Abstract
Na+/H+ antiporters influence proton or sodium motive force across the membrane. Synechocystis sp. PCC 6803 has six genes encoding Na+/H+ antiporters, nhaS1–5 and sll0556. In this study, the function of NhaS3 was examined. NhaS3 was essential for growth of Synechocystis, and loss of nhaS3 was not complemented by expression of the Escherichia coli Na+/H+ antiporter NhaA. Membrane fractionation followed by immunoblotting as well as immunogold labeling revealed that NhaS3 was localized in the thylakoid membrane of Synechocystis. NhaS3 was shown to be functional over a pH range from pH 6.5 to 9.0 when expressed in E. coli. A reduction in the copy number of nhaS3 in the Synechocystis genome rendered the cells more sensitive to high Na+ concentrations. NhaS3 had no K+/H+ exchange activity itself but enhanced K+ uptake from the medium when expressed in an E. coli potassium uptake mutant. Expression of nhaS3 increased after shifting from low CO2 to high CO2 conditions. Expression of nhaS3 was also found to be controlled by the circadian rhythm. Gene expression peaked at the beginning of subjective night. This coincided with the time of the lowest rate of CO2 consumption caused by the ceasing of O2-evolving photosynthesis. This is the first report of a Na+/H+ antiporter localized in thylakoid membrane. Our results suggested a role of NhaS3 in the maintenance of ion homeostasis of H+, Na+, and K+ in supporting the conversion of photosynthetic products and in the supply of energy in the dark.
Na+/H+ antiporters are integral membrane proteins that transport Na+ and H+ in opposite directions across the membrane and that occur in virtually all cell types. These transporters play an important role in the regulation of cytosolic pH and Na+ concentrations and influence proton or sodium motive force across the membrane (1, 2). In Escherichia coli, three Na+/H+ antiporters (NhaA, NhaB, and ChaA) have been described in detail. Of these, NhaA is the functionally best characterized transporter. The crystal structure of NhaA has been resolved (3). In addition, mutants of nhaA, nhaB, and chaA as well as the triple mutant have been generated (4). The triple mutant was shown to be hypersensitive to extracellular Na+. The genome of the cyanobacterium Synechocystis sp. PCC 6803 contains six genes encoding Na+/H+ antiporters (NhaS1–5 and sll0556). NhaS1 (slr1727) has also been designated SynNhaP (5, 6). Null mutants of nhaS1, nhaS2, nhaS4, and nhaS5 have been generated; however, a null mutant of nhaS3 could not be obtained, indicating that it is an essential gene (6–8). By heterologous expression in E. coli, Na+/H+ exchange activities could be shown for NhaS1–5 (5, 6). Inactivation of nhaS1 and nhaS2 results in retardation of growth of Synechocystis (5, 6). It has been reported that in these mutants the concentration of Na+ in cytosol and intrathylakoid space (lumen) increases and impairs the photosynthetic and/or respiratory activity of the cell (9, 10). Therefore the Na+ extrusion by Synechocystis Na+/H+ antiporters similar to E. coli NhaA, NhaB, and ChaA is essential for the adaptation to salinity stress.
In contrast to the case in E. coli, Na+ is an essential element for the growth of some cyanobacteria (11, 12). Interestingly, the Na+/H+ antiporter homolog NhaS4 was identified as an uptake system for Na+ from the medium during a screen for mutations in Synechocystis that result in lack of growth at low Na+ concentrations (7). The requirement of a Na+ uptake antiporter for cell growth is consistent with the physiology of Synechocystis. Specifically, photoautotrophic bacteria like cyanobacteria share some components (plastoquinone, cytochrome b6f, and c6) of the thylakoid membrane for electron transport for both photophosphorylation and respiratory oxidative phosphorylation. Na+/H+ antiporters therefore may coordinate both H+ and Na+ gradients across the plasma and thylakoid membranes to adapt to daily environmental changes (11). It remains to be determined whether the six Na+/H+ antiporters are localized to the plasma membrane or to the thylakoid membrane in Synechocystis. Information on the membrane localization will also provide information on the physiological role in Synechocystis. In this study, we explored the membrane localization of NhaS3, the role of specific amino acid residues for its function, and the effect of CO2 concentration and circadian rhythms on the expression pattern of nhaS3 to gain insight into the physiological role of NhaS3 in Synechocystis.
EXPERIMENTAL PROCEDURES
Synechocystis Growth Conditions
Synechocystis cells were grown at 30 °C in BG11 medium (13) containing 20 mm TES-KOH3 (pH 8.0) and bubbled with either 2% CO2 in air (v/v) or air alone (0.035% v/v CO2). Solid medium contained BG11 buffered at pH 8.0, 1.5% agar, and 0.3% sodium thiosulfate. Continuous illumination was provided by fluorescent lamps (50 μmol of photons m−2 s−1; 400–700 nm). To test the activation of expression of the nhaS3 promoter-luciferase fusions (see below), cells grown under low CO2 conditions (0.035% v/v CO2) were collected by centrifugation at 5000 × g for 10 min at 30 °C, washed with fresh growth medium to remove dissolved CO2, and inoculated into fresh medium that was aerated with air containing 2% (v/v) CO2. Control cells were kept at 0.035% (v/v CO2).
Molecular Biology Methods and Heterologous Expression in E. coli
For heterologous expression in E. coli, the nhaS3 (sll0689) gene was isolated from chromosomal DNA by PCR using KpnI site-containing forward primer 5′-ATAGGTACCAGGAGGGAAAAGAATGTTTATGAACCCAT-3′ and SalI site-containing reverse primer 5′-AAAGTCGACCTAATCTGGGGTGGGAAC-3′. The KpnI-SalI DNA fragment was ligated into the corresponding sites in pPAB404 (14), and the resulting plasmid was used to transform E. coli strain LB2003, which lacks the three K+ uptake systems (15), or E. coli strain TO114, which lacks the three Na+ extrusion type Na+/H+ antiporters (4). Growth tests of the transformed strains were carried out as described previously (16, 17). For replacement of NhaS3 with E. coli NhaA in Synechocystis, the E. coli nhaA gene was cloned behind the iron-transporter promoter in a plasmid containing the spectinomycin resistance gene (18) and inserted by homologous recombination into targeting site 44 of the chromosomal DNA of Synechocystis (16). For disruption of nhaS3, the kanamycin resistance gene was amplified by PCR using HpaI site-containing forward primer 5′-CAGTGTTAACAAAGCCACGTTGTGTCTC-3′ and HpaI site-containing reverse primer 5′-CAGTGTTAACGCGCTGAGGTCTGCCTCG-3′. The HpaI-digested DNA fragment was inserted into the EcoRV site in the middle of the nhaS3 gene of the nhaA-expressing strain. For generation of the NhaS3 knockdown strain, the same HpaI-digested DNA fragment was inserted into the EcoRV site in the middle of the nhaS3 gene of wild type Synechocystis. For generation of nhaS3-overexpressing Synechocystis, the full-length coding sequence of NhaS3 was amplified by PCR using NdeI site-containing forward primer 5′-CGGCATATGATGTTTATGAACCCATTG-3′ and SalI site-containing reverse primer 5′-AAGTCGACCTAATCTGGGGTGGGAAC-3′. The NdeI-SalI DNA fragment was ligated into the corresponding sites in p68TS4OxCm4 and inserted by homologous recombination into targeting site 4 of the chromosomal DNA of Synechocystis (16). This vector contains the inducible trc promoter. Expression of nhaS3 was induced by the addition of 0.25 mm isopropyl β-d-thiogalactopyranoside to the medium.
