Abstract
NKG2D is an important activating receptor for triggering the NK cell cytotoxic activity, although chronic engagement of specific ligands by NKG2D is also known to provoke decreased cell surface expression of the receptor and compromised NK cell function. We have studied the dynamics of surface NKG2D expression and how exposure to the specific ligand major histocompatibility complex class I chain-related molecule B (MICB) affects receptor traffic and fate. While in the NKL cell line and “resting” NK cells NKG2D was found principally at the cell surface, in activated primary NK cells an intracellular pool of receptor could also be found recycling to the plasma membrane. Exposure of NK cells to targets expressing MICB resulted in degradation of ∼50% of total NKG2D protein and lysosomal degradation of the DAP10 adaptor molecule. Consistent with these observations, confocal microscopy experiments demonstrated that DAP10 trafficked to secretory lysosomes in both transfected NKL cells and in activated primary NK cells upon interaction with MICB-expressing target cells. Interestingly, polarization to the synapse of secretory lysosomes containing DAP10 was also observed. The implications of the intracellular traffic of the NKG2D/DAP10 receptor complex for NK cell activation are discussed. We propose that the rapid degradation of NKG2D/DAP10 observed coincident with recruitment of the receptor to the cytotoxic immune synapse may explain the loss of NKG2D receptor expression after chronic exposure to NKG2D ligands.
The killer cell lectin-like receptor NKG2D is one of the best characterized NK3 cell-activating receptors. Signaling via NKG2D depends on its association with DAP10, a transmembrane adaptor molecule containing the sequence YINM, which signals via recruitment of phosphatidylinositol 3-kinase and Grb2 (growth factor receptor-bound protein 2) (1, 2). Effector cell activation mediated by NKG2D has been described as immune recognition of the “induced self,” because the cellular ligands for NKG2D (NKG2D-L): the polymorphic MHC class I chain-related molecules (MIC) A and MICB and the UL16-binding proteins are not normally expressed but instead are up-regulated on target cells after pathogen infection or tumor transformation to render these cells susceptible to NK cell lysis (3). Strikingly, however, although induced expression of NKG2D-L acts as a danger signal to provoke an immune response, a number of studies performed in mouse models have shown that chronic exposure to NKG2D-L can also lead to down-modulation of the surface expression of NKG2D and impaired NK cell cytotoxic function (4–6).
In humans, a common feature of patients with multiple different tumors is the presence in the serum of high levels of soluble MICA and -B or UL16-binding proteins, released by tumor cells, that are associated with an impairment of CTL and NK cell cytotoxic function (7–10). These observations have been interpreted as suggesting that the release of soluble NKG2D-L is a strategy of tumor immune evasion (11). However, recent data show that receptor interaction with cell membrane-anchored MICB can also lead to impaired NKG2D function. We have shown that brief cytotoxic interactions between NK cells and MICB-expressing target cells trigger a synaptic interchange of NKG2D and MICB as well as a rapid down-modulation of surface NKG2D and compromised NK cell cytotoxicity suggesting that NKG2D traffic is rapidly altered upon recognition of MICB expressed on target cell (12).
The surface level of a receptor is dictated by the relative rates of synthesis and transport to the plasma membrane and endocytosis, recycling, and degradation. The loss of cell surface NKG2D observed after NKG2D-L binding (7–10, 12) raises the question of what is the intracellular fate of the receptor on interaction with NKG2D-L. However, the traffic of this receptor has not been previously studied. Here we describe the dynamics of surface NKG2D expression and examine how cytotoxic interactions between NK cells and the MHC class I- 721.221 (here called 221) cells that express MICB (here called 221B) affect the traffic and fate of the NKG2D/DAP10 receptor complex. In NKL cells and resting primary NK cells NKG2D is mainly expressed at the cell surface; however, in activated primary NK cells an intracellular pool of receptor recycling to the cell surface is detected. During cytotoxic interactions the recognition of MICB expressed on target cells results in a rapid degradation of NKG2D/DAP10 that is associated with the traffic of DAP10 to secretory lysosomes (SLs) (13, 14). Our data provide new insights into the dynamics of NKG2D receptor expression in NK cells and suggest a plausible model to explain how chronic exposure to NKG2D-L could lead to NKG2D down-modulation and compromised NK cell function.
EXPERIMENTAL PROCEDURES
Cells and Reagents
The isolation and culture of peripheral blood mononuclear cells, human primary polyclonal NK cells, and the cell lines NKL, 721.221 (221), 221 cells stably transfected with MICB (221B) (12), RPMI-8866, and Daudi were as previously described (12, 15). NKL cells transfected with DAP10-GFP have been described (12). The DAP10(Y85F)-GFP mutant was prepared by PCR (QuikChange mutagenesis kit, Stratagene), using the DAP10-GFP plasmid as template and the oligonucleotides: For-gatggcaaagtcttcatcaacatgccaggc and Rev-gcctggcatgttgatgaagactttgccatc. The integrity of the plasmid construct was verified by DNA sequence analysis. Transfection of the plasmid in NKL cells was performed as previously described (12).
Mouse mAbs specific for 2B4, CD25, CD56 and HLA-A, -B, and -C (fluorescein isothiocyanate), CD56-PE, and CD3-PE/Cy5 were obtained from BD Pharmingen, biotinylated anti-Perforin from Ancell (Bayport, MN), anti-NKG2D (1D11) from Santa Cruz Biotechnology (Santa Cruz, CA), anti-CD71 from Immunotech, anti-EEA1, anti-GFP, and anti-GM130 from BD Biosciences, anti-β actin and IgG1 (MOPC21) from Sigma, anti-CD63 (Dr. F. Sanchez-Madrid, Hospital Universitario de la Princesa, Madrid, Spain), anti-DAP10 mAb (Dr. E. Vivier, Centre d'Immunologie de Marseille-Luminy, Université de la Méditerranée, Marseille, France), and the Fb2 phosphotyrosine-specific mAb (Dr. D. Cantrell, Division of Cell Biology and Immunology, University of Dundee, UK). The rabbit anti-DAP10 was obtained from Santa Cruz Biotechnology. Goat anti-mouse-Ig Alexa 488 and 568 (highly cross-absorbed), Streptavidin-Alexa 488, β-subunit of cholera toxin Alexa 594, and fluorescent trackers chloromethyl derivative of aminocoumarin and carboxyfluorescein diacetate succunimidyl ester were obtained from Molecular Probes (Eugene, OR). Poly-l-lysine, brefeldin A (BFA), and cycloheximide (CHX), epoxomicin, and chloroquine were obtained from Calbiochem. Mouse serum was obtained from Calbiochem (Germany).
