Abstract
Parasitic protozoa are unable to synthesize purines de novo and must import preformed purine nucleobases or nucleosides from their hosts. Leishmania major expresses two purine nucleobase transporters, LmaNT3 and LmaNT4. Previous studies revealed that at neutral pH, LmaNT3 is a broad specificity, high affinity nucleobase transporter, whereas LmaNT4 mediates the uptake of only adenine. Because LmaNT4 is required for optimal viability of the amastigote stage of the parasite that lives within acidified phagolysomal vesicles of mammalian macrophages, the function of this permease was examined under acidic pH conditions. At acidic pH, LmaNT4 acquires the ability to transport adenine, hypoxanthine, guanine, and xanthine with Km values in the micromolar range, indicating that this transporter is activated at low pH. Thus, LmaNT4 is an acid-activated purine nucleobase transporter that functions optimally under the physiological conditions the parasite is exposed to in the macrophage phagolysosome. In contrast, LmaNT3 functions optimally at neutral pH. Two-electrode voltage clamp experiments performed on LmaNT3 and LmaNT4 expressed in Xenopus oocytes revealed substrate-induced inward directed currents at acidic pH, and application of substrates induced acidification of the oocyte cytosol. These observations imply that LmaNT3 and LmaNT4 are nucleobase/proton symporters.
Leishmania species are parasitic protozoa that infect humans and animals and cause a spectrum of diseases in tropical and subtropical regions of the world (1). The life cycle consists of three principal stages: flagellated promastigotes that colonize the midgut of the sand fly vector, metacyclic promastigotes that live in the mouth parts of the sand fly and are infectious to mammalian hosts, and non-flagellated amastigotes that reside within acidified phagolysosomal vesicles of vertebrate host macrophages.
Like all other parasitic protozoa examined, Leishmania species are unable to synthesize the purine ring de novo and must import purines from both the insect and mammalian hosts (2). Salvage of these essential nutrients is initiated by uptake of nucleobases and nucleosides across the parasite plasma membrane by permeases of the ENT (equilibrative nucleoside transporter) family (SLC29 family). The Leishmania major genome (3) encodes five members of this family, NT1.1, NT1.2, and NT2, which are nucleoside transporters (4, 5), and NT3 and NT4, which are purine nucleobase permeases (6, 7). Previous functional characterization of LmaNT3 (Lma indicates that the transporter is from L. major) revealed that it mediates the uptake of hypoxanthine, adenine, guanine, and xanthine with Km values in the low micromolar range (6). In contrast, at neutral pH, LmaNT4 promotes the uptake of only adenine. Nonetheless, studies with the Δlmant4-null mutant (7) revealed that LmaNT4 was essential for optimal viability of L. major amastigotes in primary murine macrophages. In contrast, deletion of the LmaNT3 genes in the Δlmant3-null mutant was without effect on amastigote viability inside macrophages.
Because amastigotes are exposed to a pH of ∼5 in the macrophage phagolysosome (8), one potential explanation for the compromised viability of Δlmant4 amastigotes is that LmaNT4 operates optimally at acidic pH. In this case, deletion of the LmaNT4 genes would reduce the ability of amastigotes to salvage purines within the phagolysosome and would compromise their viability. If, in contrast, the LmaNT3 permease functions optimally at neutral pH, amastigotes could tolerate the loss of this transporter more easily, explaining the unaltered viability of Δlmant3 amastigotes. To test this possibility, we have studied the function of both LmaNT4 and LmaNT3 at both acidic and neutral pH. Indeed, LmaNT4, but not LmaNT3, is an acid-activated nucleobase transporter, implying that it functions optimally under the physiological conditions of the macrophage phagolysosome.
EXPERIMENTAL PROCEDURES
Parasite Culture
L. major Friedlin VI (MHOM/IL/80Friedlin) parasites were cultured at 26 °C in Medium 199 containing 15% fetal bovine serum. Generation of the Δlmant-null mutant and complementation with the LmaNT3 and LmaNT4 genes have been described previously (7).