Membrane Fractionation
Thylakoid and plasma membranes were prepared from Synechocystis cells as described previously (19). An anti-NhaS3 antibody was raised against synthetic peptides with the two sequences NH2-LAEINRLSSNEGQI-COOH and NH2-KKEEAPEKPVPTPD-COOH (Operon Biotechnologies, Japan). Polyclonal antibodies raised against the plasma membrane nitrate transporter NrtA (20) or against the thylakoid membrane proteins NdhD3 and NdhF3 (21) were used to identify they Synechocystis plasma membrane, or thylakoid membrane fractions, respectively. The proteins were separated by SDS-PAGE on 10 or 12% polyacrylamide gels and then transferred to polyvinylidene fluoride membranes. The membranes were incubated for 1 h with primary antibody (1:1000 in blocking buffer) and were then incubated for 30 min with the secondary antibody with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5000; Amersham Biosciences) and subsequently subjected to chemiluminescence detection (ECL; Amersham Biosciences).
Immunolabeling and Electron Microscopy
Synechocystis cells grown to an optical density of about 1.2 in BG11 medium were frozen and then fixed with anhydrous acetone containing 1% glutaraldehyde at −80 °C. The samples were warmed and embedded in LR White resin as described (22). Ultra-thin sections were first labeled with the IgG fraction of NhaS3 antiserum (1:20) in Tris-buffered saline and then with 12-nm colloidal gold particles coupled to goat anti-rabbit IgG. IgG fractions were purified from the NhaS3 or preimmune serum using the MelonTM Gel IgG Spin Purification Kit (Pierce). The sections were stained with uranyl acetate and examined with a 1010EX transmission electron microscope (JEOL) at 80 kV as described (22).
Measurement of Circadian Rhythm of nhaS3 Promoter Activity in Synechocystis
A 1000-bp nhaS3 promoter sequence was fused to the bacterial luciferase gene set luxAB at the BglII and NdeI sites of p68TS1ΩLuxAB(+)PLNK,4 and the construct was inserted into the TS1 region in Synechocystis chromosomal DNA (23). Bioluminescence from cells grown on the solid BG11 medium was measured as described previously (24–26). The selected cells were cultured in liquid BG11 medium at 30 °C under 91 μmol of white light illumination m−2 s−1 with bubbling of air and stirring. The optical density of the culture at 730 nm was maintained at ∼0.35 by dilution with fresh BG11 medium. To entrain the circadian clock, the culture was placed in darkness for 12 h and then kept under constant light conditions. Bioluminescence was measured every hour.
Na+/H+ Antiporter Assay
The preparation of E. coli membrane vesicles was carried out as described previously (27). Na+/H+ antiporter activity was measured by the acridine orange fluorescence quenching method (27) at 25 °C in an assay mixture (2 ml) in the buffer (10 mm Tris-HCl (pH 7.2), 140 mm choline chloride, 5 mm MgSO4, 6 mm 2-mercaptoethanol, and 10% glycerol) supplemented with 1 μm acridine orange. The membrane vesicles equal to 50 μg of protein were added to the assay mixture. Tris-dl-lactate (2 mm) was added to initiate fluorescence quenching caused by respiration. Changes in fluorescence were monitored after the addition of 5 mm NaCl. Then 20 ml of 5% Triton X-100 were added to dissipate the pH gradient across the membrane. Fluorescence emission was monitored at 530 nm with excitation at 495 nm.
Measurement of K+ Uptake in E. coli
K+ uptake by E. coli was measured at described previously (16). The cells expressing nhaS3 or containing the empty vector were incubated with 1 mm KCl in HEPES-NaOH, pH 7.5, containing 10 mm glucose, and aliquots were withdrawn at the times indicated in Fig. 3. The cellular K+ content was determined by flame photometry.
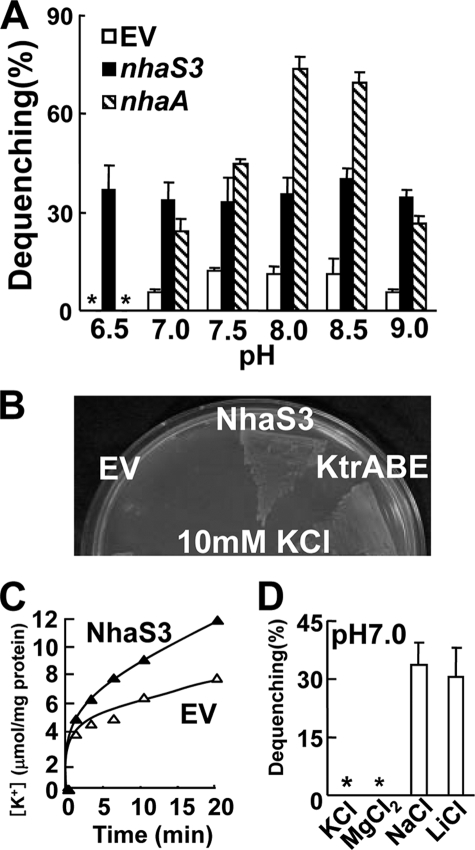
FIGURE 3.
