Abstract
Dishevelled (Dvl) is an essential protein in the Wnt signaling pathways; it uses its PDZ domain to transduce the Wnt signals from the membrane receptor Frizzled to downstream components. Here, we report identifying a drug-like small molecule compound through structure-based ligand screening and NMR spectroscopy and show the compound to interact at low micromolar affinity with the PDZ domain of Dvl. In a Xenopus testing system, the compound could permeate the cell membrane and block the Wnt signaling pathways. In addition, the compound inhibited Wnt signaling and reduced the levels of apoptosis in the hyaloid vessels of eye. Moreover, this compound also suppressed the growth of prostate cancer PC-3 cells. These biological effects suggest that by blocking the PDZ domain of Dvl, the compound identified in our studies effectively inhibits the Wnt signaling and thus provides a useful tool for studies dissecting the Wnt signaling pathways.
The Wnt signaling pathways are regulated by a family of secreted Wnt glycoproteins. The canonical Wnt pathway, which is highly conserved, is best understood. In this pathway, Wnt molecules interact with the seven-transmembrane Frizzled (Fz)2 proteins (1) by binding to an N-terminal cysteine-rich-domain (2). The signal is then transduced into the cell through an internal sequence of Fz, C-terminal to the seventh transmembrane domain, which binds directly to the PDZ (postsynaptic density-95/discs large/zonula occludens-1) domain of the cytoplasmic protein Dishevelled (Dvl) (3). Dvl then transduces the Wnt signals to downstream components (4). Three Dvl homologs (Dvl-1, -2, and -3) have been identified in humans; all are expressed in both embryonic and adult tissues, including brain, heart, lung, kidney, skeletal muscle, and others (4). Up-regulation and overexpression of Dvl proteins have been reported in many cancers, including those of breast, colon, prostate, mesothelium, and lung (non-small cell) (5–8).
The Dvl protein is made up of three conserved domains: an N-terminal DIX domain, a central PDZ domain, and a C-terminal DEP domain (9). The central PDZ domain is of particular interest because of its interaction with Fz and other Wnt pathway proteins (3, 10). The direct interaction between the PDZ domain and Fz peptides is relatively weak, and other factors may play a role to ensure the communication between the two molecules (3). For example, several studies suggest that the DEP domain of Dvl has a membrane-targeting function that may facilitate PDZ-Fz interaction (11–14). However, the weak PDZ-Fz interaction provides an opportunity to block Wnt signaling at the Dvl level by using a small molecule inhibitor. An earlier study in our laboratories used an NMR-assisted virtual ligand screening approach to identify a peptide mimic that can bind to the Dvl PDZ domain (15). We have now used an improved algorithm to conduct an additional structure-based virtual screen of the PDZ domain of Dvl and have discovered a group of drug-like compounds that bind to the PDZ domain with moderate to low micromolar affinity. One of these compounds effectively blocked Wnt signaling in vivo and reduced the growth rate of a prostate cancer cell line.
EXPERIMENTAL PROCEDURES
Ligand and Receptor Preparation
All compound data bases were obtained from the NCI (National Institutes of Health), Chemical Diversity Inc. (ChemDiv, San Diego, CA), or Sigma-Aldrich. Three-dimensional coordinates for all compounds were generated by using the Optive Research Concord program (Tripos Inc., St. Louis, MO) and stored in the Sybyl mol2 format. The Unity module of the Sybyl software package (Tripos Inc.) was used to select the compounds in the three-dimensional small molecule data base that matched the known ligand of the Dvl PDZ domain. The first Unity query was based on three-dimensional distance constraints determined by analyzing the structure of the Dapper (Dpr) peptide-PDZ complex (10), which was obtained from the Protein Data Bank (PDB code: 1L6O). The atoms of the bound Dpr peptide within hydrogen-bonding distance of suitable H-bond acceptors and donors on the backbone of the βB sheet of the PDZ domain were selected.
Docking and Scoring
The compounds selected from the Unity queries were docked to the Dpr peptide binding site of Dvl PDZ domain by using the FlexX module of the Sybyl software package. For the docking analysis, the receptor site was first defined as all of the residues in the PDZ domain within 7.0 Å of the Dpr peptide, and the core site (i.e. where the core fragment is to be placed during docking) was defined as all residues within 10.0 Å of the Thr (−2) in the Dpr peptide. In later stages of screening, the core site was redefined as a much smaller area made up of residues Ile-267, Ser-268, Ile-269, Leu-324, Arg-325, and Val-328, which make up the hydrophobic groove. This refined definition helped to prevent docked conformations that were likely to be incorrect. For example, when the larger core site was used, some docked compounds were almost entirely exposed to the solvent, although most of the molecules are hydrophobic. As many as 30 docked conformations were generated for each compound by FlexX. Docked conformations were scored by using the five consensus score functions included in the FlexX software. The Dpr peptide was used to calibrate docking parameters and to determine high (negative) values for each individual scoring function. The backbone root mean square deviation between the crystal structure conformation and the docked conformation of Dpr was 2.86 Å.
