Abstract
MicroRNA-155 (miR-155) has been involved in the response to inflammation in macrophages and lymphocytes. Here we show how miR-155 participates in the maturation of human dendritic cells (DC) and modulates pathogen binding by down-regulating DC-specific intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN), after directly targeting the transcription factor PU.1. During the maturation of DCs, miR-155 increases up to 130-fold, whereas PU.1 protein levels decrease accordingly. We establish that human PU.1 is a direct target for miR-155 and localize the target sequence for miR-155 in the 3′-untranslated region of PU.1. Also, overexpression of miR-155 in the THP1 monocytic cell line decreases PU.1 protein levels and DC-SIGN at both the mRNA and protein levels. We prove a link between the down-regulation of PU.1 and reduced transcriptional activity of the DC-SIGN promoter, which is likely to be the basis for its reduced mRNA expression, after miR-155 overexpression. Finally, we show that, by reducing DC-SIGN in the cellular membrane, miR-155 is involved in regulating pathogen binding as dendritic cells exhibited the lower binding capacity for fungi and HIV protein gp-120 when the levels of miR-155 were higher. Thus, our results suggest a mechanism by which miR-155 regulates proteins involved in the cellular immune response against pathogens that could have clinical implications in the way pathogens enter the human organism.
MicroRNAs have emerged as important regulators of key cellular processes. They consist of endogenous small, non-coding RNA molecules of about 19–22 nucleotides in length (1), which regulate mRNAs in a post-transcriptional manner. They bind to the 3′-untranslated regions of their target mRNAs and exert their function in two ways: mainly blocking the translation and also inducing their cleavage in a similar fashion to small interfering RNAs (2). MicroRNAs are initially expressed as long immature pri-microRNAs, which are processed in the nucleus into the precursor pre-microRNAs and finally matured by Dicer in the cytoplasm into the functional 19–22-nucleotide long microRNAs, which are then incorporated into the RNA-induced silencing complex (1).
The role of microRNAs is being intensively studied in many different fields such as fetal development and the immune system. One of the miRNAs that appears to play a particularly important role in the immune system is microRNA-155 (miR-155),3 the expression of which is induced by inflammatory signals such as exposure to antigen, Toll-like receptor ligands, or interferon γ stimulation in T-cells, B-cells, and macrophages, respectively (3, 4).
miR-155 knock-out mice show aberrant immune functions including defective B and T cell immunity and abnormal function of antigen-presenting cells (4, 5). These mutant mice exhibit an imbalance in the immune Th1/Th2 response, with the CD4+ T cells biased toward Th2 differentiation (4). A lack of miR-155 also leads to a failure in production of high-affinity IgG1 antibodies by murine B-cells (28). This effect has been related to its ability to target the transcription factor PU.1, a key transcription factor in human hematopoiesis, restricted to B lymphoid, granulocytic, and monocytic cells (6).
So far, the role of miR-155 in dendritic cell biology has not been studied in depth. Dendritic cells (DCs) are professional antigen presenting cells that have a pivotal role in controlling immune responses, directing them toward immune activation or tolerance (7), orchestrating an efficient and protective immune response. DCs are present as sentinels in peripheral tissues where they capture antigens that will be presented to CD4+ and CD8+ T cells in lymphoid organs. They arise either from myeloid- or lymphoid-derived precursors and exhibit an immature phenotype characterized by a high phagocytic capacity and low expression of co-stimulatory molecules (8). DCs undergo a maturation process after “sensing” pathogen-derived structures through pattern recognition receptors such as Toll-like receptors, exposure to pro-inflammatory cytokines, or after ligation of the surface receptor CD40. Upon maturation, DCs stop taking up antigens and change their pattern of homing receptors, acquiring a phenotype that allows them to migrate into the T cell compartments, where they perform their antigen presenting function. Many transcription factors such as NF-κB and PU.1 are involved in this process, although no microRNA has been implicated so far.
In addition to its role in DC maturation, PU.1 has been implicated in the differentiation of DCs from myeloid precursors. In this process, the balance between PU.1 and MafB determines the phenotype as being a dendritic cell or a macrophage (9). PU.1 controls a number of myeloid genes, and it has been shown to contribute to transcriptional expression of DC-SIGN (CD209) (10). DC-SIGN is a C-type lectin, present in myeloid dendritic cells, that binds a large array of pathogens via mannan- and Lewis oligosaccharides-dependent interactions (11–15). For example, it has been proposed that DC-SIGN binds to HIV-1, facilitating transport of the virus by DCs' migrating into the lymph nodes, thus promoting transinfection of CD4+ lymphocytes (16). Moreover, CD209 mediates transient adhesion contact with T cells through intercellular adhesion molecule 3 recognition, (17), DC transmigration across endothelium via intercellular adhesion molecule 2 interactions (18), and interaction with neutrophils via Mac-1 (19). DC-SIGN is mainly expressed in DCs and macrophages activated by interleukin-4, and is down-regulated together with PU.1 during maturation of human DCs (10).
In this report, we demonstrate that miR-155 levels increase during maturation of human monocyte-derived dendritic cells after exposure to lipopolysaccharide (LPS). We show that human miR-155 directly targets the 3′-untranslated region (3′-UTR) of PU.1 mRNA and we map the target sequence for miR-155 binding. We also present a stably transfected, inducible cell system using the THP-1 monocytic cell line. This cell line, hence forth named THP1–155, was able to overexpress miR-155 in a regulated fashion, after treatment with doxycycline. In this system, we prove that overexpression of miR-155 elicits protein level down-regulation of PU.1 and subsequently of DC-SIGN mRNA and protein. Furthermore, we demonstrate that miR-155 reduction increased DC-SIGN levels in the membrane of DCs resulting in impaired pathogen binding capacity. Thus, inhibition of miR-155 by synthetic oligonucleotides increased the binding of DCs to both Candida albicans and HIV-1 protein (gp120). Our results show how DC-SIGN, a protein of functional importance in the immune system, is regulated indirectly by miR-155 by direct targeting of the transcription factor PU.1. This has an important physiological role in the pathogen binding ability of dendritic cells, with potentially important effects on the entry and interaction of pathogens such as HIV-1.
EXPERIMENTAL PROCEDURES
Cell Culture
Dendritic cells were generated from human peripheral blood mononuclear cells as previously described (20). Monocytes were cultured for 5–7 days in complete medium (RPMI 1640 medium supplemented with 10% fetal calf serum) with 1000 units/ml granulocyte macrophage-colony stimulating factor and 1000 units/ml interleukin-4 (Immunotools) to obtain immature DCs. To mature these DCs, ultrapure LPS from Escherichia coli 0111:B4 (1 μg/ml) was used. The cell line THP1–155 was cultured in RPMI complete medium. HEK293T and HeLa cells were cultured in Dulbecco's modified Eagle's medium plus 10% fetal calf serum.