Transport activity of NhaS3. A, pH dependence of Na+/H+ antiporter activities of NhaS3 in inverted membrane vesicles prepared from nhaS3-expressing E. coli. Fluorescence from acridine orange was monitored. The percentage of fluorescence dequenching observed after the addition of 5 mm NaCl was plotted relative to that of lactate-induced quenching. EV, empty vector; *, not detected. B, complementation of an E. coli K+ uptake mutant (LB2003) by NhaS3. Growth of E. coli LB2003 expressing nhaS3 or Synechocystis K+ uptake transporter KtrABE on synthetic solid medium containing 10 mm KCl. C, K+ uptake activity by NhaS3 in E. coli. Cells containing NhaS3 or the empty vector were incubated with 1 mm KCl in HEPES-NaOH, pH 7.5, and aliquots were withdrawn at the indicated times. D, measurement of K+ or Mg2+ efflux mediated by NhaS3. K+, Mg2+, or Li+, instead of Na+ were added to membrane vesicles prepared from nhaS3-expressing E. coli cells. Antiporter activity was determined by the same procedure as in A. *, not detected.
RESULTS
E. coli NhaA Does Not Complement a Synechocystis nhaS3 Mutant
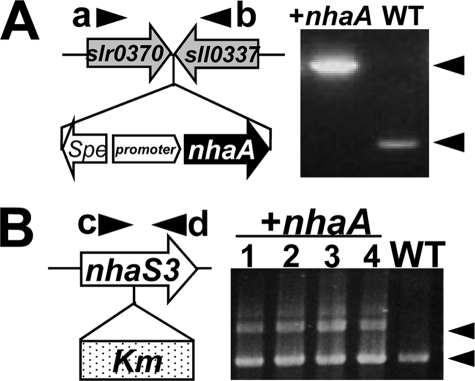
Previously it had been reported that NhaS3 is an essential gene in Synechocystis 6803 (6, 8). Likewise our attempts to construct a null mutant of nhaS3 were also not successful (see Fig. 5). In contrast, in E. coli even disruption of all three Na+/H+ antiporter genes is not lethal. Based on these observations we hypothesized that NhaS3 may have additional unknown characteristics. To test this hypothesis, we tried to replace nhaS3 in Synechocystis with the E. coli nhaA gene encoding a plasma membrane Na+/H+ antiporter (28, 29). As a first step nhaA was introduced into the Synechocystis genome (Fig. 1A). Next, nhaS3 was disrupted in this background by insertion of a kanamycin resistance cassette, and recovery of a fully segregated mutant of nhaS3 expressing E. coli nhaA was attempted. PCR analysis of kanamycin-resistant cells showed that nhaS3 was not fully disrupted by the kanamycin resistance gene because of the incomplete segregation of chromosomes (Fig. 1B). These results indicate that nhaA cannot replace nhaS3 functionally.
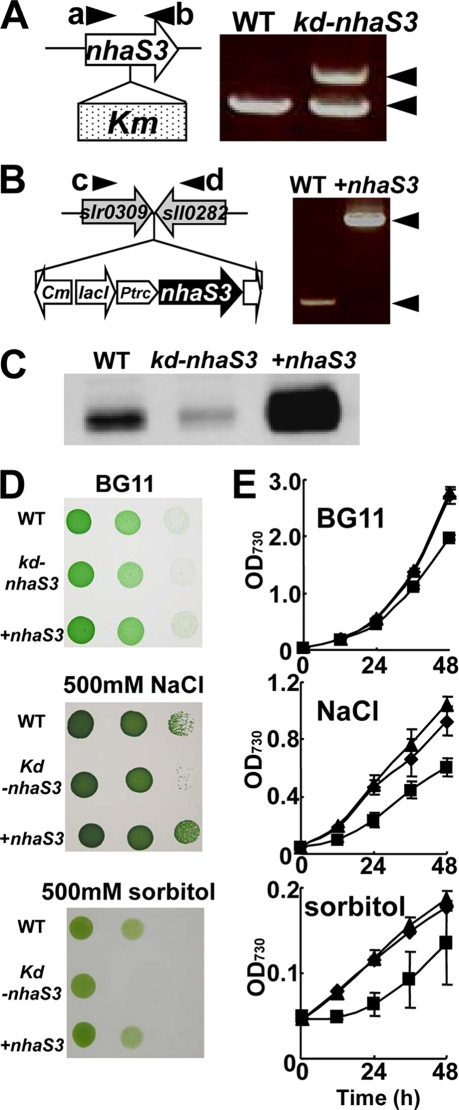
FIGURE 5.
Inhibition of growth of nhaS3 knockdown Synechocystis cells by salt stress and hyperosmotic stress. A, creation of a nhaS3 knockdown strain (kd-nhaS3). The nhaS3 gene was disrupted in Synechocystis by insertion of a kanamycin resistance gene (Km). Integration of Km was detected by PCR using primers a and b. Note that nhdS3 was not completely disrupted. B, overexpression of nhaS3 (+nhaS3). A construct containing the lacIq, Ptrc::nhaS3, and chloramphenicol resistance (Cm) gene was integrated into the TS4 sites (between slr0309 and sll0282) in the Synechocystis genome. Integration of the overexpression construct was verified by PCR using primers c and d. C, validation of both the knockdown strain (kd-nhaS3) and the overexpression strain (+nhaS3). NhaS3 protein levels were detected by Western blot analysis. D, growth of the wild type (WT), nhaS3 knockdown strain (kd-nhaS3), and nhaS3 overexpression strain (+nhaS3) on solid BG11 medium or on BG11 medium containing either 500 mm NaCl or 500 mm sorbitol. E, growth test performed in liquid culture using the same strains and conditions as in D. The OD730 was measured at the times indicated. WT, circle; kd-nhaS3, square; +nhaS3, triangle.
FIGURE 1.
Introduction of E. coli nhaA into Synechocystis. A, E. coli nhaA under control of the iron-inducible promoter was introduced at a site between slr0370 and sll0337 of Synechocystis chromosome (left panel). During growth in BG11 medium, nhaA is constitutively expressed. Correct insertion of the expression construct was confirmed by PCR on genomic DNA using specific primers a and b; the results are shown in the right panel. WT, wild type; +nhaA, nhaA-expressing cells; Spe, spectinomycin resistance gene. B, disruption of nhaS3 in the strain expressing E. coli nhaA was performed by insertion of the kanamycin resistance gene (Km) into nhaS3. Correct integration of the kanamycin gene was tested by PCR on genomic DNA using specific primers c and d. The results for four independent clones and WT are shown in the right panel. Note that nhaA could not replace nhaS3.
