Abstract
The G protein βγ subunit dimer (Gβγ) and the Gβ5/regulator of G protein signaling (RGS) dimer play fundamental roles in propagating and regulating G protein pathways, respectively. How these complexes form dimers when the individual subunits are unstable is a question that has remained unaddressed for many years. In the case of Gβγ, recent studies have shown that phosducin-like protein 1 (PhLP1) works as a co-chaperone with the cytosolic chaperonin complex (CCT) to fold Gβ and mediate its interaction with Gγ. However, it is not known what fraction of the many Gβγ combinations is assembled this way or whether chaperones influence the specificity of Gβγ dimer formation. Moreover, the mechanism of Gβ5-RGS assembly has yet to be assessed experimentally. The current study was undertaken to directly address these issues. The data show that PhLP1 plays a vital role in the assembly of Gγ2 with all four Gβ1–4 subunits and in the assembly of Gβ2 with all twelve Gγ subunits, without affecting the specificity of the Gβγ interactions. The results also show that Gβ5-RGS7 assembly is dependent on CCT and PhLP1, but the apparent mechanism is different from that of Gβγ. PhLP1 seems to stabilize the interaction of Gβ5 with CCT until Gβ5 is folded, after which it is released to allow Gβ5 to interact with RGS7. These findings point to a general role for PhLP1 in the assembly of all Gβγ combinations and suggest a CCT-dependent mechanism for Gβ5-RGS7 assembly that utilizes the co-chaperone activity of PhLP1 in a unique way.
Eukaryotic cells utilize receptors coupled to heterotrimeric GTP-binding proteins (G proteins)3 to mediate a vast array of responses ranging from nutrient-induced migration of single-celled organisms to neurotransmitter-regulated neuronal activity in the human brain (1). Ligand binding to a G protein-coupled receptor (GPCR) initiates GTP exchange on the G protein heterotrimer (composed of Gα, Gβ, and Gγ subunits), which in turn causes the release of Gα-GTP from the Gβγ dimer (2–4). Both Gα-GTP and Gβγ propagate and amplify the signal by interacting with effector enzymes and ion channels (1, 5). The duration and amplitude of the signal is dictated by receptor phosphorylation coupled with arrestin binding and internalization (6) and by regulators of G protein signaling (RGS) proteins, which serve as GTPase-activating proteins for the GTP-bound Gα subunit (7, 8). The G protein signaling cycle is reset as the inactive Gα-GDP reassembles with the Gβγ dimer and Gαβγ re-associates with the GPCR (5).
To fulfill its essential role in signaling, the G protein heterotrimer must be assembled post-translationally from its nascent polypeptides. Significant progress has been made recently regarding the mechanism by which this process occurs. It has been clear for some time that the Gβγ dimer must assemble first, followed by subsequent association of Gα with Gβγ (9). What has not been clear was how Gβγ assembly would occur given the fact that neither Gβ nor Gγ is structurally stable without the other. An important breakthrough was the finding that phosducin-like protein 1 (PhLP1) functions as a co-chaperone with the chaperonin containing tailless complex polypeptide 1 (CCT) in the folding of nascent Gβ and its association with Gγ (10–15). CCT is an important chaperone that assists in the folding of actin and tubulin and many other cytosolic proteins, including many β propeller proteins like Gβ (16). PhLP1 has been known for some time to interact with Gβγ and was initially believed to inhibit Gβγ function (17). However, several recent studies have demonstrated that PhLP1 and CCT work together in a highly orchestrated manner to form the Gβγ dimer (10–15).
Studies on the mechanism of PhLP1-mediated Gβγ assembly have focused on the most common dimer Gβ1γ2 (10, 13, 14), leaving open questions about the role of PhLP1 in the assembly of the other Gβγ combinations. These are important considerations given that humans possess 5 Gβ genes and 12 Gγ genes with some important splice variants (18, 19), resulting in more than 60 possible combinations of Gβγ dimers. Gβ1–4 share between 80 and 90% sequence identity and are broadly expressed (18, 19). Gβ5, the more atypical isoform, shares only ∼53% identity with Gβ1, carries a longer N-terminal domain, and is only expressed in the central nervous system and retina (20). The Gγ protein family is more heterogeneous than the Gβ family. The sequence identity of the 12 Gγ isoforms extends from 10 to 70% (21), and the Gγ family can be separated into 5 subfamilies (21–23). All Gγ proteins carry C-terminal isoprenyl modifications, which contribute to their association with the cell membrane, GPCRs, Gαs, and effectors (9). Subfamily I Gγ isoforms are post-translationally farnesylated, whereas all others are geranylgeranylated (22, 24).
There is some inherent selectivity in the assembly of different Gβγ combinations, but in general Gβ1–4 can form dimers with most Gγ subunits (25). The physiological purpose of this large number of Gβγ combinations has intrigued researchers in the field for many years, and a large body of research indicates that GPCRs and effectors couple to a preferred subset of Gβγ combinations based somewhat on specific sequence complementarity, but even more so on cellular expression patterns, subcellular localization, and post-translational modifications (18).
In contrast to Gβ1–4, Gβ5 does not interact with Gγ subunits in vivo, but it instead forms irreversible dimers with RGS proteins of the R7 family, which includes RGS proteins 6, 7, 9, and 11 (26). All R7 family proteins contain an N-terminal DEP (disheveled, Egl-10, pleckstrin) domain, a central Gγ-like (GGL) domain, and a C-terminal RGS domain (8, 26). The DEP domain interacts with the membrane anchoring/nuclear shuttling R7-binding protein, and the GGL domain binds to Gβ5 in a manner similar to other Gβγ associations (27, 28). Like Gβγs, Gβ5 and R7 RGS proteins form obligate dimers required for their mutual stability (26). Without their partner, Gβ5 and R7 RGS proteins are rapidly degraded in cells (26, 29). Gβ5-R7 RGS complexes act as important GTPase-accelerating proteins for Gi/oα and Gqα subunits in neuronal cells and some immune cells (26).
It has been recently shown that all Gβ isoforms are able to interact with the CCT complex, but to varying degrees (15). Gβ4 and Gβ1 bind CCT better than Gβ2 and Gβ3, whereas Gβ5 binds CCT poorly (15). These results suggest that Gβ1 and Gβ4 might be more dependent on PhLP1 than the other Gβs, given the co-chaperone role of PhLP1 with CCT in Gβ1γ2 assembly. However, another report has indicated that Gγ2 assembly with Gβ1 and Gβ2 is more PhLP1-dependent than with Gβ3 and Gβ4 (30). Thus, it is not clear from current information whether PhLP1 and CCT participate in assembly of all Gβγ combinations or whether they contribute to the specificity of Gβγ dimer formation, nor is it clear whether they or other chaperones are involved in Gβ5-R7 RGS dimer formation. This report was designed to address these issues.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK 293T cells were cultured in Dulbecco's modified Eagle's medium/F-12 (50/50 mix) growth media containing l-glutamine and 15 mm HEPES supplemented with 10% fetal bovine serum (Sigma-Aldrich). The cells were subcultured regularly to maintain growth, but were not used beyond 25 passages.
Preparation of cDNA Constructs
The pcDNA3.1 vectors containing N-terminally FLAG-tagged human Gβs 1–4, Gβ5short, N-terminally HA-tagged Gγs 1–5 and 7–13, and 3× HA-tagged RGS7 (S2), were obtained from the Missouri University of Science and Technology cDNA Resource Center. Wild-type human PhLP1 and the PhLP1 Δ1–75 N-terminal truncation variant each with a 3′ c-Myc and His6 tag were constructed in pcDNA3.1/Myc-His B vector using PCR as described (14, 31).
RNA Interference Experiments
Short interfering RNAs (siRNAs) were chemically synthesized (Dharmacon) to target nucleotides 608–628 of human lamin A/C (14), nucleotides 345–365 of human PhLP1 (14), and nucleotides 172–192 of human CCTζ-1 (32). HEK 293T cells were grown in 12-well plates to 50–70% confluency at which point they were transfected with siRNA at 100 nm final concentration using Oligofectamine reagent (Invitrogen) as described previously (14). 24 h later, the cells were transfected with 0.5 μg each of FLAG-Gβ and HA-Gγ or HA-RGS7 in pcDNA3.1(+) using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). The cells were harvested for subsequent immunoprecipitation experiments 72 h later. 10 μg of cell lysate was immunoblotted with an anti-PhLP1 antibody (33) to assess the percent PhLP1 knockdown, and 20 μg was immunoblotted with anti-CCTζ and anti-CCTϵ antibodies (Santa Cruz Biotechnology) to determine the percent CCT knockdown.
Dominant Interfering Mutant Experiments
HEK 293T cells were plated in 6-well plates and grown to 70–80% confluency. The cells were then transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Each well was transfected with 1.0 μg of the empty vector control, wild-type PhLP1-Myc, or PhLP1 Δ1–75-Myc along with 1.0 μg each of the indicated FLAG-Gβ and HA-Gγ or HA-RGS7 cDNAs. The cells were harvested for immunoprecipitation 48 h after transfection.