Protein Preparation
Different forms of the Dvl PDZ domain protein were synthesized by using an Escherichia coli system as described (3, 15, 16). To prepare TMR-labeled PDZ domain, a cysteine located outside of the binding site was mutated to alanine to increase solubility of the PDZ domain. A construct without a T328C mutation was made for use in fluorescence spectroscopy experiments. The fluorescent label 2-((5(6)-tetramethylrhodamine)carboxyamino)ethyl methanethiosulfonate (TMR) was covalently bound to this Cys residue, which is the only Cys in this PDZ domain. The PDZ solution was dialyzed overnight at 4 °C against 100 mm potassium phosphate buffer at pH 7.5 to remove dithiothreitol, which had been added to prevent disulfide bond formation. A 10-fold excess of TMR dissolved in DMSO was added dropwise to the PDZ solution while stirring. After 2 h at room temperature, the unbound TMR was removed from the solution by dialysis against 100 mm potassium phosphate buffer, pH 7.5, at 4 °C.
NMR Spectroscopy
All NMR studies used either a 600-MHz Varian INOVA spectrometer or a 600-MHz Bruker Avance spectrometer. 15N-labeled PDZ samples were prepared at a concentration of 0.3 mm in 100 mm KHPO4, 0.5 mm EDTA, and 10% D2O at pH 7.5. The compounds were dissolved either in the same buffer as the PDZ domain or in DMSO, depending on the aqueous solubility of the individual compound. Titration was carried out by adding small amounts of the compound to the PDZ domain and taking 15N-HSQC spectra of the mixture. Compound concentrations varied from 0.3 to 6.0 mm during the course of these titrations. All NMR spectra were processed with NMRPipe (17) software and analyzed with the Sparky program(18).
Fluorescence Spectroscopy
All fluorescence measurements were obtained by using a Jobin-Yvon Fluorolog-3 spectrofluorometer (HORIBA Jobin-Yvon Inc., Edison, NJ) (15). Fluorescence anisotropy measurements of TMR-labeled PDZ domain were obtained for binding affinity calculations. Titrations of compounds to the solution of TMR-labeled PDZ domain were performed at 25 °C in 100 mm KHPO4, 0.5 mm EDTA buffer, at pH 7.5. The excitation wavelength for TMR-labeled PDZ domain was 551 nm with an entrance slit width of 5 nm. The maximum fluorescence emission wavelength was 578 nm with an exit slit width of 5 nm. The compounds were prepared to a concentration of ∼1.0–20.0 mm in the same buffer as that used for the protein. During titration, the range of concentrations of the compound was ∼100 nm–1.0 mm, depending on binding affinity for the particular compound. The anisotropy data were analyzed by fitting the data to the standard ligand binding curve in the program Prism (GraphPad Software Inc., San Diego, CA). Best-fit curves were obtained by using a global, non-linear regression model that assumed that the law of mass action was followed. Although changes of fluorescence anisotropy due to the compounds binding were small, we were able to obtain binding affinity values for the small molecule compounds. Under the same conditions, fluorescence polarization measurements of the ROX-labeled Dpr peptide (ROX-N-butyric-SGSLKMTTV-COOH) were also performed. The excitation wavelength for the ROX-labeled Dpr peptide was 578 nm with an entrance slit width of 5 nm. The maximum fluorescence emission wavelength was 605 nm with an exit slit width of 5 nm. By titrating the PDZ domain into 50 nm ROX-Dpr peptide solution in the absence and presence of 6 μm compound 3289-8625, respectively, we obtained the binding affinity (Kd) of Dpr and the apparent binding affinity (Kdapp) of the peptide in the presence of 3289-8625. The competition binding constant (Ki) between PDZ and 3289-8625 was calculated by the equation Kdapp = Kd (1 + [I]/Ki).