Uptake Assays
Uptake of 3H-labeled nucleobases into parasites and Xenopus laevis oocytes were performed as described previously (7). The buffer for measurement of nucleobase uptake in parasites at different pH values was prepared by mixing solutions of 0.1 m Na2HPO4 and 0.1 m NaH2PO4 to obtain the desired pH value. Concentrated parasites in phosphate-buffered saline, pH 7.4, were diluted ∼5-fold into the sodium phosphate buffer and assayed immediately for uptake using radiolabeled ligand resuspended in the same buffer. Substrate saturation curves were fitted to the Michaelis-Menten equation by non-linear regression using Prism 4.0b (GraphPad Software, Inc.).
Electrophysiology and pH Measurements
cRNA transcripts were synthesized from linearized inserts containing the LmaNT3 and LmaNT4 open reading frames subcloned into the pL2.5 vector (9) using the mMESSAGE mMACHINE T7 ultra kit (Ambion). Defolliculated X. laevis oocytes were microinjected with 23 nl (∼10 ng) cRNA and maintained in ND96 buffer (96 mm NaCl, 2.0 mm KCl, 1.8 mm CaCl2, and 5 mm HEPES, pH 7.5) for 3 days at 16 °C. Water-injected oocytes were used as controls. Two-electrode voltage clamp experiments were performed in ND96 buffer at room temperature using GeneClamp 500B (Axon Instruments, Inc.). For measurements performed at pH 5.5, ND96 buffer was supplemented with 40 mm MES3 buffer adjusted to that pH with HCl. Charge-flux ratios were determined by integrating the charge induced by 25 μm [3H]hypoxanthine or [3H]adenine over 5 min and by measuring the radioactivity accumulated by the same isolated oocytes over that time. Background uptake by uninjected oocytes was subtracted.
For fluorescence measurements, oocytes were preloaded with 30 μm BCECF for 30 min in ND96 buffer at room temperature. Fluorescence was measured using a Leica upright microscope equipped with a 20× objective, an Oregon Green Filter cube (exciter, HQ495/30X; dichroic, Q515LP; and emitter, HQ545/50m), and a photodiode (pin-020A, UCT Sensors, Inc.) coupled to an Axopatch 200B patch clamp amplifier (Axon Instruments, Inc.). The objective was focused on the animal pole, and fluorescence was monitored with Clampex 8.2 (Axon Instruments, Inc.).
RESULTS
Activation of LmaNT4 Nucleobase Permease at Acidic pH
Functional characterization of LmaNT3 in Xenopus oocytes (6) established that it transports the purine nucleobases hypoxanthine, adenine, guanine, and xanthine with Km values in the low micromolar range. Furthermore, this transporter is the principal nucleobase permease in promastigotes because deletion of the LmaNT3 genes in the Δlmant3-null mutant abrogated uptake of hypoxanthine and guanine, greatly reduced uptake of xanthine, but left residual transport of adenine (7) that is probably due to the intact LmNT4 transporter gene. Consequently, the Δlmant3-null mutant provided an opportune genetic background in which nucleobase transporters could be expressed from episomal expression vectors and functionally characterized. Subtraction of background uptake of any nucleobase by the Δlmant3-null mutant from the uptake measured for the null mutant complemented with LmaNT4, Δlmant3[pNT4], provided a quantitative measurement of uptake due to LmaNT4 alone. These experiments revealed that LmaNT4 transports adenine with low affinity, being non-saturable at 1 mm adenine but does not transport hypoxanthine, guanine, or xanthine at pH 7.4 (7).
