Abstract
It has previously been shown that highly invasive MDA-MB231 human breast cancer cells express vacuolar proton-translocating ATPase (V-ATPases) at the cell surface, whereas the poorly invasive MCF7 cell line does not. Bafilomycin, a specific V-ATPase inhibitor, reduces the in vitro invasion of MB231 cells but not MCF7 cells. Targeting of V-ATPases to different cellular membranes is controlled by isoforms of subunit a. mRNA levels for a subunit isoforms were measured in MB231 and MCF7 cells using quantitative reverse transcription-PCR. The results show that although all four isoforms are detectable in both cell types, levels of a3 and a4 are much higher in MB231 than in MCF7 cells. Isoform-specific small interfering RNAs (siRNA) were employed to selectively reduce mRNA levels for each isoform in MB231 cells. V-ATPase function was assessed using the fluorescent indicators SNARF-1 and pyranine to monitor the pH of the cytosol and endosomal/lysosomal compartments, respectively. Cytosolic pH was decreased only on knockdown of a3, whereas endosome/lysosome pH was increased on knockdown of a1, a2, and a3. Treatment of cells with siRNA to a4 did not affect either cytosolic or endosome/lysosome pH. Measurement of invasion using an in vitro transwell assay revealed that siRNAs to both a3 and a4 significantly inhibited invasion of MB231 cells. Immunofluorescence staining of MB231 cells for V-ATPase distribution revealed extensive intracellular staining, with plasma membrane staining observed in ∼18% of cells. Knockdown of a4 had the greatest effect on plasma membrane staining, leading to a 32% reduction. These results suggest that the a4 isoform may be responsible for targeting V-ATPases to the plasma membrane of MB231 cells and that cell surface V-ATPases play a significant role in invasion. However, other V-ATPases affecting the pH of the cytosol and intracellular compartments, particularly those containing a3, are also involved in invasion.
The leading cause of mortality from cancer is metastasis, making inhibition of metastasis an important strategy in controlling cancer progression. The metastatic cascade involves a series of steps that include escape of cells from the site of the primary tumor into the circulation or lymphatic system and the extravasation of cells from the circulation or lymphatic system into secondary sites (1, 2). Both of these processes require the tumor cells to display an invasive phenotype in which they degrade extracellular matrix of the surrounding tissue. The vacuolar H+-ATPases (or V-ATPases)2 are a family of ATP-dependent proton pumps that have been implicated in tumor cell invasion (3). V-ATPases are up-regulated in tissue samples from highly invasive pancreatic carcinomas (4), and treatment of a human cancer cell line with antisense oligonucleotides to the V-ATPase c subunit decreased invasion in vitro (5). Inhibition of V-ATPase expression in hepatocellular carcinoma cells using siRNAs reduces invasiveness of these cells in vitro and metastasis in vivo (6). In addition, treatment of highly invasive MDA-MB231 human breast cancer cells with the specific V-ATPase inhibitors bafilomycin or concanamycin inhibits the invasion of these cells in vitro (7).
How might V-ATPases function in tumor cell invasion? Because V-ATPases are present in both intracellular compartments and the plasma membrane, there are a number of possibilities. Plasma membrane V-ATPases are poised to acidify the extracellular environment and have been shown to be active in transport of protons across the plasma membrane of a number of tumor cell types, including MB231 cells (7–9). Extracellular acidification is required for activation of secreted cathepsins, proteases that normally reside in lysosomes (10), but which have been shown to be secreted by a variety of cancer cells and to be important for tumor cell invasion (11). Once in the extracellular space, activated cathepsins can both degrade extracellular matrix proteins and activate other secreted proteases involved in invasion, such as matrix metalloproteases (11, 12). Intracellular V-ATPases may also function in invasion by either aiding in the proteolytic activation of cathepsins or matrix metalloproteases within lysosomes or secretory vesicles or by assisting in the trafficking of vesicles containing these proteases to the cell surface.
To begin to unravel the role of V-ATPases in tumor cell invasion, we have investigated the role of particular isoforms of the V-ATPase in the invasive phenotype of a human breast cancer cell line. V-ATPases are multisubunit complexes composed of a peripheral domain (V1) responsible for ATP hydrolysis and an integral domain (V0) that carries out proton transport (3). The core V0 domain in mammals contains five subunits (a, d, c, c″, and e), with the information necessary to target the V-ATPase to different cellular membranes located in the a subunit (13–19). Subunit a is a 100-kDa integral membrane protein containing a 50-kDa hydrophilic N-terminal domain located on the cytoplasmic side of the membrane and a 50-kDa C-terminal domain containing multiple membrane-spanning segments (20). Targeting information is localized to the cytoplasmic N-terminal domain (21). In mammals subunit a exists in four isoforms (a1–a4) (15–19, 22–24). The a1 and a2 isoforms appear to localize primarily to intracellular compartments, with a1 present in synaptic vesicles (25) and clathrin-coated vesicles in brain, whereas a2 is present in Golgi and endosomal compartments (18, 26). The a3 isoform is responsible for targeting the V-ATPase to the plasma membrane of osteoclasts, where it plays an essential role in bone resorption (18, 19, 23, 27), although a3 has also been localized to lysosomes and insulin-containing secretory vesicles in pre-osteoclasts (18) and pancreatic islet cells (17), respectively. The a4 isoform functions to target V-ATPases to the apical membrane of renal intercalated cells, where they are responsible for acid secretion into the urine (15, 16, 24). a4 has also been localized to the apical membrane of epididymal clear cells (28).
The purpose of this study was to determine the a subunit isoform expression profile of the human breast cancer cell line MDA-MB231 and to determine the effect of reducing expression of each of the four a subunit isoforms on V-ATPase function and the invasiveness of these cells in vitro. The results suggest distinct functions of a subunit isoforms in control of cytosolic and endosome/lysosome pH and a role for both intracellular and plasma membrane V-ATPases in tumor cell invasion.
EXPERIMENTAL PROCEDURES
Cell Culture
Human breast cancer cell lines, MDA-MB231 and MCF7, were purchased from American Type Culture Collection (ATCC). Cells were grown in FalconTM T-75 flasks in Dulbecco's modified Eagle's medium (DMEM) with phenol, 4.5 g/liter d-glucose, 4 mm (584 mg/liter) l-glutamine, and 110 mg/liter sodium pyruvate (Invitrogen) supplemented with 10% FBS (Invitrogen), minimum Eagle's medium nonessential amino acids (Invitrogen), 60 μg/ml penicillin, and 125 μg/ml streptomycin (Invitrogen). Cells were grown in a 95% air, 5% CO2 humidified environment at 37 °C.