NhaS3 Is Localized to the Thylakoid Membrane
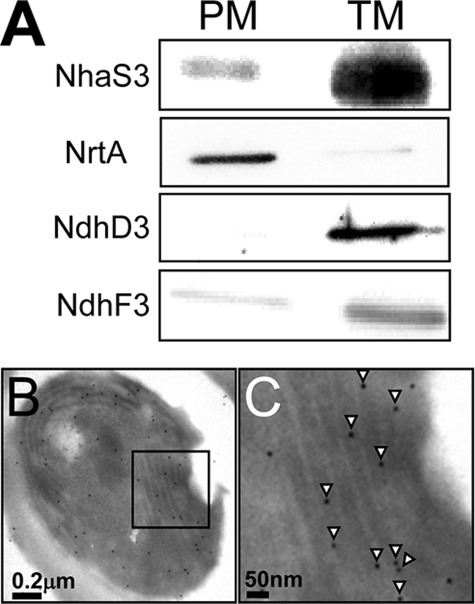
The finding that E. coli nhaA cannot substitute for nhaS3 (Fig. 1) suggested that NhaS3 might differ from E. coli NhaA with respect to its activity and/or subcellular localization. We therefore performed immunolocalization experiments using anti-NhaS3 antibodies to determine the subcellular localization of NhaS3. Membrane fractions of thylakoid and plasma membranes were prepared by aqueous polymer two-phase partitioning followed by sucrose density gradient centrifugation (19). As shown in Fig. 2A, a single protein band of the corresponding molecular mass of NhaS3 was found in the thylakoid membrane fraction, which was identified by the presence of the thylakoid membrane marker proteins NdhD3 and NdhF3 (21, 30). Only a weak signal for NhaS3 was found in the plasma membrane fraction, which was identified by the presence of the nitrate transporter NrtA (20). This indicated that NhaS3 was localized in the thylakoid membrane in Synechocystis. In addition, the subcellular localization of the NhaS3 protein was analyzed by immunogold labeling followed by electron microscopy. A cross-section of wild type Synechocystis cells grown under standard conditions (see “Experimental Procedures”) showed gold particles decorating the thylakoid membrane when the IgG fraction of NhaS3 antiserum was used (Fig. 2, B and C). Only a small amount of the label was found on the plasma membrane or in other locations. Control experiments with IgG fraction of preimmune serum did not show any significant labeling (data not shown). These results indicate that NhaS3 is associated with the thylakoid membrane fraction (Fig. 2A).
FIGURE 2.
Membrane localization of NhaS3. A, membrane fractions were prepared by aqueous polymer two-phase partitioning and sucrose density gradient and separated by SDS-PAGE. NhaS3, NrtA, NdhD3, and NdhF3 proteins were detected on Western blots using the corresponding antibodies. PM, plasma membrane; TM, thylakoid membrane. B and C, cross-section of a Synechocystis cell immunolabeled using an anti-NhaS3 antibody. C is an enlarged section of B. NhaS3 protein, indicated by the presence of gold particles (arrowheads in C), was detected in the thylakoid membrane.
NhaS3 Has Na+/H+ Antiporter Activity and Induces K+ Uptake in E. coli
The activity of NhaS3 may be influenced by the proton gradient formed through respiration or photosynthesis across the thylakoid membrane. In E. coli, three Na+/H+ antiporters, NhaA, NhaB, and ChaA, show different pH dependence from each other and also have different physiological roles (31, 32). In Synechocystis it has been reported that NhaS3 has antiporter activity at pH 8.5 (6), but its detailed properties remain to be studied. To measure the pH dependence of NhaS3, E. coli strain TO114, which possesses low Na+/H+ exchange activity, was used as a host (4). Inverted membrane vesicles were prepared from E. coli TO114 cells transformed with plasmids encoding NhaS3 or NhaA or the empty plasmid pPAB404. Transport activities were assessed by measuring the dequenching of acridine orange fluorescence upon the addition of 5 mm NaCl at different pH values (Fig. 3A). E. coli NhaA had a peak of activity at pH 8.0 and 8.5, which is consistent with the pH profile of NhaA previously reported (33). In contrast, NhaS3 showed a similar level of activity across the entire pH spectrum tested (pH 6.5–9.0). This indicates that NhaS3 activity is pH-independent.
Na+/H+ antiporter activities affect K+ transport across the membrane and the balance of the cytosolic Na+/K+ concentration ratio (34–37). To test the influence of NhaS3 on K+ uptake, it was expressed in an E. coli strain, LB2003, lacking three major K+ uptake systems and unable to grow at low K+ concentrations (15). NhaS3 restored growth of the LB2003 strain at low K+ concentrations (10 mm) (Fig. 3B). Under the same conditions, expression of the Synechocystis K+ uptake system, KtrABE (16), which was used as a positive control, rescued the mutant. No growth was observed with the empty vector alone. Although the initial net K+ uptake of the cells expressing NhaS3 was higher than that of the cells containing the empty vector (Fig. 3C), NhaS3 did not show K+/H+ exchange activity (Fig. 3D). The measurement was performed under conditions where NhaS3 showed Li+/H+ as well as Na+/H+ antiporter activities, but no Mg2+/H+ antiporter activity (Fig. 3D). These data indicate that NhaS3 either functions as a K+ uptake transporter or that Na+ extrusion mediated by NhaS3 may indirectly increase the influx of K+ into the cells. NhaS3 may contribute to maintaining the balance of the cytosolic Na+/K+ ratio.