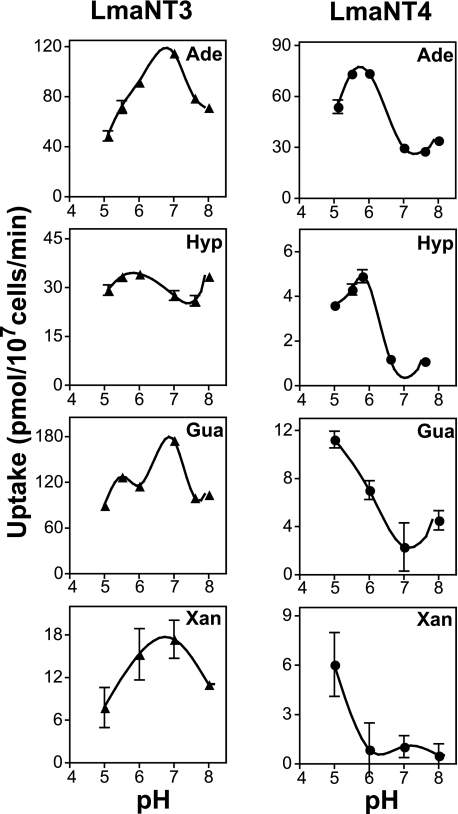
To determine whether LmaNT4 might be a nucleobase permease that is optimized to function in the acid environment that amastigotes experience in the macrophage phagolysosome (8), uptake of 25 μm purine nucleobases by Δlmant3[pNT4] parasites was measured over a range of pH values from 5 to 8 (Fig. 1). Because the concentration of nucleobases in plasma and other extracellular fluids is typically in the low micromolar range (10), uptake properties at this concentration are likely to be relevant to the roles of these permeases during an infection of a mammalian host. Indeed, LmaNT4 exhibited a pH maximum between 5 and 6 for transport of each nucleobase. In contrast, uptake by LmaNT3, measured using the Δlmant3[pNT3] cell line (Fig. 1), revealed maximum uptake activity at pH 7 for uptake of each of the purine nucleobases, except for hypoxanthine, whose uptake was not very responsive to changes in pH.
FIGURE 1.
pH profiles for uptake of nucleobases by LmaNT3 and LmaNT4. Δlmant3-null mutants complemented with the LmaNT3 or LmaNT4 gene on an episomal expression vector were assayed for uptake of each nucleobase (25 μm) over a range of pH values from 5.0 to 8.0. Uptake values for Δlmant3 parasites, measured in parallel, were subtracted from each data point to provide uptake specific for LmaNT3 or LmaNT4 permeases. Data represent the mean ± S.D. (error bars) of triplicate measurements. Hyp, hypoxanthine.
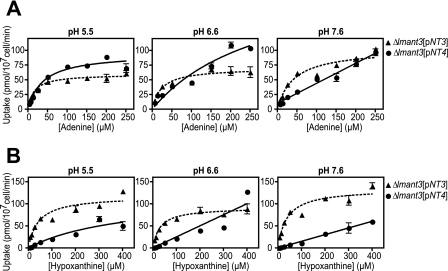
To determine the apparent affinity for uptake of nucleobases, substrate response curves for adenine (Fig. 2A) and hypoxanthine (Fig. 2B) were obtained for LmaNT3 and LmaNT4 at pH 7.6, 6.6, and 5.5. LmaNT3 exhibited Km values in the low micromolar range for both substrates at all three pH values (Table 1). LmaNT4 revealed non-saturable transport for adenine at pH 7.6, but the apparent affinity increased with decreasing pH to exhibit a Km value of ∼40 μm at pH 5.5. For hypoxanthine, transport was non-saturable at pH 7.5 and 6.5, but transport began to saturate at pH 5.5 (estimated Km ∼ 400 μm).
FIGURE 2.
Substrate dependence curves at pH 5.5, 6.6, and 7.6 for uptake of adenine (A) and hypoxanthine (B) by LmaNT3 and LmaNT4. Δlmant3-null mutants complemented with the LmaNT3 (dotted lines) or LmaNT4 (solid lines) gene (Δlmant3[pNT3] and Δlmant3[pNT4], respectively) on an episomal expression vector were assayed for uptake (1-min assays) of [3H]adenine and [3H]hypoxanthine over a range of substrate concentrations. Uptake values for Δlmant3 parasites, measured in parallel, were subtracted from each data point to provide uptake specific for LmaNT3 or LmaNT4 permeases. Data represent the mean ± S.D. (error bars) of triplicate measurements.
TABLE 1.