FACS Experiments
Mixtures of cells were incubated for the time indicated at 37 °C in 96-well plates (105 NK cells plus 2 × 105 target cells), disrupted with ice-cold phosphate-buffered saline/0.5 mm EDTA, fixed with 4% paraformaldehyde, permeabilized, or not with 0.1% saponin in phosphate-buffered saline/0.1% fetal calf serum and stained as indicated. Target cells were excluded from the analysis by labeling them with carboxyfluorescein diacetate succunimidyl ester following the manufacturer's instructions.
To study the dynamics of cell surface NKG2D, cells were cultured in 96-well plates (in triplicates for each time point) in the presence of either BFA (10 μg/ml) or CHX (50 μg/ml). After the time points indicated in the experiments cells were harvested and stained for the indicated molecules. % of modulation was calculated as follows: (geometric mean fluorescence intensity (gmfi) of treated cells/gmfi of untreated cells) − 1.
Immunoprecipitation, Cell Fractionation, and Western Blot
For immunoprecipitation experiments 20 × 106 NKL (DAP10-GFP) cells alone or mixed with 221 or 221B cells for 5 min were lysed (20 mm Tris.HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 1% Nonidet P-40, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mm NaVO3, and 1 mm NaF) and, after centrifugation to remove nuclei, immunoprecipitated with the FB2 anti-phosphotyrosine mAb coupled to Protein A-Sepharose. Immunoprecipitations were run in 12% SDS-PAGE and analyzed by Western blot for DAP10. NK cells cultured alone or in the presence of 221B target cells were purified by cell sorting, lysed as before, and 100 μg of total protein was resolved in 15% SDS-PAGE and analyzed for DAP10 expression by Western blot. The indicated amounts of chloroquine or epoxomicin were included in the incubations in certain samples.
Cell fractionation was carried out as previously described (16). Briefly, 5 × 107 primary activated NK cells were cultured alone, or mixed with an equal number of 221B cells for 15 min. The cells were then washed three times in cold phosphate-buffered saline, washed once in 50 mm HEPES, pH 7.2, 90 mm KCl, resuspended in 6 ml of cold HEPES/KCl, and disrupted by passage through a ball-bearing cell homogenizer 20 times using a ball bearing 8.006 mm in diameter. Nuclear material was spun out at 2100 rpm for 4 min at 4 °C, and the pellet was washed once in 4 ml of HEPES/KCl buffer to make a final volume of 10 ml. Using a stock of 90 ml of Percoll (Amersham Biosciences) with 10 ml of 500 mm HEPES, pH 7.2, and 900 mm KCl, a Percoll “step” gradient was formed by underlaying 15.9 ml of 39% Percoll with 12.1 ml of 90% Percoll. All dilutions of Percoll were made in 50 mm HEPES, pH 7.2, 90 mm KCl. The soluble cell lysate was then overlaid onto this gradient and centrifuged at 40,000 × g for 30 min at 4 °C. The top 10 ml of soluble lysate that did not enter the gradient were removed and discarded, and 1-ml fractions were collected from the top of the gradient. Samples of each fraction were assayed for hexosaminidase activity: 20-μl samples of each fraction were diluted in 200 mm citric acid, pH 4.5, 0.2% Triton X-100, and incubated in the presence of 8 mm p-nitrophenyl-N-acetyl-β-d-glucosaminide (Sigma) for 30 min at 37 °C. The reaction was stopped by the addition of 1 ml of 67 mm NaCl, 83 mm Na2CO3, and 33 mm glycine, pH 10.5, and the absorbance was measured at 405 nm. The lytic granules corresponded to fractions 23–26 with a peak of hexosaminidase activity of 1, compared with a background of 0.2 absorbance units. Fractions corresponding to SL were made to 0.5% deoxycholic acid, centrifuged to remove Percoll, and then analyzed for expression of DAP10 and CD63 by Western blot.
Confocal Microscopy
Target cells were labeled with chloromethyl derivative of aminocoumarin following the manufacturer's instructions. Conjugates were formed by incubation at 37 °C during 15 min and processed for immunofluorescence as described (12). NKL Cells were loaded with cholera toxin Alexa Fluor 594 by incubation at 37 °C for 15 min (17). Series of optical sections were obtained from the different samples with a Leica DM IRE2 confocal scanning laser microscope and the Leica confocal software. Series of at least 10 confocal sections through the whole cell were analyzed using the ImageJ software (National Institutes of Health) and then Manders' coefficients (R) and two-dimensional co-localization histograms were obtained. Time-lapse experiment procedures are detailed in the video 1 legend.
Cytotoxicity Assays
Cytotoxic assays were performed as described (12).