Immunoprecipitation Experiments
Transfected HEK 293T cells were washed with phosphate-buffered saline (Fisher) and solubilized in immunoprecipitation buffer (phosphate-buffered saline, pH 7.4, 2% Nonidet P-40 (Sigma), 0.6 mm phenylmethylsulfonyl fluoride, 6 μl/ml protease inhibitor mixture per ml of buffer (Sigma P8340)). The lysates were passed through a 25-gauge needle 10 times and centrifuged at maximum speed for 10–12 min at 4 °C in an Eppendorf microcentrifuge. The protein concentration for each sample was determined using the DC Protein Assay Kit II (Bio-Rad), and equal amounts of protein were used in the subsequent immunoprecipitations. Approximately 150 μg of total protein was used in immunoprecipitations from cells in 12-well plates and 450 μg from cells in 6-well plates. The clarified lysates were incubated for 30 min at 4 °C with 2.5 μg of anti-FLAG antibody (clone M2, Sigma), 1 μg of anti-CCTϵ (clone PK/29/23/8d Serotec), 1.75 μg of anti-Myc or 1.5 μg of anti-HA (clone 3F10, Roche Applied Science) for lysates from 12-well plates or with 6.25 μg of anti-FLAG for lysates from 6-well plates. Next, 30 μl of a 50% slurry of Protein A/G Plus-agarose (Santa Cruz Biotechnology) was added, and the mixture was incubated for 30 min at 4 °C as described (14). The immunoprecipitated proteins were solubilized in SDS sample buffer and resolved on a 10% Tris-glycine-SDS gel or a 16.5% Tris-Tricine-SDS gel for Gγ. The proteins were transferred to nitrocellulose and immunoblotted using an anti-FLAG (clone M2, Sigma), anti-c-Myc (BioMol), anti-HA (Roche Applied Science), or an anti-PhLP1 antibody (14). Immunoblots were incubated with the appropriate anti-rabbit, anti-mouse, anti-goat (Li-Cor Biosciences), or anti-rat (Rockland) secondary antibody conjugated with an infrared dye. Blots were scanned using an Odyssey Infrared Imaging System (Li-Cor Biosciences), and protein band intensities were quantified using the Odyssey software. The data are presented as the mean value ± S.E. from at least three experiments.
Radiolabel Pulse-Chase Assays
Radiolabel pulse-chase assays were preformed as previously described (14). Briefly, siRNA-treated or transfected HEK 293T cells in 12-well plates were washed once with 1.5 ml of methionine-free Dulbecco's modified Eagle's medium media (Mediatech) and then incubated in 1 ml of this same media at 37 °C for 1 h. The media was discarded, and 400 μl of media supplemented with 200 μCi/ml radiolabeled l-[35S]methionine (PerkinElmer Life Sciences) was added. The cells were incubated in the radiolabeled media for 10 min at 23 °C. Subsequently, the cells were washed once with 1.6 ml of Dulbecco's modified Eagle's medium media supplemented with 4 mm non-radiolabeled l-methionine (Sigma) and 4 μm cycloheximide to remove the radiolabeled media and then incubated in 0.8 ml of this same media at 23 °C for the chase times indicated. After the chase periods, the cells were harvested for immunoprecipitation. Radiolabeled gels were visualized with a Storm 860 PhosphorImager, and the band intensities were quantified using ImageQuant software (Amersham Biosciences). The molar ratios were determined by normalizing the band intensities to the number of methionine residues found in FLAG-Gβ1, HA-Gγ, FLAG-Gβ5, or HA-RGS7 and then calculating the ratios. The molar ratios were consistently substoichiometric, with the HA-Gγ2/FLAG-Gβ1 ratio reaching ∼0.4 by 60 min of chase (Fig. 9B) and the HA-RGS7/Gβ5 ratio reaching ∼0.1 by 60-min chase (Figs. 8 and 9A). The lower HA-RGS7/Gβ5 ratio probably results from less efficient synthesis and folding of the nascent RGS7 compared with nascent Gβ5. The rate data for Gβγ and Gβ5-RGS7 assembly were fit to a first-order rate equation with background correction to determine the rate constant for assembly.
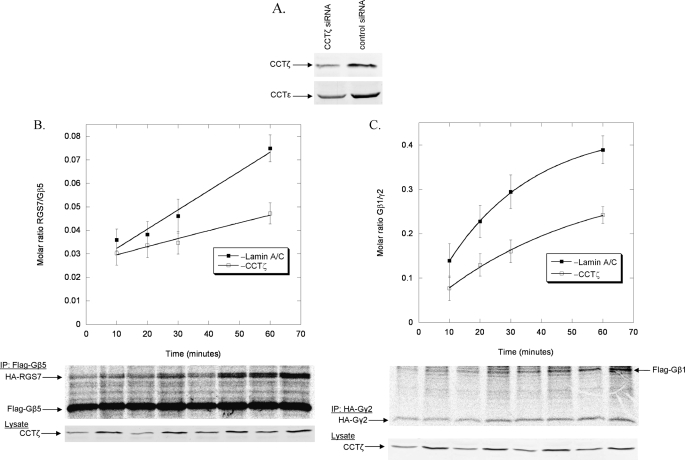
FIGURE 9.
Effects of CCT on the rate of Gβ5-RGS7 dimer formation. A, HEK-293T cells were treated with CCTζ or lamin A/C siRNA for 96 h and the expression of CCTζ and CCTϵ was measured by immunoblotting 20 μg of whole cell lysate. Representative blots are shown. B, the rate of Gβ5-RGS7 dimer assembly was measured in HEK-293T cells with or without CCTζ knockdown as in Fig. 8A. The data are from three separate experiments. C, the rate of Gβ1γ2 dimer assembly was measured in HEK-293T cells with or without CCTζ knockdown as in Fig. 8A. The data are from three separate experiments.
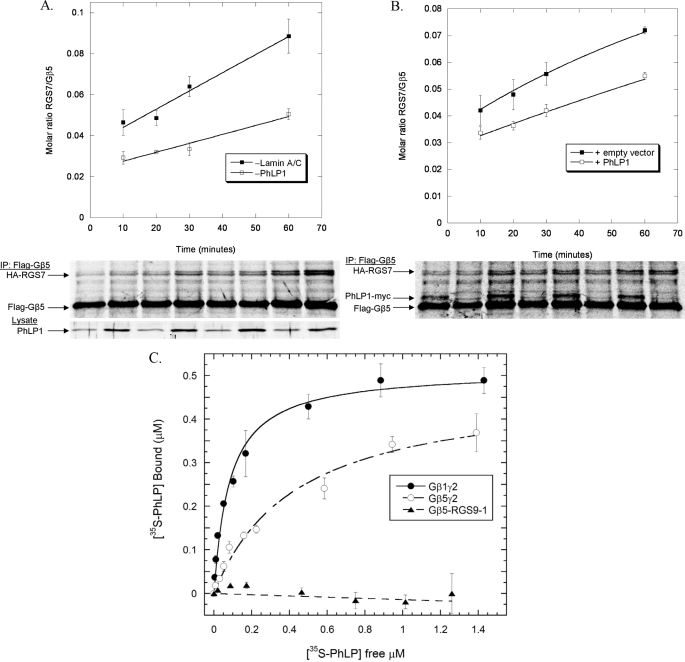
FIGURE 8.
Effects of PhLP1 on the rate of Gβ5-RGS7 dimer formation. A, the rate of Gβ5-RGS7 dimer assembly was measured in HEK-293T cells with or without PhLP1 knockdown. Cells were treated with PhLP1 or lamin A/C siRNAs as indicated. Twenty-four hours later, the cells were transfected with FLAG-Gβ5 and HA-RGS7 cDNAs. After 72 additional h, nascent polypeptides were labeled for 10 min with [35S]methionine and then chased with unlabeled methionine and cycloheximide. At the chase times indicated, the FLAG-Gβ5 was immunoprecipitated and the proteins were separated by SDS-PAGE. The radioactive bands were visualized and quantified using a PhosphorImager, and the molar ratio of Gβ5 to RGS7 was calculated. The data points represent the average ± S.E. from three separate experiments, and lines represent fits of the data to a first order rate equation. A representative gel is shown below the graph as is a PhLP1 immunoblot of 10 μg of whole cell lysate showing the degree of siRNA knockdown. B, HEK-293 cells were transfected with FLAG-Gβ5 and HA-RGS7 with and without PhLP1-Myc cDNAs for 48 h, and the rate of Gβ5-RGS7 assembly was measured using the pulse-chase assay as in panel A. The data are from three separate experiments. C, the binding of the indicated concentrations of 35S-PhLP1 to 0.5 μm purified Gβ1γ2 (○), Gβ5γ2 (○), or Gβ5-RGS9-1 (▴) was measured by in vitro co-immunoprecipitation (see “Experimental Procedures”). Symbols represent the average ± S.E. from three separate experiments. Lines represent non-linear least squares fits of the data to a one-to-one binding equation. The fits yielded Kd values of 83 ± 13 nm for Gβ1γ2, 440 ± 70 nm for Gβ5γ2, and no measurable value for Gβ5-RGS9-1.
Protein Purifications
Gβ1γ2, Gβ5γ2, and Gβ5-RGS9-1 were expressed and purified from insect cells. Recombinant baculovirus constructs were generously provided by Narasimhan Gautam of Washington University (Gβ1(34)), James Garrison from the University of Virginia (Gβ5 and Gγ2 (35)), and Ching-Kang Chen from Virginia Commonwealth University (RGS9-1 (36)). The Gγ2 subunit contained both a His6 tag and a FLAG epitope tag on the N terminus. The RGS9-1 subunit contained only a FLAG epitope tag on the C terminus. Sf9 cells (Amersham Biosciences) were grown to a density of 2 × 106 cell/ml and then co-infected with a Gβ and Gγ2 or RGS9-1 baculovirus at an multiplicity of infection of 5 for each virus type. Cells were grown in shaker culture for ∼60 h and then pelleted by centrifugation at 250 × g for 10 min at 4 °C. The supernatant was discarded, and the cell pellet was snap frozen in liquid nitrogen and stored at −80 °C.