Estimation ofKm and Vmaxvalues for uptake of adenine and hypoxanthine by LmaNT3 (NT3) and LmaNT4 (NT4). The data inFig. 2 were fitted to the Michaelis-Menten equation by non-linear regression using Prism 4.0b software (GraphPad Software, Inc.). The numerical values forKm and Vmaxare reported as best fit values and “asymptotic” S.E. ((17)). ND indicates that kinetic values were not determined due to non-saturable substrate dependence curves. TheVmax(pmol/107cell/min)/Km(μm) values for data whereKm and Vmaxwere not determined and for hypoxanthine uptake by NT4 at pH 5.5 whereKm and Vmaxvalues exhibited high S.E. were calculated from the slope of the initial linear portion of theVversus[S] curve, where [S] ≪Km and the slope = Vmax/Km.
| Adenine |
Hypoxanthine |
|||||
|---|---|---|---|---|---|---|
| Km | Vmax | Vmax/Km | Km | Vmax | Vmax/Km | |
| μm | pmol/107cell/min | μm | pmol/107cell/min | |||
| NT3 | ||||||
| pH 7.6 | 42 ± 10 | 102 ± 7 | 2.4 | 38 ± 12 | 134 ± 11 | 3.5 |
| pH 6.6 | 24 ± 7 | 70 ± 5 | 2.9 | 31 ± 5 | 92 ± 4 | 2.9 |
| pH 5.5 | 16 ± 3 | 60 ± 2 | 3.7 | 42 ± 16 | 118 ± 13 | 2.8 |
| NT4 | ||||||
| pH 7.6 | ND | ND | 0.35 | ND | ND | 0.15 |
| pH 6.6 | ND | ND | 0.40 | ND | ND | 0.22 |
| pH 5.5 | 39 ± 6 | 94 ± 5 | 2.4 | 420 ± 370 | 119 ± 64 | 0.43 |
These experiments reveal that at 25 μm substrate, LmaNT3 activity is optimal at neutral pH (Fig. 1). In contrast, transport of purine nucleobases by LmaNT4 is strongly activated by acidic pH (Fig. 1), and apparent substrate affinity is markedly increased at acidic pH values (Fig. 2 and Table 1). Hence, LmaNT4 functions as an adenine transporter at neutral pH but acquires the ability to transport all four purine nucleobases at acidic pH.
In addition, the Vmax/Km values, a measure of transporter efficiency, were calculated for LmaNT3 and LmaNT4 employing both adenine and hypoxanthine at the three pH values investigated (Table 1). For LmaNT3, transporter efficiency was not strongly dependent upon pH. In contrast, LmaNT4 exhibited an ∼7-fold (adenine) and ∼3-fold (hypoxanthine) increase in transporter efficiency as the pH was lowered from 7.6 to 5.5. This result underscores the observation that transport by LmaNT4 is activated by acidic pH conditions.
Transport of the Drug Allopurinol by LmaNT4 Is Also Activated by Acidic pH
Allopurinol is a hypoxanthine analog that is employed in the treatment of leishmaniasis (11) and is taken up by parasite nucleobase permeases including LmaNT3 (7). To determine whether LmaNT4 could also provide a route for drug uptake in the acidified phagolysosomes in which amastigotes reside, we measured the uptake of [3H]allopurinol by both nucleobase transporters at pH 5.5 and 7.6 (Fig. 3, A and B). At a 1 μm concentration of allopurinol (Fig. 3A), LmaNT3 promoted the uptake of allopurinol at both pH values. In contrast, LmaNT4 did not induce transport of 1 μm allopurinol at pH 7.6 above the level already observed in Δlmant3-null mutants that represent the host cells for expression of LmaNT4. However, LmaNT4 did mediate drug uptake at pH 5.5. LmaNT3 and LmaNT4 promote allopurinol uptake to similar degrees at pH 5.5, at least at early time points, suggesting that drug import in amastigotes may be mediated by both nucleobase permeases. LmaNT3 and LmaNT4 mRNAs are expressed in both promastigotes and amastigotes (7), implying that both permeases are likely to be expressed in amastigotes. However, direct measurements of permease levels in amastigotes would require the development of transporter-specific antibodies.
FIGURE 3.