Electron Microscopy
Cell mixtures (E:T ratio 1:1) centrifuged (1200 rpm 1 min) and incubated 20 min at 37 °C before fixation with 0.1 m PIPES buffer, 2% formaldehyde, and 0.05% glutaraldehyde were processed for electron microscopy as described previously (18), stained with rabbit anti-GFP antibodies, washed twice with Tris-buffered saline, incubated with a goat anti-rabbit immunoglobulins conjugated to 10 nm gold particles (British Biocell, UK), and observed in a Philips CM100 transmission electron microscope (FEI-Philips, Netherlands). In control experiments (not shown), the anti-GFP antibody stained specifically intracellular organelles in cells transfected with sets of markers for specific organelles fused to GFP.4
RESULTS
Dynamics of Surface NKG2D on NK Cells
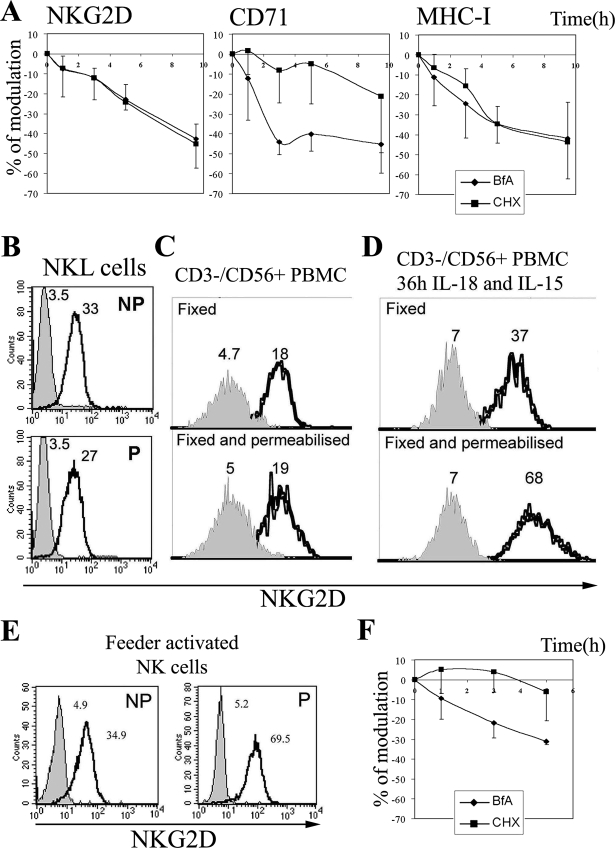
The dynamics of NKG2D expression at the cell surface was first studied in the NKL cell line, which mediates cytotoxic activity mainly via this receptor (12). Treatment of cells with BFA blocks anterograde transport of proteins from the endoplasmic reticulum to the Golgi complex and causes tubulation and fusion of early endosomes with the trans-Golgi network (19). Blockade, in parallel, of protein synthesis with CHX permits discrimination between the effects of BFA on the prevention of endocytic versus endoplasmic reticulum-Golgi transport. In control experiments we verified that the BFA and CHX were working as expected, because surface expression of CD71 (transferrin receptor), which is known to be rapidly recycled (20), was significantly reduced (40–50% from the control) after 3–5 h of BFA treatment, and only some of this reduction was due to prevention of egress of newly synthesized protein, as shown by CHX treatment (Fig. 1A). In contrast, the loss of surface expression of MHC-I over time was due to degradation and replacement with newly synthesized molecules, because the effects of BFA and CHX were comparable. NKG2D surface expression decreased at a similar rate after treatment with either BFA or CHX, suggesting that NKG2D molecules do not recycle between an intracellular compartment and the cell surface in the NKL cell line. FACS analysis and confocal microscopy (not shown) of NKL cells stained for NKG2D confirmed that this receptor was mainly expressed on the cell surface (Fig. 1B). “Resting” NK cells freshly isolated from peripheral blood mononuclear cell also mainly expressed NKG2D on the surface (Fig. 1C); however, after stimulation with IL-15 and IL-18 (Fig. 1D) or exposure to irradiated Daudi and RPMI8866 feeder cells (Fig. 1E) an increase in surface expression of NKG2D, as well as the appearance of intracellular receptor was found. Importantly, treatment with these cytokines did not induce an up-regulation of MICA/B in the peripheral blood mononuclear cells (data not shown) suggesting that these changes in the behavior of the NKG2D receptor were not driven by exposure to ligand. Interestingly, BFA treatment of purified primary polyclonal activated NK cells produced a more rapid down-regulation of NKG2D than after exposure to CHX (Fig. 1F), which suggested that a proportion of NKG2D molecules recycled between the cell surface and intracellular compartments in activated primary NK cells.
FIGURE 1.
Traffic of NKG2D in NK cells. A and F, dynamics of NKG2D, CD71, and MHC-I surface expression. NKL cells (A) or cultured activated NK cells (F) were treated with BFA or CHX and at different time points stained as indicated. The mean ± S.D. of the % of modulation of four experiments is shown. B–E, FACS analysis for NKG2D expression under permeabilizing (P) or non-permeabilizing (NP) conditions. The gray histogram represents the isotype control. Numbers indicate the geometric mean fluorescence intensity (gmfi). Representative experiments, out of five per panel, are shown.
These data indicate that, in the NKL cell line and primary resting NK cells, NKG2D is stably expressed at the cell surface and does not undergo any significant level of recycling after internalization. However priming of NK cells by exposure to feeder cells or cytokines induced NKG2D up-regulation, and the appearance of intracellular receptor accompanied by recycling of NKG2D between the cell surface and intracellular compartments.
MICB Expressed on Target Cells Induces the Rapid Degradation of NKG2D and DAP10
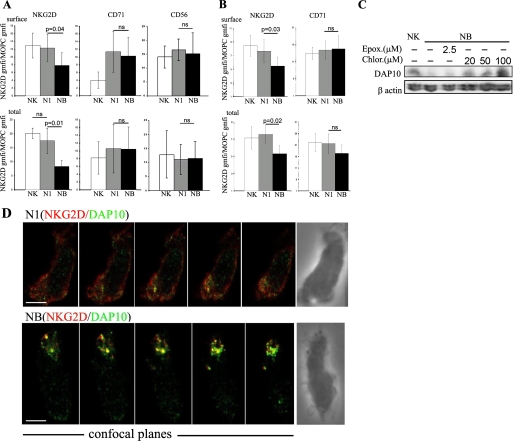
Because activation by exposure to feeder cells was able to induce recycling of the NKG2D receptor to the cell surface (Fig. 1), and NKG2D-mediated cytotoxicity has been correlated with receptor down-modulation and compromised NK cell function (4, 12), it was of interest to study the fate of surface and intracellular NKG2D after binding to the specific ligand MICB. Primary activated NK cells were incubated for 1 h at 37 °C alone, or in the presence of 221 or 221B cells and then analyzed by FACS to assess the levels of surface and total NKG2D (Fig. 2A). As expected, a clear reduction in cell surface NKG2D was found in NK cells cultured with 221B cells (NB samples). Importantly, a marked reduction of around 50% in total cellular NKG2D was also found. A small loss in total NKG2D was also noted in NK cells co-cultured with 221 cells (N1 samples), but these differences were not statistically significant. The levels of CD56 did not change significantly, however, there was an increase in cell surface expression of CD71 on both N1 and NB samples reminiscent of the polarization of the recycling endosomal compartment toward the contact site seen at the T cell synapse (21, 22). Similar results were obtained in experiments done with the NKL cell line except that no significant changes in cell surface expression of CD71 were noted in these cells (Fig. 2B), probably reflecting the lower cytotoxic efficiency mediated by the NKL cell line against N1 samples compared with primary human NK cells. These data indicated that, although treatment with feeder cell lines stimulated recycling of NKG2D between intracellular compartments and the plasma membrane (Fig. 1F), interaction with ligand-bearing target cells was associated with receptor degradation. Interestingly the loss of total cellular NKG2D was more pronounced than the decrease in surface expression, which indicated that traffic of the intracellular pool of NKG2D is altered so that the receptor is delivered for degradation routes upon 221B encounter.