Real Time Reverse Transcription-PCR
Cells were harvested and lysed, and RNA was isolated using RNeasy® mini kit (Qiagen). After RNA isolation, mRNA was isolated with the MicroPoly(A) PuristTM kit from Ambion®. Total RNA or mRNA concentration was quantified using Quant-iT RiboGreen® RNA reagent (Molecular Probes). One-step quantitative RT-PCR was performed in a 96-well format on a Stratagene MX-3000P® QPCR system using Brilliant® SYBR® Green QRT-PCR master mix kit (Stratagene). The PCR cycling sequence consisted of 30 min at 50 °C to allow for reverse transcription and then a heat inactivation and denaturation step for 10 min at 95 °C; this was followed by 40 cycles of 30 s at 95 °C, 1 min at 55 °C, and 30 s at 72 °C to allow for denaturation, annealing, and extension, respectively. To quantitate the results, cDNA clones of the a1, a2 (ATCC), a3, and a4 (Open Biosystems) isoforms were purchased. The cDNA sequences were verified by sequencing. Plasmid DNA for each isoform isolated from Escherichia coli was quantitated by measuring the absorbance at 260 nm; serial dilutions were made, and these DNA standards were used during the quantitative RT-PCRs to facilitate quantification of the initial mRNA levels for each experimental sample by use of a standard curve.
RNA Interference
siRNA pools specific for the a1, a2, a3, or a4 isoform were purchased from Dharmacon. Each pool contained four siRNAs specific for the appropriate a subunit isoform. MB231 cells were plated in 60-mm dishes at 4 × 105 cells/dish in the media described above without antibiotics and incubated overnight. Cells were transfected with 20 nm siRNA directed against a1 or a2, 100 nm siRNA directed against a3, or 10 nm siRNA directed against a4 according to the manufacturer's directions. Briefly, the siRNA was mixed 1:1 with Opti-MEMTM (Invitrogen), allowed to incubate for 5 min, and then mixed with transfection reagent (Dharmafect2TM, Dharmacon). The siRNA/transfection reagent mix was incubated for 20 min at room temperature and then added to the appropriate volume of DMEM + serum, and 4 ml was added to each dish. After incubation of cells with siRNA for 24 h, the media were changed to siRNA-free media, and cells were incubated for an additional time (depending upon the assay) prior to harvesting. For all experiments, data were collected 96 h post-transfection. To quantitate reduction in the a subunit isoform mRNA levels, quantitative RT-PCR was performed as described above using RNA isolated from cells after siRNA treatment. To confirm the specificity of knockdown, we used primers specific for the isoforms that were not targeted by the siRNA treatment.
Loading Cells with Fluorescent pH Indicators
To evaluate the localization of fluorescent pH indicators, cells were grown onto round coverslips (25 mm) in DMEM supplemented with FBS. After 48 h in culture, cells were incubated for 12 h with 1 mm 8-hydroxypyrene-1,3,6-trisulfonic acid (pyranine), a pH fluorescent indicator, in DMEM supplemented with FBS. Cells take up pyranine by pinocytosis, which effectively labels endosomal and lysosomal compartments (29). The cytosol was loaded with 7 μm SNARF-1, a pH fluoroprobe. This probe is internalized by incubating cells for 30 min with the acetoxymethyl ester form of SNARF-1 (30). Cellular esterases cleave the acetoxymethyl groups leaving the charged form of SNARF-1 in the cytosol. Fluorescence images of cells were obtained using ×20 objective and a digital camera (Orca, Hamamatsu). Cells loaded with SNARF-1 were excited at 534 nm, and the fluorescence emission signal was collected at 590 nm (long bandpass filter). Cells loaded with pyranine were excited at 465 nm, and the emission was collected at 514 nm (20 nm bandpass). To co-label cells loaded with pyranine with a lysosomal marker, pyranine-loaded cells were washed, to remove extracellular pyranine, and further incubated with 2 μm LysoTracker® Red DND-99 for 3 min. LysoTracker® Red labels acidic compartments corresponding to endosomes and lysosomes (E/L). Cells were transferred to the microscope chamber, and the fluorescence of pyranine was excited at 458 nm (argon laser 200 milliwatts), and the emission was collected from 514 to 540 nm using the λ mode in the Zeiss confocal microscope (LSM510 Meta System). The fluorescence of LysoTracker® Red was excited at 543 nm (helium-neon laser, 5 milliwatts), and the emission was collected from 580 to 650 nm. Images were acquired using a ×63 water immersion microscope objective (0.95 N.A.). Images were analyzed using Physiology software (Zeiss).
Cytosolic and Endosomal/Lysosomal pH Measurements
To verify the usefulness of pyranine and SNARF for in vivo pH measurements, 1 μm pyranine and 2 μm SNARF-1 (free acid) were dissolved in high K+ buffers of increasing pH (from 5.5 to 8.5), and their spectral properties were evaluated using a fluorometer. The excitation spectra of pyranine were collected using an emission wavelength of 514 nm, and the emission spectra of SNARF-1 were acquired using 534 nm as excitation wavelength. The significant properties of pyranine are the increase in the excitation peak at 465 nm and the decrease at 405 nm as pH is increased from 5.5 to 8.5. The significant properties of SNARF-1 are the increase in the emission signal at 644 nm and the decrease at 584 nm as pH is increased. Consequently, the fluorescence ratios at 465/405 nm and 644/584 nm can be used to monitor pH in endosomes/lysosomes and cytosol, respectively, using a ratiometric approach that allows us to quantitate pH.
To evaluate the effect of siRNA treatment on steady state cytosolic and endosome/lysosome pH, cells growing on rectangular coverslips (8 × 22 mm) were treated with siRNA directed against the different a isoforms for 24 h as described above. 84 h post-transfection, cells were incubated with pyranine and/or SNARF-1 for 12 h as described above. Following incubation, for each experiment two coverslips containing cells were washed, to remove external dye, and then transferred to a thermostated fluorescence chamber kept at 37 °C and continuously perfused at 3 ml/min. The excitation spectra of pyranine in endosomes/lysosomes was acquired using an emission wavelength of 514 nm, and the emission spectra of SNARF-1 were acquired using 534 nm as excitation wavelength, and the conversion of ratio values to pHcyt and pHE/L was performed.