Vectors Generated
pCDNA3.1.BIC
The genomic region encompassing miR-155 was amplified and cloned into HindIII/XhoI of the pCDNA3.1 multicloning site. Primers employed were BIC-FOR, AAGCTTTATGCCTCATCCTCTGAGTGC and BIC-REV, CTCGAGACGAAGGTTGAACATCCCAGTGACC.
pLVTHM_BIC
The same fragment as above was first cloned in pSUPER in the HindIII/XhoI sites, from where it was removed using EcoRI and MluI sites, and then subcloned into pLVTHM. pRLTK_WT_3′UTR_PU1 was generated by cloning the 3′-UTR of human PU.1 into XbaI and NotI sites of the pRLTK vector (Promega). PU.1 3′UTR was amplified from genomic DNA by PCR amplification following the protocol established by Ralser et al. (21) using the following primers: 3′-UTR PU.1 FOR2 (TCT AGA TAC GAC TTC AGC GGC GAA GTG CTG) and 3′-UTR PU.1 REV BamHI (GGA TCC GGA TTG AGA ATA ACT TTA CTT G). pRLTK_MUT_ 3′UTR_PU1 was generated by site-directed mutagenesis on pRLTK_WT_3′UTR_PU1. This vector was mutated in the putative miR-155 binding site, using the following primers: 3′UTR_Mut_PU.1 FOR (GCC TCC CCG CTG GCC TGA ATT CGA AGC CCT CGC CCG GCC CGG) and 3′UTR_Mut_PU.1 REV (CCG GGC CGG GCG AGG GCT TCG AAT TCA GGC CAG CGG GGA GGC). Mutagenesis was done using the QuikChange® Site-directed Mutagenesis Kit (Stratagene) and following the manufacturer's instructions.
Transfections
To generate the THP1–155 cell line, HEK293 T cells were transfected with Superfect (Qiagen) following the manufacturer's protocol with 5 μg of pLVTHM_BIC (or pLV/tTR_KRAB_Red in the case of generating the repressor lentiviral particles), 3.75 μg of pPAX2 and 1.5 μg of pMD2G. Supernatant from these cells, containing lentiviral particles, was added to THP-1 cells that were preincubated in the presence of 8 μg/ml of Polybrene (Sigma) for 30 min at 37 °C. Infection was checked 4 days after infection, and positive cells were sorted. All the vectors used in the lentiviral system were kindly provided by Prof. Didier Trono (Ecole Polytechnique Fédérale de Lausanne, Switzerland).
For the luciferase promoter assays, we used the pCD209–468 pXP2 reporter plasmid, which contains the proximal region of the DC-SIGN promoter and pCDNA3.1-PU.1 encoding for full-length PU.1 (10). These plasmids were kindly provided by Prof. A. L. Corbi (Centro de Investigaciones Biologicas, CSIC, Spain). pCDNA3.1_PU.1_3UTR was generated by inserting the 3′-UTR of PU.1 after the coding region of pCDNA3.1 employing a blunt end ligation strategy. THP1–155 cells were electroporated following standard procedures. Electroporation results were normalized by co-transfection with the pRLTK-Renilla luciferase plasmid. To assess direct targeting of miR-155, pRLTK, pRLTK_WT_3′UTR_PU1, or pRLTK_MUT_3′UTR_ PU1 were transfected into HeLa cells employing Superfect (Qiagen) following the manufacturer's instructions. pCDNA3.1.BIC or pCDNA3.1 empty vector was co-transfected. Normalization was achieved with co-transfection of pGL3 (Promega). All luciferase/Renilla luciferase assays were measured employing the Dual-Glo kit from Promega. The experiments were performed three times in triplicates. Statistical differences were determined using Student's t test.
Flow Cytometry
For the pathogen binding experiments, mature DCs were transfected with 100 nm of oligonucleotides anti-miR-155 or anti-miR-Control, and 50 nm Cy3-premiR-control-1 (Ambion) at day 5 of culture. Cy3 fluorescence was used both to assess transfection efficiency and to specifically select the transfected population of DCs. DC-SIGN surface expression was checked by flow cytometry, using 0.3 ng/μl APC-anti-human DC-SIGN antibody or APC Rat IgG2a as Isotype Control (eBiosciences). To perform the binding assays, C. albicans was resuspended in phosphate-buffered saline, inactivated at 90 °C for 20 min, and stained using propidium iodide (1 mg/ml) for 1 h at 4 °C with shaking. Blocking of DC-SIGN was done using 20 μg/ml of AZND1 (Beckman Coulter) or a matched isotype, incubating cells for 20 min at room temperature. Cells were then fixed with CellFix (BD Bioscience) for 30 min at 4 °C, and labeled Candida conidia or gp120-fluorescein isothiocyanate (Trinity Biotech) were added. Binding was performed in binding buffer (20 mm Tris, pH 7.5, 150 mm NaCl, 1 mm CaCl2, 2 mm MgCl2, and 1% bovine serum albumin), after which cells were washed and analyzed by flow cytometry (FACSAria, BD Biosciences). Data were processed with the program FlowJo.
RT and qPCR Analysis
RNA samples were obtained using the TRIzol isolation (Invitrogen) method. Real time PCR using Applied Biosystems TaqMan® MicroRNA Assays was used to detect both mature miR-155 and the housekeeping RNU6B, which was used as normalizing control. These assays were performed following the manufacturer's instructions. Briefly, a stem loop primer was used for reverse transcription of 2 ng of total RNA in two steps (30 min, 16 °C; 30 min, 37 °C), followed by qPCR employing the FAM-TaqMan probe and primers provided. Perfectprobe, from PrimerDesign (Southampton SO15 0DJ), was employed to detect DC-SIGN (For, TGTAGGAATGGTCTGGACTAGG; Rev, CAAGGGGAGAGAGAGGATGG) and PU.1 (For, TGCCCTATGACACGGATCTATA; Rev, GTAATGGTCGCTATGGCTCTC) mRNA.
Western Blotting
Cells were lysed in Nonidet P-40 (1%), 2 mm Pefablock, and 2 μg/ml aprotinin, leupeptin, and pepstatin, at 4 °C for 10 min. 30 μg of cell lysates were subjected to SDS-PAGE under reducing conditions and transferred onto an Immobilon polyvinylidene difluoride membrane (Millipore Corp., Bedford, MA). Protein detection was performed using the SuperSignal West Pico chemiluminescent system (Pierce). Antibodies employed were from Santa Cruz Biotechnology, Inc.: α-PU.1 (sc-352); α-DC-SIGN (sc-20081), and α-tubulin (sc-58667). Quantity One program was used to quantify PU.1 in Fig. 3.