Xenopus Embryo Studies
Xenopus eggs were obtained from females that had been injected with 500 IU of human chorionic gonadotropin (Sigma-Aldrich) and had been artificially fertilized. Synthesis and microinjection of mRNAs were carried out as described previously (15, 19). Briefly, for the luciferase assay, the siamois promoter-driven reporter DNA construct (3, 15) (400 pg) was injected alone or co-injected with Wnt3A mRNA (1 pg) into the animal pole region at the two-cell stage. Injected embryos were cultured in the absence or presence of the compound 3289-8625 at different concentrations, and animal cap explants were dissected at late blastula stage. For secondary axis assay, Wnt3A mRNA (1 pg) was injected in the ventro-vegetal region at the four-cell stage, and injected embryos were cultured in the absence or presence of the compound 3289-8625 until larval stage.
Cell Proliferation Assay
The effect of the compound 3289-8625 on the PC-3 cells was examined by using the MTT cell proliferation assay (Promega, Madison, WI). In the experiments, PC-3 cells were plated in 24-well culture plates at a density of 1 × 104 cells/well and cultured overnight in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum plus penicillin and streptomycin at 37 °C. On the second day, compound 3289-8625 was added to the cells at different concentrations. Before being added to the cells, the compound was dissolved in DMSO at 200 mm. While adding the compound to the cells, the same amounts of DMSO were added to the control cells. After 68–69 h of treatment, 10% volume of MTT stock solution (5 mg/ml) was added to the cell culture. The cells were incubated at 37 °C until the total treatment time reached 72 h. The converted dye was then solubilized, and the absorbance was measured at 570 nm. Three sets of independent experiments were performed, and each data point was normalized against the control cells.
The β-Catenin Level in PC-3
PC-3 cells were seeded in 100-mm tissue culture dish, cultured in completed Dulbecco's modified Eagle's medium overnight, and then treated with 3289-8625 compound (final concentration 80 μm) or DMSO vehicle for 72 h. For the extraction of membrane and cytosolic proteins (20, 21), cells were collected in TES suspension buffer and homogenized on ice. The lysate was centrifuged for 10 min at 500 × g. The crude supernatant was then fractionated at 100,000 × g for 90 min at 4 °C to generate a supernatant or cytosolic fraction and a membrane-rich pellet fraction. The membrane-enriched pellet was dissolved in phosphate-buffered saline buffer containing 1% Triton X-100 and 1% Nonidet P-40. Equal volumes of Laemmli buffer were added to protein solutions, and the samples were boiled for 5 min. The proteins were then separated by 10% SDS-PAGE electrophoresis under reducing condition, transferred to nitrocellulose membranes, blocked with 5% nonfat dry milk in phosphate-buffered saline with Tween, probed with antibody against β-catenin (Santa Cruz Biotechnology Inc., Santa Cruz, CA) and horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody (Sigma-Aldrich), and visualized by a chemiluminescence detection system (PerkinElmer Life Sciences). Membranes were then stripped and reprobed with antibody against integrin α3 protein (22) as equal loading control.
RESULTS
Screening of Dvl PDZ Binding Compounds
To identify additional scaffolds of Dvl PDZ domain inhibitors, we carried out several rounds of new computational screening on the basis of our earlier studies (15, 23, 24). To identify possible PDZ binding compounds, we first used the UNITY module in the Sybyl software (Tripos Inc.) to screen data bases of drug-like compounds from the NCI (National Institutes of Health), ChemDiv, and Sigma-Aldrich. Hits returned from these searches were docked to the protein receptor site, and the conformations of the complexes were scored by using the FlexX module in Sybyl. The overall procedure of the computational screening was similar to that used in our earlier work (15), but many details were further refined in this study.
The first Unity query was based on three-dimensional distance constraints determined by analyzing the structure of the Dpr peptide-PDZ complex (10); we chose the ligand-based query approach, which was not used in our earlier study (15). The atoms of the bound Dpr peptide within hydrogen-bonding distance of suitable H-bond acceptors and donors on the backbone of the βB sheet of the PDZ domain were selected. Distance and angle constraints between those atoms were used to run a three-dimensional flexible screen of the NCI (National Institutes of Health) and ChemDiv data bases; in addition, the Sigma-Aldrich data bases were searched by using the Sigma-Aldrich online two-dimensional search utility. The initial Unity query generated a list of several thousand compounds as potential hits.