Invasion Assay
Assays for in vitro invasion were performed using ChemoTx® membranes (Neuroprobe) with an 8-μm pore size membrane and coated with MatrigelTM (BD Biosciences). A chemoattractant (FBS) was added to the trans-side of the membrane to induce invasive cells to digest the coating and migrate through the pores to the trans-side. MatrigelTM was diluted in PBS to a final concentration of 0.3 μg/μl, and a total of 4 μg was coated onto the membrane in each well. The membrane was allowed to dry overnight under vacuum at room temperature. MatrigelTM-coated membrane was re-hydrated with 50 μl of DMEM + 4.5 g/liter d-glucose, without phenol, l-glutamine, or sodium pyruvate (Invitrogen) (termed Media) for at least two h. 30 μl of the Media containing 5% FBS were added to the wells of the complementary ChemoTx® plate to act as the chemoattractant. 72 h post-transfection, cells were harvested to set up the invasion assay. To ensure that only viable cells were loaded onto the membrane, cells were washed to remove unattached cells and then harvested via trypsinization. Viability of the harvested cells was assessed by trypan blue exclusion; less than 1% of the harvested cells stained with trypan blue. Cells were next brought to a concentration of 2 × 105 cells/ml in Media that did not contain siRNA. For inhibitor-treated samples, cells were resuspended in Media containing either DMSO or 100 nm concanamycin A in DMSO and allowed to incubate for 15 min at 37 °C. 5 × 104 cells were seeded onto the rehydrated membrane, which was then fitted onto the wells containing chemoattractant. The invading cells were incubated under normal growth conditions. After 24 h, the noninvaded cells and Matrigel® were scraped from the top of the membrane using a cell scraper, and the membrane with the invaded cells was fitted onto wells containing 30 μl of 4% paraformaldehyde and incubated for 10 min at room temperature to fix the cells. The membrane was then fitted onto wells containing 30 μl of 4 μg/ml propidium iodide in warm PBS to stain the cells. The cells were incubated with propidium iodide for 30 min at 37 °C and then moved to a Zeiss Axiovert 10 fluorescence microscope for analysis. Thirteen images per well were taken of the trans-side of the membrane. The number of invaded cells per well was counted from the images. Each condition was performed in triplicate, and the number of invading cells was averaged over the three wells. Percent invasion for treated cells was normalized to nontreated cells.
Immunocytochemistry
A polyclonal antibody against the V-ATPase subunit E (provided by Dr. Reuveni) was used to determine the in situ localization of V-ATPases (31). MB231 cells were plated onto 60-mm dishes containing poly-d-lysine-coated 12-mm round coverslips. Approximately 24 h later, the cells were treated with siRNA for 24 h. The media were then changed to siRNA-free media, and the cells were allowed to incubate for an additional 72 h. 96 h post-transfection with siRNA, the cells on coverslips were retrieved. Using a 200-μl pipette tip, the confluent monolayer was scratched to create a wound to induce migration. Wounding was performed because it has previously been shown that cells utilizing V-ATPases for pH regulation and migration target them to the plasma membrane at the leading edge (31). Cells were then incubated for an additional 4 h, washed, fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100. The cells were blocked with 1% bovine serum albumin in PBS for 1 h and then incubated with anti-E antibody at a 1:500 dilution overnight. The cells were next rinsed with PBS, and then Alexa Fluor® 488-conjugated goat anti-rabbit secondary antibody (1:500) and Alexa Fluor® 594 phalloidin (to stain F-actin, 1:250) (Invitrogen) in 1% bovine serum albumin/PBS were added to the cells. After 1 h of incubation at room temperature, the cells were rinsed with PBS. The cells were prepared for viewing using ProLong® Gold (Invitrogen) mounting medium that was allowed to cure at room temperature for 24 h. The fluorescent cells were imaged on either the Zeiss Axiovert 10 fluorescence microscope or a Leica TCS SP2 confocal microscope. To quantitate the appearance of plasma membrane staining, 30–40 consecutive images were taken along the wound. For each image, the total number of cells was counted, and the number showing a distinct line of staining at the plasma membrane was counted. The ratio of cells with plasma membrane staining versus the total number of cells was calculated.
Cathepsin L Secretion
Cells plated in 60-mm dishes were treated with siRNA and allowed to incubate for 24 h at 37 °C. Media were replaced with siRNA-free media, and cells were incubated for an additional 48 h The media were then replaced with serum-free media, and cells were incubated for an additional 24 h. 96 h post-transfection the conditioned media from each dish was recovered and concentrated 80-fold using an Amicon® Ultra (4 ml, 10,000 cutoff, Millipore). Samples of the concentrated media for each condition were resolved by SDS-PAGE and blotted for cathepsin L using a monoclonal antibody from Zymed Laboratories Inc.
Statistical Analysis
All results are expressed as means ± S.D. The significant differences were determined by analysis of variance for each experiment, followed by pairwise t test to compare individual treatments to nontreated. All statistical tests were considered significant at p < 0.05.
RESULTS
mRNA Levels of a Subunit Isoforms in MDA-MB231 and MCF7 Cells as Measured Using Quantitative RT-PCR
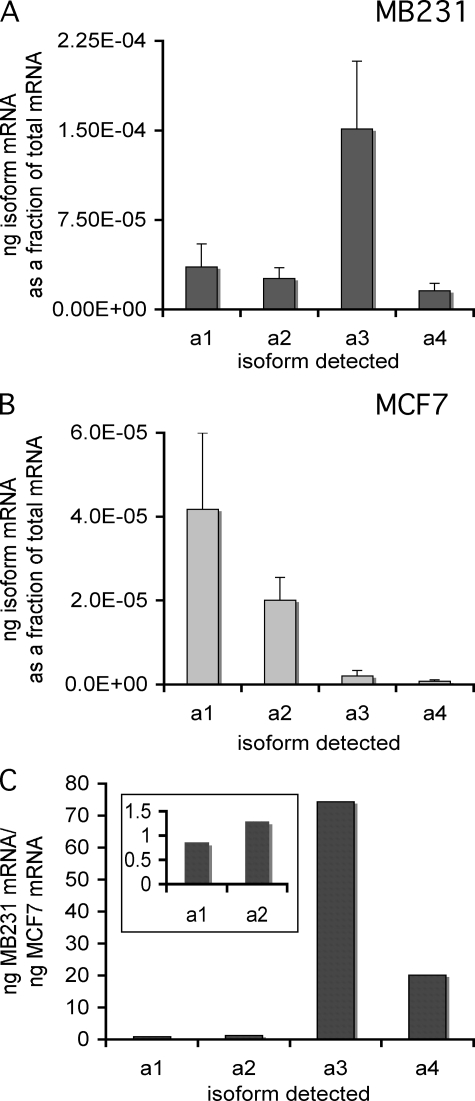
Because of the role V-ATPases have been reported to play in invasiveness of breast cancer cells, it was of interest to compare the mRNA expression profiles for a subunit isoforms in the highly invasive breast cancer cell line MDA-MB231 with those of the poorly invasive cell line MCF7. Quantitative RT-PCR was performed with isoform-specific primers on mRNA isolated from each cell line, using plasmid-borne cDNAs encoding each isoform as standards. As can be seen in Fig. 1, although all four a subunit isoforms can be detected in both cell lines, their relative abundance differs dramatically between MB231 and MCF7 cells. Thus the predominant a subunit isoform in MB231 cells is a3, whereas the predominant isoform in MCF7 cells is a1, followed closely by a2. Comparing the ratio of expression of each isoform between the two cell lines (Fig. 1C) reveals that both the a3 and a4 isoforms are expressed at much higher levels in MB231 than MCF7 cells.