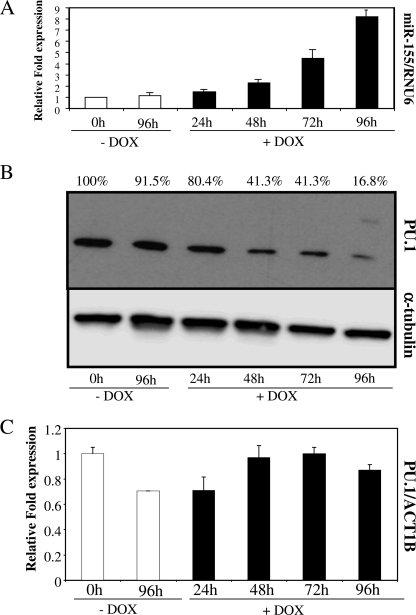
FIGURE 3.
THP1–155 cells overexpressing miR-155 showed down-regulation of PU.1 protein expression. A, THP1–155 cells were treated or not with doxycycline (DOX) to de-repress or not the expression of the miR-155 transgene, respectively. Cells were collected at different time points, and subjected to RNA extraction. RNA from these samples was subjected to specific microRNA RT qPCR, and miR-155 levels were quantified. RNU6B was employed for normalization purposes. B, protein extracts from the same samples were subjected to Western blotting, and PU.1 levels were determined. As a control, α-tubulin was also detected in the same blot. Values represent percentage of PU.1 normalized against α-tubulin and compared with control. C, the same RNA extracts were also used to perform standard RT qPCR, to detect PU.1 mRNA levels. Values were normalized against ACT1B. This figure shows one representative experiment of three. Data in A and C represent mean ± S.D. (error bars). Differences in B (+Dox versus −Dox), not significant.
RESULTS
MiR-155 Increases during Maturation of DCs
DC maturation can be induced by several stimuli such as components of bacteria, viruses, parasites, and cytokines. Lipopolysaccharides, peptidoglycans, flagellin, CpG motifs, and viral nucleic acids induce Toll-like receptor signaling, which triggers dendritic cell maturation. It has been shown previously that most of these inflammatory stimuli up-regulate miR-155 levels in macrophages by activating the NF-κB signaling pathway (3, 22). To determine whether this up-regulation occurs during dendritic cell maturation, we exposed monocyte-derived DC to LPS, which is widely reported to drive DC maturation (23) (supplemental Fig. S1). We then followed the changes in miR-155 levels over time in these cells, by use of reverse transcription followed by stem loop-specific quantitative PCR. During LPS-induced maturation, we could detect increased differential expression of miR-155 by 6 h, whereas a maximum value of 136-fold increase was reached by 48 h; no significant change occurred in the immature DCs over that time (Fig. 1A).
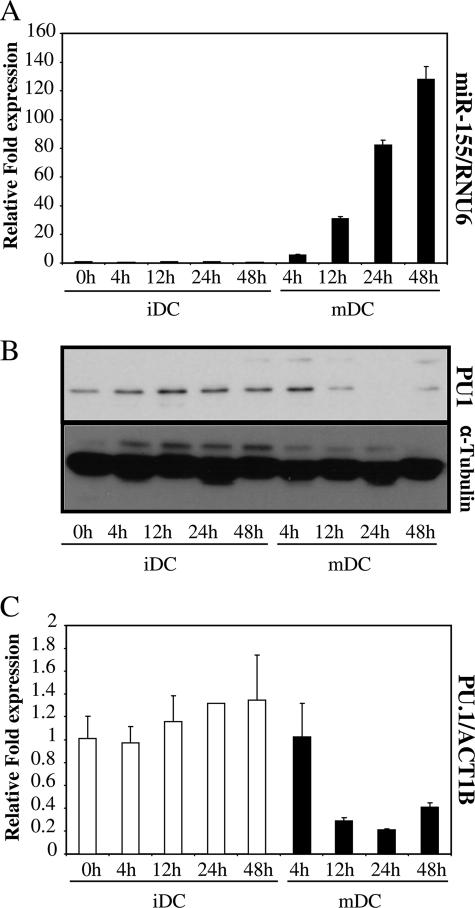
FIGURE 1.
miR-155 increases during maturation of DCs. A, RNA from LPS matured dendritic cells (mDC) and untreated DCs (iDC) was collected at different time points and mature miR-155 was determined by microRNA-specific RT qPCR. Data were normalized with RNU6B. B, protein extracts of the same samples were subjected to Western blot. PU.1 expression was determined and protein concentration controlled with α-tubulin. C, RNA extracted for A was also subjected to standard RT qPCR and PU.1 mRNA levels were quantified, normalizing with ACT1B. Data in A and C represent mean ± S.D. (error bars). These experiments were done on cells from three different donors, with similar results. A representative one is shown.
It has been described that PU.1 protein levels decrease during DC maturation (10). From computer analyses using different programs that predicted PU.1 as a putative target of miR-155 (24, 25), we wondered whether there was a correlation between the increase in miR-155 levels and the pattern of expression of PU.1 over the maturation process. PU.1 protein expression in maturing DCs was assessed by Western blotting, and we observed that PU.1 levels decreased over time; levels were reduced at 12 h, whereas they remained constant in immature DCs over this time (Fig. 1B). Also, the levels of PU.1 mRNA in these cells were determined by qPCR. LPS stimulation also reduced the levels of PU.1 mRNA (Fig. 1C). Thus, miR-155 levels increased during DC maturation, whereas PU.1 protein and mRNA levels decreased. These data suggested that miR-155 might play a role during DC maturation, and it raised the possibility that PU.1 might be targeted by miR-155 during this process.
miR-155 Directly Targets Human PU.1
Using bioinformatic databases, miR-155 was predicted to target human PU.1 (24–26). The core binding sequence (seeding region) for this microRNA in the 3′-UTR of PU.1 has a perfect 9-base Watson-Crick match, above the usual target-microRNA matches, and is also widely conserved across several species. To date, there have been several reports unveiling direct targets of miR-155: angiotensin II, several NF-κB pathway gene transcripts (Ripk1, IKKϵ, and FADD), MAF and, more recently, miR-155 was demonstrated to target the 3′-UTR of murine PU.1 (4, 22, 27, 28).
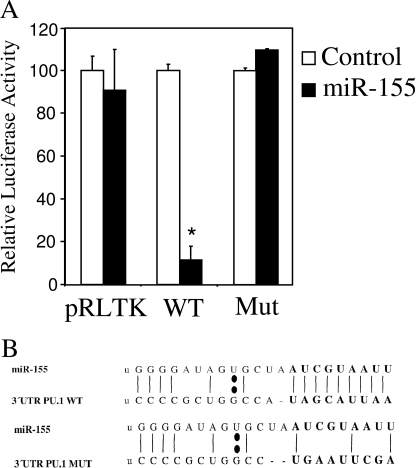
To prove, for the first time, a direct link between miR-155 and human PU.1, we performed luciferase reporter assays in HeLa cells. For this assay, we generated a reporter construct that expressed a fusion protein between the Renilla luciferase mRNA and the 3′-UTR of PU.1 (pRLTK-WT-PU.1). Also, we generated an alternative construct in which the predicted seeding region for miR-155 in the 3′-UTR of PU.1 was mutated (pRLTK-MUT-PU.1) (shown in Fig. 2B). This mutation was decided upon by the abrogation of the match between miR-155 and the 3′-UTR of PU.1, as predicted by the RNA-Hybrid program (29). HeLa cells were co-transfected with a plasmid that expressed miR-155 and one of the Renilla luciferase fusion plasmids described, or the empty vector. It was found that co-transfection of the plasmid expressing miR-155 reduced the activity of the wild type 3′-UTR of the PU.1 reporter by ∼85%. The reporters that did not contain the seeding sequence for miR-155, both control and mutant exhibited no significant reduction in their Renilla luciferase activity, when co-transfected with the miR-155 expressing vector (Fig. 2A).