Analysis of Conserved Residues in NhaS3
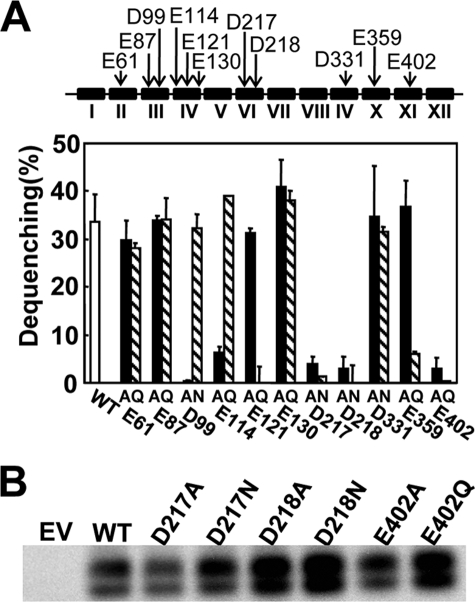
In the NhaA antiporter protein negatively charged residues in the hydrophobic transmembrane domains play a crucial role for Na+ or H+ electrostatic interaction and ion transport function (3). Two aspartates, corresponding to Asp217 and Asp218 in NhaS3, are proposed to be the ion binding site in NhaS3 (3). Another negatively charged residue, glutamate (Glu402), is located in the eleventh hydrophobic domain of NhaS3 but is not conserved in the other Synechocystis Na+/H+ antiporters or in E. coli NhaA (supplemental Fig. S1). To test the role of these charged residues in the transport function of NhaS3, mutant versions of NhaS3 were generated. In these mutant proteins single negatively charged residues (Asp or Glu) were replaced by either a neutral nonpolar residue (alanine) or a neutral polar residue (asparagine or glutamine). The mutant proteins were then expressed in E. coli, and their antiporter activity was determined by fluorescence dequenching in membrane vesicles (Fig. 4A). Amino acid substitutions at Asp217, Asp218, and Glu402 abolished the transport activities of NhaS3 regardless of the replacing amino acid. The protein expression levels of these NhaS3 variants that caused loss of transport activity were verified by Western blotting analysis (Fig. 4B). There was no difference in the amount of protein present, indicating that the loss of activity was not caused by a lack of protein. The lower bands seen on the blots are most likely degradation products of the full-length protein (upper band). Based on data available for NhaA (3), Asp217, Asp218, and Glu402 are likely to be part of the structure for translocation of Na+/H+. Replacement of Asp99, Glu114, Glu121, and Glu359 caused a marked decrease in the activities only in the case of one of the substituted amino acids (Fig. 4B). In these cases the replacement of the residues affected the transport activity, but the results suggest that they may not be essential for binding or translocation of cations. These results show that the conserved negative residues Asp217 and Asp218 in NhaS3 have the same function as their counterparts in NhaA, which provides further evidence that NhaS3 functions as a Na+/H+ antiporter.
FIGURE 4.
Na+/H+ antiporter activities in the inverted membrane vesicles of E. coli expressing wild type NhaS3 or NhaS3 variants. A, antiporter activities of the NhaS3 variants. The locations of the negatively charged residues (Asp or Glu) that were replaced with Ala or Asn/Gln are indicated with arrows (top). The Roman numerals designate the numbers of the putative transmembrane spanning domains predicted by the TopPredII algorithm and counted from the N terminus of NhaS3. Na+/H+ antiporter activity was determined in inverted membrane vesicles by measuring the percentage of fluorescence dequenching observed after addition of 5 mm NaCl (bottom). B, immunological detection of NhaS3 variants containing mutations at Asp217, Asp218, and Glu402 that showed no transport activities in A. The membrane proteins isolated from the strains expressing the nhaS3 variants were separated by SDS-polyacrylamide gel electrophoresis (12% acrylamide) and blotted, and the blots were probed using anti-NhaS3 antibodies. WT, wild type; EV, empty vector.
NhaS3 Contributes to Na+ Homeostasis and Na+ Tolerance in Synechocystis
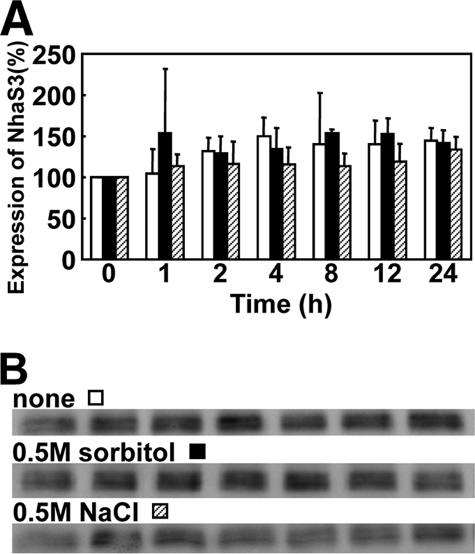
Based on the above data on NhaS3-mediated Na+ transport activities, we examined whether NhaS3 contributes to the cellular ion homeostasis in response to environmental stress. It was tested whether the amount of NhaS3 affects the stress tolerance of Synechocystis. An nhaS3 knockdown strain was obtained by insertion of kanamycin resistance gene into nhaS3 (Fig. 5A). The decrease in NhaS3 protein was confirmed by Western blotting (Fig. 5C). A Synechocystis strain overexpressing nhaS3 was generated by integration of a copy of nhaS3 under control of the inducible trc promoter (Fig. 5B). Overexpression of nhaS3 and the increased presence of the NhaS3 protein was confirmed by Western blot (Fig. 5C). Both knockdown cells and cells overexpressing nhaS3 were grown under conditions of salinity stress and hyperosmotic stress (Fig. 5, D and E). The nhaS3 knockdown strain grew less well on solid or liquid medium supplemented with either 500 mm NaCl or 500 mm sorbitol when compared with the growth of wild type or overexpressor. This suggests that NhaS3 contributes to the osmoadaptation of Synechocystis. Overexpression of NhaS3 did not increase the salt or osmotolerance of the cells. In wild type cells the amount of NhaS3 protein increased only slightly (1.1–1.6-fold) during salt stress or osmotic stress compared with the untreated control (Fig. 6), indicating that both stress conditions had only a small effect on NhaS3 expression at the translational level.
FIGURE 6.
Effect of salt stress or hyperosmotic stress on the expression of NhaS3. A, wild type cells cultured in liquid BG11 medium (OD730 = 0.5) were transferred to the same medium (white bars) or to hyperosmotic medium containing either 500 mm NaCl (black bars) or 500 mm sorbitol (hatched bars) and cultured for the time indicated. B, cells samples taken from the cultures in A at the times indicated were subjected to Western blot analysis using anti-NhaS3 antibodies.