FIGURE 1.
mRNA levels of subunit a isoforms in MB231 and MCF7 cells. Using mRNA isolated from cells, quantitative RT-PCR was used to determine the mRNA levels of the different subunit a isoforms, as described under “Experimental Procedures.” Plasmids expressing the cDNA for each isoform were used to construct a standard curve. All values are normalized to total mRNA loaded as determined by Quant-iT RiboGreen® reagent. The values reported are the ratio of nanograms of isoform-specific mRNA to the total nanograms of mRNA. A, a subunit isoform-specific mRNA levels in MB231 cells. B, a subunit isoform-specific mRNA levels in MCF7 cells. C, ratio of a subunit isoform-specific mRNAs in MB231 versus MCF7 cells. Inset shows data for a1 and a2 on an expanded scale. n = 7, error bars indicate standard deviation.
Selective Knockdown of Each Subunit a Isoform mRNA Using siRNA
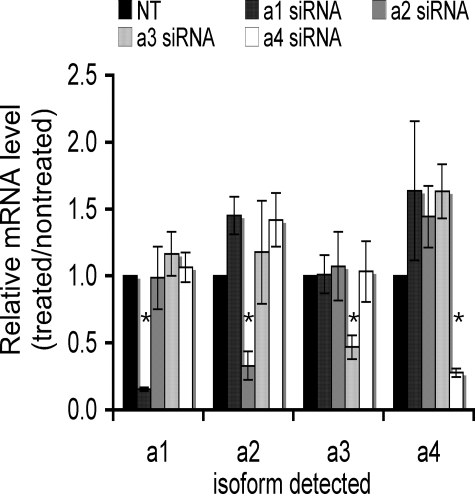
To evaluate the role of a subunit isoforms in V-ATPase function and invasiveness, we employed isoform-specific siRNAs to reduce the level of each a subunit isoform mRNA in MB231 cells. The effect of siRNA treatment on mRNA levels for each isoform was assessed using quantitative RT-PCR. mRNA was isolated from cells treated with siRNA for 24 h followed by 72 h of post-treatment incubation. As shown in Fig. 2, greater than 50% reduction in mRNA levels was observed for each a subunit isoform. Moreover, reductions in mRNA levels were isoform-specific, although it should be noted that there does appear to be some compensatory up-regulation of the a1, a2, and a3 mRNA levels upon treatment with siRNA to a4. These results indicate that the siRNA treatment is effective at selectively knocking down mRNA levels for each a subunit isoform.
FIGURE 2.
Isoform specificity of mRNA knockdown using a subunit isoform-specific siRNAs. siRNA pools from Dharmacon were used to reduce the mRNA levels of each isoform in MB231 cells as described under “Experimental Procedures.” The level of knockdown was quantitated by performing QRT-PCR on RNA from cells harvested 96 h siRNA post-treatment. Knockdown is reported as the ratio of the nanograms of mRNA in treated cells versus the nanograms of mRNA in nontreated (NT) cells as determined using the standard curve after QRT-PCR. Values are means; error bars indicate standard deviation, n = 5. *, p < 0.01 versus nontreated.
Effect of a Subunit Isoform Reduction on Cytoplasmic and Endosomal/Lysosomal pH in MB231 Cells
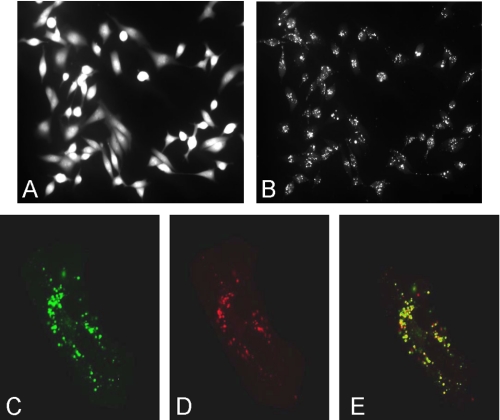
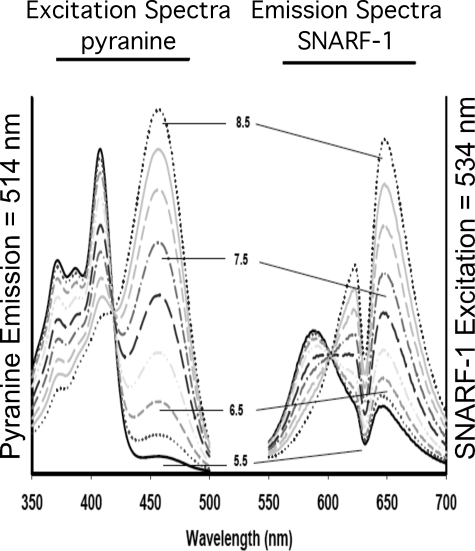
To investigate the effect of reducing each subunit a isoform on V-ATPase function in MB231 cells, the fluorescence indicators SNARF-1 and pyranine were employed to monitor cytosolic and endosome/lysosome pH, respectively. Cells loaded with SNARF-1 show a homogeneous distribution of fluorescence consistent with a cytoplasmic distribution (Fig. 3A). By contrast, cells allowed to endocytose the membrane-impermeant pH indicator pyranine show punctate staining (Fig. 3B). Cells loaded with both pyranine and the lysosomal marker LysoTracker® Red (Fig. 3, C–E) show extensive co-localization of these two probes (Fig. 3E), indicating that pyranine localizes to the endosome/lysosome compartment. The pH dependence of the spectral properties of pyranine and SNARF-1 are shown in Fig. 4. For pyranine, the excitation spectra at an emission wavelength of 514 nm shows an increase in the excitation peak at 465 nm and a decrease at 405 nm with increasing pH. The ratio of fluorescence at 465 and 405 nm can therefore be used to quantitate endosome/lysosome pH. For SNARF-1 the emission spectra at an excitation wavelength of 534 nm shows an increase at 644 nm and a decrease at 584 nm with increasing pH. Cytosolic pH can therefore be determined from the ratio of fluorescence at 644 and 584.
FIGURE 3.
Fluorescent pH indicators in cytosol and endosomes/lysosomes. MB231 cells were loaded with 1 mm 8-hydroxypyrene-1,3,6-risulfonic acid (pyranine), to label endosomes/lysosomes or 7 μm of the acetoxymethyl ester form of SNARF-1 to label the cytosol as described under “Experimental Procedures.” A, fluorescence image of cells loaded with SNARF-1, excited at 534 nm, and the fluorescence emission signal collected at 590 nm (long bandpass filter). B, same field as for A, except that pyranine was excited at 465 nm and its emission collected at 514 nm (20 nm bandpass). C–E, cells incubated with pyranine as in A were washed to remove extracellular pyranine and further incubated with 2 μm LysoTracker® Red DND-99 for 3 min to label endosomes/lysosomes. C, cells were transferred to the microscope chamber, and the fluorescence of pyranine was excited at 458 nm and the emission collected from 514 to 540 nm. D, fluorescence of LysoTracker® Red was excited at 543 nm, and the emission was collected from 580 to 650 nm. E shows the merge of C and D and indicates that these fluoroprobes exhibit significant co-localization.
FIGURE 4.