FIGURE 2.
Effect of cytotoxic interactions on NKG2D expression. A and B, FACS analysis for NKG2D protein levels in NK (A) and NKL (B) cells cultured alone or with either 221 (N1) or 221B targets (NB). The mean ± S.D. of the ratio between the gmfi for NKG2D and isotype control staining obtained in seven (A) or five (B) different experiments is shown. Statistical significance was assessed by the Mann Whitney test. p > 0.05 was considered non significant (ns). Target cells, labeled with carboxyfluorescein diacetate succunimidyl ester, were excluded from the analysis. C, degradation of DAP10 upon MICB engagement. NK cells or NK cells purified by cell sorting after 1-h incubation with 221B targets (NB) were analyzed by Western blot for DAP10 expression. The effect of the lysosomal activity inhibitor chloroquine or the proteasome inhibitor epoxomicin was evaluated in the indicated samples. One experiment, out of three performed is shown. D, N1 or NB samples, like in A, were stained for NKG2D and DAP10. Series of confocal sections through the z axel of the merged channels and the differential interference contrast images (right panels) are shown. Scale bars, 2.5 μm.
NKG2D and the signaling adaptor DAP10 form a functional receptor complex for NK cell activation (1, 23). Therefore, we next studied if the adaptor molecule DAP10 was also degraded after cytotoxic interactions with 221B cells. Cell lysates of NK cells, purified by cell sorting after culture with 221B cells, were analyzed by Western blot for DAP10 expression. As shown in Fig. 2C a clear loss of DAP10 was observed. Increasing concentrations of chloroquine, but not the proteasome inhibitor epoxomicin, inhibited the degradation of DAP10 provoked by exposure to the NKG2D ligand MICB (Fig. 2C), strongly suggesting that the degradation of DAP10 occurred in a lysosomal compartment.
The effect of cytotoxic interactions on the intracellular traffic of NKG2D and DAP10 in primary cultured NK cells was then studied by confocal microscopy. Consistent with the FACS experiments, 15 min of co-culture with 221B targets, but not 221 cells, produced a down-modulation of NKG2D from the NK cell surface (Fig. 2D). Intracellular vesicles containing NKG2D and DAP10 were observed in NK cells incubated 15 min with 221B cells in different confocal sections acquired every 0.3 μm through the z axel. This clear-cut co-localization of NKG2D and DAP10 in vesicular structures was not observed in NK cells incubated with 221 cells (Fig. 2D).
These results showed that interaction with MICB expressing targets induces the internalization of NKG2D/DAP10 complexes. Importantly, endocytosed DAP10 and NKG2D are both rapidly degraded after MICB engagement.
Traffic of DAP10 to SL in NK Cells
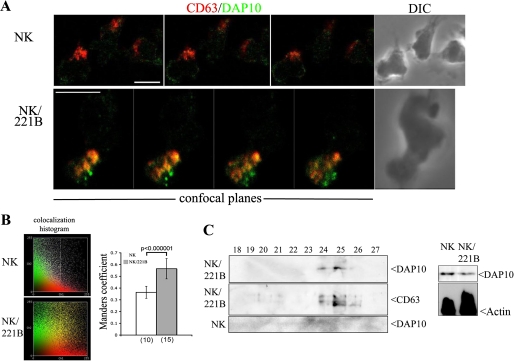
The overlapping, vesicular pattern of staining seen for NKG2D and DAP10 plus the evidence for lysosomal degradation of DAP10 (Fig. 2C) led us to investigate the hypothesis that DAP10 could traffic to SL after the formation of NKG2D/DAP10 endocytic complexes (Fig. 2D). NK cells incubated for 15 min alone or with 221B cells were fixed and stained for both DAP10 and CD63, a classic marker of SL (SL). Although DAP10 was barely detected in this organelle in purified NK cells, it was clearly found in SL in NB samples (Fig. 3A). This co-localization was quantified by calculation of Manders' coefficient (24) (Fig. 3B). To confirm the recruitment of DAP10 to SL we isolated these organelles by fractionation in Percoll gradients from primary cultured activated NK cells and NK cells cultured for 15 min with 221B cells and looked for DAP10 protein by Western blot (Fig. 3C). In NB samples, DAP10 was found in the peak of SL fractions, defined by assay for activity of the lysosomal hydrolase hexosaminidase (not shown), co-fractionating with CD63. No DAP10 was found in SL isolated from NK cells cultured without exposure to 221B cells. Thus, efficient recruitment of DAP10 to SL in primary cultured activated NK cells was observed on encounter with target cells expressing MICB. Interestingly, Western blot analysis of unfractionated lysates of these samples already showed some loss of DAP10 at this 15-min time point.
FIGURE 3.
Traffic of the DAP10 adaptor molecule in cultured activated NK cells. A, NK and NB samples were stained for NKG2D and CD63. The merged channels of 3 (NK) or 4 (NK/221B) confocal planes obtained from representative cells and the differential interference contrast (DIC) images (right panels) are shown. Scale bars, 5 μm. B, co-localization histogram and Manders' coefficient (R) calculated at least from 10 confocal sections in the number of cells indicated in parenthesis. Samples were statistically compared by the Student's t test. C, expression of DAP10 in SL purified by subcellular fractionation of NK and NB samples. Fractions 18–27 were analyzed for DAP10 and CD63 by Western blot. DAP10 protein could be detected in both sets of lysates prior to fractionation (left panels), thus the absence of DAP10 in fractions 24–26 of the NK cell alone is consistent with the idea that the large scale recruitment of the receptor complex to SL is consequent on this cytotoxic interactions.