Uptake of allopurinol by LmaNT3 and LmaNT4 as a function of pH. A, time courses for uptake of 1 μm [3H]allopurinol by Δlmant3-null mutants and by the null mutant complemented with either the LmaNT3 or LmaNT4 gene (Δlmant3[pNT3] and Δlmant3[pNT4], respectively). Uptake data in A and B represent the mean ± S.D. of three independent measurements. B, substrate dependence curves for uptake of [3H]allopurinol by LmaNT3 and LmaNT4 at pH 5.5 and 7.6. Uptake measurements (1 min) were performed on Δlmant3-null mutants, and the mutant was complemented with each permease gene. Values for uptake of Δlmant3 parasites were subtracted from those for the complemented lines to obtain the velocity due to each permease alone. C, sensitivity of promastigotes of wild type L. major, the Δlmant4-null mutant, and the complemented null mutant Δlmant4[pNT4] to allopurinol. Parasites were cultured (initial inoculum of 1 × 106 cells/ml) for 72 h in RPMI 1640 medium, supplemented with 40 mm MES buffer adjusted to pH 5.5 and 20% dialyzed fetal bovine serum, containing 100 μm xanthosine as the sole purine and increasing concentrations of allopurinol. Parasites were counted in triplicate on a hemacytometer grid, and data represent the mean ± S.D.
Measurement of allopurinol uptake revealed that at sufficiently high concentrations (Fig. 3B), both permeases can mediate the uptake of this drug at both pH values. Nonetheless, LmaNT4 transports the drug more efficiently than LmaNT3 at pH 5.5, whereas both transporters exhibit similar velocities of transport at pH 7.6.
To further explore uptake by LmaNT4 of allopurinol at pH 5.5, allopurinol killing curves were obtained for promastigotes of (i) wild type L. major, (ii) the previously generated Δlmant4-null mutant in which both copies of the LmaNT4 gene have been ablated by targeted gene replacement (7), and (iii) the complemented null mutant, Δlmant4[pNT4], in which the LmaNT4 open reading frame is expressed from an episomal expression vector (7). As revealed in Fig. 3C, at pH 5.5 the EC50 for killing of the Δlmant4-null mutant was ∼10-fold higher (115 μm) than that for either the wild type or the complemented null mutant Δlmant4[pNT4] (11 μm and 17 μm, respectively). In contrast, no difference in drug sensitivity was observed at pH 7.6 for the wild type compared with the Δlmant4-null mutant. This result confirms that LmaNT4 plays a significant role in uptake of allopurinol at acidic pH, as sensitivity to the drug depends upon expression of this permease.
To observe killing by allopurinol in vitro, it was necessary to use xanthosine as the sole purine source (Fig. 3C) and to exclude other purines from the medium, such as hypoxanthine or adenine, which have been shown to interfere with allopurinol toxicity in Leishmania species (12). For this reason, allopurinol killing curves were not carried out in macrophages infected with the three strains used above, as allopurinol cytotoxicity would likely be abrogated by the presence of even low concentrations of natural purine nucleobases within the host cells.
Expression of LmaNT3 and LmaNT4 in Xenopus Oocytes Reveals Substrate-dependent Currents in Two-electrode Voltage Clamp Experiments
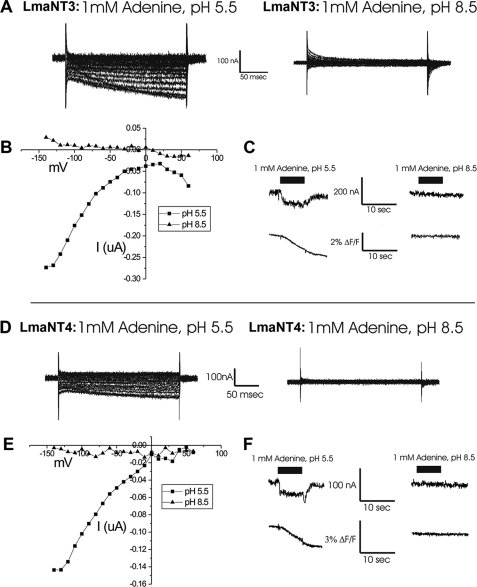
The LdNT1.1, LdNT1.2, and LdNT2 nucleoside transporters from Leishmania donovani are electrogenic proton symporters that exhibit substrate-induced currents when expressed in Xenopus oocytes (13). To determine whether LmaNT3 and LmaNT4 might also be proton symporters and to examine potential proton symport as a function of extracellular pH, both nucleobase permeases were expressed in Xenopus oocytes. Two-electrode voltage clamp experiments performed at pH 5.5 in the presence of 1 mm adenine using oocytes expressing LmaNT3 (Fig. 4A) and LmaNT4 (Fig. 4D) revealed inward directed currents that increased with increasing negative inward polarization (Fig. 4, B and E) for both transporters. In contrast, neither permease exhibited significant currents when monitored at either pH 8.5 (Fig. 4, A, B, D, and E) or at pH 7.5 (data not shown). Hence, currents were strongly pH-sensitive. Furthermore, currents were dependent upon application of the substrate adenine (Fig. 4, C and F) and hence were substrate-induced.