Gβ1γ2 and Gβ5γ2 were purified by a modification of a previously described protocols (35, 37). The cell pellet from 1 liter of cells was thawed and resuspended in 100 ml of homogenization buffer (20 mm HEPES, pH 7.5, 3 mm MgCl2, 150 mm NaCl, 2 μg/ml aprotinin, 2 μg/ml leupeptin, 2 μg/ml pepstatin, 20 μg/ml benzamidine, and 0.1 mm phenylmethylsulfonyl fluoride). The suspension was sonicated with a tip sonicator on ice and centrifuged at 100,000 × g for 1 h. The pellet was homogenized in 100 ml of extraction buffer (homogenization buffer + 0.1% polyoxyethylene 10 laurel ether) using a Dounce homogenizer and stirred on ice for 1 h. The suspension was centrifuged again at 100,000 × g for 1 h. The supernatant was collected and applied to a 5-ml M2 FLAG-agarose column (Sigma-Aldrich) equilibrated in extraction buffer. The column was washed with 30 ml of extraction buffer, and the Gβγ dimers were eluted with 15 ml of FLAG elution buffer (extraction buffer plus 250 μg/ml FLAG peptide). Fractions containing the purified dimers were combined and applied to a 2 ml nickel-nitrilotriacetic acid column (Novagen) equilibrated in extraction buffer plus 30 mm imidazole. The column was washed with 20 ml of this buffer, and then eluted with 10 ml of extraction buffer plus 500 mm imidazole. Fractions containing Gβγ were combined and dialyzed in extraction buffer plus 50% glycerol, which caused a 4-fold increase in protein concentration. Gβ5-RGS9-1 was purified the same way except the nickel-nitrilotriacetic acid column was skipped because the RGS9-1 protein did not contain an His6 tag. This procedure generally resulted in ∼1 ml of ∼1 mg/ml protein that was 90% pure.
Metabolically labeled 35S-PhLP1 was prepared by transforming DE3 Escherichia coli cells with a PhLP1 pET15b vector (38) and inoculating 100 ml of M9 minimal media with a single colony of cells. The culture was incubated ∼20 h at 37 °C until the absorbance at 600 nm reached 0.6–0.7. The cells were collected by centrifugation and resuspended in 100 ml of reduced Na2SO4 M9 minimal media. At this point, 12 mg of isopropyl-β-d-thiogalactopyranoside was added along with 500 μl of 2 mCi/ml [35S]H2SO4. The culture was grown for 3.5 h at 37 °C to an absorbance at 600 nm of ∼1.0. The labeled PhLP1 was then purified as described previously (38).
In Vitro Binding Assays
The binding of 35S-PhLP1 to Gβγ or Gβ5-RGS9-1 dimers was determined by mixing 35 μl of a 50% slurry of M2 FLAG-agarose beads equilibrated in assay buffer (extraction buffer without protease inhibitors) with purified Gβγ or Gβ5-RGS9-1 (final concentration, 0.5 μm). The 35S-PhLP1 was then added to the reaction mixture at final concentrations ranging from 0.01 μm to 2 μm in a total reaction volume of 150 μl. The reaction mixture was incubated on a rotator at 4 °C for 1 h. Each reaction was briefly vortexed, and 50 μl of the mixture was counted in a scintillation counter to obtain the total amount of PhLP1 added. Each reaction was then centrifuged for 1 min at 1000 × g to separate the bound from the free 35S-PhLP1. A 50-μl aliquot of the supernatant was then counted as described above to obtain the free counts. The free counts were subtracted from the total counts to determine the counts of bound 35S-PhLP1. Nonspecific binding was determined by running the assay in parallel with FLAG-glutathione S-transferase in place of Gβγ or Gβ5-RGS9-1. The specific binding was determined by subtracting the nonspecific binding from the total binding. Counts were converted into concentration units using the specific activity of the 35S-PhLP1. The concentration of specifically bound 35S-PhLP1 was then plotted versus the free PhLP1 concentration, and the Kd for the interaction was determined by fitting the data to a one-to-one binding equation, B = Bmax/(1 + Kd/[PhLP1]), where B is the amount of PhLP1 bound to the beads, Bmax is the maximal binding of PhLP1, and Kd is the dissociation constant for the interaction.
RESULTS
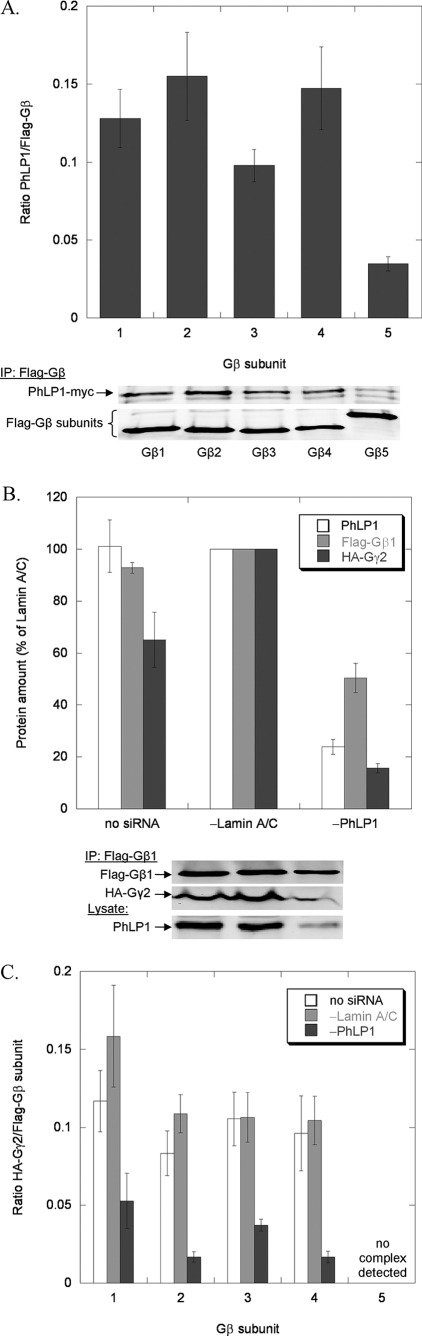
It has been shown previously that, to mediate Gβγ assembly, PhLP1 must bind Gβγ with high affinity (13, 14). As a first step toward determining the ability of PhLP1 to catalyze Gβγ dimer formation with the five Gβ subunits, we measured the interaction of PhLP1 with each Gβ subunit in complex with Gγ2 by co-immunoprecipitation. Equal amounts of Myc-tagged PhLP1, Gγ2, and FLAG-tagged Gβ1–5 were overexpressed in HEK-293T cells. After incubation, cells were harvested and immunoprecipitated with an anti-FLAG antibody and immunoblotted with anti-Myc and anti-FLAG antibodies. Protein band intensities were quantified, and the ratio of the PhLP1-Myc band to each FLAG-Gβ band was determined (Fig. 1A). The data show that Gβs 1–4 all co-immunoprecipitated similar amounts of PhLP1 while Gβ5 co-immunoprecipitated significantly less, indicating that PhLP1 binds Gβ5 complexes with a lower affinity than it does Gβ1–4 complexes. All five Gβs expressed equally well under these conditions, so the differences in binding cannot be attributed to different Gβ expression levels (Fig. 1A). These results suggest that PhLP1 may be involved in Gβγ assembly of Gβ1–4, but perhaps not Gβ5.
FIGURE 1.
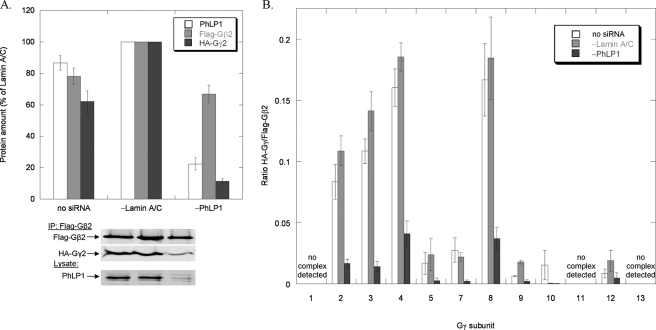
Effects of PhLP1 siRNA knockdown on the assembly of all Gβ subunits with Gγ2. HEK 293T cells were treated as follows. A, cells were transfected with PhLP1-Myc, HA-Gγ2, and the indicated FLAG-Gβ cDNAs. After 48 h, cells were lysed, immunoprecipitated with an anti-FLAG antibody, and immunoblotted with anti-Myc or anti-FLAG antibodies. The graph represents the ratio of the PhLP1-Myc/FLAG-Gβ band intensities for all 5 Gβs. Bars represent the average ± S.E. from three separate experiments. A representative blot is shown below the graph. B and C, cells were treated with siRNA against PhLP1, lamin A/C, or no siRNA as indicated. Twenty-four hours later, cells were transfected with the indicated FLAG-Gβ subunit and HA-Gγ2 cDNAs. After 72 additional hours, cells were lysed, immunoprecipitated with an anti-FLAG antibody, and immunoblotted with anti-FLAG or anti-HA antibodies. Bands were quantified and expressed as a percentage of the lamin A/C control for Gβ1γ2 in B or as the ratio of HA-Gγ2/FLAG-Gβ for all five Gβs in C. PhLP1 knockdown was measured by quantifying the PhLP1 band intensity in immunoblots of 10 μg of whole cell lysate. The average PhLP1 knockdown was between 60 and 76% compared with the lamin A/C control. Bars represent the average ± S.E. from 3–5 separate experiments. A representative blot for Gβ1γ2 is shown below the graph in B. The ncd indicates no complex detected under these conditions. This same abbreviation is also used in Figs. 2–4.
