In an effort to gain insight into the functional role of H3T45ph, we performed chromatin immunoprecipitations linked to direct DNA sequencing (ChIP-Seq; data not shown). However, we did not observe significant enrichment of any region of the genome. We believe that this is due to the fact that the anti-H3T45ph antibody immunoprecipitates nucleosomes very poorly, presumably because H3T45 is located close to the DNA gyres, and consequently, recognition of the epitope may be sterically hindered. Nevertheless, our data indicate that PKCδ is the kinase responsible for phosphorylation of H3T45 during the latter stages (i.e. downstream of caspase-3 activation) of neutrophil apoptosis.
A role for PKCδ in apoptosis is well documented (24). Indeed, sustained nuclear accumulation of activated PKCδ following cleavage by caspase-3 appears to be highly important in a number of other apoptotic systems (Ref. 24 and references therein). It is difficult to determine the actual reason for phosphorylation of H3T45 by PKCδ, but we can speculate based on the fact that H3T45 occupies a structurally interesting position within the nucleosome. It is positioned at the N terminus of the first H3 α-helix (αN1-helix), and is an important determinant for the exit of the H3 N-terminal tail from the nucleosome. This position is consistent with H3T45, and perhaps its modification, having a role in nucleosome structure and stability. Indeed, mutation of H3T45 to alanine has recently been shown to promote ATP-dependent nucleosomal remodeling in vitro, directly demonstrating that H3T45 does play an important role in nucleosome structure (26). We suggest that phosphorylation of this residue, with the resulting introduction of negative charge close to the DNA, would affect the binding energies of the nucleosome. Perhaps the altered nucleosomal/chromatin structure favors DNA processing in late apoptosis, such as DNA nicking and/or fragmentation, explaining the abundance of the mark at this time. Alternatively, but not mutually exclusively, phosphorylation of H3T45 may provide a binding site for a protein in a similar manner to that occurring at the c-jun gene where phosphorylation of H3S10 provides a binding site for a 14-3-3 protein (27). However, we have been unable to identify such binding proteins in H3T45ph peptide pull-down experiments coupled to mass spectroscopy (data not shown). It is of course possible, and perhaps likely given the nucleosomal location of H3T45, that such binding studies need to be performed in a nucleosomal context.
Neutrophil lifespan is critically important in vivo; a tightly regulated extension in neutrophil survival is necessary for efficient protection from invading pathogens, whereas an unregulated increase in neutrophil lifespan is linked to a number of pathologies including inflammatory and immunological diseases (reviewed in Ref. 21). Likewise, an unregulated decrease in neutrophil lifespan has also been linked to numerous diseases. Thus, the tight control of neutrophil survival is essential for normal homeostasis. PKCδ is certainly involved in this control, and our data provide evidence that H3T45ph, at least in part, is also intimately involved. Understanding the exact role of this histone modification in neutrophil apoptosis remains to be established, but it is quite possible that it will have implications in the treatment/screening of certain human pathological conditions that are linked to neutrophil survival.
Supplementary Material
This work was supported by grants from Cancer Research UK and the 6th Research Framework Programme of the European Union (EPIgenetic TReatment Of Neoplastic disease (Epitron) and Small Modulators of Gene Activation and Repression by Targeting Epigenetic Regulators (SMARTER)) (to T. K.). We declare competing financial interests; T. K. is a director of Abcam Ltd.
This article was selected as a Paper of the Week.
- PTM
- post-translational modification
- PKCδ
- protein kinase C-δ
- CA
- calyculin A
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- ELISA
- enzyme-linked immunosorbent assay
- DMSO
- dimethyl sulfoxide
- MOPS
- 3-(N-morpholino) propanesulfonic acid
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- bis-tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- Fam
- 6-carboxyfluorescein
- Tamra
- 6-carboxytetramethylrhodamine
- JNK
- c-Jun N-terminal kinase.