FIGURE 4.
Adenine-dependent currents generated in Xenopus oocytes expressing LmaNT3 or LmaNT4. Oocytes expressing LmaNT3 (A) or LmaNT4 (D) were voltage clamped, and 1 mm adenine was applied at pH 5.5 or 8.5. Voltage steps were applied from a holding potential of −30 mV to command potentials between −140 and +60 mV, and currents obtained in the absence of adenine were subtracted from those applied in the presence of adenine. Current/voltage curves were generated for oocytes expressing LmaNT3 (B) or LmaNT4 (E) at pH 5.5 and 8.5. Data represent recordings from individual oocytes, but curves were representative of those performed on at least three different oocytes with at least four repetitions/oocyte. Oocytes expressing LmaNT3 (C) or LmaNT4 (F) were pre-loaded with the pH-sensitive dye BCECF, clamped at a holding potential of −80 mV, and superfused with 1 mm adenine at pH 5.5 or 8.5. Current (top) and fluorescence (bottom) changes were measured for each oocyte before, during, and after application of adenine.
To determine whether the elicited currents represented inward flux of protons, oocytes were loaded with the pH-sensitive dye BCECF. Upon application of 1 mm adenine at pH 5.5, a negative fluorescence change (ΔF/F) occurred for oocytes expressing either LmaNT3 (Fig. 4C) or LmaNT4 (Fig. 4F), indicating acidification of the oocyte cytosol. At pH 8.5, no such acidification was detected for either permease. These results reveal that inward flux of protons occurs at pH 5.5, where substrate-induced currents are observed, but not at pH 8.5, where no such currents are apparent.
To determine the approximate stoichiometry of proton versus nucleobase flux, oocytes expressing LmaNT3 and LmaNT4 were clamped at a membrane potential of −80 mV and incubated with either 25 μm [3H]adenine or [3H]hypoxanthine for 5 min at pH 5.5. The substrate-induced current flux over this time period was measured. Following current measurements, the same oocytes were lysed, and the pmol of labeled nucleobase taken up was determined by scintillation spectrometry. Comparison of current to nucleobase flux, measured in four oocytes in each case, revealed a stoichiometry for charge/adenine of 4.08 ± 0.4 for LmaNT3 and 7.4 ± 0.3 for LmaNT4 and charge/hypoxanthine of 6.5 ± 0.7 for LmaNT3 and 8.5 ± 0.4 for LmaNT4.
DISCUSSION
Activation of LmaNT4 Transport at Acidic pH
The principal conclusion of the current studies is that the transport activity of the LmaNT4 nucleobase permease is activated at acidic pH. When measured at 25 μm concentrations, this transporter mediates the uptake of only adenine at neutral pH but acquires the ability to take up hypoxanthine, guanine, and xanthine as well at pH 5–6 (Fig. 1). The pH activation is due, at least in part, to increased apparent affinity of LmaNT4 for nucleobases at acidic pH values compared with neutrality (Table 1). Thus, adenine transport by LmaNT4 was non-saturable up to 250 μm substrate at pH 7.6 but exhibited a Km value of ∼40 μm at pH 5.5 (Fig. 2 and Table 1). Similarly, uptake of hypoxanthine by LmaNT4 did not saturate up to 400 μm substrate at pH 7.6 or 6.6 but exhibited an estimated Km value of ∼400 μm at pH 5.5. In contrast to LmaNT4, the related nucleobase permease LmaNT3 had optimal transport activity for all four purine nucleobases at neutral pH (Fig. 1), and apparent affinity for substrates was not strongly pH-dependent (Table 1).
These observations demonstrate a clear functional difference between the two nucleobase permeases of L. major and suggest that LmaNT4 is optimized to function in the low pH environment of the phagolysosome to which the infectious amastigote forms are exposed. This interpretation is consistent with the observation (7) that deletion of the LmaNT4 genes compromises the survival of amastigotes within murine macrophages, whereas deletion of the LmaNT3 genes does not affect amastigote viability.