To directly measure the contribution of PhLP1 to the assembly of the five Gβ isoforms with Gγ, the effect of siRNA-mediated PhLP1 knockdown on Gβγ dimer formation was measured by co-immunoprecipitation of Gγ2 with the Gβs. We chose Gγ2 because it is a common isoform that associates to some extent with all Gβ subunits in vitro (25). HEK 293T cells were treated with PhLP1 siRNA, a control siRNA to lamin A/C or a mock treatment with no siRNA and then co-expressed with HA-Gγ2 and one of the five FLAG-tagged Gβ subunits. Cell lysates were immunoprecipitated with an anti-FLAG antibody, and the precipitate was immunoblotted with anti-HA and anti-FLAG antibodies to detect the amount of Gγ2 bound to each Gβ subunit. Fig. 1B shows the levels of PhLP1 in the cell extract and the amounts of FLAG-Gβ1 and HA-Gγ2 in the immunoprecipitate relative to the lamin A/C siRNA control. A 75% knockdown of PhLP1 resulted in a 50% decrease in Gβ1 and a striking 85% decrease in Gγ2 compared with the lamin A/C control. This pattern was consistent among all the Gβ subunits except for Gβ5, which had no detectable Gγ2 bound under these conditions (Fig. 1C). To more directly compare the effects of PhLP1 knockdown, the Gγ2/Gβ1–4 band intensity ratios in the immunoprecipitates were determined for the three siRNA conditions (Fig. 1C). In each case, much less Gγ2 was associated with Gβ when PhLP1 was knocked down. The Gγ2/Gβ ratio decreased between 65 and 84% compared with the lamin A/C control. These results indicate that PhLP1 does assist in the formation of Gβγ complexes containing Gβs 1–4 with Gγ2.
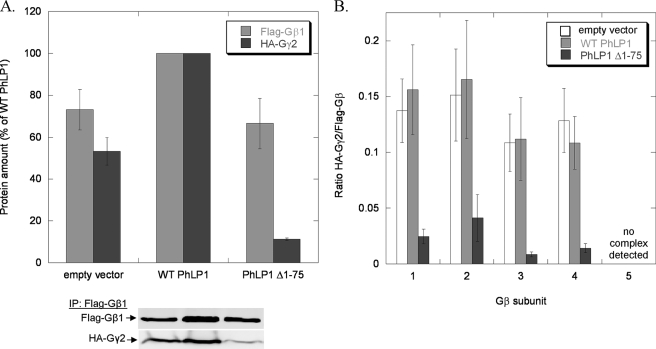
To further examine the role of PhLP1 in Gβγ assembly with the different Gβ subunits, an alternative method to block PhLP1 function was employed. It has been shown previously that an N-terminally truncated PhLP1 variant in which the first 75 amino acids have been removed (PhLP1 Δ1–75) acts in a dominant interfering manner to block Gβγ assembly by forming a stable PhLP1 Δ1–75-Gβ-CCT ternary complex that does not release Gβ from CCT for association with Gγ (13, 14). Co-expression of PhLP1 Δ1–75 with FLAG-Gβ1 and HA-Gγ2 resulted in a dramatic reduction in the amount of Gγ2 in the Gβ1 immunoprecipitate compared with wild-type PhLP1 (expressed at comparable levels) or to an empty vector control (Fig. 2A). This pattern was similar among Gβs 1–4. PhLP1 Δ1–75 decreased the Gγ2/Gβ ratios by 75–92% in the Gβ1–4 immunoprecipitates (Fig. 2B). For Gβ5, again very little Gγ2 was associated with it under these conditions. Interestingly, co-expression of wild-type PhLP1 increased the amount of both Gβ and Gγ2 in the FLAG-Gβ immunoprecipitate by 30–50% for all five Gβ isoforms (see Figs. 2A, 4A, and 6B). This observation is consistent with a PhLP1-mediated enhancement of Gβγ formation, resulting in a stabilization of Gβ and Gγ expression. Together, these findings confirm the siRNA knockdown results by showing that PhLP1 is important in the assembly of each of the Gβs 1–4 with Gγ2.
FIGURE 2.
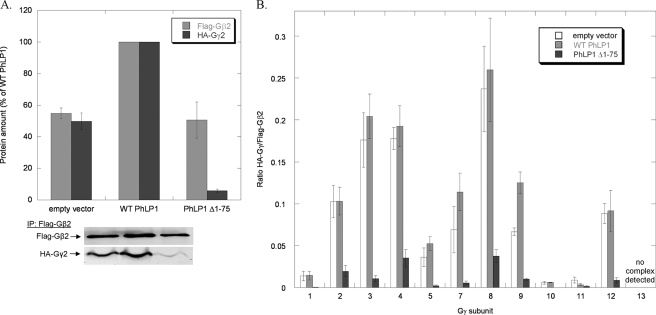
Effects of PhLP1 Δ1–75 expression on the assembly of all Gβ subunits with Gγ2. HEK 293T cells were transfected with either wild-type PhLP1, PhLP1 Δ1–75, or an empty vector control along with the indicated FLAG-Gβ subunit and HA-Gγ2 cDNAs. After 48 h, cells were lysed, immunoprecipitated with an anti-FLAG antibody, and immunoblotted with anti-FLAG or anti-HA antibodies. Bands were quantified and expressed as a percentage of the wild-type PhLP1 control for Gβ1γ2 in A or as the ratio of HA-Gγ2/FLAG-Gβ for all five Gβs in B. Bars represent the average ± S.E. from 3–5 separate experiments. A representative blot for Gβ1γ2 is shown below the graph in A.
FIGURE 4.
Effects of PhLP1 Δ1–75 expression on the assembly of all Gγ subunits with Gβ2. HEK 293T were transfected with either WT PhLP1, PhLP1 Δ1–75, or an empty vector control along with the indicated HA-Gγ subunit and FLAG-Gβ2 cDNAs. After 48 h, cells were lysed, immunoprecipitated with an anti-FLAG antibody and immunoblotted with anti-FLAG or anti-HA antibodies. Bands were quantified and expressed as a percentage of the wild-type PhLP1 control for Gβ2γ2 in A or as the relative ratio of HA-Gγ/FLAG-Gβ2 for all 12 Gγs in B. Bars represent the average ± S.E. from 3–6 separate experiments. A representative blot for Gβ2γ2 is shown below the graph in A.
FIGURE 6.
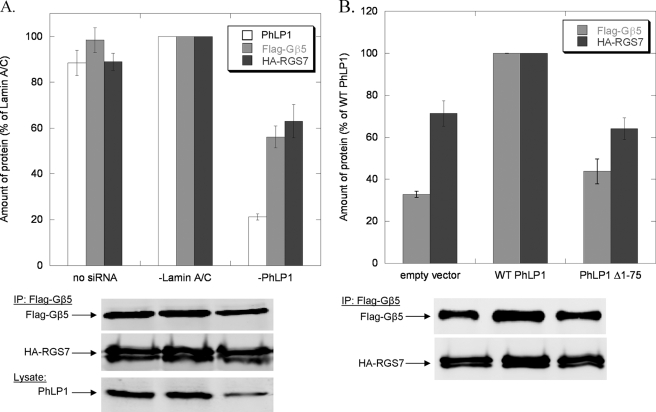
Effects of PhLP1 on the assembly of RGS7 with Gβ5. A, HEK 293T cells were treated with siRNA against PhLP1, lamin A/C, or no siRNA as indicated. Twenty-four hours later, cells were transfected with HA-RGS7 and FLAG-Gβ5 cDNAs. After 72 additional hours, cells were lysed, immunoprecipitated with an anti-FLAG antibody, and immunoblotted with anti-FLAG or anti-HA antibodies. Bands were quantified and expressed as a percentage of the lamin A/C control. PhLP1 knockdown was measured by quantifying the PhLP1 band intensity in immunoblots of 10 μg of whole cell lysate. B, cells were transfected with either WT PhLP1, PhLP1 Δ1–75, or an empty vector control along with HA-RGS7 and FLAG-Gβ5 cDNAs. After 48 h, cells were lysed, immunoprecipitated with an anti-FLAG antibody, and immunoblotted with anti-FLAG or anti-HA antibodies. Bands were quantified and expressed as a percentage of the wild-type PhLP1 control. Bars represent the average ± S.E. from three separate experiments. Representative blots are shown below the graphs.
A second question regarding the scope of PhLP1-mediated Gβγ assembly is whether all 12 Gγ subunits or just a subset require PhLP1 to associate with Gβ. To address this question, the effects of siRNA-mediated PhLP1 knockdown and PhLP1 Δ1–75 overexpression on the association of the twelve Gγ subunits with Gβ2 were measured. Gβ2 was chosen because it forms dimers with most Gγ isoforms, yet it shows selectivity between the different Gγs (25). The siRNA knockdown experiments followed the same format as those in Fig. 1. HEK 293T cells were treated with PhLP1 siRNA, a control siRNA to lamin A/C or no siRNA and then co-expressed with FLAG-Gβ2 and each of the 12 HA-tagged Gγ subunits. Cell lysates were immunoprecipitated with an anti-FLAG antibody, and the precipitate was immunoblotted with anti-HA and anti-FLAG antibodies to detect the amount of each Gγ subunit bound to Gβ2. Fig. 3A shows the levels of PhLP1 in the cell extract and FLAG-Gβ2 and HA-Gγ2 in the immunoprecipitate relative to the lamin A/C siRNA control. The results were similar to the Gβ1γ2 experiment. The PhLP1 knockdown was 80%, which resulted in a 30% decrease in Gβ2 and a 90% decrease in Gγ2 compared with the lamin A/C control. This pattern was consistent among all the Gβ2Gγ combinations that formed dimers. Gβ2 decreased by 20–50% while the co-immunoprecipitating Gγs decreased by 80–95% (data not shown). Fig. 3B compares the Gγ/Gβ2 band intensity ratios for the three siRNA conditions. In each case, much less Gγ was associated with Gβ when PhLP1 was knocked down. The Gγ/Gβ2 ratios decreased between 74 and 91% compared with the lamin A/C control, except for Gγs 1, 11, and 13, which did not form dimers with Gβ2. These results indicate that all Gβ2Gγ dimers depend upon PhLP1 for their assembly.