The program FlexX (25) was then used in the docking studies. This program uses five scoring functions to evaluate docking results: the standard FlexX scoring function F_score (26–28), the Chemscore function (29), the knowledge-based proton motive force (PMF) score (based entirely on protein-ligand atom pairs and their distances (30)), the G_score function (calculated from ligand-protein atom pair interactions based on values from the Tripos force field (31)), and the D_score (based on the atom charges and the Van der Waals interactions between the ligand and protein (32)). Data from our earlier studies (15) and from the additional experiments in this study revealed that the F_score and Chemscore functions were the most reliable in predicting binding to the PDZ domain. Therefore, in this study, we used only the F_score and Chemscore rather than the consensus score of all five scoring functions. The G_score function was used as an additional reference because in our experience, G_score can help to identify internal steric hindrance in a docked conformation. Approximately 50 high scoring compounds were then obtained from the NCI (National Institutes of Health) and from Sigma-Aldrich and further screened by using NMR spectroscopy. Before the NMR studies, the FlexX docked complexes of all selected compounds were visually inspected to confirm that the ligands were in the peptide binding groove of the PDZ domain and that there was no internal steric hindrance.
To validate docking results, we performed 1H-15N correlated NMR spectroscopy. We obtained the 15N-HSQC spectra (15) by titrating various concentrations of the small molecules into samples of 15N-labeled mDvl1 PDZ domain. Examination of the spectra for chemical shift perturbations (33) revealed several small molecules that bound to the conventional C-terminal peptide binding groove of the PDZ domain; many of those NMR-confirmed hits showed a butyric acid substructure at one end. On the basis of this butyric acid group, we used the program Unity to perform a substructure search of the NCI (National Institutes of Health), Sigma-Aldrich, and ChemDiv compound data bases. The program FlexX was then used to dock and score the hits returned from the Unity screen. Several of the highest scoring compounds in the FlexX docking had a similar core structure that was predicted by FlexX to bind in the traditional binding groove of the PDZ domain. Those compounds were obtained from ChemDiv, Inc. and were tested by NMR spectroscopy.
Characterization of Compound 3289-8625, a PDZ Domain Inhibitor
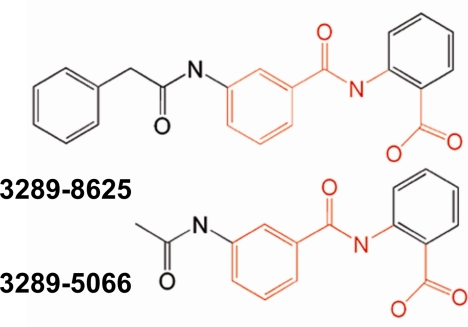
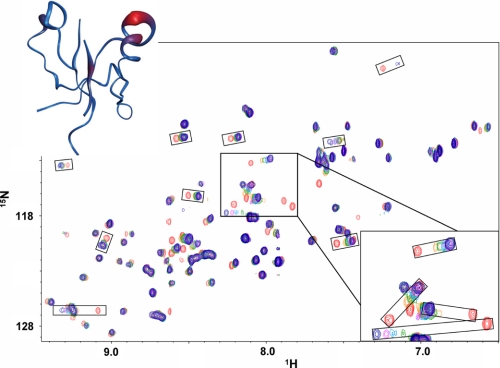
In the NMR experiments, compound 3289-5066 and compound 3289-8625 (Fig. 1) displayed the most significant chemical shift perturbations when titrated into the solution of 15N-labled Dvl PDZ domain. Their chemical shift patterns were generally similar and closely resembled that seen with Dpr and Fz binding (3). Compound 3289-8625 bound more strongly to the Dvl PDZ domain, as judged by the chemical shift perturbations due to ligand binding (Fig. 2). When the weighted sums of the chemical shift data were plotted onto a tube representation of the mDvl1 backbone, compound 3289-8625 was seen to bind in the groove between the βB sheet and the αB helix (Fig. 2).
FIGURE 1.
Chemical structures of the compounds identified in the substructure search. The chemical structures of compounds 3289-8625 and 3289-5066 are shown. Parts shown in red share the common scaffold.
FIGURE 2.
Interaction between the Dvl PDZ domain and compound 3289-8625. Shown are 15N-HSQC spectra of free PDZ domain (red) and PDZ domain with increasing concentrations of compound 3289-8625 (orange, green, cyan, purple, and blue, ligand:protein ratios of 1, 3, 5, 7, and 15). Upper inset, tube diagram of the PDZ domain with the weighted chemical shift intensities from the overlaid NMR spectra shown as regions of differing width and color. The widest (and red) regions contain the residues showing the greatest chemical shift. Lower inset, detail view of several peaks showing large chemical shift.