FIGURE 2.
MiR-155 targets PU.1 3′-UTR. Dual luciferase assay was performed on HeLa cells transfected with pGL3 (normalizing control) and pRLTK (Control), pRLTK_WT_3′UTR_PU1 (WT), or pRLTK_MUT_3′UTR_PU1 (Mut). These plasmids were co-transfected with pCDNA3.1 (Control) or pCDNA3.1-BIC (miR-155). Normalized Renilla luciferase values were represented relative to the control in each case. Data represent mean ± S.D. (error bars). *, statistically significant, ≤0.05.
As a conclusion, our experimental results confirm the target prediction given by different bioinformatic databases and establish that human PU.1 is indeed a direct functional target of miR-155. We also localized the binding region for miR-155 on the 3′-UTR of PU.1, because mutation of the predicted seeding region abrogated the down-regulation exerted by miR-155.
miR-155-induced Overexpression Down-regulates PU.1 Protein Levels
During DC maturation, after LPS stimulation, there is a correlation between the increase in miR-155 levels and the decrease in PU.1 (Fig. 1). Because of the profound influence of pro-inflammatory stimuli on gene expression, LPS may trigger a number of direct or indirect cellular responses that could lead to a decrease of PU.1 during DC maturation. To dissect whether there is a link between the presence of increased levels of miR-155 and the down-regulation of PU.1, we generated a cellular system in which we could rule out an effect due to other signaling pathways activated by LPS. To achieve this, we employed lentiviral vectors containing a sequence that could be processed into mature miR-155, under the transcriptional control of the Tetracycline TeT repressor motif, based on a Tet-on system (30). We stably transduced THP-1 monocytic cells, thus generating a cell line in which the expression of miR-155 is induced after addition of doxycycline, a tetracycline derivative, to the culture medium. Therefore, this cell line, THP1–155, is able to express miR-155 in the absence of LPS or other inflammatory stimuli. This allowed us to elucidate more clearly the effect of miR-155 on the regulation of PU.1 in a myeloid cell line.
We compared THP1–155 cells treated or untreated with doxycycline over the course of 96 h. Then, we determined the levels of miR-155 during this period, using RT qPCR, as described above. The results showed an increase in the levels of miR-155 (Fig. 3A) over time, reaching a maximum of 8-fold induction 96 h after transgene induction, when compared with the doxycycline untreated, non-induced control.
We then interrogated the system for the expression of PU.1 to establish a link between the expression of miR-155 and the previously observed reduction in PU.1 protein and mRNA levels in DCs (Fig. 1). We performed PU.1 protein detection by Western blot and, as expected, when miR-155 was overexpressed following doxycycline treatment, the levels of PU.1 were clearly reduced, reaching the minimum expression in the doxycycline-treated THP1–155 after 96 h (Fig. 3B). PU.1 protein levels were reduced by approximately 85% at this time point, which correlated with the maximum value of miR-155, an 8-fold induction, both compared with time 0 h. To learn more about the mechanism of action of miR-155 on the regulation of PU.1, we determined the mRNA levels of PU.1 over the same time course. We quantified the expression of PU.1 mRNA by RT qPCR, comparing doxycycline-treated with untreated THP1–155 cells. Fig. 3C shows that the increase of miR-155 in THP1–155 cells does not affect the expression levels of PU.1 mRNA. This result suggests that miR-155 might be blocking the translation of PU.1 mRNA, a common mechanism of action of microRNAs.
Thus, aspects of DC maturation that are related to miR-155 are mimicked by THP1–155 cells. In this regard, an increase of miR-155 over time results in a decrease in PU.1 protein levels. These data demonstrate a direct link between the expression of miR-155 and the protein expression levels of the transcription factor PU.1.
miR-155 Expression Down-regulates DC-SIGN Levels in THP1–155 Cells
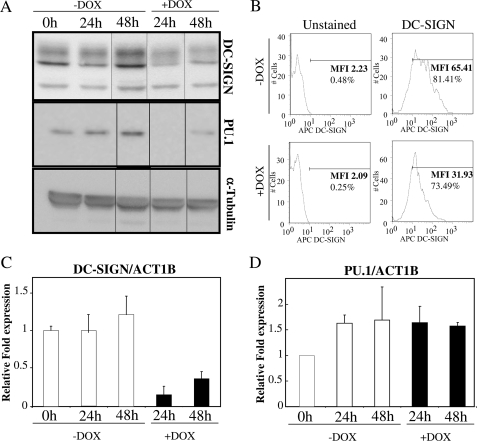
DC-SIGN is a C-type lectin that mediates binding and internalization of viral, bacterial, and fungal pathogens by myeloid dendritic cells. This is an important role of immature DCs, as they act as immune sentinels sampling their surroundings in search for pathogen antigens. Also, it has been reported that DC-SIGN triggers intracellular signals that modulate dendritic cell maturation (31). DC-SIGN is reported to be down-regulated during the maturation of dendritic cells (10). After phagocytosis and activation, mature dendritic cells show a significantly reduced capacity to detect and ingest antigens, which is reflected by down-regulation of DC-SIGN. The transcriptional down-regulation of DC-SIGN in DC maturation seems to depend on the concurrent decrease of PU.1 (10). Taking these previous studies into consideration, we investigated whether miR-155 could be regulating DC-SIGN levels during the maturation process by targeting PU.1.