FIGURE 3.
Effects of PhLP1 knockdown on the assembly of all Gγ subunits with Gβ2. HEK 293T cells were treated with siRNA against PhLP1, lamin A/C, or no siRNA as indicated. Twenty-four hours later, cells were transfected with the indicated HA-Gγ subunit and FLAG-Gβ2 cDNAs. After 72 additional hours, cells were lysed, immunoprecipitated with an anti-FLAG antibody, and immunoblotted with anti-FLAG or anti-HA antibodies. Bands were quantified and expressed as a percentage of the lamin A/C control for Gβ2γ2 in A or as the ratio of HA-Gγ/FLAG-Gβ2 for all 12 Gγs in B. PhLP1 knockdown was measured by quantifying the PhLP1 band intensity in immunoblots of 10 μg of whole cell lysate. The average PhLP1 knockdown was between 66 and 90% compared with the lamin A/C control. Bars represent the average ± S.E. from 3–14 separate experiments. A representative blot for Gβ2γ2 is shown below the graph in A.
The dominant interference experiments with PhLP1 Δ1–75 followed a similar pattern. Co-expression of PhLP1 Δ1–75 with FLAG-Gβ2 and HA-Gγ2 resulted in a 50% reduction in the amount of Gβ2 and a 95% reduction in the amount of Gγ2 in the FLAG immunoprecipitate when compared with the wild-type PhLP1 control (Fig. 4A). Moreover, co-expression of wild-type PhLP1 increased both Gβ2 and Gγ2 levels by 50%, similarly to Gβ1γ2 (Fig. 2A). The effect of PhLP1 Δ1–75 on the Gγ/Gβ2 ratio was the same for all the Gγs that formed dimers with Gβ2. The ratios were drastically reduced by amounts ranging from 81–100%. Together with the PhLP1 knockdown data, these results clearly demonstrate that all Gγ subunits that interact with Gβ2 require PhLP1 for dimer formation.
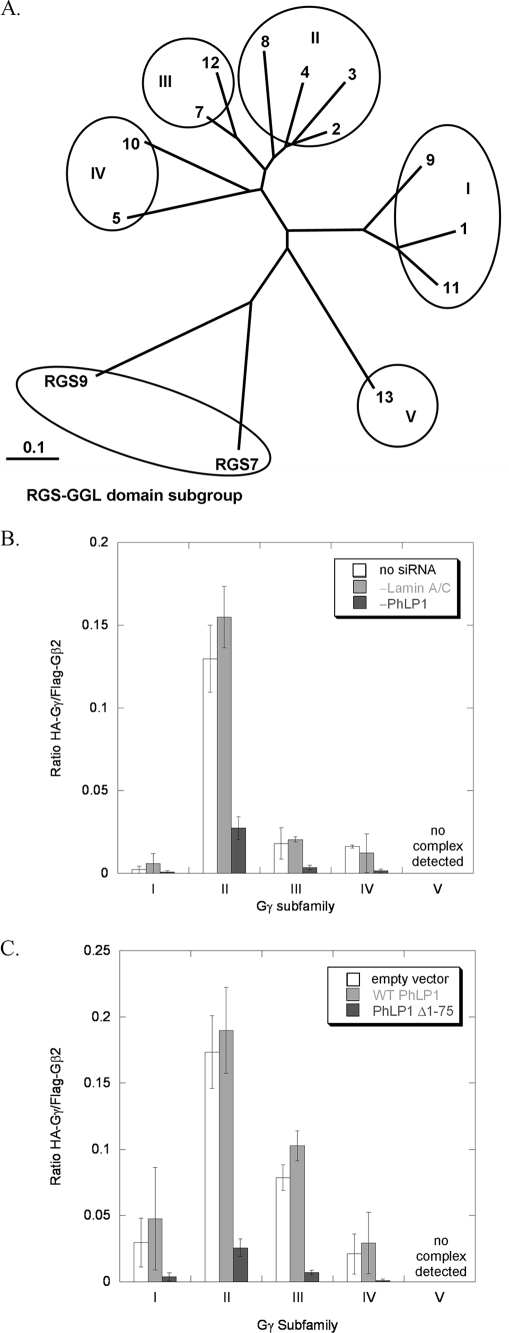
Another interesting observation that can be made from the data in Figs. 3B and 4B concerns the effect of PhLP1 on the specificity of Gβ2γ dimer formation. The Gγ subunits can be divided genetically into five subfamilies as shown in the phylogenetic tree of Fig. 5A. Members of subfamily II form dimers with Gβ2 readily, whereas members of the other subfamilies interact weakly with Gβ2 or not at all (Fig. 5, B and C). The order of dimer formation of the Gγ subfamilies with Gβ2 is II > III > I/IV with no dimer formation found with subgroup V. This pattern of Gβ2γ dimer specificity is similar to in vitro data reported previously (25). Importantly, PhLP1 does not appear to influence the specificity of Gβ2γ dimer formation. The specificity pattern is the same no matter the level of PhLP1 activity. For example, when PhLP1 is siRNA-depleted, Gβ2γ formation with Gγ subfamily II is greater than with subfamilies III, I, and IV by a similar factor as when PhLP1 is at endogenous levels. Similarly, when PhLP1 function is blocked by the PhLP1 Δ1–75 variant, Gβ2γ formation with subgroup II is greater than the other subgroups by a similar factor as when PhLP1 is overexpressed (Fig. 5, B and C). Thus, it appears that PhLP has no effect on which Gγ subunit will interact with Gβ2.
FIGURE 5.
Effects of PhLP1 knockdown on the specificity of Gβ2 dimerization with Gγ subfamilies. A, the phylogenetic relationship between human Gγ subunits and RGS7 and 9 is depicted. An unrooted dendrogram was made using TreeView from a Gγ family sequence alignment created with ClustalX. The Gγ family can be separated into five subfamilies as indicated. The scale bar represents a substitution rate of 0.1 per amino acid. B, the Gγ/Gβ2 ratios within each Gγ subfamily under the different siRNA conditions from Fig. 3B were averaged and plotted to show the effects of PhLP1 knockdown on the subfamily specificity of Gβ2Gγ dimer formation. Error bars represent the S.E. of the mean within each subfamily. C, a similar average of the Gβ2Gγ ratios for each subfamily under the different PhLP1 overexpression conditions from Fig. 4B was calculated and plotted.
A third important question regarding the scope of PhLP1-mediated dimer assembly is whether PhLP1 assists in the formation of Gβ5-R7 RGS protein complexes. Gβ5 binds both CCT (15) and PhLP1 (Fig. 1) weakly compared with the other Gβ subunits, suggesting that CCT and PhLP1 may not be required for Gβ5-R7 RGS dimer assembly. To begin to address this issue, the effects of PhLP1 knockdown and PhLP1 Δ1–75 overexpression on the interaction of Gβ5 with RGS7 were assessed by co-immunoprecipitation as in Fig. 1. PhLP1 knockdown decreased both Gβ5 expression and RGS7 co-immunoprecipitation with Gβ5 by 40% (Fig. 6A). This result is in contrast to the Gβγ co-immunoprecipitation data, which showed a similar 40% decrease in Gβ1 and Gβ2 expression but exhibited a much greater decrease (80–90%) in the amount of Gγ co-immunoprecipitating with Gβ upon PhLP1 knockdown (Figs. 1B and 3A). The results were similar in the dominant interference experiments (Fig. 6B). Overexpression of wild-type PhLP1 increased Gβ5 expression by ∼2-fold over the empty vector control, as was observed with Gβ1 and Gβ2 (Figs. 2A and 4A). However, the proportional increase in co-immunoprecipitation seen with Gγ2 (Figs. 2A and 4A) was not observed with RGS7, which showed a much smaller increase. Moreover, overexpression of PhLP1 Δ1–75 did not cause the dramatic decrease in RGS7 co-immunoprecipitation that was observed with Gγ2 (Fig. 6B). These findings suggest that the effect of PhLP1 on the expression of Gβ5 is similar to the other Gβs but that PhLP1 may not be as important in Gβ5-RGS7 assembly as it is in Gβγ assembly.