Increased CO2 Concentrations Induce nhaS3 Expression
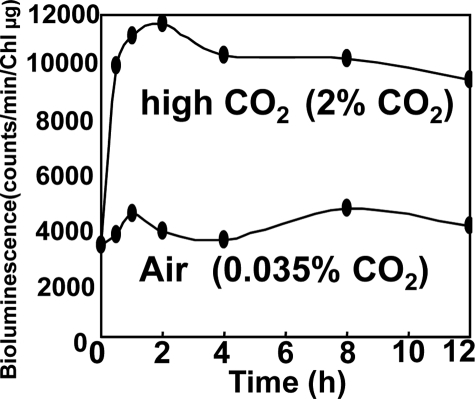
The proton gradient across the thylakoid membrane generated by the electron transfer machinery during respiration and photosynthesis may be influenced by NhaS3-mediated H+ transport activities and vice versa. Therefore the effect of CO2 concentration on the expression of nhaS3 was examined. Cells carrying a reporter construct consisting of the luciferase gene under control of the NhaS3 promoter (PnhaS3::luxAB) were grown under low CO2 conditions (0.035% CO2 in air), transferred to fresh medium, and incubated under high CO2 conditions (2% in air). Luciferase activity was monitored during growth at high CO2 concentrations and compared with the activity in cells grown at low CO2 concentrations. Luciferase activity was increased by more than 2.5-fold after transfer of the cells to high CO2 conditions (Fig. 7). These results suggest that nhaS3 expression is induced in response to increased CO2 concentrations.
FIGURE 7.
Induction of nhaS3 expression under high CO2 conditions. Synechocystis cells containing a luciferase reporter gene under control of the nhaS3 promoter (PnhaS3::luxAB) were grown under low CO2 conditions (0.035% CO2 in air) and transferred to high CO2 conditions (2% CO2 in air). Control cells were left at low CO2 conditions (0.035% in air). The time scale represents the time after transfer. At the times indicated the luciferase activity was determined.
Expression of Nha3S Is Controlled by the Circadian Clock in Synechocystis
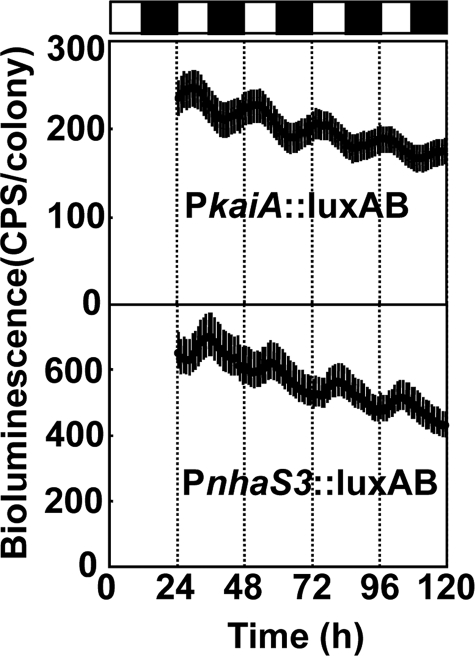
In cyanobacteria, genes involved in respiration, photosynthesis, carbohydrate synthesis, cell division, and nitrogen fixation are under circadian control (38), and their expression changes during light and dark cycles. The same promoter-luciferase construct used above (PnhaS3::luxAB) was used to determine whether expression of nhaS3 is regulated by the circadian clock. Synechocystis cells were entrained by a 12-h dark period and then placed under continuous light (24–26). Luciferase activity was determined every hour (Fig. 8). Cells expressing luciferase driven by the kaiA promoter were used as controls. The circadian period (wave length of the cosine curve) of the cells containing PnhaS3::luxAB was 22.8 h, which is very close to the standard circadian period (22.4 h) of Synechocystis 6803 (39) and also to the period length of kaiA expression (22.9 h). The peak of the expression of nhaS3 was at 12.5 h, which corresponds to the beginning of subjective night, whereas for kaiA the peak of expression was at 6.4 h (corresponding to the middle of the day). It has been shown before that the expression of most cycling genes related to respiration peaks around the time of transition from subjective day to subjective night (39). Subjective early night is also the time when the intracellular CO2 concentration increases because of the lack of photosynthesis. Hence, the result that nhaS3 expression peaks at the beginning of subjective night (Fig. 8) is consistent with the fact that nhaS3 expression increases under high CO2 conditions (Fig. 7). It also suggests that NhaS3 may have a role in mediating the conversion of photosynthetic products and in the supply of energy in the dark.
FIGURE 8.
Circadian rhythm of nhaS3 expression. Synechocystis cells containing a luciferase reporter gene under control of the nhaS3 promoter (PnhaS3::luxAB) were entrained by a 12-h dark period and then shifted to continuous light. The PkaiA::luxAB reporter strain, which shows circadian bioluminescence rhythms,4 was used as control. The time scale represents the actual time after transfer to continuous light conditions. The bioluminescence was determined every hour. Each point indicates the average ± standard deviation from seven replicates. The white and black boxes above the graphs represent subjective day and night, respectively.
DISCUSSION
Although the identity of the Na+/H+ antiporter in the thylakoid membrane of cyanobacteria or of chloroplasts in plant cells had been unknown, our results presented here suggest that NhaS3 functions as a Na+/H+ antiporter in the thylakoid membrane (Fig. 2). These findings account for the inability of E. coli nhaA to replace NhaS3 despite the fact that NhaS3 showed typical Na+/H+ antiporter activity like NhaA (Fig. 4); because NhaA is localized to the plasma membrane in E. coli, it is likely also to be targeted to the plasma membrane in Synechocystis. The amino acid alignment of Na+/H+ antiporters (supplemental Fig. S1) shows that NhaS3 contains a unique sequence, GGLAPEDSLVIQLLMGSADLSPEAAQSVFSAQSEV, in its N terminus. Interestingly, this sequence is not found in the homologous Na+/H+ antiporter in the cyanobacterium Gloeobacter violaceus, which lacks a thylakoid membrane or in other Synechocystis Na+/H+ antiporters. The amino acid sequence of NhaS3 shows high similarity to that of the Gloeobacter Na+/H+ antiporter (40% identity). It is possible that the unique sequence in NhaS3 acts as a signal for targeting to the thylakoid membrane. We therefore constructed a plasmid containing an hemagglutinin epitope-tagged NhaS3 lacking the putative targeting sequence. At this time we have not yet obtained cells expressing this construct. The role of the putative targeting sequence therefore remains to be elucidated.
The finding that nhaS3 is an essential gene in Synechocystis (Fig. 1) supports the interpretation that NhaS3 plays a crucial role in cytosolic and luminal pH regulation or ion homeostasis. The nhaS3 knockdown mutant showed increased sensitivity to elevated Na+ concentrations in the medium (Fig. 5). The most likely transport function for NhaS3 is to sequester Na+ into the thylakoid lumen by utilizing the transmembrane pH gradient and thereby alleviating the toxic effect of sodium on carbon fixation in the cytoplasm (9, 10). It has been reported that the Na+/H+ antiporter sequesters Na+ into the vacuole from the cytosol to avoid Na+ inhibition of the metabolism taking place in the cytosol or to partially substitute for K+ or to balance the osmolarity (40–42). When Na+ increases to toxic levels or Synechocystis cells encounter Na+ stress, NhaS3-mediated accumulation of Na+ in the thylakoid lumen would result in the inactivation of the oxygen evolving activity of photosystem II to prevent over-reduction of the photosynthetic electron transport chain and formation of active oxygen, while dissipating the pH gradient across the thylakoid membrane (9, 10, 43). NhaS3 may function as a putative uncoupler of the electrochemical proton gradient generated by photosynthesis.