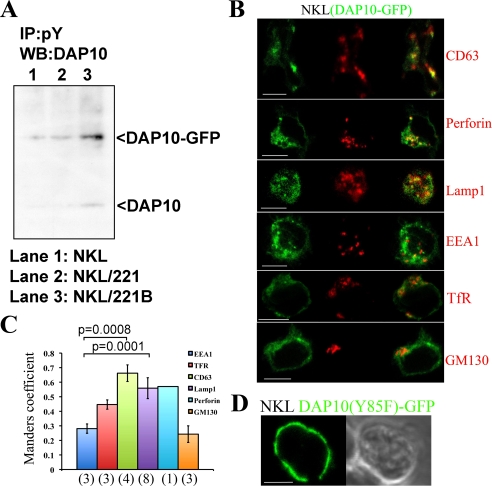
The intracellular traffic of DAP10 was also studied in NKL cells stably transfected with DAP10-GFP. These transfectants displayed levels of cell surface NKG2D and cytotoxic activity similar to the parental cells (supplemental Fig. S1, A and B), and confocal microscopy indicated a very high degree of overlap between DAP10 staining and the GFP signal (supplemental Fig. S1C). Importantly, although some basal phosphorylation of DAP10-GFP was noted, immunoprecipitation and Western blot experiments demonstrated a marked induction of tyrosine phosphorylation of DAP10-GFP after exposure to MICB (Fig. 4A). These data demonstrated that addition of the GFP tag to DAP10 did not significantly affect the ability of this protein to respond to engagement of NKG2D by the natural ligand MICB. Time-lapse confocal microscopy showed that DAP10-GFP was located not only at the cell surface, but also accumulated intracellularly in dynamic vesicles (video 1) positive for cholera toxin (supplemental Fig 1D), strongly suggesting the traffic of overexpressed DAP10-GFP through endosomes. Confocal microscopy analysis (Fig. 4, B and C) showed that endosomal DAP10-GFP extensively co-localized with the SL markers CD63, Lamp1, and Perforin. Some co-localization was also noted with transferrin receptor (CD71), while co-localization was not observed with the Golgi marker GM130 or the early endosome marker EEA1. Substitution of the tyrosine residue of the Y85XXM motif present in the cytoplasmic tail of DAP10 with a phenylalanine led to the accumulation of the mutant DAP10(Y85F)-GFP on the NKL cell surface suggesting that this mutation abrogates internalization of this molecule (Fig. 4D). These data demonstrated the traffic of overexpressed DAP10-GFP to SL in NKL cells, which supported the findings obtained in primary activated NK cells (Fig. 3, A–C).
FIGURE 4.
Traffic of DAP10-GFP in the NKL cell line. A, phosphorylation of DAP10-GFP molecule upon MICB stimulation. NKL cells were stimulated for 5 min with 221 and 221B cells and lysates were immunoprecipitated with an anti-phosphotyrosine Ab. Immunoprecipitates were analyzed by Western blot with an anti-DAP10 Ab for phosphorylated DAP10-GFP and DAP10 detection. B, NKL (DAP10-GFP) stable transfectants were stained as indicated. The green, red, and merged channels of one equatorial confocal plane are shown. Scale bars, 5 μm. C, histograms (mean ± S.D.) represent Manders' coefficients obtained for quantification of the co-localization. Samples were statistically compared by the Student's t test. Numbers in parenthesis indicate the number of cells analyzed. D, localization of the mutant DAP10(Y85F)-GFP at the cell surface in the NKL cell line. The green channel and the differential interference contrast image are shown. Scale bar, 5 μm.
NKG2D and DAP10 Accumulate at the cNK-IS in Organelles of the Endosomal Compartment
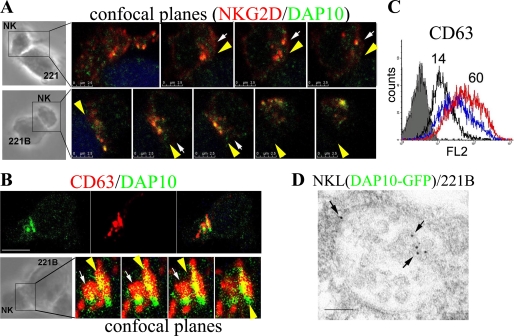
Given that the recognition of MICB expressing targets leads to the localization of DAP10 in SL (Fig. 3), it was of interest to study the recruitment of NKG2D/DAP10 endocytosed complexes to the cNK-IS. As shown in Fig. 5A, NKG2D/DAP10 containing vesicles polarized to the cNK-IS formed between cultured primary NK cells and both 221 and 221B cells. Interestingly however, NKG2D/DAP10 clearly clustered at the cell surface at the contact site only when the target cells expressed MICB. Moreover, conjugates formed between cultured primary NK cells and 221B cells showed that DAP10 co-localized with CD63 in both intracellular vesicles and at the surface interface of the cNK-IS (Fig. 5B and supplemental Fig. S2), which suggested the presence of DAP10 in degranulating SL. Degranulation during the cytotoxic process was then studied. Expression of CD63 at the NK cell surface during degranulation was 4-fold greater on NK cells co-cultured with 221B target cells than in NK cells cultured alone. Likewise culture with 221 cells also enhanced surface expression of CD63, although to a lesser degree, suggesting that culture with 221 cells was a less efficient trigger of exocytosis of SL (Fig. 5C). To give more experimental support to the presence of DAP10 in SL, this phenomenon was further confirmed by ultra-structural analysis. Immunogold labeling was performed with an anti-GFP antibody in NKL(DAP10-GFP)/221B cell conjugates processed for electron microscopy. Interestingly, gold particles were found at the outer membrane and at the membrane of internal vesicles of multivesicular structures typical of SL (13) (Fig. 5D, arrows). Accumulation of SL containing DAP10-GFP at the cNK-IS was also observed by electron microscopy in these conjugates (supplemental Fig. S3). Importantly, gold particles were observed in polarized SL (arrowheads) and in tight contact with the target cell membrane (arrows). Therefore, together these data are consistent with polarized traffic of SL containing DAP10 toward the cNK-IS, although our data do not exclude the possibility that the accumulation of the receptor complex at the site of target cell contact is influenced by interactions with specific ligands that affect the internalization of NKG2D and DAP10.
FIGURE 5.