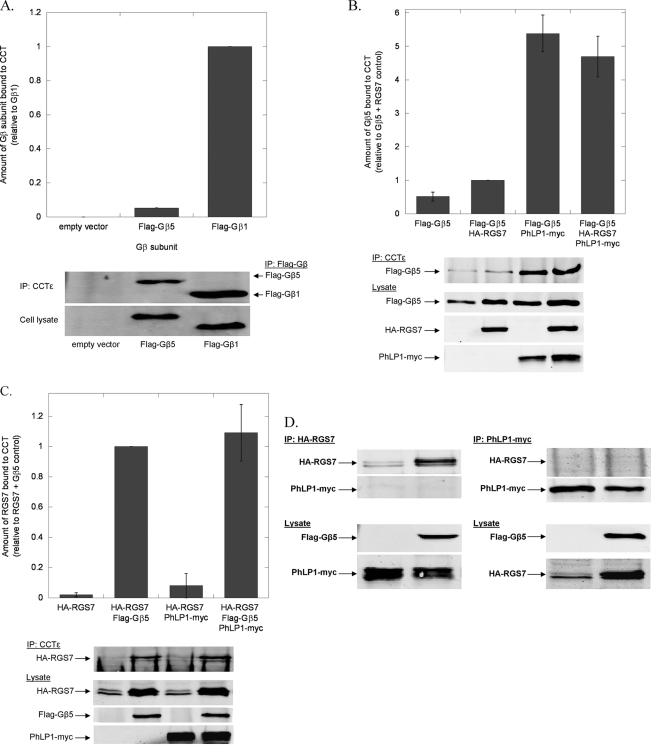
The findings of Fig. 6 point to potentially significant differences between the mechanisms of Gβ5-RGS7 assembly and Gβγ assembly. As a result, Gβ5-RGS7 assembly was further investigated to better understand the role of PhLP1 and CCT in this process. If CCT were involved in Gβ5 folding, the two would have to interact, yet Gβ5 has been reported to bind CCT poorly in vitro (15). To further test the ability of Gβ5 to interact with CCT, the co-immunoprecipitation of overexpressed Gβ5 with endogenous CCT in HEK-293T cells was measured. Gβ5 was readily detected in the CCT immunoprecipitate, but the amount was 20-fold less than that of Gβ1 (Fig. 7A), confirming the finding that Gβ5 binds CCT much less than other Gβs. Importantly, co-expression of PhLP1 increased Gβ5 binding to CCT by nearly 10-fold (Fig. 7B), indicating that PhLP1 stabilizes the interaction of Gβ5 with CCT considerably. In contrast, co-expression of RGS7 had no effect on Gβ5 binding to CCT. These results are very different from the effect of PhLP1 and Gγ2 on Gβ1 binding to CCT in which both PhLP1 and Gγ2 contributed significantly to the release of Gβ1 from the CCT complex (13). Thus, it appears that the role of PhLP1 in the binding of Gβ1 and Gβ5 to CCT are opposite, with PhLP assisting in the release of a tightly binding Gβ1 while stabilizing the weak interaction of Gβ5.
FIGURE 7.
Effects of PhLP1 and RGS7 on the binding of Gβ5 to CCT. A, binding of Gβ5 to CCT was compared with that of Gβ1 by co-immunoprecipitation. HEK 293T cells were transfected with cDNAs for FLAG-Gβ1, FLAG-Gβ5 or an empty vector control as indicated. After 48 h, cells were lysed, immunoprecipitated with an anti-CCTϵ antibody, and immunoblotted with anti-FLAG antibodies. Bands were quantified and the binding of Gβ5 to CCT was expressed relative to that of Gβ1. Bars represent the average ± S.E. from three separate experiments, and representative blots are shown below the graphs. (The Gβ5 error bar is very small.) For all experiments A–D, the expression of each transfected cDNA was confirmed by immunoblotting 5 μg of whole cell lysate with the antibodies indicated. B, the effect of PhLP1 and RGS7 on the binding of Gβ5 to CCT was measured by co-immunoprecipitation as in panel A. Cells were transfected with the indicated cDNAs, and CCT was immunoprecipitated and immunoblotted for FLAG-Gβ5. Bands were quantified and expressed relative to the FLAG-Gβ5/HA-RGS7 sample. Data are from eight separate experiments. C, the effects of PhLP1 and Gβ5 on RGS7 binding to CCT was measured by co-immunoprecipitation as in panel A. Cells were transfected with the indicated cDNAs, and CCT was immunoprecipitated and immunoblotted for HA-RGS7. Bands were quantified and expressed relative to the FLAG-Gβ5/HA-RGS7 sample. Data are from three separate experiments. D, the ability of PhLP1 and RGS7 to co-exist in CCT or other complexes was tested by co-immunoprecipitation. Cells were transfected with cDNAs to PhLP1-Myc and HA-RGS7 with or without FLAG-Gβ5, immunoprecipitated with anti-Myc or anti-HA antibodies and immunoblotted with these same antibodies as indicated. The resulting blots are shown.
To complete the investigation of CCT interacting partners in the Gβ5-RGS7 dimer, the binding of RGS7 was also measured by co-immunoprecipitation with CCT. No RGS7 bound CCT when RGS7 was overexpressed alone, but co-expression of Gβ5 caused a detectible amount of RGS7 to co-immunoprecipitate with CCT (Fig. 7C). In contrast, co-expression of PhLP1 with RGS7 did not cause RGS7 to bind CCT and co-expression of PhLP1 together with Gβ5 and RGS7 did not increase RGS7 co-immunoprecipitation with CCT. The total amount of RGS7 in the cell lysate also increased significantly upon Gβ5 co-expression, consistent with the fact that R7 RGS proteins require Gβ5 for stable expression in the cell (29). These results suggest that in the process of Gβ5-RGS7 assembly Gβ5 recruits RGS7 to CCT. The lack of effect of PhLP1 on RGS7 binding to CCT suggests that PhLP1 does not play a role in this recruitment. To further test this notion, RGS7 and PhLP1 were co-expressed with and without Gβ5, and their ability to co-immunoprecipitate each other was measured. Neither protein was found in the immunoprecipitate of the other in the presence or absence of Gβ5 (Fig. 7D), indicating that RGS7 and PhLP1 do not exist in any complexes together. From these binding experiments, it appears that PhLP1 stabilizes the interaction of Gβ5 with CCT and that Gβ5 recruits RGS7 to CCT, but only after PhLP1 has been released from the complex.
The data from Figs. 6 and 7 suggest that PhLP1 may be involved in the folding of Gβ5 by stabilizing its interaction with CCT but that PhLP1 may not participate in Gβ5-RGS7 assembly. This concept was further tested by measuring the effect of PhLP1 knockdown or overexpression on the rate of Gβ5-RGS7 dimerization. In these experiments, PhLP1 was either siRNA-depleted or overexpressed in HEK-293T cells, and the rate of Gβ5-RGS7 dimer formation was measured in a pulse-chase experimental format (14). PhLP1 knockdown resulted in a 2-fold decrease in the rate of Gβ5-RGS7 dimerization compared with a control siRNA (Fig. 8A), which is somewhat less than the 5-fold decrease in the rate of Gβ1γ2 dimerization observed with a similar PhLP1 knockdown (14). In contrast, the effects of PhLP1 overexpression on the rate of Gβ5-RGS7 assembly were strikingly different than what was observed for Gβ1γ2 assembly. PhLP1 overexpression actually caused a small decrease in the rate of Gβ5-RGS7 assembly (Fig. 8B), whereas it resulted in a 4-fold increase in the rate of Gβ1γ2 assembly (14). Interestingly, PhLP overexpression increased the amount of Gβ5 produced during the 10-min pulse by 40%, which in turn caused a small increase in RGS7 co-immunoprecipitation. However, the net effect was a decrease in the RGS7/Gβ5 ratio, indicating an inhibition of RGS7/Gβ5 dimer formation despite the fact that more Gβ5 was available for assembly. It is clear from these results that the role of PhLP1 in Gβ5-RGS7 assembly is much different than its role in Gβγ assembly. It appears that endogenous levels of PhLP1 may contribute to Gβ5-RGS7 assembly by stabilizing the interaction of Gβ5 with CCT, but that excess PhLP1 inhibits Gβ5-RGS7 assembly, possibly by interfering with the Gβ5-RGS7 interaction.
The recently published structure of the Gβ5-RGS9 complex (39) suggests a possible reason for the observed inhibition of Gβ5-RGS7 assembly by excess PhLP1. In the structure, the Gγ-like domain interacts along the expected Gγ binding surface of Gβ5, opposite the predicted PhLP1 binding site (39). However, the N-terminal lobe of RGS9 interacts with Gβ5 on the same surface as PhLP1 (39, 40). This overlap may preclude the formation of a PhLP1-Gβ5-RGS7 complex analogous to the PhLP1-Gβγ complex that is believed to be an intermediate in Gβγ assembly (13, 14). To test this possibility, the binding of PhLP1 to the Gβ5-RGS9-1 complex was measured. An in vitro assay was preformed in which Gβ5-RGS9-1 was immobilized on FLAG antibody-linked agarose beads via a FLAG tag on the RGS9-1. Increasing concentrations of metabolically labeled 35S-PhLP1 were added to the beads and allowed to reach equilibrium. The beads were pelleted, and the amount of bound and free 35S-PhLP1 was determined. The results show that indeed there was no measurable binding of PhLP1 to Gβ5-RGS9-1 (Fig. 8C). In contrast, PhLP1 readily bound Gβ1γ2 and to a lesser extent Gβ5γ2 in this assay. The dissociation constants for the interactions were 83 ± 13 nm for Gβ1γ2 and 440 ± 72 nm for Gβ5γ2. The Kd for Gβ1γ2 binding is similar to the 107 nm Kd reported previously for the PhLP1-Gβ1γ1 interaction using surface plasmon resonance methods (38), so the assay appears to be measuring the binding accurately. The inability of Gβ5-RGS9-1 to bind PhLP1 suggests that excess PhLP1 interferes with Gβ5-RGS7 dimer formation, because it binds Gβ5 in a manner that does not allow RGS7 to simultaneously interact.