Spectral properties and pH dependence of fluorescence of SNARF-1 and pyranine. 1 μm pyranine and 2 μm SNARF-1 (free acid) were dissolved in high K+ buffer and spectral properties evaluated as follows. The excitation spectra of pyranine were collected using an emission wavelength of 514 nm, whereas the emission spectra of SNARF-1 were acquired using 534 nm as the excitation wavelength. Note that these fluoroprobes do not exhibit spectral overlap and do not exhibit fluorescence resonance energy transfer that could hamper the interpretation of the data. For pyranine, an increase in the excitation peak at 465 nm and a decrease at 405 nm as pH is increased from 5.5 to 8.5 is observed. 415 nm represents the isoexcitation wavelength and is used to evaluate dye concentration and/or quenching artifacts. For SNARF-1, the emission signal at 644 nm decreases and at 584 nm increases as pH is decreased. 600 nm represents the isoemissive wavelength. Consequently, the fluorescence ratios at 465/405 and 644/584 nm can be used to monitor pH in endosomes/lysosomes and cytosol, respectively, using a ratiometric approach that allows quantitation of pH.
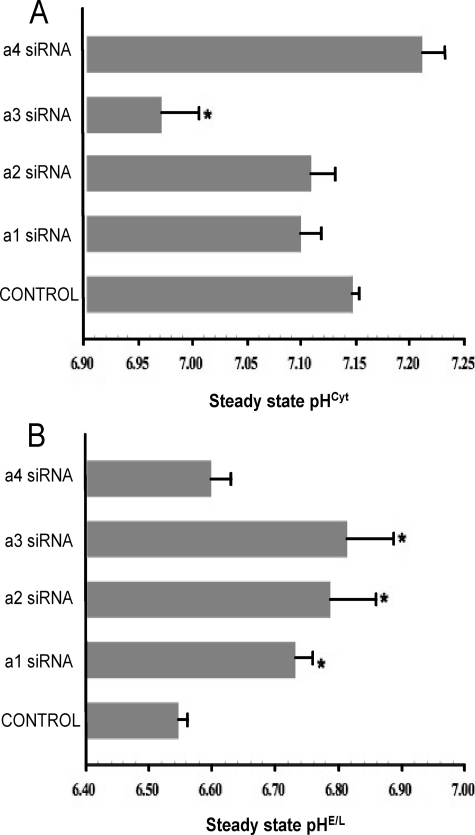
Using this ratiometric approach, cytosolic and endosome/lysosome pH were quantitated following treatment of MB231 cells with isoform-specific siRNAs. As shown in Fig. 5A, treatment with siRNA specific for a3 significantly reduced the cytosolic pH by nearly 0.2 unit, whereas siRNA specific for a1, a2, or a4 had little effect. By contrast, treatment of cells with siRNA for a1, a2, and a3 all significantly increased the pH of endosomes/lysosomes by 0.2–0.25 unit, whereas a4 siRNA had no significant effect (Fig. 5B). These results suggest that under steady state conditions the a3 isoform plays a role in controlling both cytosolic and endosome/lysosome pH, whereas the a1 and a2 isoforms function in regulation of only endosome/lysosome pH.
FIGURE 5.
Steady state pH of cytosol and endosomes/lysosomes in MB231 cells treated with a subunit isoform-specific siRNAs. Cells that had been treated with siRNA directed against a1, a2, a3, or a4 or no siRNA (Control) were incubated as described under “Experimental Procedures” followed by loading of endosomes/lysosomes with pyranine or cytosol with SNARF-1, as described. 96 h post-transfection, simultaneous measurements of pHcyt and pHE/L were performed in cells co-loaded with pyranine and SNARF-1 as described under “Experimental Procedures.” The conversion of ratio values to pHcyt and pHE/L were performed as described previously. Values are means, and error bars indicate standard deviation, n = 6. *, p < 0.05 versus control.
Effect of Reduction of a Subunit mRNA Levels on in Vitro Invasion of MB231 Cells
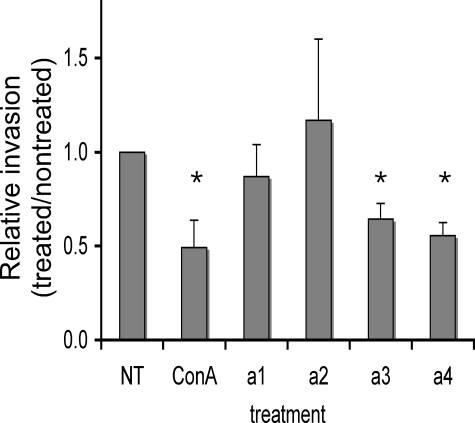
To assess the effects of reducing mRNA levels for a subunit isoforms on the invasiveness of MB231 cells, an in vitro invasion assay was employed. Cells were plated on MatrigelTM-coated wells and induced to migrate to the trans-side by the presence of a chemoattractant (serum). After 24 h, cells on the cis-side were removed, and cells on the trans-side were fluorescently stained with propidium iodide and counted as described under “Experimental Procedures.” As shown in Fig. 6, treatment of MB231 cells with 100 nm concanamycin inhibited invasion by ∼50%. No effect of concanamycin treatment on the invasiveness of MCF7 cells was observed (data not shown), as reported previously (7). Treatment of MB231 cells with siRNAs specific for either a3 or a4 inhibited invasion by 40–50%. By contrast, treatment with siRNAs specific for a1 or a2 had no significant effect on the invasiveness of MB231 cells. Under the conditions employed, neither treatment with concanamycin nor siRNAs affected cell viability or proliferation (data not shown). These results suggest that both the a3 and a4 isoforms function in invasion of MB231 cells.
FIGURE 6.
In vitro invasion of MB231 cells after siRNA treatment. In vitro invasion was assayed using MatrigelTM-coated ChemoTx® membranes as described under “Experimental Procedures.” Cells treated with either concanamycin (ConA) or siRNA were allowed to invade and then stained with propidium iodide. Fourteen images were taken of the trans-side of the membrane in each well, and the number of cells per well were counted, and the average over three wells was calculated. Invasion is reported as the ratio of the amount of invasion observed for treated cells divided by the amount of invasion observed for nontreated cells. For concanamycin experiments, nontreated (NT) samples include an equivalent volume of the solvent (DMSO). Values are means, and error bars indicate standard deviation, n = 5. *, p < 0.01 versus nontreated.