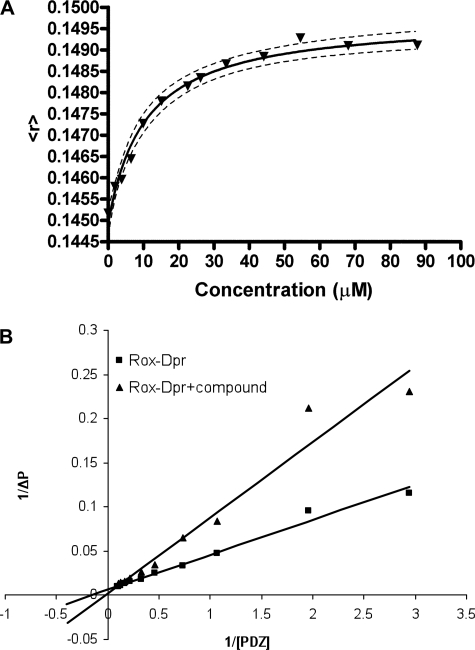
To further assess the binding characteristics of the molecules identified in the virtual screen, we used fluorescence anisotropy to measure the binding affinities of the identified inhibitors to the Dvl PDZ domain. In these experiments, each small molecule inhibitor was titrated into a solution of TMR-labeled PDZ domain, and the anisotropy change due to ligand binding was used to determine the binding affinity of the inhibitor to the Dvl PDZ domain. To verify the accuracy of this method, we first determined the binding affinity between the PDZ domain and the Dpr peptide; this value (11.0 ± 1.4 μm) was consistent with the value obtained by other methods (3, 15). The fluorescence method yielded a measured binding affinity (Kd) value of 10.6 ± 1.7 μm between the PDZ domain and compound 3289-8625; the fluorescence data are plotted in Fig. 3. We also measured the binding of other, similar compounds identified in this study to the Dvl PDZ domain. As suggested by the NMR studies, all of these compounds had weaker binding affinities than did compound 3289-8625; for example, measured by the fluorescence anisotropy method, the binding affinity between compound 2372-2393 and the Dvl PDZ domain was 18.9 ± 2.1 μm.
FIGURE 3.
Binding of compound 3289-8625 to the Dvl PDZ domain. A, binding was followed by monitoring the changing of fluorescence anisotropy of the TMR-labeled Dvl PDZ domain by binding compound 3289-8625. In the plot of the fluorescence anisotropy of the TMR-labeled Dvl PDZ domain with increasing concentrations of compound 3289-8625, the y axis is fluorescence anisotropy, and the x axis is the concentration of the compound. The Kd value was determined from the fitted curve. B, polarization change was monitored during titration of the PDZ domain into 50 nm ROX-Dpr peptide solution in the absence and presence of 6 μm compound 3289-8625, respectively; the reciprocal of the polarization change (1/ΔP) was plotted against the reciprocal of the PDZ protein concentration (1/[PDZ]). Data were analyzed by the program Prism (GraphPad Software Inc.).
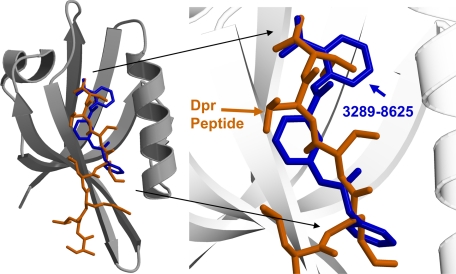
The binding affinity between compound 3289-8625 and the Dvl PDZ domain is comparable with the binding affinity of the Dpr peptide to the Dvl PDZ domain. To further demonstrate that compound 3289-8625 can compete with the Dpr peptide, we also measured the binding affinity between the Dpr peptide and the PDZ domain by monitoring the change of fluorescence polarization during titration of unlabeled Dvl PDZ into a solution of the fluorescent ROX-labeled Dpr peptide. This method yielded a measured binding affinity of 6.1 ± 0.4 μm., which is in good agreement with the value obtained by using TMR-labeled PDZ domain as described above (the slight difference may due to the effects of fluorescence tag labeling or experimental errors). By using the same assay, we showed that compound 3289-8625 inhibited the interaction between the Dpr peptide and the PDZ domain in the manner of classical competitive inhibition (KI values, 4.9 ± 1.7 μm, Fig. 3B). The data clearly show that compound 3289-8625 and the Dpr peptide competed for the same site on the surface of Dvl PDZ domain. Indeed, the conformation of PDZ-bound compound 3289-8625 as calculated by FlexX lies in this same region and closely resembles the crystal structure conformation of the MTTV motif of the Dpr peptide (10). When overlaid, the conformations of Dpr and this compound are seen to be very similar. The benzene ring on one end of the compound mimics the Val side chain of Dpr, and the benzene ring on the other end of the compound lies in the same region as the Met side chain. The C-terminal carboxylate ion of Dpr and that of the compound lie in the same region, where they can interact similarly with the PDZ domain (Fig. 4).