We used THP1–155 in which we could control miR-155 expression and down-regulate PU.1. It has already been reported that THP-1 cells could be differentiated into dendritic-like cells, sharing some inherent functions such as pathogen binding, T cell stimulation, and DC-SIGN down-regulation after LPS treatment (32). Also, THP1 cells have been shown to overexpress miR-155 in response to LPS treatment (3). To determine whether the presence of miR-155 and the subsequent down-regulation of PU.1 could affect the expression of DC-SIGN, we cultured THP1–155 cells, in the presence or absence of doxycycline, and determined the levels of DC-SIGN protein by Western blot after 24 and 48 h. In concordance with the expected outcome, the induction of miR-155 by doxycycline reduced the levels of DC-SIGN to ∼50% when compared with untreated cells (Fig. 4A, upper panel). This result was further confirmed by flow cytometry, where the membrane expression of DC-SIGN was similarly reduced when cells overexpressed miR-155 (Fig. 4B). DC-SIGN mRNA levels were quantified to elucidate whether the effect of miR-155 was taking place at the mRNA expression level. Cells overexpressing miR-155 expressed lower levels of DC-SIGN mRNA than the untreated control (Fig. 4C). A control experiment was performed to show that this effect was indeed due to miR-155. THP1–155 cells were transfected with oligonucleotide anti-miR-155 (or an irrelevant anti-miR control). This was expected to block the effects of miR-155 overexpression, which would establish that they were due solely to the presence of miR-155. In this experiment (supplemental Fig. S1) we confirmed that we could rescue the expression of DC-SIGN with an antagonist to miR-155. We confirmed in THP-155 cells that when miR-155 was overexpressed PU.1 protein levels were down-regulated, and that PU.1 expression was not affected at the mRNA level (Fig. 4, A, lower panel, and D).
FIGURE 4.
DC-SIGN levels are down-regulated by miR-155 expression in THP1–155 cells. A, THP1–155 cells were treated or not with doxycycline (DOX), to de-repress miR-155 inducible expression, respectively. Cells were collected at different time points and protein extracts subjected to Western blot. DC-SIGN and PU.1 protein expression was determined, as well as α-tubulin, as control. B, these cells were subjected to flow cytometry and DC-SIGN and graphs show percentage of positive population (%) and mean fluorescence intensity (MFI). C and D, RNA from the same samples was subjected to standard RT qPCR. DC-SIGN (C) and PU.1 (D) mRNA levels were quantified and normalized against ACT1B. Shown is one representative experiment of three. Data in C and D represent mean ± S.D. (error bars). Differences in D (+Dox versus −Dox), not significant.
Overall, we have demonstrated that THP1–155 cells are a good model in which to investigate the effect of miR-155 on DC-SIGN and PU.1 regulation. We have shown that, as in maturation of monocyte-derived DCs (10), so in THP-155 cells, an increase in miR-155 correlates with down-regulation of DC-SIGN, at both the mRNA and protein levels.
The Transcriptional Activation of DC-SIGN Promoter Is Regulated by miR-155 via PU.1
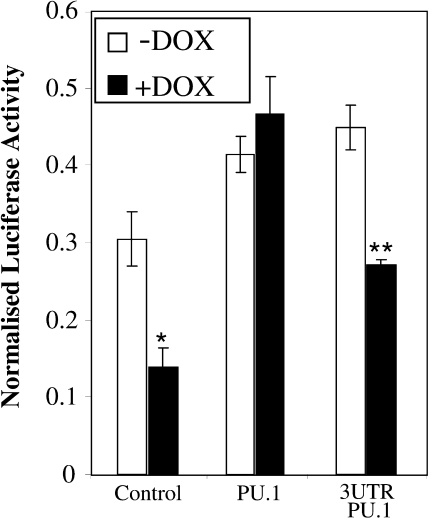
Both PU.1 and DC-SIGN appear to be regulated by miR-155 in THP1–155 cells. However, DC-SIGN mRNA is down-regulated by the presence of miR-155, whereas PU.1 mRNA remains unaltered. These results suggest a different regulatory mechanism for both proteins; whereas DC-SIGN could be affected at the transcriptional level, PU.1 seems to be regulated by miR-155 at the translational level. Interestingly, PU.1 has been shown to regulate DC-SIGN expression through binding of two motifs in its promoter region, both in dendritic cells and THP1 cells (10). Based on this finding, we hypothesized that miR-155 was affecting DC-SIGN levels indirectly through down-regulation of PU.1 and the subsequent decrease in the transcriptional activity of the DC-SIGN promoter. To test this, we employed a luciferase-based reporter construct in which transcriptional activity is controlled by the proximal DC-SIGN promoter (10). We transfected this construct into THP1–155 cells, which overexpressed miR-155 when treated with doxycycline. The results showed that overexpression of miR-155 reduced the activity of the DC-SIGN-luciferase reporter; the decrease was similar to that found in previous assays of DC-SIGN protein and mRNA levels (Fig. 5). To investigate the role of PU.1, we co-transfected the cells with expression vectors coding for PU.1. In both doxycycline-treated and untreated cells, overexpression of PU.1 induced a similar relative luciferase activity in the DC-SIGN reporters, which was clearly independent of the presence of miR-155. Thus, the miR-155-mediated suppression of the DC-SIGN promoter was effectively rescued by the presence of PU.1. Of note, the PU.1 cDNA sequence cloned into the expression vector does not contain the 3′-UTR of PU.1 and therefore does not contain the target seeding region for miR-155 (Fig. 2). This presumably renders the expressed PU.1 mRNA resistant to the blocking activity of miR-155. Interestingly, co-transfection of a PU.1 construct containing the 3′-UTR was not able to efficiently rescue expression of the DC-SIGN promoter. Altogether, these results indicate that miR-155 regulates the expression of DC-SIGN at the transcriptional level, and that this regulation is related to the 3′-UTR-dependent down-regulation of PU.1.
FIGURE 5.
The transcriptional activity of DC-SIGN promoter decreases when miR-155 is overexpressed. THP1–155 cells were treated or not with doxycycline (DOX) for 96 h, to allow for miR-155 to be expressed or to maintain the repression, respectively. Cells were then transfected with pRLTK-Renilla, as control for transfection, and a reporter containing the proximal promoter of DC-SIGN (pCD209–468 pXP2). These cells were co-transfected with expression vectors pCDNA3.1 (Control), pCDNA3.1_PU.1 (PU.1), or pCDNA3.1_PU.1_3UTR. Luciferase values were determined and normalized against Renilla luciferase. Data represent mean ± S.D. (error bars). *, statistically significant ≤0.05 compared with −Dox control. **, statistically significant (≤0.05) when compared with −Dox/pCDNA3,1_PU.1_3UTR-transfected.
Pathogen Binding Capacity Is Affected by miR-155 Overexpression
Finally, the functional consequences of the expression of miR-155 and subsequent down-regulation of DC-SIGN were investigated. DC-SIGN recognizes pathogens by binding to pathogen-specific carbohydrate residues and is mainly expressed in DCs and alternatively activated macrophages (32). Importantly, the binding activity of DC-SIGN seems to be involved in determining the immune response triggered by the presence of certain pathogens (33, 34). To determine the effect of miR-155 on the expression of DC-SIGN in the membrane of DCs, we transfected mature DCs with anti-miR-155 to inhibit the activity of miR-155. Then, we incubated the DCs transfected with anti-miR-155 or anti-miR-Control with a specific anti-DC-SIGN antibody and analyzed the cells by flow cytometry. Fig. 6A shows that the expression of DC-SIGN on DCs increases when miR-155 is inhibited by anti-miR-155. This result is in accordance with the decrease in total and membrane-expressed protein as well as in the mRNA expression observed previously in THP1–155 cells, when miR-155 was overexpressed (Fig. 4). A parallel control experiment revealed that transfection with anti-miR-155 significantly reduced the level of miR-155 in LPS-treated DCs (supplemental Fig. S3).