REFERENCES
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. ( 1997) Nature 389, 251– 260 [DOI] [PubMed] [Google Scholar]
- 2.Fischle W., Wang Y., Allis C. D. ( 2003) Curr. Opin. Cell Biol. 2, 172– 183 [DOI] [PubMed] [Google Scholar]
- 3.Strahl B. D., Allis C. D. ( 2000) Nature 403, 41– 45 [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T. ( 2007) Cell 128, 693– 705 [DOI] [PubMed] [Google Scholar]
- 5.Rea S., Eisenhaber F., O'Carroll D., Strahl B. D., Sun Z. W., Schmid M., Opravil S., Mechtler K., Ponting C. P., Allis C. D., Jenuwein T. ( 2000) Nature 406, 593– 599 [DOI] [PubMed] [Google Scholar]
- 6.Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., Kouzarides T. ( 2001) Nature 410, 120– 124 [DOI] [PubMed] [Google Scholar]
- 7.Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. ( 2001) Nature 410, 116– 120 [DOI] [PubMed] [Google Scholar]
- 8.Wallace J. A., Felsenfeld G. ( 2007) Curr. Opin. Genet. Dev. 17, 400– 407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton A. L., Rose S., Barratt M. J., Mahadevan L. C. ( 2000) EMBO J. 19, 3714– 3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomson S., Clayton A. L., Mahadevan L. C. ( 2001) Mol. Cell 8, 1231– 1241 [DOI] [PubMed] [Google Scholar]
- 11.Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., Brinkley B. R., Bazett-Jones D. P., Allis C. D. ( 1997) Chromosoma 106, 348– 360 [DOI] [PubMed] [Google Scholar]
- 12.Bannister A. J., Kouzarides T. ( 2004) Methods Enzymol. 376, 269– 288 [DOI] [PubMed] [Google Scholar]
- 13.Dignam J. D., Lebovitz R. M., Roeder R. G. ( 1983) Nucleic Acids Res. 11, 1475– 1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen M. R., Thingholm T. E., Jensen O. N., Roepstorff P., Jørgensen T. J. ( 2005) Mol. Cell. Proteomics 4, 873– 886 [DOI] [PubMed] [Google Scholar]
- 15.Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. ( 2005) Mol. Cell. Proteomics 4, 2010– 2021 [DOI] [PubMed] [Google Scholar]
- 16.Schroeder M. J., Shabanowitz J., Schwartz J. C., Hunt D. F., Coon J. J. ( 2004) Anal. Chem. 76, 3590– 3598 [DOI] [PubMed] [Google Scholar]
- 17.Collins S. J., Ruscetti F. W., Gallagher R. E., Gallo R. C. ( 1978) Proc. Natl. Acad. Sci. U. S. A. 75, 2458– 2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes P. J., Twist L. E., Durham J., Choudhry M. A., Drayson M., Chandraratna R., Michell R. H., Kirk C. J., Brown G. ( 2001) Biochem. J. 355, 361– 371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olins A. L., Herrmann H., Lichter P., Kratzmeier M., Doenecke D., Olins D. E. ( 2001) Exp. Cell Res. 268, 115– 127 [DOI] [PubMed] [Google Scholar]
- 20.Choi M. R., Groot M., Drexler H. C. ( 2007) Apoptosis 12, 2025– 2035 [DOI] [PubMed] [Google Scholar]
- 21.Luo H. R., Loison F. ( 2008) Am. J. Hematol. 83, 288– 295 [DOI] [PubMed] [Google Scholar]
- 22.Khwaja A., Tatton L. ( 1999) Blood 94, 291– 301 [PubMed] [Google Scholar]
- 23.Pongracz J., Webb P., Wang K., Deacon E., Lunn O. J., Lord J. M. ( 1999) J. Biol. Chem. 274, 37329– 37334 [DOI] [PubMed] [Google Scholar]
- 24.Reyland M. E. ( 2007) Biochem. Soc. Trans. 35, 1001– 1004 [DOI] [PubMed] [Google Scholar]
- 25.Simon H. U. ( 2003) Immunol. Rev. 193, 101– 110 [DOI] [PubMed] [Google Scholar]
- 26.Ferreira H., Somers J., Webster R., Flaus A., Owen-Hughes T. ( 2007) Mol. Cell. Biol. 27, 4037– 4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macdonald N., Welburn J. P., Noble M. E., Nguyen A., Yaffe M. B., Clynes D., Moggs J. G., Orphanides G., Thomson S., Edmunds J. W., Clayton A. L., Endicott J. A., Mahadevan L.C. ( 2005) Mol. Cell 20, 199– 211 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.