FIGURE 4.
The complex structures. The crystal structure of the Dvl PDZ domain-bound Dpr peptide is shown overlaid with the highest scoring predicted conformation (from FlexX) of compound 3289-8625 in the binding groove. The carbon atoms of the bound Dpr peptide are shown in brown, the carbon atoms of the bound compound 3289-8625 are shown in blue, and the carbon atoms of the PDZ domain are shown in gray.
Inhibition of Wnt Signaling by Compound 3289-8625
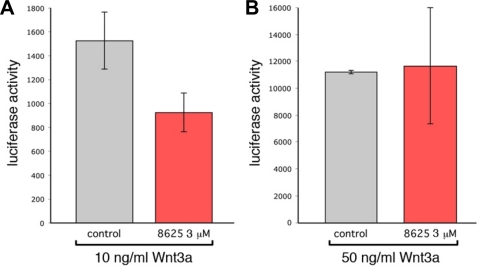
The PDZ domain of Dvl interacts directly with the conserved sequence C-terminal to the seventh transmembrane helix of the Wnt receptor Fz (3), and Wnt signaling can be inhibited by blocking this interaction (3, 15). To test whether compound 3289-8625 inhibits Wnt signaling in the cell, we evaluated the ability of compound 3289-8625 to block the canonical Wnt pathway using a 293 cell line stably transfected with a luciferase reporter under the control of lymphoid enhancer factor/T-cell factor binding sites (SuperTopflash) (34). To activate the Wnt pathway, we applied recombinant Wnt3a to the cells at either 10 ng/ml or 50 ng/ml (Fig. 5). When used at 3 μm, 3289-8625 was effective at reducing luciferase activity by about 2-fold (Fig. 5A). However, at 50 ng/ml Wnt3a 3 μm, compound 3289-8625 was not effective (Fig. 5B). These observations clearly indicated that the compound 3289-8625 competitively inhibits the Wnt signaling.
FIGURE 5.
Compound 3289-8625 blocks Wnt signaling. SuperTopflash 293 cells were treated with either 10 ng/ml (A) or 50 ng/ml (B) recombinant Wnt3a. The level of the Wnt pathway response according to luciferase activity was measured in the absence (gray bars) or presence (red bars) of 3289-8625 inhibitor at 3 μm. These data indicated that 3289-8625 could inhibit the Wnt pathway response at 10 ng/ml Wnt3a but that inhibition could be eliminated by increasing the level of Wnt pathway stimulation. Significance values are: in A, p = 0.05; in B, not significant. Error bars indicate S.E.
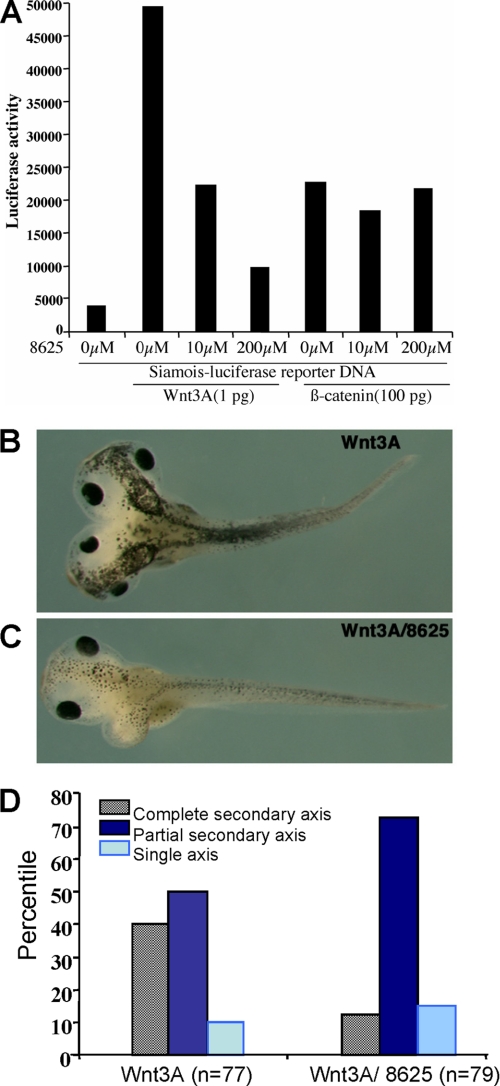
To further assess the ability of the compound to inhibit the Wnt signaling pathway in vivo, we used a previously established Xenopus system (15, 19). Xenopus Wnt target gene siamois promoter-driven luciferase reporter construct (35) was injected alone or co-injected with synthetic Wnt3A mRNA (1 pg) or β-catenin RNA (100 pg) in the animal pole region at the two-cell stage. Groups of 10 injected embryos were cultured in the absence or presence of different concentrations of the compound 3289-8625. Ectodermal explants were dissected at the late blastula stage and assayed for luciferase activity. Because the molecular mass of compound 3289-8625 is 374 daltons and its calculated octanol-water partition (logP) value and polar surface are 3.15 and 116 Å2, respectively, it is likely that the compound can penetrate the cell membrane. Therefore, instead of co-injecting the compound with Wnt3A