Endosomal compartment polarizes NKG2D/DAP10 to the synapse. A, the indicated conjugates were stained for NKG2D and DAP10. Differential interference contrast images and merged channels of four (NK/221) or five (NK/221B) confocal planes at the synapse are shown. B, NK/221B conjugates were stained for CD63 and DAP10. The green, red, and merged channels of one equatorial confocal section as well as four confocal planes at the synapse are shown. A and B, vesicles are labeled by arrows, and synapse surface interfaces are indicated by yellow arrowheads. Target cells were labeled with chloromethyl derivative of aminocoumarin. At least ten cell conjugates, from three different donors, were analyzed in each condition. Scale bars 2.5 μm (A) and 5 μm (B). C, degranulation of SL. NK cells (black histogram line), NK/221 (blue histogram line), or NK/221B samples (red histogram line) were stained for surface CD63 and analyzed by FACS. Target cells, labeled with carboxyfluorescein diacetate succunimidyl ester, were excluded from the analysis. The gray histogram represents the isotype control staining for the NK cells alone (similar to the NK/221 and NK/221B control staining). Numbers indicate the gmfi of NK and NK/221B samples. D, distribution of DAP10-GFP by immunogold labeling at external and internal membranes (arrows) of a SL. Gold particle, 10 nm. Scale bar, 100 nm.
DISCUSSION
In this study, we have investigated the traffic of the NKG2D/DAP10 receptor complex during NK cell activation and upon interaction with the specific ligand MICB. In NKL and resting NK cells NKG2D is principally found stably expressed at the cell surface. Activation of resting NK cells, by exposure to either cytokines or feeder cells, is associated with an increased expression of NKG2D at the cell surface and the appearance of an intracellular pool of receptor that recycles to the cell surface. Strikingly, however, engagement with the natural ligand MICB induces a rapid and significant endocytosis and degradation of both, NKG2D and DAP10. The internalized DAP10 is recruited to SL where part polarizes toward the cNK-IS, but a significant proportion is rapidly degraded. Thus these findings confirm previous studies describing a down-modulation of surface NKG2D after exposure to NKG2D-L and provide a mechanism for this phenomenon.
Although treatment with cytokines such as IL-2 and IL-15 had been shown to induce an increased expression of NKG2D at the surface of both T and NK cells (25, 26), the dynamics of surface expression of NKG2D had not been studied before. In NKL cells surface NKG2D molecules are long lived, and no intracellular NKG2D is found in either the NKL cell line or resting primary NK cells. In contrast, ex vivo cytokine activated and primary cultured NK cells display increased levels of cell surface NKG2D and an intracellular pool of recycling receptor (Fig. 1). It is not clear how cytokine activation of primary cells leads to this altered traffic of NKG2D, but it is interesting to note that IL-15 as well as “priming” NK cells for cytotoxicity via trans-presentation on dendritic cells (27), also appears to be able to induce the appearance of a population of intracellular receptors (Fig. 1) perhaps as preparation for polarization toward the cNK-IS. It is tempting to speculate that this dual effect of IL-15 is related to the observation that the signaling pathways initiated by the interleukin 15 receptor are coupled to NKG2D/DAP10 signaling (28). It is interesting to note that mutation of the critical residue Tyr-85 in DAP10 blocks the localization of DAP10 in CD63+ endosomes in NKL cells. It can be possible that the basal level of DAP10 phosphorylation observed can be due to the intracellular traffic of the overexpressed protein.
Having seen that activation of primary NK cells leads to the induction of a continuous process of internalization and recycling of NKG2D to and from the cell surface, it was of interest to study how contact with target cells expressing specific ligands of NKG2D would affect receptor traffic. Our experiments show that recognition of MICB leads to the formation of NKG2D/DAP10 endocytic complexes and the localization of DAP10 in vesicles with features of SL. Three sets of experiments show that these vesicles are closely related to SL: confocal microscopy showing intracellular DAP10 co-localizing extensively with CD63, electron microscopy showing the presence of DAP10-GFP in multivesicular structures typical of SL, and lastly biochemical isolation on Percoll gradients of the SL compartment showing that DAP10 co-fractionates with CD63.
SL form an intracellular compartment with dual function: secretion and degradation (13). They can switch function from an intracellular organelle, in which proteins can be degraded, to a secretory organelle with proteins that become functional as they are released from the cell. In this context it is easy to imagine that, although molecules normally resident in lysosomes such as CD63 and CD107a can recycle between this compartment and the cell surface, NKG2D and DAP10 may be considerably more susceptible to lysosomal degradation, and this may explain why, although NK recognition of 221B cells is associated with increased surface expression of CD63 (Fig. 5C), the expression of both cell surface and total NKG2D/DAP10 actually decreases (Fig. 2). It is thus plausible to speculate that contact with target cells expressing MICB might alter the pattern of intracellular traffic of NKG2D/DAP10 to simultaneously enhance recruitment of NKG2D/DAP10 to SL and polarized secretion, but also the degradation of NKG2D/DAP10 recruited to these structures. SL containing DAP10 are seen to polarize toward the cNK-IS (Fig. 5 and supplemental Figs. S2 and S3) and cluster at the cell surface in the cNK-IS. Importantly, the DAP10-GFP molecule is clearly observed by electron microscopy at the outer and exosome membranes of SL and in tight contact with the target cell membrane at the synaptic cleft in NKL(DAP10-GFP)/221B conjugates (Fig. 5D and supplemental Fig. S3). This is reminiscent of previous observations of exosomes conveying the TCR, perforin and granzyme degranulated to the synaptic cleft formed at the cytotoxic T lymphocyte synapse (29), and supports the hypothesis that the presence of recognition molecules such as the TCR/CD3 or NKG2D/DAP10 in these vesicles could help the unidirectional delivery of lytic substances during the cytotoxic process (30, 31). Although NKG2D/DAP10 appear to move to the cNK-IS via polarized traffic of SL, our data do not exclude the possibility that the accumulation of the receptor complex at the site of target cell contact depends on stable interactions with specific ligands to avoid re-internalization. Live cell imagining to monitor the trafficking of DAP10-GFP in NKL cells in complex with MICB expressing 221 cells to clarify these issues will be an important objective for future research.