The binding of Gβ5 to CCT and the Gβ5-dependent recruitment of RGS7 to CCT suggest an important role for CCT in the Gβ5-RGS7 assembly process. This possibility was tested further by measuring the effect of CCT knockdown on the rate of Gβ5-RGS7 dimerization using the pulse-chase assay. An siRNA to CCTζ that results in a substantial knockdown of CCT complexes has been reported (32, 41). Using this siRNA, CCTζ expression was decreased by 50% in HEK-293T cells (Fig. 9A). In addition, expression of the CCTϵ subunit was also decreased by a similar amount (Fig. 9A), indicating that expression of the entire CCT complex was reduced by 50%. This reduction in CCT resulted in a proportional decrease in the rate of Gβ5-RGS7 assembly of 50% (Fig. 9A), suggesting that Gβ5-RGS7 assembly is very dependent on CCT. For comparison, the effect of this CCT knockdown on Gβγ assembly, which is expected to be CCT-dependent (13, 15), was also measured. The 50% reduction in CCT caused a similar 50% decrease in the rate of Gβγ assembly (Fig. 9B), confirming the importance of CCT in Gβγ formation. The striking similarity of these effects of CCT knockdown on the rates of both Gβ5-RGS7 and Gβγ dimerization show that Gβ5-RGS7 assembly is just as dependent on CCT as Gβγ assembly. Together, the data in Figs. 8 and 9 indicate a similar role for CCT in both Gβ5-RGS7 and Gβγ assembly, but a much less critical role for PhLP1 in Gβ5-RGS7 assembly compared with its essential role in Gβγ assembly.
DISCUSSION
Post-translational assembly of stable G protein heterotrimers is a fundamental prerequisite for G protein signaling, yet the mechanism by which the assembly process occurs had been an enigma for more than two decades since the G protein heterotrimer was initially discovered. The most puzzling issue has been how the Gβ and Gγ subunits could come together to form a stable dimer when the individual polypeptides were structurally unstable. Recent studies have shed considerable light on the assembly process and have outlined a mechanism by which CCT and PhLP1 work as co-chaperones to fold Gβ and present it to Gγ for dimerization to occur (10–15, 42, 43). Gγ itself appears to be held by another chaperone DRiP78 (30) until it can interact with PhLP1-Gβ. Mechanistic studies have thus far focused on the most common Gβ1γ2 dimer combination and have not addressed whether this assembly mechanism was general to the many other Gβγ or Gβ5-RGS protein dimers, or specific to only a subset. All the Gβ subunits have been recently shown to interact with CCT, but the interaction of Gβ5 with CCT was much weaker that Gβ1–4 (15). The current study has addressed the scope of PhLP1-mediated dimer assembly for many Gβγ combinations. The results clearly show that PhLP1 is a general co-chaperone for Gβγ assembly. All Gβ subunits required PhLP1 for association with Gγ2 (Figs. 1 and 2), and all Gγ subunits that form dimers with Gβ2 required PhLP1 for association with Gβ2 (Figs. 3 and 4). It seems very likely that the other possible Gβγ dimer combinations would also require PhLP1 for their assembly as well. Thus, it appears that all Gβγs follow a similar mechanism of dimer formation.
Understanding the reasons why some Gβγ combinations form dimers and other do not has been of interest in the field for some time (18). Apparent differences in Gβγ specificity between in vitro assays and cell-based assays have suggested that cellular factors that are involved in the assembly process such as PhLP1 might influence Gβγ specificity (25). However, this does not appear to be the case. As noted above, the specificity of Gβγ dimer formation was not changed by increases or decreases in PhLP1 activity. Thus, it appears that PhLP1 is acting as a true catalyst in Gβγ assembly by not influencing which Gβs can bind which Gγs but by simply facilitating the association of Gβγ combinations that are intrinsically stable. In the case of the Gβ2γ combinations investigated here, dimer stability appears to be determined by sequence specificity, because Gγ binding segregated along subfamily lines according to sequence homology (Fig. 5). Hence, the major factors that determine Gβ2γ specificity appear to be limited to complementarity of the binding surfaces as determined by specific amino acid interactions, the expression of the complementary Gβγ combinations in the same cell types, and the subcellular localization within the cell (18).
It is interesting to note that inhibition of PhLP1 activity through siRNA-mediated knockdown or overexpression of the PhLP1 Δ1–75 dominant negative variant resulted in a surprisingly small decrease in Gβ expression (∼50%), despite the fact that very little of this residual Gβ was associated with Gγ (Figs. 1–4). This finding indicates that Gβ can exist in the cell unassociated with Gγ. It is likely that this pool of undimerized Gβ is associated with CCT, because it has been previously shown that Gβ-CCT complexes are relatively stable in the absence of PhLP1 and Gγ (13). Thus, it appears that the role of CCT is to fold Gβ and protect it from aggregation or proteolytic degradation until it can be released by PhLP1 to interact with Gγ.
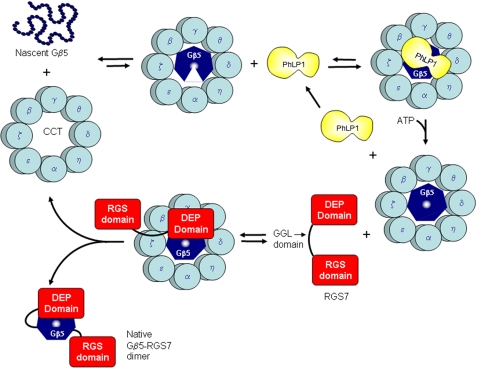
In the case of Gβ5-RGS7 dimers, the data suggest a very different assembly mechanism than that of Gβγ. An outline of a possible mechanism for Gβ5-RGS7 assembly that is consistent with the data presented is depicted in Fig. 10. The decrease in the rate of Gβ5-RGS7 assembly upon siRNA-mediated CCT knockdown (Fig. 9A) indicates that CCT is involved in the assembly process, most likely by folding the nascent Gβ5 despite the weak interaction of Gβ5 with CCT. Likewise, the decrease in the rate of Gβ5-RGS7 assembly upon PhLP1 knockdown (Fig. 8A) shows that PhLP1 also contributes to the assembly process, possibly by increasing the efficiency of Gβ5 folding by increasing the binding of Gβ5 to CCT through the formation of a stable PhLP1-Gβ5-CCT ternary complex (Fig. 7B). However, the decrease in the rate of Gβ5-RGS7 assembly upon overexpression of PhLP1 (Fig. 8B) indicates that excess PhLP1 interferes with the assembly process. A logical explanation of this effect is that PhLP1 must be released from Gβ5 prior to its interaction with RGS7 and that excess PhLP1 blocks the association of RGS7 with Gβ5. Once PhLP1 is released, it appears that RGS7 can associate with Gβ5 while still bound to CCT, given the fact that Gβ5 initiates the co-immunoprecipitation of RGS7 with CCT (Fig. 7C). Once formed, the Gβ5-RGS7 complex would be expected to readily release from CCT because of the relatively weak interaction of the complex with CCT (Fig. 7, B and C). The folded and assembled Gβ5-RGS7 complex would then be able to interact with its R7 anchoring protein and with its Gα targets.
FIGURE 10.
Proposed model of Gβ5-RGS7 assembly. A speculative model of the mechanism of Gβ5-RGS7 assembly that is consistent with current data is depicted. In this model, Gβ5 binds CCT, but is unable to fold into its seven-bladed β-propeller structure (illustrated by the gap in the Gβ5 heptagon) without PhLP1. PhLP1 binding increases the interaction of Gβ5 with CCT, allowing folding to occur (Fig. 7B). PhLP1 is then released, perhaps by ATP binding to CCT. The folded Gβ5 is then able to interact with RGS7 on CCT. The initial interaction is most likely via its N-terminal DEP/DHEX domain, because this domain binds the same face of Gβ5 as PhLP1 (39). Once formed, the Gβ5-RGS7 can release from CCT as a functionally active dimer.
The unique roles for PhLP1 in Gβγ versus Gβ5-RGS7 dimer formation can be understood by examining the structures of the complexes. In the case of Gβγ, the Gγ binding surface is on the opposite side of Gβ from the principal PhLP1 binding surface (40), allowing PhLP1 and Gγ to interact with Gβ simultaneously. It has been proposed that this configuration allows nascent Gγ to associate with Gβ while the Gβ β-propeller is being stabilized by PhLP1 (13). In the case of Gβ5-RGS9, the N-terminal lobe of RGS9 covers a 2600-Å2 area on the same face of Gβ5 (39) predicted to bind PhLP1, based on the phosducin-Gβ1γ1 structure (40). In fact, several residues of Gβ5 that contact the N-terminal lobe of RGS9 are also expected to contact PhLP1 (39, 40). Because of this overlap, assembly of the Gβ5-RGS complex apparently cannot proceed through a PhLP1-Gβ5-RGS intermediate.
A question that is not clear from the structures is how PhLP1 assists in the release of Gβ1 from CCT, whereas it stabilizes the binding of Gβ5 to CCT. More structural information on the PhLP1-Gβ-CCT complexes for both the Gβ1 and Gβ5 complexes would be required to understand the underling molecular basis for these disparate binding properties. Perhaps the differences lie more in the interactions of the Gβ subunits with CCT, with Gβ1 making high affinity contacts and Gβ5 making only low affinity contacts in the absence of PhLP1. Upon PhLP1 binding, it is possible that both Gβ1 and Gβ5 form a similar complex with CCT in which the high affinity contacts of Gβ1 have been lost but indirect contacts with CCT through PhLP1 have been gained, thereby increasing the binding of Gβ5 to CCT.