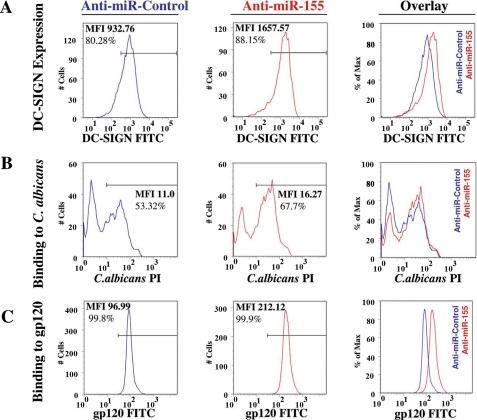
FIGURE 6.
Pathogen binding capacity of DCs is affected by miR-155. DCs were transfected with either oligonucleotide anti-miR-155 or anti-miR-Control, and Cy3 labeled pre-miR-Control. A, DC-SIGN membrane expression in transfected cells was assessed by flow cytometry and presented as graphs showing percentage of positive population (%) and mean fluorescence intensity (MFI). Overlay of both graphs is also shown. Cell number is normalized as percentage of maximum (FlowJo). B, these cells were assayed for their binding capacity to labeled C. albicans conidia. The binding ability of the cells was determined by flow cytometry and presented as in A. C, DCs transfected as previously described were subjected to binding assays employing HIV-1 recombinant gp120-fluorescein isothiocyanate (FITC). Binding capacity was determined through flow cytometry and expressed as in A. The flow cytometry data in this figure correspond to a representative experiment out of three independent repeats. PI, propidium iodide.
We then hypothesized that the increase in DC-SIGN observed in the membrane of DCs when miR-155 levels were reduced, could affect their capacity to bind pathogens. To test this hypothesis, we employed C. albicans as a model pathogen bound by DC-SIGN (13), and performed a pathogen binding assay. The fungi were heat inactivated, labeled with propidium iodide as described under “Experimental Procedures,” and co-incubated with DCs (transfected with anti-miR-155 or anti-miR-Control). After several washes, cells binding labeled C. albicans were analyzed using flow cytometry. DCs transfected with anti-miR-155 showed an increased binding capacity for C. albicans (Fig. 6B) when compared with anti-miR-Control-transfected DCs. Furthermore, blocking DC-SIGN with a specific anti-DC-SIGN antibody reduced binding to C. albicans, suggesting the participation of DC-SIGN in this process (supplemental Fig. S4). We then performed binding experiments with labeled HIV-1 protein gp120. gp120 protein has been shown to bind DC-SIGN, which is crucial in the trans-infection of HIV-1 (16). As expected, the anti-miR-155 augmented the binding capacity of mature DCs for gp120 (Fig. 6C). Taken together, these data show that the increase of miR-155 during DC maturation reduces the capacity of the cells to bind pathogens by DC-SIGN down-regulation.
DISCUSSION
In their normal peripheral location, the main function of DCs is to sample their surroundings, detecting pathogens and foreign molecules. They express a variety of pathogen binding molecules such as DC-SIGN and are actively endocytic. After activation, the DC migrates to the lymph nodes to present the foreign antigen to T cells. During this migration the DC undergoes maturation, down-regulating endocytic activity and pathogen binding molecules, whereas up-regulating molecules such as major histocompatibility complex class II that will be involved in antigen presentation. In this study we have shown that miR-155 regulates the levels of DC-SIGN at the transcriptional level, indirectly, through direct targeting of transcription factor PU.1.
The reciprocal relationship between increased miR-155 levels and reduced PU.1 expression during LPS induced DC maturation (Fig. 1) suggested a link or interaction between miR-155 and PU.1. Searches of computer databases predicted that the recognition sequence for miR-155 was present in the 3′-UTR of PU.1. By use of a reporter assay in which expression of Renilla luciferase was driven by the 3′-UTR of PU.1 (Fig. 2) we have shown that human PU.1 is indeed a direct target for miR-155. A similar observation has been made in murine systems although the sequences differ between the species. Moreover, employing the program RNA Hybrid (29), we show a difference in the predicted secondary structure of the complex of miR-155:PU.1 (supplemental Fig. S5), which is known to be key in microRNA targeting. We also mapped the binding region for miR-155, which coincides with the one predicted using several bioinformatic tools. It consists of a seeding region of 9 bases, which is stronger than those of other targets studied so far, which are in the range of 6–8 bases (4, 27). This region is likely to play an important role in PU.1 regulation in different contexts such as hematopoietic development and myeloid disorders (9, 35–37). Intriguingly, it has recently been demonstrated that miR-155 overexpression is linked to a myeloid disorder (38), which could be related to the ability of miR-155 to target PU.1.
Apart from its role in hematopoietic development, PU.1 plays an important role in dendritic cells, as described in several reports (9, 10, 39). Furthermore, it is well established that PU.1 is able to regulate the levels of DC-SIGN (10). PU.1 is down-regulated during maturation of DCs, and this is associated with reduced levels of DC-SIGN. To investigate a possible link between miR-155 and the maturation-related decrease in DC-SIGN expression, we generated a system in which THP1 cells were able to express miR-155 in an inducible way, isolating its effect from other pathways activated during DC maturation. We chose THP1 cells as they can regulate DC-SIGN and miR-155 in a similar way to DCs (3, 32), and it has been described as a cellular model that shares characteristics with DCs (32). In the newly established cell line, THP1–155, increased levels of miR-155 lead to reduced expression of the PU.1 protein (Fig. 3). This result mimics that in DCs, where PU.1 decreased during maturation, whereas miR-155 increased (Fig. 1). However, there appears to be a difference in the regulation of PU.1 mRNA in maturing DCs and induced THP1–155 cells. In THP1–155, PU.1 mRNA levels are stable (Fig. 3), which suggests that miR-155 is blocking PU.1 mRNA translation, rather than promoting its cleavage. During DC maturation, however, PU.1 mRNA is down-regulated (Fig. 1). A possible explanation could lie in the different levels of miR-155 expression attained by the two cell types (with a similar basal expression of miR-155); the expression of miR-155 in THP1–155 reaches a maximum of 8-fold over the basal levels, whereas mature DCs have ∼130 times more miR-155 than the immature ones. Thus, it is conceivable that when miR-155 reaches certain levels of overexpression, it could promote cleavage of PU.1 mRNA as well as blocking its translation. Other explanations, not exclusive of the previous one, could involve either other microRNAs, or possibly, yet to be described pathways triggered by LPS, which might affect PU.1 mRNA expression at the transcriptional level.