The binding of MICB by NKG2D on activated NK cells triggers both cytotoxicity and down-modulation of the surface expression of NKG2D (Fig. 2) reminiscent of the TCR down-modulation on T cells after antigen recognition. It has been proposed that the mechanism responsible for the loss of cell surface TCR/CD3 complexes is an increase in degradation, whereas the endocytosis rate is not changed (32). It is then plausible to think that the recycling of NKG2D may also be affected by raising its degradation. How the rate of endocytosis of NKG2D is affected by MICB engagement is not known, but it is clear that ligand recognition is associated with a marked increase in degradation. Although the destruction of TCR/CD3 complexes is known to depend on both the lysosomal compartment and the proteasome, our data show that DAP10 degradation can be abrogated by inhibitors of the lysosomal activity but not by the proteasome inhibitor epoxomicin (Fig. 2C). This observation supports the involvement of SL in this process, but whether NKG2D degradation depends on lysosomal activity, the proteasome, or both still remains to be determined.
Some degradation of total NKG2D, although not statistically significant, was also observed in NK cells cultured for 1 h with 221 cells (Fig. 2A), and, consistent with this observation, some loss of DAP10 was also noted in these samples (data not shown). Because this is a cytotoxic interaction mainly mediated by NKp46 and NKp44 (33, 34), a possible explanation for this phenomenon may be the existence of cross-talk between the signaling pathways triggered through different activating receptors. An idea consistent with the observation that engagement of a single natural cytotoxicity receptor can trigger the activation of signaling cascades associated with other activating receptors (35).
The mechanism by which DAP10 is recruited to the SL is not yet known, although it has been shown that the Y85XXM motif present in the cytoplasmic tail of DAP10 is necessary for NKG2D internalization (36), and DAP10 targeting to the cNK-IS (37). A glycine in the −1 position with respect to the Tyr in this motif (GYXXM) is a common feature of proteins, such as CD63 or Lamp1, that traffic directly from the trans-Golgi network to lysosomes. The absence of this residue in the YXNM motif at the DAP10 cytoplasmic tail suggests an indirect route for the traffic of this adaptor molecule to SL in NK cells. Antibody uptake experiments (data not shown) suggest that NKG2D/DAP10 can be endocytosed by a clathrin-mediated pathway in the NKL cell line. Our data support the idea that mutation of the Tyr-85 for Phe abrogates receptor internalization (Fig. 4D), however, protein sorting to SL after internalization depends on distinct mechanisms such as the mannose 6-phosphate pathway used to sort lysosomal hydrolases and mono-ubiquitylation used to sort proteins to multivesicular bodies (38). For example, sorting of Fas ligand to SL is regulated by mono-ubiquitylation and phosphorylation (39), and it will be important to study the roles of these different post-translational modifications in influencing the intracellular traffic of NKG2D and DAP10. The significance of the intracellular traffic of NK cell-activating receptors during the cytotoxic process clearly deserves future research.
The simultaneous polarization to the cNK-IS and degradation of NKG2D/DAP10 receptor complexes observed upon MICB engagement lead to a model to explain the observation that both in vitro and in vivo, chronic exposure to NKG2D-L induces down-regulation of NKG2D on human CD8+ T cells and murine NK cells (4–6, 11, 36). The degradation of NKG2D and DAP10 is clearly associated with a dramatic change in the traffic of NKG2D after ligand engagement; from a recycling molecule, to a molecule mainly directed to degradation routes. This altered traffic seems to be important for establishment of the cNK-IS for NK cell cytotoxicity, but the concomitant receptor degradation may also explain the impairment in cytotoxic capacity observed after chronic exposure to NKG2D ligands. The down-regulation of NKG2D and DAP10 could simply be a consequence of sustained stimulation triggering continued recruitment of these molecules to SL where, although some receptor polarizes to the cNK-IS, much is degraded in a process that is considerably more rapid than the replenishment of cell surface NKG2D/DAP10 by de novo synthesis.
Supplementary Material
Acknowledgments
We thank Dr. F. Sanchez-Madrid, Dr. E. Vivier, and Dr. D. Cantrell for kind gifts of mAbs; Dr. N. Miller for assistance with cell sorting; Prof. J. Trowsdale for helpful advice and support; and Dr. M. Vales-Gomez for critical reading of the manuscript.
This work was supported in part by the Leukaemia Research Fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Video 1.
J. N. Skepper, manuscript in preparation.
- NK
- natural killer
- NKG2D
- natural killer cell lectin-like receptor gene 2D
- MHC
- major histocompatibility complex
- MIC
- MHC class I chain-related molecule
- cNK-IS
- cytotoxic NK cell immune synapse
- SL
- secretory lysosome
- BFA
- brefeldin A
- CHX
- cycloheximide
- GFP
- green fluorescent protein
- mAb
- monoclonal antibody
- FACS
- fluorescence-activated protein kinase
- gmfi
- geometric mean fluorescence intensity
- IL-2
- interleukin-2
- DIC
- differential interference contrast.