In conclusion, this work expands the role of PhLP1 as an essential co-chaperone in the assembly of all Gβγ combinations and outlines a mechanism for Gβ5-RGS7 dimer formation. This mechanism is similar to Gβγ assembly in its CCT dependence but differs significantly in its PhLP1 dependence. The data provide additional insight into the intricate means by which the cell utilizes its molecular chaperones to bring the unstable β-propeller fold of Gβ subunits together with their complementary Gγ-like domains to create stable Gβγ and Gβ5-RGS7 dimers to perform their vital functions in G protein signaling.
This work was supported, in whole or in part, by National Institutes of Health Grants GM078550 and EY012287 (to B. M. W.).
- G protein
- heterotrimeric GTP-binding protein
- PhLP1
- phosducin-like protein 1
- CCT
- cytosolic chaperonin containing tailless complex polypeptide 1
- RGS
- regulator of G protein signaling
- R7 RGS
- RGS proteins of the R7 subfamily
- GPCR
- G protein-coupled receptor
- DRiP78
- dopamine receptor-interacting protein 78
- DEP domain
- disheveled, Egl-10, pleckstrin homology domain
- GGL domain
- Gγ-like domain
- HEK
- human embryonic kidney cell
- HA
- hemagglutinin
- siRNA
- short interfering RNA
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1.Wettschureck N., Offermanns S. ( 2005) Physiol. Rev. 85, 1159– 1204 [DOI] [PubMed] [Google Scholar]
- 2.Farrens D. L., Altenbach C., Yang K., Hubbell W. L., Khorana H. G. ( 1996) Science 274, 768– 770 [DOI] [PubMed] [Google Scholar]
- 3.Li J., Edwards P. C., Burghammer M., Villa C., Schertler G. F. ( 2004) J. Mol. Biol. 343, 1409– 1438 [DOI] [PubMed] [Google Scholar]
- 4.Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. ( 2000) Science 289, 739– 745 [DOI] [PubMed] [Google Scholar]
- 5.Cabrera-Vera T. M., Vanhauwe J., Thomas T. O., Medkova M., Preininger A., Mazzoni M. R., Hamm H. E. ( 2003) Endocr. Rev. 24, 765– 781 [DOI] [PubMed] [Google Scholar]
- 6.Reiter E., Lefkowitz R. J. ( 2006) Trends Endocrinol. Metab. 17, 159– 165 [DOI] [PubMed] [Google Scholar]
- 7.Ross E. M., Wilkie T. M. ( 2000) Annu. Rev. Biochem. 69, 795– 827 [DOI] [PubMed] [Google Scholar]
- 8.Willars G. B. ( 2006) Semin. Cell Dev. Biol. 17, 363– 376 [DOI] [PubMed] [Google Scholar]
- 9.Marrari Y., Crouthamel M., Irannejad R., Wedegaertner P. B. ( 2007) Biochemistry 46, 7665– 7677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humrich J., Bermel C., Büunemann M., Häarmark L., Frost R., Quitterer U., Lohse M. J. ( 2005) J. Biol. Chem. 280, 20042– 20050 [DOI] [PubMed] [Google Scholar]
- 11.Knol J. C., Engel R., Blaauw M., Visser A. J., Van Haastert P. J. ( 2005) Mol. Cell. Biol. 25, 8393– 8400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubota S., Kubota H., Nagata K. ( 2006) Proc. Natl. Acad. Sci. U. S. A. 103, 8360– 8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lukov G. L., Baker C. M., Ludtke P. J., Hu T., Carter M. D., Hackett R. A., Thulin C. D., Willardson B. M. ( 2006) J. Biol. Chem. 281, 22261– 22274 [DOI] [PubMed] [Google Scholar]
- 14.Lukov G. L., Hu T., McLaughlin J. N., Hamm H. E., Willardson B. M. ( 2005) EMBO J. 24, 1965– 1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wells C. A., Dingus J., Hildebrandt J. D. ( 2006) J. Biol. Chem. 281, 20221– 20232 [DOI] [PubMed] [Google Scholar]
- 16.Valpuesta J. M., Martín-Benito J., Gómez-Puertas P., Carrascosa J. L., Willison K. R. ( 2002) FEBS Lett. 529, 11– 16 [DOI] [PubMed] [Google Scholar]
- 17.McLaughlin J. N., Thulin C. D., Bray S. M., Martin M. M., Elton T. S., Willardson B. M. ( 2002) J. Biol. Chem. 277, 34885– 34895 [DOI] [PubMed] [Google Scholar]
- 18.Robishaw J. D., Berlot C. H. ( 2004) Curr. Opin. Cell Biol. 16, 206– 209 [DOI] [PubMed] [Google Scholar]
- 19.Gautam N., Downes G. B., Yan K., Kisselev O. ( 1998) Cell Signal 10, 447– 455 [DOI] [PubMed] [Google Scholar]
- 20.Watson A. J., Katz A., Simon M. I. ( 1994) J. Biol. Chem. 269, 22150– 22156 [PubMed] [Google Scholar]
- 21.Downes G. B., Gautam N. ( 1999) Genomics 62, 544– 552 [DOI] [PubMed] [Google Scholar]
- 22.Ray K., Hansen C. A., Robishaw J. D. ( 1996) Trends Cardiovasc. Med. 6, 115– 121 [DOI] [PubMed] [Google Scholar]
- 23.Myung C. S., Lim W. K., DeFilippo J. M., Yasuda H., Neubig R. R., Garrison J. C. ( 2006) Mol. Pharmacol. 69, 877– 887 [DOI] [PubMed] [Google Scholar]
- 24.Jones M. B., Siderovski D. P., Hooks S. B. ( 2004) Mol. Interv. 4, 200– 214 [DOI] [PubMed] [Google Scholar]
- 25.Dingus J., Wells C. A., Campbell L., Cleator J. H., Robinson K., Hildebrandt J. D. ( 2005) Biochemistry 44, 11882– 11890 [DOI] [PubMed] [Google Scholar]
- 26.Witherow D. S., Slepak V. Z. ( 2003) Receptors Channels 9, 205– 212 [PubMed] [Google Scholar]
- 27.Hu G., Wensel T. G. ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 9755– 9760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martemyanov K. A., Yoo P. J., Skiba N. P., Arshavsky V. Y. ( 2005) J. Biol. Chem. 280, 5133– 5136 [DOI] [PubMed] [Google Scholar]
- 29.Chen C. K., Burns M. E., He W., Wensel T. G., Baylor D. A., Simon M. I. ( 2000) Nature 403, 557– 560 [DOI] [PubMed] [Google Scholar]
- 30.Dupré D. J., Robitaille M., Richer M., Ethier N., Mamarbachi A. M., Hébert T. E. ( 2007) J. Biol. Chem. 282, 13703– 13715 [DOI] [PubMed] [Google Scholar]
- 31.Carter M. D., Southwick K., Lukov G., Willardson B. M., Thulin C. D. ( 2004) J. Biomol. Tech. 15, 257– 264 [PMC free article] [PubMed] [Google Scholar]
- 32.Grantham J., Brackley K. I., Willison K. R. ( 2006) Experimental cell research 312, 2309– 2324 [DOI] [PubMed] [Google Scholar]
- 33.Thulin C. D., Howes K., Driscoll C. D., Savage J. R., Rand T. A., Baehr W., Willardson B. M. ( 1999) Mol. Vis. 5, 40. [PubMed] [Google Scholar]
- 34.Kisselev O., Gautam N. ( 1993) J. Biol. Chem. 268, 24519– 24522 [PubMed] [Google Scholar]
- 35.Fletcher J. E., Lindorfer M. A., DeFilippo J. M., Yasuda H., Guilmard M., Garrison J. C. ( 1998) J. Biol. Chem. 273, 636– 644 [DOI] [PubMed] [Google Scholar]
- 36.He W., Lu L., Zhang X., El-Hodiri H. M., Chen C. K., Slep K. C., Simon M. I., Jamrich M., Wensel T. G. ( 2000) J. Biol. Chem. 275, 37093– 37100 [DOI] [PubMed] [Google Scholar]
- 37.Jones M. B., Garrison J. C. ( 1999) Anal. Biochem. 268, 126– 133 [DOI] [PubMed] [Google Scholar]
- 38.Savage J. R., McLaughlin J. N., Skiba N. P., Hamm H. E., Willardson B. M. ( 2000) J. Biol. Chem. 275, 30399– 30407 [DOI] [PubMed] [Google Scholar]
- 39.Cheever M. L., Snyder J. T., Gershburg S., Siderovski D. P., Harden T. K., Sondek J. ( 2008) Nat. Struct. Mol. Biol. 15, 155– 162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaudet R., Bohm A., Sigler P. B. ( 1996) Cell 87, 577– 588 [DOI] [PubMed] [Google Scholar]
- 41.Kunisawa J., Shastri N. ( 2003) Mol. Cell 12, 565– 576 [DOI] [PubMed] [Google Scholar]
- 42.Martín-Benito J., Bertrand S., Hu T., Ludtke P. J., McLaughlin J. N., Willardson B. M., Carrascosa J. L., Valpuesta J. M. ( 2004) Proc. Natl. Acad. Sci. U. S. A. 101, 17410– 17415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLaughlin J. N., Thulin C. D., Hart S. J., Resing K. A., Ahn N. G., Willardson B. M. ( 2002) Proc. Natl. Acad. Sci. U. S. A. 99, 7962– 7967 [DOI] [PMC free article] [PubMed] [Google Scholar]