Effect of siRNA Treatment on Cellular Distribution of V- ATPases as Assessed Using Immunofluorescence
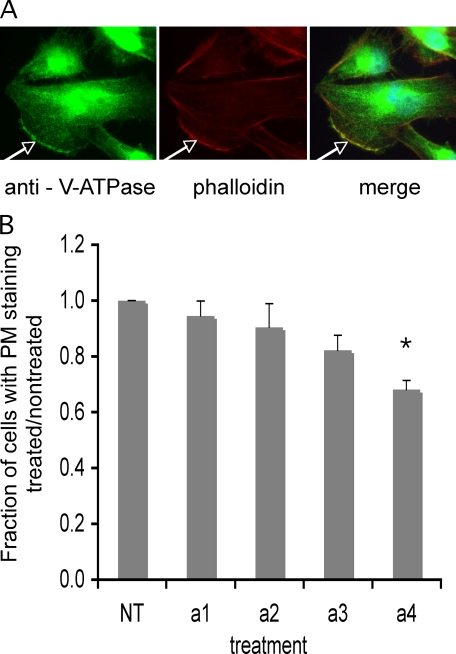
To determine the effect of treatment of cells with siRNAs specific for a subunit isoforms on the cellular distribution of V-ATPases, immunofluorescence studies were performed using an antibody directed against the E subunit of the V-ATPase (part of the peripheral V1 domain). This antibody was employed because of the lack of suitable antibodies capable of recognizing the a subunits in an isoform-specific manner in these cells. Cells were induced to migrate by creation of a “wound” in the cell monolayer. Representative immunofluorescence images of cells at the border of such “wounds” are shown in Fig. 7A. As can be seen, extensive intracellular staining was observed, indicative of V-ATPase localization to intracellular compartments (endosomes, lysosomes, and Golgi). In addition, plasma membrane staining was observed in ∼18% of cells, with staining primarily localized to the leading edge, consistent with previous reports (7, 31). Next, the effect of treatment with isoform-specific siRNAs on cellular distribution of V-ATPase was assessed. No detectable effect on the intracellular distribution of V-ATPases was observed following treatment with any of the siRNAs (data not shown). Treatment of cells with siRNAs specific for a3 or a4 but not a1 or a2 resulted in a reduction in the fraction of cells showing plasma membrane staining (Fig. 7B), with a4 resulting in the largest decrease (32%). These results suggest that the a4 isoform may be responsible for targeting of the V-ATPase to the plasma membrane in these cells.
FIGURE 7.
Immunostaining of MB231 cells with antibody against the V-ATPase and quantitation of plasma membrane staining. MB231 cells were grown as a monolayer on coverslips inside 60-mm dishes. A wound was made through the monolayer to polarize the cells after which they were immunostained with an antibody directed against the E subunit of V-ATPase as described under “Experimental Procedures.” A, three panels show fluorescence using anti-E (left), phalloidin to stain actin filaments (middle), and the merge (right), respectively. White arrows indicate the leading edge. B, cells grown as described above were treated with siRNA against the four a subunit isoforms, allowed to reach confluence, wounded, and immunostained as described under “Experimental Procedures.” Thirty to 40 consecutive images were captured along the wound. For each image, the total number of cells was counted, and the number showing a distinct line of staining at the plasma membrane (as indicated by the white arrow) was counted. The data are presented as a fraction of cells with plasma membrane staining relative to nontreated. Values are means; error bars indicate standard deviation, n = 3. *, p < 0.01 versus nontreated.
Effect of siRNA Treatment on Secretion of Pro-cathepsin L
To determine whether treatment with any of the isoform-specific siRNAs had effects on protein trafficking and secretion, the levels of a secreted protein (pro-cathepsin L) in the media were compared for cells treated with siRNAs and untreated cells. As shown by the representative blot in Fig. 8, none of the treatments with the isoform-specific siRNAs had a significant effect on secretion of pro-cathepsin L.
FIGURE 8.
Secretion of cathepsin L by MB231 cells. MB231 cells were treated with siRNA against the four a subunit isoforms or no siRNA (nontreated) for 24 h, and the media were then replaced with media without siRNA. The cells were incubated for an additional 48 h in serum-containing media and 24 h in serum-free media, and then conditioned media from these cells were collected and analyzed for the presence of pro-cathepsin L by Western blotting using a commercially available antibody as described under “Experimental Procedures.” Shown is a representative result.
DISCUSSION
Although V-ATPases have been implicated in tumor cell invasion, nothing is known concerning which populations of V-ATPase are functioning in this process. Thus, inhibitors like bafilomycin and concanamycin, although specific for V- ATPases, do not distinguish between V-ATPases containing different subunit isoforms or localized to intracellular versus plasma membranes. This latter problem is because of the membrane permeability of bafilomycin and concanamycin. Similarly, treatment of cells using siRNAs to subunits common to all V-ATPases (such as subunit c) (6) results in inhibition of all V-ATPases in the cell.
Targeting of V-ATPases to different cellular compartments is controlled by isoforms of subunit a, part of the integral V0 domain (15–19, 21–24, 28). In mammals, there are four isoforms of subunit a, with a3 and a4 responsible for plasma membrane targeting in osteoclasts and renal intercalated cells, respectively (15–19, 22–24). To begin to understand the role of different populations of V-ATPases in tumor cell invasiveness, we wished to compare the expression profile of subunit a isoforms in two human breast cancer cell lines with different metastatic potential. MDA-MB231 are highly invasive cells that display plasma membrane V-ATPases and show invasion that is inhibited by bafilomycin and concanamycin (7, 30, 31). By contrast, MCF7 cells are poorly invasive, show little evidence of plasma membrane V-ATPases, and display invasion that is not sensitive to V-ATPase inhibitors at the concentrations that affect MB231 cells (7). The results using quantitative RT-PCR demonstrate that although mRNA encoding all four a subunit isoforms can be detected in both cell types, both a3 and a4 are expressed at much higher levels in MB231 cells as compared with MCF7 cells. It is interesting to note that although the a3 isoform is expressed in a variety of cell types and tissues in addition to osteoclasts (19, 22), the a4 isoform has thus far been detected almost exclusively in renal cells (15, 24). This result suggests that MB231 cells have up-regulated an atypical isoform of subunit a.
To determine the role that different isoforms of subunit a play in V-ATPase function and tumor cell invasion, isoform-specific siRNAs were employed to selectively reduce mRNA levels for each of these isoforms in MB231 cells. These siRNAs were shown to reduce mRNA levels for the appropriate a subunit species in an isoform-specific manner. To assess the effects of siRNA treatment on V-ATPase function, the pH of the cytosolic and endosome/lysosome compartments was quantitated using the fluorescent pH indicators SNARF and pyranine, respectively. SNARF is taken up by cells as the acetoxymethyl ester and becomes trapped in the cytosol following cleavage by cytosolic esterases (30). As a result, cells labeled with SNARF show a diffuse, cytoplasmic staining pattern (Fig. 3A). Pyranine is a membrane-impermeant probe that is internalized by cells via fluid phase endocytosis (29). The localization of pyranine to the endosome/lysosome compartment was confirmed by co-localization with the lysosomal marker Lysotracker® Red (Fig. 3E). The results show that treatment of MB231 cells with siRNA specific for a3 both acidifies the cytosol and alkalinizes the endosome/lysosome compartment. This result is consistent with the presence of a3 in the endosome/lysosome compartment, where inhibition of V-ATPase activity prevents transport of protons out of the cytosol as well as into the lumen of endosomes/lysosomes. Similar conclusions can be drawn for the a1 and a2 isoforms with regard to their effect on endosome/lysosome pH; however, no significant effect on the steady state pH of the cytosol is observed (Fig. 5A). This may be related to the much lower levels of expression of a1 and a2 relative to a3 in MB231 cells (Fig. 1A). It is also possible that a1 or a2 are involved in the targeting of V-ATPases to the endosome/lysosome compartment such that their reduction secondarily results in effects on endosome/lysosome acidification. The fact that no significant effect on the intracellular staining pattern was observed by immunofluorescence following treatment with any of the isoform-specific siRNAs, however, argues against this possibility.