mRNA as in earlier studies (15), we added the compounds to the culture medium at different concentrations. As shown in Fig. 6A, compound 3289-8625 was able to penetrate the keratin-rich outer embryonic cell membrane and to block Wnt signaling in a concentration-dependent fashion. At 10 μm, it inhibited Wnt3A-induced luciferase activity by 2-fold. However, the compound had no effect if the Wnt signal was activated by β-catenin, indicating that the compound inhibited Wnt signaling upstream of β-catenin. Furthermore, ventro-vegetal injection of Wnt3A mRNA at the four-cell stage induced a complete secondary axis. When these Wnt3A-injected embryos were incubated with compound 3289-8625 at 25 μm, the formation of a complete secondary axis was significantly reduced (Fig. 6, B and C). This further suggests that compound 3289-8625 was efficient at inhibiting Wnt signaling in vivo.
FIGURE 6.
Compound 3289-8625 penetrates the cell membrane and blocks Wnt signaling in Xenopus. A, the siamois reporter construct and synthetic mRNAs corresponding to Wnt3A (1 pg) and β-catenin (100 pg) were injected into the animal-pole region of two-cell stage Xenopus embryos, and ectodermal explants were dissected at late blastula stage for a luciferase assay. B, an embryo that received an injection of Wnt3A mRNA (1 pg) developed a complete secondary axis. C, an embryo that received injections of Wnt3A mRNA (1 pg) and was treated with compound 3289-8625 (10 μm) developed a partial secondary axis. D, summary of the effect of compound 3289-8625 on the formation of the secondary axis induced by Wnt3A. All results are the mean value from three independent experiments.
Compound 3289-8625 Suppresses Wnt Signaling in the Hyaloid Vessel System
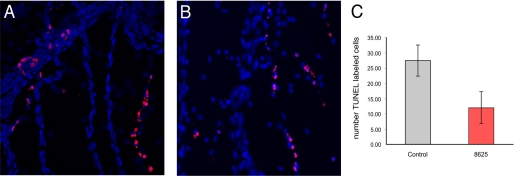
In the mouse eye, the canonical Wnt pathway is critical for scheduled vascular regression. In this system, resident macrophages produce Wnt7b and, through close contact with vascular endothelial cells of the temporary hyaloid vessels, Wnt7b activates the Wnt pathway to promote programmed cell death (36). To further examine the effects of the compound 3289-8625 on the Wnt signaling pathway in vivo, we took advantage of the earlier studies (36) and examined whether the compound was capable to interfere with the canonical Wnt pathway and reduce programmed cell death and vascular regression in the hyaloid vessel system. In the experiments, 120 nl of 12 μm compound 3289-8625 was injected into the vitreous of the eye, where the hyaloid vessels reside. Based on an estimated vitreous volume of 780 nl, we anticipated an immediate 6.5-fold dilution to give an initial concentration no less than 2.5 μm. After 24 h, hyaloid vessel networks were removed by dissection, and a TUNEL analysis (37) was performed for detection of apoptotic cells (Fig. 7). Quantification revealed that compound 3289-8625 could suppress apoptosis consistent with inhibition of the Wnt pathway.
FIGURE 7.