There are no direct data concerning the concentrations of purine nucleobases within Leishmania-infected phagolysomes. However, nucleobases and nucleosides in plasma and extracellular fluids are typically in the range of 0.4–6 μm (10). This observation suggests that amastigotes are probably exposed to purine nucleobases in the low micromolar range. Hence, the reduction in Km values of LmaNT4 as a function of decreasing pH will make this permease a more efficient carrier for purines under the physiological conditions in which amastigotes likely reside. In contrast, LmaNT4 probably plays a less significant role in purine uptake in promastigotes that reside in conditions closer to neutrality (14). This suggestion is consistent with the observation that LmaNT3 is the principal nucleobase transporter of promastigotes and that the Δlmant3-null mutant exhibits very low levels of purine nucleobase uptake, with the exception of residual adenine transport (7).
It is noteworthy that the human nucleoside transporters hENT3 (15) and hENT4 (16), display a similar activation at acidic pH, exhibiting adenosine transport at pH 5.5 but not at pH 7.4. It has been suggested that the hENT4 permease may play an important role in adenosine uptake under ischemic conditions in the heart, where pH may drop as low as 6.6. Thus, at least three members of the ENT family in organisms as diverse as protozoa and humans exhibit activation at acidic pH values.
Electrogenic Transport by LmaNT3 and LmaNT4
The observation of inward directed substrate-induced currents mediated by LmaNT3 and LmaNT4 (Fig. 4) suggests that these permeases likely function as proton symporters under low pH conditions. These currents increased with increasing negative membrane polarization, as expected for proton-mediated currents. Furthermore, application of substrate to oocytes expressing these permeases resulted in acidification of the cytosol, also indicating an inward flux of protons. The measured stoichiometry of ∼4–8 for charge/nucleobase could have different interpretations. Thus, there may be some component of the current that is directly coupled to nucleobase flux and another that is substrate-induced but not directly coupled. Alternatively, a stoichiometry of >1 for proton flux would generate a strong thermodynamic driving force to concentrate purine nucleobases inside the parasite. Currents were not coupled to Na+ flux, as identical currents were elicited by substrate in buffers containing Na+ or in which Na+ was replaced by choline (data not shown).
The currents mediated by LmaNT3 and LmaNT4 were only observed at pH 5.5 and not at pH 7.5 or 8.5. Thus, the proton transport pathways of both permeases are pH-dependent. These observations suggest that proton symport is likely to occur for both permeases under the acidic conditions of the phagolysosome. In contrast, the high affinity transport by LmaNT3 and lower affinity transport by LmaNT4 at neutral pH appears to be uncoupled from proton flux. Hence, LmaNT3 may function as a symporter at low pH and as a facilitative transporter at neutral pH. In principle, the same shift in mechanism may apply to LmaNT4. However, the reduced transport capacity of LmaNT4 at neutral pH and at physiological concentrations of nucleobases implies that facilitative transport by this permease may not be of biological significance.
This work was supported, in whole or in part, by National Institutes of Health Grants AI44138 (to S. M. L.) and NS051169 (to H. P. L.).
- MES
- 4-morpholineethanesulfonic acid
- BCECF
- 2′,7′-bis(2-carboxyethyl)-5(6)-carboxyfluorescein.