Arabidopsis thaliana has 8 genes belonging to the NHX family encoding Na+/H+ antiporters and 28 genes belonging to the CHX family encoding cation/H+ antiporters. Bioinformatic analyses revealed that many AtCHX genes including AtCHX23, which is one of the closest homologs of NhaS3, are preferentially expressed in pollen, suggesting a potential role of AtCHX in pollen function (44). Song et al. (45) reported that AtCHX23 is localized in the chloroplast envelope. They also reported that RNA interference inhibition of AtCHX23 led to physiological malfunction and morphological changes in the chloroplasts. The essentiality of NhaS3 in Synechocystis may reflect a physiological function of members of the AtCHX family in vegetative tissues in Arabidopsis.
It has been suggested that Na+ is an essential element for cell division, photosynthesis, and pH regulation in cyanobacteria (11, 46). Consistent with this, it has been shown that Na+ is required for some types of membrane transport proteins for the uptake of K+ or bicarbonate into Synechocystis cells (16, 47, 48). Moreover, one of the six Na+/H+ transporters in Synechocystis, NhaS4, was reported to function as a Na+ uptake system, most likely located in the plasma membrane (7). NhaS3-mediated thylakoid membrane ion transport may participate in the intracellular circulation of Na+.
In the light, a relatively high proton gradient occurs across the thylakoid membrane, and in the dark, this difference in pH between cytosol and thylakoid lumen is decreased (49, 50). The transition between these two states is achieved by proton diffusion across the thylakoid membrane, which is accompanied by a counter movement of cations to balance both osmolarity and electrochemical gradient. The dissipation of transmembrane pH gradients upon addition of Na+ was demonstrated by expression in E. coli (Fig. 3A). NhaS3 did allow E. coli strain LB2003 to grow on low K+, because of the K+ uptake activity or likely through an indirect mechanism (Fig. 3, B and C). This ability to control the transport of H+, Na+, and K+ indicates that NhaS3 has the potential to control thylakoid membrane transport, which leads to maintenance of ion homeostasis in the cytoplasm and the thylakoid lumen.
Expression of nhaS3 was up-regulated by shifting the cells from low CO2 to high CO2 (Fig. 7), and nhaS3 expression peaked at the beginning of subjective night (Fig. 8). Because of special limitations in the unicellular bacterium Synechocystis, each physiological process, such as light-dependent reaction of photosynthesis, carbon fixation (i.e. light-independent reaction of photosynthesis), respiration, or nitrogen fixation occurs at a different phase in the cell division cycle (51). In the dark, nitrogenase activity for nitrogen fixation increases and glucose is consumed by conversion into other metabolites (51). The main role of the circadian clock in Synechocystis seems to be to adjust the physiological state of the cell to the upcoming dark period (39). Consistent with this hypothesis, the respiration-related genes in cyanobacteria show peak expression at the beginning of subjective night, which probably helps provide energy and carbon source during the night (39, 51). It can be anticipated that NhaS3 may influence thylakoid membrane transport of photosynthetic products and/or energy conversion by regulating the electrochemical potential across the thylakoid membrane in the dark.
Supplementary Material
Acknowledgments
We thank Eva-Mari Aro (University of Turku, Turku, Finland) and Teruo Ogawa (Nagoya University, Nagoya, Japan) for generously supplying antibodies for NdhD3 and NdhF3.
This work was supported by Grants-in-aid for Scientific Research 17078005 and 20-08103 (to N. U.) from MEXT and JSPS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 .
K. Onai and M. Ishiura, unpublished observations.
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid.
REFERENCES
- 1.Padan E., Schuldiner S. ( 1994) Biochim. Biophys. Acta 1185, 129– 151 [DOI] [PubMed] [Google Scholar]
- 2.Padan E., Schuldiner S. ( 1994) Biochim. Biophys. Acta 1187, 206– 210 [DOI] [PubMed] [Google Scholar]
- 3.Hunte C., Screpanti E., Venturi M., Rimon A., Padan E., Michel H. ( 2005) Nature 435, 1197– 1202 [DOI] [PubMed] [Google Scholar]
- 4.Ohyama T., Igarashi K., Kobayashi H. ( 1994) J. Bacteriol. 176, 4311– 4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamada A., Hibino T., Nakamura T., Takabe T. ( 2001) Plant Physiol. 125, 437– 446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inaba M., Sakamoto A., Murata N. ( 2001) J. Bacteriol. 183, 1376– 1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikkat S., Milkowski C., Hagemann M. ( 2000) Plant Cell Environ. 23, 549– 559 [Google Scholar]
- 8.Elanskaya I. V., Karandashova I. V., Bogachev A. V., Hagemann M. ( 2002) Biochemistry 67, 432– 440 [DOI] [PubMed] [Google Scholar]
- 9.Allakhverdiev S. I., Nishiyama Y., Suzuki I., Tasaka Y., Murata N. ( 1999) Proc. Natl. Acad. Sci. U. S. A. 96, 5862– 5867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allakhverdiev S. I., Sakamoto A., Nishiyama Y., Inaba M., Murata N. ( 2000) Plant Physiol. 123, 1047– 1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller A. G., Turpin D. H., Canvin D. T. ( 1984) J. Bacteriol. 159, 100– 106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan A., Scherer S., Lerner M. ( 1989) Plant Physiol. 89, 1220– 1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y. ( 1979) J. Gen. Microbiol. 111, 1– 61 [Google Scholar]