To evaluate the role of a subunit isoforms in the invasiveness of MB231 cells, an in vitro invasion assay was employed. The results show that only siRNAs specific for a3 and a4 significantly inhibit invasion. This result is somewhat surprising in light of the absence of any effect of a4-specific siRNA treatment on either cytosolic or endosome/lysosome pH. To assess the effect of siRNA treatment on V-ATPase localization, immunofluorescence was performed using an antibody that recognizes the E subunit (part of the V1 domain). The results demonstrate the presence of V-ATPase at the plasma membrane in a significant fraction of MB231 cells induced to migrate by creation of a wound in the cell monolayer. In addition, V-ATPase tends to localize to the leading edge of the cell, as indicated by co-localization with F-actin. Treatment of cells with siRNA to a4 reduces the fraction of cells showing plasma membrane staining by 32% relative to untreated controls. Inhibition of a3 expression also reduced plasma membrane staining, although to a lesser degree (18%). These results suggest that the a4 isoform (and perhaps to a lesser degree a3) are involved in targeting the V-ATPase to the plasma membrane of MB231 cells and that this cell surface V-ATPase plays a role in the invasiveness of these cells. It should be noted that although the decrease in plasma membrane staining with a4-siRNA treatment is consistent with the a4 isoform being part of the plasma membrane V-ATPase, it does not eliminate the possibility that a4 is involved in the trafficking to the plasma membrane of V-ATPase complexes containing a different isoform. The fact that all of the other siRNA treatments lead to smaller effects on plasma membrane staining, however, argues against this possibility. The absence of any effect of a4 knockdown on cytosolic pH suggests that the cell surface V-ATPases containing a4 do not contribute significantly to cytosolic pH homeostasis in these cells, or that their function in this context can to some degree be compensated by a partial up-regulation of other a subunit isoforms (Fig. 2). It is interesting to note in this regard that a1, a3, and a4 have all been reported to be present in the plasma membrane of renal proximal tubule cells, so that a subunit isoforms may have partially overlapping functions (26).
We speculate that a4-containing V-ATPases at the cell surface may promote the invasiveness of MB231 cells by locally acidifying the extracellular environment (i.e. the space between the cell surface and the extracellular matrix). This extracellular acidification may promote invasion by activation of secreted proteases, such as cathepsins, which require an acidic environment for optimal activity. Because the a4 isoform is typically expressed only in renal cells (15, 16, 24), we suggest that MB231 cells have up-regulated an atypical form of the a subunit for the purpose of enhancing their invasive potential. Additional experiments measuring the effect of siRNA treatment on proton secretion into the extracellular space and looking at whether a4 expression levels in other tumor cell types correlates with their invasive properties will be required to test these ideas.
With respect to inhibition of invasion by knockdown of a3, it seems most likely that this is due, at least in part, to inhibition of intracellular V-ATPases, because the effect on invasion is more significant than the effect observed for plasma membrane staining. Moreover, the effects of a3 knockdown on cytoplasmic and endosome/lysosome pH are most consistent with a3 having an intracellular role. There are a number of possible functions that intracellular V-ATPases may play in promoting invasion, including intracellular activation of secreted proteases involved in degradation of extracellular matrix and trafficking of these proteases to the cell surface. Preliminary experiments monitoring protein secretion in MB231 cells have not detected any significant changes following siRNA treatment (Fig. 8), although this does not rule out more selective effects on particular trafficking pathways. Additional experiments will be required to elucidate the role of a subunit isoforms in protein trafficking in mammalian cells. It should be noted that up-regulation of intracellular and plasma membrane V-ATPases may play additional roles in cancer progression beyond promotion of tumor cell invasion. For example, increased V-ATPase activity in intracellular compartments may aid in the drug resistance of tumor cells by driving uptake into intracellular vesicles via proton-coupled drug transporters (32–35). Intracellular and plasma membrane V-ATPases may also function in cytoplasmic pH homeostasis. This is particularly important in tumor cells that, because of their high glycolytic rates (32), often require additional aid in acid secretion from the cytoplasm. In summary, this study represents the first examination of the role of V-ATPase a subunit isoforms in pH regulation and invasiveness of breast cancer cells, and suggests the possibility that V-ATPases may represent an effective and potentially selective therapeutic target in controlling tumor cell metastasis.
Acknowledgments
We thank Drs. Daniel Cipriani, Yanru Wang, Kevin Jefferies, Jie Qi, and Masashi Toei for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM34478 (to M. F.), post-doctoral support (to A. H.) and pre-doctoral support (to S. B.) from National Institutes of Health Ruth L. Kirschstein Institutional Research Training Grant T32DK007542, and post-doctoral support (to A. H.) from National Institutes of Health Training in Education and Critical Research Skills Grant K12GM074869. This work was also supported by American Heart Association Grants SD 0435197N (to S. R. S.) and GIA 0855168 (to R. M. Z.).