REFERENCES
- 1.Herwaldt B. L. ( 1999) Lancet 354, 1191– 1199 [DOI] [PubMed] [Google Scholar]
- 2.Carter N. S., Rager N., Ullman B. ( 2003) in Molecular and Medical Parasitology ( Marr J. J., Nilsen T., Komuniecki R. eds) pp. 197– 223, Academic Press, London [Google Scholar]
- 3.Ivens A. C., Peacock C. S., Worthey E. A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M. A., Adlem E., Aert R., Anupama A., Apostolou Z., Attipoe P., Bason N., Bauser C., Beck A., Beverley S. M., Bianchettin G., Borzym K., Bothe G., Bruschi C. V., Collins M., Cadag E., Ciarloni L., Clayton C., Coulson R. M., Cronin A., Cruz A. K., Davies R. M., De Gaudenzi J., Dobson D. E., Duesterhoeft A., Fazelina G., Fosker N., Frasch A. C., Fraser A., Fuchs M., Gabel C., Goble A., Goffeau A., Harris D., Hertz-Fowler C., Hilbert H., Horn D., Huang Y., Klages S., Knights A., Kube M., Larke N., Litvin L., Lord A., Louie T., Marra M., Masuy D., Matthews K., Michaeli S., Mottram J. C., Müller-Auer S., Munden H., Nelson S., Norbertczak H., Oliver K., O'neil S., Pentony M., Pohl T. M., Price C., Purnelle B., Quail M. A., Rabbinowitsch E., Reinhardt R., Rieger M., Rinta J., Robben J., Robertson L., Ruiz J. C., Rutter S., Saunders D., Schäfer M., Schein J., Schwartz D. C., Seeger K., Seyler A., Sharp S., Shin H., Sivam D., Squares R., Squares S., Tosato V., Vogt C., Volckaert G., Wambutt R., Warren T., Wedler H., Woodward J., Zhou S., Zimmermann W., Smith D. F., Blackwell J. M., Stuart K. D., Barrell B., Myler P. J. ( 2005) Science 309, 436– 442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasudevan G., Carter N. S., Drew M. E., Beverley S. M., Sanchez M. A., Seyfang A., Ullman B., Landfear S. M. ( 1998) Proc. Natl. Acad. Sci. U. S. A. 95, 9873– 9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter N. S., Drew M. E., Sanchez M., Vasudevan G., Landfear S. M., Ullman B. ( 2000) J. Biol. Chem. 275, 20935– 20941 [DOI] [PubMed] [Google Scholar]
- 6.Sanchez M. A., Tryon R., Pierce S., Vasudevan G., Landfear S. M. ( 2004) Mol. Membr. Biol. 21, 11– 18 [DOI] [PubMed] [Google Scholar]
- 7.Ortiz D., Sanchez M. A., Pierce S., Herrmann T., Kimblin N., Archie Bouwer H. G., Landfear S. M. ( 2007) Mol. Microbiol. 64, 1228– 1243 [DOI] [PubMed] [Google Scholar]
- 8.Antoine J. C., Prina E., Jouanne C., Bongrand P. ( 1990) Infect Immun. 58, 779– 787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arriza J. L., Kavanaugh M. P., Fairman W. A., Wu Y. N., Murdoch G. H., North R. A., Amara S. G. ( 1993) J. Biol. Chem. 268, 15329– 15332 [PubMed] [Google Scholar]
- 10.Traut T. W. ( 1994) Mol. Cell. Biochem. 140, 1– 22 [DOI] [PubMed] [Google Scholar]
- 11.Martinez S., Marr J. J. ( 1992) N. Engl. J. Med. 326, 741– 744 [DOI] [PubMed] [Google Scholar]
- 12.Ullman B. ( 1984) Pharmaceut. Res. 1, 194– 203 [DOI] [PubMed] [Google Scholar]
- 13.Stein A., Vaseduvan G., Carter N. S., Ullman B., Landfear S. M., Kavanaugh M. P. ( 2003) J. Biol. Chem. 278, 35127– 35134 [DOI] [PubMed] [Google Scholar]
- 14.Gontijo N. F., Almeida-Silva S., Costa F. F., Mares-Guia M. L., Williams P., Melo M. N. ( 1998) Exp. Parasitol. 90, 212– 219 [DOI] [PubMed] [Google Scholar]
- 15.Baldwin S. A., Yao S. Y., Hyde R. J., Ng A. M., Foppolo S., Barnes K., Ritzel M. W., Cass C. E., Young J. D. ( 2005) J. Biol. Chem. 280, 15880– 15887 [DOI] [PubMed] [Google Scholar]
- 16.Barnes K., Dobrzynski H., Foppolo S., Beal P. R., Ismat F., Scullion E. R., Sun L., Tellez J., Ritzel M. W., Claycomb W. C., Cass C. E., Young J. D., Billeter-Clark R., Boyett M. R., Baldwin S. A. ( 2006) Circ. Res. 99, 510– 519 [DOI] [PubMed] [Google Scholar]
- 17.Motulsky H. ( 1999) Analyzing Data with GraphPad Prism, GraphPad Software Inc., San Diego, CA [Google Scholar]