- 14.Buurman E. T., Kim K. T., Epstein W. ( 1995) J. Biol. Chem. 270, 6678– 6685 [DOI] [PubMed] [Google Scholar]
- 15.Stumpe S., Bakker E. P. ( 1997) Arch. Microbiol. 167, 126– 136 [PubMed] [Google Scholar]
- 16.Matsuda N., Kobayashi H., Katoh H., Ogawa T., Futatsugi L., Nakamura T., Bakker E. P., Uozumi N. ( 2004) J. Biol. Chem. 279, 54952– 54962 [DOI] [PubMed] [Google Scholar]
- 17.Uozumi N., Nakamura T., Schroeder J. I., Muto S. ( 1998) Proc. Natl. Acad. Sci. U. S. A. 95, 9773– 9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katoh H., Hagino N., Grossman A. R., Ogawa T. ( 2001) J. Bacteriol. 183, 2779– 2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norling B., Zak E., Andersson B., Pakrasi H. ( 1998) FEBS Lett. 436, 189– 192 [DOI] [PubMed] [Google Scholar]
- 20.Omata T. ( 1995) Plant Cell Physiol. 36, 207– 213 [DOI] [PubMed] [Google Scholar]
- 21.Zhang P., Battchikova N., Jansen T., Appel J., Ogawa T., Aro E. M. ( 2004) Plant Cell 16, 3326– 3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toyooka K., Moriyasu Y., Goto Y., Takeuchi M., Fukuda H., Matsuoka K. ( 2006) Autophagy 2, 96– 106 [DOI] [PubMed] [Google Scholar]
- 23.Kucho K., Aoki K., Itoh S., Ishiura M. ( 2005) Genes Genet. Syst. 80, 19– 23 [DOI] [PubMed] [Google Scholar]
- 24.Onai K., Morishita M., Itoh S., Okamoto K., Ishiura M. ( 2004) J. Bacteriol. 186, 4972– 4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto K., Onai K., Furusawa T., Ishiura M. ( 2005) Plant Cell Environ. 28, 1305– 1315 [Google Scholar]
- 26.Okamoto K., Onai K., Ishiura M. ( 2005) Anal. Biochem. 340, 193– 200 [DOI] [PubMed] [Google Scholar]
- 27.Kuroda T., Shimamoto T., Inaba K., Tsuda M., Tsuchiya T. ( 1994) J. Biochem. 116, 1030– 1038 [DOI] [PubMed] [Google Scholar]
- 28.Goldberg E. B., Arbel T., Chen J., Karpel R., Mackie G. A., Schuldiner S., Padan E. ( 1987) Proc. Natl. Acad. Sci. U. S. A. 84, 2615– 2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karpel R., Olami Y., Taglicht D., Schuldiner S., Padan E. ( 1988) J. Biol. Chem. 263, 10408– 10414 [PubMed] [Google Scholar]
- 30.Ohkawa H., Price G. D., Badger M. R., Ogawa T. ( 2000) J. Bacteriol. 182, 2591– 2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shijuku T., Saito H., Kakegawa T., Kobayashi H. ( 2001) Biochim. Biophys. Acta 1506, 212– 217 [DOI] [PubMed] [Google Scholar]
- 32.Shijuku T., Yamashino T., Ohashi H., Saito H., Kakegawa T., Ohta M., Kobayashi H. ( 2002) Biochim. Biophys. Acta 1556, 142– 148 [DOI] [PubMed] [Google Scholar]
- 33.Inoue H., Sakurai T., Ujike S., Tsuchiya T., Murakami H., Kanazawa H. ( 1999) FEBS Lett. 443, 11– 16 [DOI] [PubMed] [Google Scholar]
- 34.Serrano R., Rodriguez-Navarro A. ( 2001) Curr. Opin. Cell Biol. 13, 399– 404 [DOI] [PubMed] [Google Scholar]
- 35.Shi H., Quintero F. J., Pardo J. M., Zhu J. K. ( 2002) Plant Cell 14, 465– 477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venema K., Belver A., Marin-Manzano M. C., Rodríguez-Rosales M. P., Donaire J. P. ( 2003) J. Biol. Chem. 278, 22453– 22459 [DOI] [PubMed] [Google Scholar]
- 37.Yamaguchi T., Apse M. P., Shi H., Blumwald E. ( 2003) Proc. Natl. Acad. Sci. U. S. A. 100, 12510– 12515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Golden S. S., Ishiura M., Johnson C. H., Kondo T. ( 1997) Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 327– 354 [DOI] [PubMed] [Google Scholar]
- 39.Kucho K., Okamoto K., Tsuchiya Y., Nomura S., Nango M., Kanehisa M., Ishiura M. ( 2005) J. Bacteriol. 187, 2190– 2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apse M. P., Aharon G. S., Snedden W. A., Blumwald E. ( 1999) Science 285, 1256– 1258 [DOI] [PubMed] [Google Scholar]
- 41.Zhang H. X., Blumwald E. ( 2001) Nat. Biotechnol. 19, 765– 768 [DOI] [PubMed] [Google Scholar]
- 42.Apse M. P., Sottosanto J. B., Blumwald E. ( 2003) Plant J. 36, 229– 239 [DOI] [PubMed] [Google Scholar]
- 43.Nowaczyk M. M., Hebeler R., Schlodder E., Meyer H. E., Warscheid B., Rögner M. ( 2006) Plant Cell 18, 3121– 3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sze H., Padmanaban S., Cellier F., Honys D., Cheng N. H., Bock K. W., Conéjéro G., Li X., Twell D., Ward J. M., Hirschi K. D. ( 2004) Plant Physiol. 136, 2532– 2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song C. P., Guo Y., Qiu Q., Lambert G., Galbraith D. W., Jagendorf A., Zhu J. K. ( 2004) Proc. Natl. Acad. Sci. U. S. A. 101, 10211– 10216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J. D., Brand J. J. ( 1988) Arch. Biochem. Biophys 264, 657– 664 [DOI] [PubMed] [Google Scholar]
- 47.Shibata M., Katoh H., Sonoda M., Ohkawa H., Shimoyama M., Fukuzawa H., Kaplan A., Ogawa T. ( 2002) J. Biol. Chem. 277, 18658– 18664 [DOI] [PubMed] [Google Scholar]
- 48.Berry S., Esper B., Karandashova I., Teuber M., Elanskaya I., Rögner M., Hagemann M. ( 2003) FEBS Lett. 548, 53– 58 [DOI] [PubMed] [Google Scholar]
- 49.Jagendorf A. T., Uribe E. ( 1966) Proc. Natl. Acad. Sci. U. S. A. 55, 170– 177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schönknecht G., Neimanis S., Katona E., Gerst U., Heber U. ( 1995) Proc. Natl. Acad. Sci. U. S. A. 92, 12185– 12189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitsui A., Kumazawa S., Takahashi A., Ikemoto H., Cao S., Arai T. ( 1986) Nature 323, 720– 722 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.