- V-ATPase
- vacuolar proton-translocating ATPase
- MB231
- MDA-MB231
- RT
- reverse transcription
- DMSO
- dimethyl sulfoxide
- QRT
- quantitative RT
- pHcyt
- cytosolic pH
- pHE/L
- endosomes/lysosomes pH
- siRNA
- small interfering RNA
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Gupta G. P., Massagué J. ( 2006) Cell 127, 679– 695 [DOI] [PubMed] [Google Scholar]
- 2.Pantel K., Brakenhoff R. H. ( 2004) Nat. Rev. Cancer 4, 448– 456 [DOI] [PubMed] [Google Scholar]
- 3.Forgac M. ( 2007) Nat. Rev. Mol. Cell Biol. 8, 917– 929 [DOI] [PubMed] [Google Scholar]
- 4.Ohta T., Numata M., Yagishita H., Futagami F., Tsukioka Y., Kitagawa H., Kayahara M., Nagakawa T., Miyazaki I., Yamamoto M., Iseki S., Ohkuma S. ( 1996) Br. J. Cancer 73, 1511– 1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kubota S., Seyama Y. ( 2000) Biochem. Biophys. Res. Commun. 278, 390– 394 [DOI] [PubMed] [Google Scholar]
- 6.Lu X., Qin W., Li J., Tan N., Pan D., Zhang H., Xie L., Yao G., Shu H., Yao M., Wan D., Gu J., Yang S. ( 2005) Cancer Res. 65, 6843– 6849 [DOI] [PubMed] [Google Scholar]
- 7.Sennoune S. R., Bakunts K., Martínez G. M., Chua-Tuan J. L., Kebir Y., Attaya M. N., Martínez-Zaguilán R. ( 2004) Am. J. Physiol. Cell Physiol. 286, C1443– 1452 [DOI] [PubMed] [Google Scholar]
- 8.Martínez-Zaguilán R., Raghunand N., Lynch R. M., Bellamy W., Martinez G. M., Rojas B., Smith D., Dalton W. S., Gillies R. J. ( 1999) Biochem. Pharmacol. 57, 1037– 1046 [DOI] [PubMed] [Google Scholar]
- 9.Philippe J. M., Dubois J. M., Rouzaire-Dubois B., Cartron P. F., Vallette F., Morel N. ( 2002) Glia 37, 365– 373 [PubMed] [Google Scholar]
- 10.Ghosh P., Dahms N. M., Kornfeld S. ( 2003) Nat. Rev. Mol. Cell Biol. 4, 202– 212 [DOI] [PubMed] [Google Scholar]
- 11.Gocheva V., Joyce J. A. ( 2007) Cell Cycle 6, 60– 64 [DOI] [PubMed] [Google Scholar]
- 12.Joyce J. A., Hanahan D. ( 2004) Cell Cycle 3, 1516– 1619 [DOI] [PubMed] [Google Scholar]
- 13.Kawasaki-Nishi S., Nishi T., Forgac M. ( 2001) J. Biol. Chem. 276, 17941– 17948 [DOI] [PubMed] [Google Scholar]
- 14.Manolson M. F., Wu B., Proteau D., Taillon B. E., Roberts B. T., Hoyt M. A., Jones E. W. ( 1994) J. Biol. Chem. 269, 14064– 14074 [PubMed] [Google Scholar]
- 15.Oka T., Murata Y., Namba M., Yoshimizu T., Toyomura T., Yamamoto A., Sun-Wada G. H., Hamasaki N., Wada Y., Futai M. ( 2001) J. Biol. Chem. 276, 40050– 40054 [DOI] [PubMed] [Google Scholar]
- 16.Smith A. N., Finberg K. E., Wagner C. A., Lifton R. P., Devonald M. A., Su Y., Karet F. E. ( 2001) J. Biol. Chem. 276, 42382– 42388 [DOI] [PubMed] [Google Scholar]
- 17.Sun-Wada G. H., Toyomura T., Murata Y., Yamamoto A., Futai M., Wada Y. ( 2006) J. Cell Sci. 119, 4531– 4540 [DOI] [PubMed] [Google Scholar]
- 18.Toyomura T., Murata Y., Yamamoto A., Oka T., Sun-Wada G. H., Wada Y., Futai M. ( 2003) J. Biol. Chem. 278, 22023– 22030 [DOI] [PubMed] [Google Scholar]
- 19.Toyomura T., Oka T., Yamaguchi C., Wada Y., Futai M. ( 2000) J. Biol. Chem. 275, 8760– 8765 [DOI] [PubMed] [Google Scholar]
- 20.Wang Y., Toei M., Forgac M. ( 2008) J. Biol. Chem. 283, 20696– 20702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawasaki-Nishi S., Bowers K., Nishi T., Forgac M., Stevens T. H. ( 2001) J. Biol. Chem. 276, 47411– 47420 [DOI] [PubMed] [Google Scholar]
- 22.Nishi T., Forgac M. ( 2000) J. Biol. Chem. 275, 6824– 6830 [DOI] [PubMed] [Google Scholar]
- 23.Frattini A., Orchard P. J., Sobacchi C., Giliani S., Abinun M., Mattsson J. P., Keeling D. J., Andersson A. K., Wallbrandt P., Zecca L., Notarangelo L. D., Vezzoni P., Villa A. ( 2000) Nat. Genet. 25, 343– 346 [DOI] [PubMed] [Google Scholar]
- 24.Smith A. N., Skaug J., Choate K. A., Nayir A., Bakkaloglu A., Ozen S., Hulton S. A., Sanjad S. A., Al-Sabban E. A., Lifton R. P., Scherer S. W., Karet F. E. ( 2000) Nat. Genet. 26, 71– 75 [DOI] [PubMed] [Google Scholar]
- 25.Morel N., Dedieu J. C., Philippe J. M. ( 2003) J. Cell Sci. 116, 4751– 4762 [DOI] [PubMed] [Google Scholar]
- 26.Hurtado-Lorenzo A., Skinner M., El Annan J., Futai M., Sun-Wada G. H., Bourgoin S., Casanova J., Wildeman A., Bechoua S., Ausiello D. A., Brown D., Marshansky V. ( 2006) Nat. Cell Biol. 8, 124– 136 [DOI] [PubMed] [Google Scholar]
- 27.Hu Y., Nyman J., Muhonen P., Väänänen H. K., Laitala-Leinonen T. ( 2005) FEBS Lett. 579, 4937– 4942 [DOI] [PubMed] [Google Scholar]
- 28.Pietrement C., Sun-Wada G. H., Silva N. D., McKee M., Marshansky V., Brown D., Futai M., Breton S. ( 2006) Biol. Reprod. 74, 185– 194 [DOI] [PubMed] [Google Scholar]
- 29.Gillies R. J., Martinez-Zaguilan R. ( 1991) J. Biol. Chem. 266, 1551– 1556 [PubMed] [Google Scholar]
- 30.Martínez-Zaguilán R., Martínez G. M., Lattanzio F., Gillies R. J. ( 1991) Am. J. Physiol. 260, C297– 307 [DOI] [PubMed] [Google Scholar]
- 31.Rojas J. D., Sennoune S. R., Maiti D., Bakunts K., Reuveni M., Sanka S. C., Martinez G. M., Seftor E. A., Meininger C. J., Wu G., Wesson D. E., Hendrix M. J., Martínez-Zaguilán R. ( 2006) Am. J. Physiol. Heart Circ. Physiol. 291, H1147– 1157 [DOI] [PubMed] [Google Scholar]
- 32.Murakami T., Shibuya I., Ise T., Chen Z. S., Akiyama S., Nakagawa M., Izumi H., Nakamura T., Matsuo K., Yamada Y., Kohno K. ( 2001) Int. J. Cancer 93, 869– 874 [DOI] [PubMed] [Google Scholar]
- 33.Raghunand N., Martínez-Zaguilán R., Wright S. H., Gillies R. J. ( 1999) Biochem. Pharmacol. 57, 1047– 1058 [DOI] [PubMed] [Google Scholar]
- 34.Sennoune S. R., Luo D., Martínez-Zaguilán R. ( 2004) Cell Biochem. Biophys. 40, 185– 206 [DOI] [PubMed] [Google Scholar]
- 35.Torigoe T., Izumi H., Ishiguchi H., Uramoto H., Murakami T., Ise T., Yoshida Y., Tanabe M., Nomoto M., Itoh H., Kohno K. ( 2002) J. Biol. Chem. 277, 36534– 36543 [DOI] [PubMed] [Google Scholar]