We found that the levels of DC-SIGN mRNA and protein were down-regulated in THP1–155 cells that overexpressed miR-155 (Fig. 4). These results were consistent with our initial hypothesis that DC-SIGN is regulated at the transcriptional level by PU.1, and that down-regulation of PU.1 by miR-155 would lead to a decrease in the transcriptional expression of DC-SIGN. By using a reporter containing the DC-SIGN promoter fused to the luciferase gene, we showed that the promoter activity of DC-SIGN is down-regulated when miR-155 is overexpressed. Furthermore, this down-regulation is rescued when a vector encoding for PU.1 but lacking the 3′-UTR, is co-transfected into the cells (Fig. 5). The experiments shown in demonstrate the indirect effect of miR-155 on the transcriptional regulation of DC-SIGN, through targeting PU.1.
Having shown how, as DCs mature, miR-155 down-regulates expression of PU.1, which in turn results in reduced expression of DC-SIGN, we then predicted that this should result in impaired recognition and binding of pathogens by DCs. Our data (Fig. 6) show that miR-155 levels are correlated with the ability of the cell to bind pathogens (C. albicans and HIV-1 gp120 protein). To test this hypothesis we employed mature DCs and showed that blocking the activity of miR-155 by transfecting an anti-miR-155 oligonucleotide, levels of DC-SIGN were increased and there was augmented binding of pathogens by DCs. Thus, the increase in miR-155 levels has a functional consequence, with an important physiological role in DCs.
The link between miR-155 and the pathogen binding ability of DC-SIGN could have a role in determining the immune response against certain pathogens or in the ability of certain pathogens to infect the humans. It has been reported that SIGNR1 (human DC-SIGN orthologue in mouse) knock-out mice have a more Th1-dominated immune response against Mycobacterium tuberculosis when compared with WT mice, suggesting a role for SIGNR1 in the Th1/Th2 balance of the immune response. In agreement with this, there is also reported evidence for an important role of DC-SIGN during tuberculosis in humans (40). Thus, DC-SIGN-mediated pathogen binding may have important consequences on the immune responses against the tubercle bacilli in the infected host.
The dramatic increase of the levels of miR-155 (up to 130-fold, Fig. 1) during DC maturation, not previously described, adds this process to others regulated by miR-155 (4, 5). Importantly, DCs are implicated in the delicate balance of T cell polarization and the maturing DC in response to different microbial products could have a decisive influence (41). It is tantalizing to hypothesize that the effect of miR-155 on DC-SIGN and the PU.1 expression described here could affect the balance of the immune response against pathogens. Thus, miR-155 in DCs could contribute to driving Th1 polarization, hence playing an important role in the initial steps of infection. In addition to effects on the immune response, miR-155 could have a more obvious impact on the infection process of certain pathogens. Our results could have important consequences in infection by HIV-1, and might suggest a role for miR-155 in making subjects more or less susceptible to infection. Interestingly, there are several studies reporting the possible use of DC-SIGN blocking agents that aim to stop HIV-1 infection (42–45). Thus, miR-155 could be a new therapeutic target that could help prevent entrance of HIV-1 through binding of DC-SIGN.
In conclusion, our study reveals that miR-155 has an important role during DC maturation, inhibiting the expression of the transcription factor PU.1 and thus decreasing the levels of DC-SIGN and the pathogen binding ability of the cells. miR-155 could be of importance in several infectious diseases, and may contribute to susceptibility to infection and invasion by a range of pathogens. Furthermore, our findings suggest an additional explanation for how miR-155 is involved in modulating the Th1/Th2 balance, namely through controlling the maturation of DCs.
Supplementary Material
Acknowledgments
We thank Prof. Angel L. Corbi for being the most helpful critic and Dr. Chris Pickard for reviewing the manuscript.
This work was supported by Wessex Medical Research Grant M14.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- miR-155
- microRNA-155
- UTR
- untranslated region
- DC-SIGN
- dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin
- DC
- dendritic cell
- HIV-1
- human immunodeficiency virus, type 1
- LPS
- lipopolysaccharide
- WT
- wild type
- RT
- reverse transcriptase
- qPCR
- quantitative PCR.
REFERENCES
- 1.Bartel D. P. ( 2004) Cell 116, 281– 297 [DOI] [PubMed] [Google Scholar]
- 2.Stefani G., Slack F. J. ( 2008) Nat. Rev. Mol. Cell Biol. 9, 219– 230 [DOI] [PubMed] [Google Scholar]
- 3.O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. ( 2007) Proc. Natl. Acad. Sci. U. S. A. 104, 1604– 1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez A., Vigorito E., Clare S., Warren M. V., Couttet P., Soond D. R., van Dongen S., Grocock R. J., Das P. P., Miska E. A., Vetrie D., Okkenhaug K., Enright A. J., Dougan G., Turner M., Bradley A. ( 2007) Science 316, 608– 611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thai T. H., Calado D. P., Casola S., Ansel K. M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J. L., Schmidt-Supprian M., Rajewsky N., Yancopoulos G., Rao A., Rajewsky K. ( 2007) Science 316, 604– 608 [DOI] [PubMed] [Google Scholar]
- 6.Fisher R. C., Scott E. W. ( 1998) Stem Cells 16, 25– 37 [DOI] [PubMed] [Google Scholar]
- 7.Banchereau J., Steinman R. M. ( 1998) Nature 392, 245– 252 [DOI] [PubMed] [Google Scholar]
- 8.Wallet M. A., Sen P., Tisch R. ( 2005) Clin. Med. Res. 3, 166– 175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakri Y., Sarrazin S., Mayer U. P., Tillmanns S., Nerlov C., Boned A., Sieweke M. H. ( 2005) Blood 105, 2707– 2716 [DOI] [PubMed] [Google Scholar]
- 10.Domínguez-Soto A., Puig-Kröger A., Vega M. A., Corbí A. L. ( 2005) J. Biol. Chem. 280, 33123– 33131 [DOI] [PubMed] [Google Scholar]