REFERENCES
- 1.Wu J., Song Y., Bakker A. B., Bauer S., Spies T., Lanier L. L., Phillips J. H. ( 1999) Science 285, 730– 732 [DOI] [PubMed] [Google Scholar]
- 2.Upshaw J. L., Arneson L. N., Schoon R. A., Dick C. J., Billadeau D. D., Leibson P. J. ( 2006) Nat. Immunol. 7, 524– 532 [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez S., Groh V., Spies T. ( 2006) Curr. Top. Microbiol. Immunol. 298, 121– 138 [DOI] [PubMed] [Google Scholar]
- 4.Coudert J. D., Zimmer J., Tomasello E., Cebecauer M., Colonna M., Vivier E., Held W. ( 2005) Blood 106, 1711– 1717 [DOI] [PubMed] [Google Scholar]
- 5.Oppenheim D. E., Roberts S. J., Clarke S. L., Filler R., Lewis J. M., Tigelaar R. E., Girardi M., Hayday A. C. ( 2005) Nat. Immunol. 6, 928– 937 [DOI] [PubMed] [Google Scholar]
- 6.Wiemann K., Mittrucker H. W., Feger U., Welte S. A., Yokoyama W. M., Spies T., Rammensee H. G., Steinle A. ( 2005) J. Immunol. 175, 720– 729 [DOI] [PubMed] [Google Scholar]
- 7.Salih H. R., Antropius H., Gieseke F., Lutz S. Z., Kanz L., Rammensee H. G., Steinle A. ( 2003) Blood 102, 1389– 1396 [DOI] [PubMed] [Google Scholar]
- 8.Salih H. R., Goehlsdorf D., Steinle A. ( 2006) Hum. Immunol. 67, 188– 195 [DOI] [PubMed] [Google Scholar]
- 9.Salih H. R., Rammensee H. G., Steinle A. ( 2002) J. Immunol. 169, 4098– 4102 [DOI] [PubMed] [Google Scholar]
- 10.Waldhauer I., Steinle A. ( 2006) Cancer Res. 66, 2520– 2526 [DOI] [PubMed] [Google Scholar]
- 11.Groh V., Wu J., Yee C., Spies T. ( 2002) Nature 419, 734– 738 [DOI] [PubMed] [Google Scholar]
- 12.Roda-Navarro P., Vales-Gomez M., Chisholm S. E., Reyburn H. T. ( 2006) Proc. Natl. Acad. Sci. U. S. A. 103, 11258– 11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkhardt J. K., Hester S., Lapham C. K., Argon Y. ( 1990) J. Cell Biol. 111, 2327– 2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blott E. J., Griffiths G. M. ( 2002) Nat. Rev. Mol. Cell Biol. 3, 122– 131 [DOI] [PubMed] [Google Scholar]
- 15.Perussia B., Ramoni C., Anegon I., Cuturi M. C., Faust J., Trinchieri G. ( 1987) Nat. Immun. Cell Growth Regul. 6, 171– 188 [PubMed] [Google Scholar]
- 16.Bossi G., Griffiths G. M. ( 1999) Nat. Med. 5, 90– 96 [DOI] [PubMed] [Google Scholar]
- 17.Nichols B. J., Kenworthy A. K., Polishchuk R. S., Lodge R., Roberts T. H., Hirschberg K., Phair R. D., Lippincott-Schwartz J. ( 2001) J. Cell Biol. 153, 529– 541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skepper J. N. ( 2000) J. Microsc. 199, 1– 36 [DOI] [PubMed] [Google Scholar]
- 19.Lippincott-Schwartz J., Glickman J., Donaldson J. G., Robbins J., Kreis T. E., Seamon K. B., Sheetz M. P., Klausner R. D. ( 1991) J. Cell Biol. 112, 567– 577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klausner R. D., Van Renswoude J., Ashwell G., Kempf C., Schechter A. N., Dean A., Bridges K. R. ( 1983) J. Biol. Chem. 258, 4715– 4724 [PubMed] [Google Scholar]
- 21.Das V., Nal B., Dujeancourt A., Thoulouze M. I., Galli T., Roux P., Dautry-Varsat A., Alcover A. ( 2004) Immunity 20, 577– 588 [DOI] [PubMed] [Google Scholar]
- 22.Batista A., Millan J., Mittelbrunn M., Sanchez-Madrid F., Alonso M. A. ( 2004) J. Immunol. 172, 6709– 6714 [DOI] [PubMed] [Google Scholar]
- 23.Bauer S., Groh V., Wu J., Steinle A., Phillips J. H., Lanier L. L., Spies T. ( 1999) Science 285, 727– 729 [DOI] [PubMed] [Google Scholar]
- 24.Manders E. M., Stap J., Brakenhoff G. J., van Driel R., Aten J. A. ( 1992) J. Cell Sci. 103, 857– 862 [DOI] [PubMed] [Google Scholar]
- 25.Roberts A. I., Lee L., Schwarz E., Groh V., Spies T., Ebert E. C., Jabri B. ( 2001) J. Immunol. 167, 5527– 5530 [DOI] [PubMed] [Google Scholar]
- 26.Maasho K., Opoku-Anane J., Marusina A. I., Coligan J. E., Borrego F. ( 2005) J. Immunol. 174, 4480– 4484 [DOI] [PubMed] [Google Scholar]
- 27.Lucas M., Schachterle W., Oberle K., Aichele P., Diefenbach A. ( 2007) Immunity 26, 503– 517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horng T., Bezbradica J. S., Medzhitov R. ( 2007) Nat. Immunol. 8, 1345– 1352 [DOI] [PubMed] [Google Scholar]
- 29.Peters P. J., Geuze H. J., Van der Donk H. A., Slot J. W., Griffith J. M., Stam N. J., Clevers H. C., Borst J. ( 1989) Eur. J. Immunol. 19, 1469– 1475 [DOI] [PubMed] [Google Scholar]
- 30.Peters P. J., Geuze H. J., van der Donk H. A., Borst J. ( 1990) Immunol. Today 11, 28– 32 [DOI] [PubMed] [Google Scholar]
- 31.Roda-Navarro P., Reyburn H. T. ( 2007) FASEB J. 21, 1636– 1646 [DOI] [PubMed] [Google Scholar]
- 32.Liu H., Rhodes M., Wiest D. L., Vignali D. A. ( 2000) Immunity 13, 665– 675 [DOI] [PubMed] [Google Scholar]
- 33.Sivori S., Vitale M., Morelli L., Sanseverino L., Augugliaro R., Bottino C., Moretta L., Moretta A. ( 1997) J. Exp. Med. 186, 1129– 1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitale M., Bottino C., Sivori S., Sanseverino L., Castriconi R., Marcenaro E., Augugliaro R., Moretta L., Moretta A. ( 1998) J. Exp. Med. 187, 2065– 2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Augugliaro R., Parolini S., Castriconi R., Marcenaro E., Cantoni C., Nanni M., Moretta L., Moretta A., Bottino C. ( 2003) Eur. J. Immunol. 33, 1235– 1241 [DOI] [PubMed] [Google Scholar]
- 36.Ogasawara K., Hamerman J. A., Hsin H., Chikuma S., Bour-Jordan H., Chen T., Pertel T., Carnaud C., Bluestone J. A., Lanier L. L. ( 2003) Immunity 18, 41– 51 [DOI] [PubMed] [Google Scholar]
- 37.Giurisato E., Cella M., Takai T., Kurosaki T., Feng Y., Longmore G. D., Colonna M., Shaw A. S. ( 2007) Mol. Cell. Biol. 27, 8583– 8599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stinchcombe J. C., Griffiths G. M. ( 2007) Annu. Rev. Cell Dev. Biol. 23, 495– 517 [DOI] [PubMed] [Google Scholar]
- 39.Zuccato E., Blott E. J., Holt O., Sigismund S., Shaw M., Bossi G., Griffiths G. M. ( 2007) J. Cell Sci. 120, 191– 199 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.