- 11.Serrano-Gómez D., Sierra-Filardi E., Martínez-Nuñez R. T., Caparrós E., Delgado R., Muñoz-Fernández M. A., Abad M. A., Jimenez-Barbero J., Leal M., Corbí A. L. ( 2008) J. Biol. Chem. 283, 3889– 3903 [DOI] [PubMed] [Google Scholar]
- 12.Arrighi J. F., Pion M., Garcia E., Escola J. M., van Kooyk Y., Geijtenbeek T. B., Piguet V. ( 2004) J. Exp. Med. 200, 1279– 1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cambi A., Gijzen K., de Vries J. M., Torensma R., Joosten B., Adema G. J., Netea M. G., Kullberg B. J., Romani L., Figdor C. G. ( 2003) Eur. J. Immunol. 33, 532– 538 [DOI] [PubMed] [Google Scholar]
- 14.Alvarez C. P., Lasala F., Carrillo J., Muñiz O., Corbí A. L., Delgado R. ( 2002) J. Virol. 76, 6841– 6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colmenares M., Puig-Kröger A., Pello O. M., Corbí A. L., Rivas L. ( 2002) J. Biol. Chem. 277, 36766– 36769 [DOI] [PubMed] [Google Scholar]
- 16.Geijtenbeek T. B., Kwon D. S., Torensma R., van Vliet S. J., van Duijnhoven G. C., Middel J., Cornelissen I. L., Nottet H. S., KewalRamani V. N., Littman D. R., Figdor C. G., van Kooyk Y. ( 2000) Cell 100, 587– 597 [DOI] [PubMed] [Google Scholar]
- 17.Geijtenbeek T. B., Torensma R., van Vliet S. J., van Duijnhoven G. C., Adema G. J., van Kooyk Y., Figdor C. G. ( 2000) Cell 100, 575– 585 [DOI] [PubMed] [Google Scholar]
- 18.Geijtenbeek T. B., Krooshoop D. J., Bleijs D. A., van Vliet S. J., van Duijnhoven G. C., Grabovsky V., Alon R., Figdor C. G., van Kooyk Y. ( 2000) Nat. Immunol. 1, 353– 357 [DOI] [PubMed] [Google Scholar]
- 19.van Gisbergen K. P., Sanchez-Hernandez M., Geijtenbeek T. B., van Kooyk Y. ( 2005) J. Exp. Med. 201, 1281– 1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sallusto F., Lanzavecchia A. ( 1994) J. Exp. Med. 179, 1109– 1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ralser M., Querfurth R., Warnatz H. J., Lehrach H., Yaspo M. L., Krobitsch S. ( 2006) Biochem. Biophys. Res. Commun. 347, 747– 751 [DOI] [PubMed] [Google Scholar]
- 22.Tili E., Michaille J. J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., Croce C. M. ( 2007) J. Immunol. 179, 5082– 5089 [DOI] [PubMed] [Google Scholar]
- 23.Cella M., Sallusto F., Lanzavecchia A. ( 1997) Curr. Opin. Immunol. 9, 10– 16 [DOI] [PubMed] [Google Scholar]
- 24.Griffiths-Jones S., Saini H. K., van Dongen S., Enright A. J. ( 2008) Nucleic Acids Res. 36, D154– D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. ( 2003) Cell 115, 787– 798 [DOI] [PubMed] [Google Scholar]
- 26.Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. ( 2007) Nat. Genet. 39, 1278– 1284 [DOI] [PubMed] [Google Scholar]
- 27.Martin M. M., Lee E. J., Buckenberger J. A., Schmittgen T. D., Elton T. S. ( 2006) J. Biol. Chem. 281, 18277– 18284 [DOI] [PubMed] [Google Scholar]
- 28.Vigorito E., Perks K. L., Abreu-Goodger C., Bunting S., Xiang Z., Kohlhaas S., Das P. P., Miska E. A., Rodriguez A., Bradley A., Smith K. G., Rada C., Enright A. J., Toellner K. M., Maclennan I. C., Turner M. ( 2007) Immunity 27, 847– 859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krüger J., Rehmsmeier M. ( 2006) Nucleic Acids Res. 34, W451– 454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulliard Y., Wiznerowicz M., Barde I., Trono D. ( 2006) J. Biol. Chem. 281, 35742– 35746 [DOI] [PubMed] [Google Scholar]
- 31.Caparrós E., Munoz P., Sierra-Filardi E., Serrano-Gómez D., Puig-Kröger A., Rodríguez-Fernández J. L., Mellado M., Sancho J., Zubiaur M., Corbí A. L. ( 2006) Blood 107, 3950– 3958 [DOI] [PubMed] [Google Scholar]
- 32.Puig-Kröger A., Serrano-Gómez D., Caparrós E., Domínguez-Soto A., Relloso M., Colmenares M., Martínez-Muñoz L., Longo N., Sánchez-Sánchez N., Rincon M., Rivas L., Sánchez-Mateos P., Fernández-Ruiz E., Corbí A. L. ( 2004) J. Biol. Chem. 279, 25680– 25688 [DOI] [PubMed] [Google Scholar]
- 33.Lanoue A., Clatworthy M. R., Smith P., Green S., Townsend M. J., Jolin H. E., Smith K. G., Fallon P. G., McKenzie A. N. ( 2004) J. Exp. Med. 200, 1383– 1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wieland C. W., Koppel E. A., den Dunnen J., Florquin S., McKenzie A. N., van Kooyk Y., van der Poll T., Geijtenbeek T. B. ( 2007) Microbes Infect. 9, 134– 141 [DOI] [PubMed] [Google Scholar]
- 35.Dahl R., Walsh J. C., Lancki D., Laslo P., Iyer S. R., Singh H., Simon M. C. ( 2003) Nat. Immunol. 4, 1029– 1036 [DOI] [PubMed] [Google Scholar]
- 36.Anderson K. L., Smith K. A., Conners K., McKercher S. R., Maki R. A., Torbett B. E. ( 1998) Blood 91, 3702– 3710 [PubMed] [Google Scholar]
- 37.Rosenbauer F., Wagner K., Kutok J. L., Iwasaki H., Le Beau M. M., Okuno Y., Akashi K., Fiering S., Tenen D. G. ( 2004) Nat. Genet. 36, 624– 630 [DOI] [PubMed] [Google Scholar]
- 38.O'Connell R. M., Rao D. S., Chaudhuri A. A., Boldin M. P., Taganov K. D., Nicoll J., Paquette R. L., Baltimore D. ( 2008) J. Exp. Med. 205, 585– 594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laiosa C. V., Stadtfeld M., Xie H., de Andres-Aguayo L., Graf T. ( 2006) Immunity. 25, 731– 744 [DOI] [PubMed] [Google Scholar]
- 40.Tailleux L., Pham-Thi N., Bergeron-Lafaurie A., Herrmann J. L., Charles P., Schwartz O., Scheinmann P., Lagrange P. H., de Blic J., Tazi A., Gicquel B., Neyrolles O. ( 2005) PLoS Med. 2, e381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapsenberg M. L. ( 2003) Nat. Rev. Immunol. 3, 984– 993 [DOI] [PubMed] [Google Scholar]
- 42.Spear G. T., Zariffard M. R., Xin J., Saifuddin M. ( 2003) Immunology 110, 80– 85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deiva K., Khiati A., Hery C., Salim H., Leclerc P., Horellou P., Tardieu M. ( 2006) AIDS Res. Hum. Retroviruses 22, 1152– 1161 [DOI] [PubMed] [Google Scholar]
- 44.Rappocciolo G., Piazza P., Fuller C. L., Reinhart T. A., Watkins S. C., Rowe D. T., Jais M., Gupta P., Rinaldo C. R. ( 2006) PLoS Pathog. 2, e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S. K., Liang P. H., Astronomo R. D., Hsu T. L., Hsieh S. L., Burton D. R., Wong C. H. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 3690– 3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.