Compound 3289-8625 inhibits Wnt pathway responses in culture and in vivo. A and B, control-treated (A) or inhibitor-treated (B) hyaloid vessels labeled with Hoechst 33258 (blue nuclei) and with the TUNEL technique for apoptotic cells (red nuclei). Quantification of the number of TUNEL-labeled cells (C) reveals that injections of 6 μm 3289-8625 reduced the level of cell death by about 2-fold. Significance values are: p = 0.0002. Error bars indicate S.E.
Compound 3289-8625 Slows the Growth of Prostate Cancer PC-3 Cells
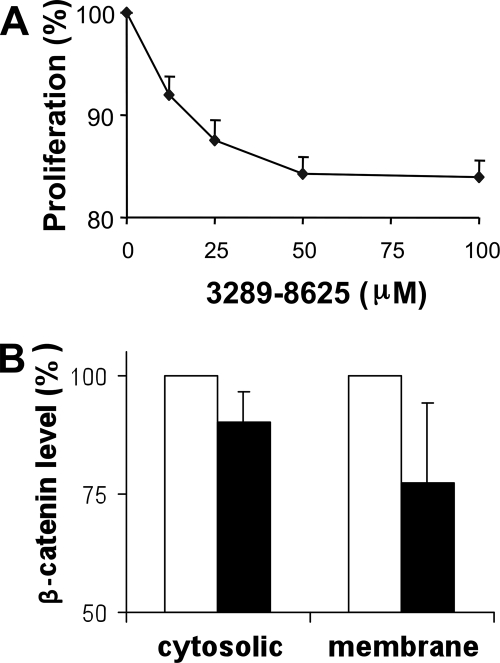
Amplification, up-regulation, and overexpression of Dvl have been reported in many cancers, including prostate cancer (6–8, 38, 39). Therefore, compounds that inhibit Wnt signaling by blocking the Fz-Dvl interaction may lead to the development of anticancer therapeutic agents (40). To test this premise, we experimentally assessed the effect of compound 3289-8625 on PC-3 prostate cancer cells, whose growth can be suppressed by inhibition of Wnt signaling (20). Fig. 8A shows the effect of the addition of compound 3289-8625 to the culture medium of PC-3 cells. At the time point of 72 h after treatment, the compound suppressed the growth of the PC-3 cells about 16% with a IC50 value about 12.5 μm, which is very close to the value of the binding affinity (10.6 ± 1.7 μm) between the Dvl PDZ domain and compound 3289-8625. To confirm that the suppression of PC-3 cells was due to inhibition of the Wnt signaling, we examined the β-catenin level in the compound 3289-8625-treated cells. Fig. 8B shows that, as expected, in the cells treated with 100 μm compound 3289-8625 for 72 h, the levels of β-catenin were decreased in both cytosolic fraction and membrane fraction, indicating that compound 3289-8625 indeed inhibited the Wnt signaling in the cells.
FIGURE 8.
Compound 3289-8625 suppresses cell proliferation and reduces β-catenin level in prostate cancer PC-3 cells. A, PC-3 cells were treated with compound 3289-8625 at different concentrations or with DMSO as a control (the same amount of DMSO was used to dissolve the compound). Cells were examined with the Promega MTT cell proliferation assay, and at the data points, cell proliferation was normalized against the control cells. Three sets of independent experiments were performed, and each data point represents the measurements from the three experiments. Error bars indicate S.E. B, the levels of β-catenin protein in the PC-3 cells treated with 80 μm compound 3289-8625 for 72 h (close bar). The β-catenin proteins isolated from cell membrane and cytosolic fractions were analyzed in Western blots as described under “Experimental Procedures.” The β-catenin protein levels were calibrated with integrin α3, a housekeeping protein that served as loading control, and normalized with the β-catenin levels in the cells that were treated with DMSO, i.e. 100% (open bar). The densitometries of protein bands presented here were quantified using the Scion Image software (Scion Corp., Frederick, MD). Each bar represents the mean of three Western blot results. Error bars indicate S.E.
DISCUSSION
Because of advances in technology, it is now possible to identify small molecules that can block the active site of a specific protein by screening an extensive small molecule data base in silico (41, 42). However, the current technology is far from perfect. Frequently, a large percentage of the potential ligands identified by in silico screening yields false positive results, and additional methods are needed to evaluate these screening results. In our studies, NMR spectroscopy was used to perform such a final validation to identify small molecules that yield true positive results. This approach has proved to be very effective in our study. The aim of the current study was to identify and evaluate small molecule inhibitors of the PDZ domain of Dvl, and using the approach of NMR-assisted virtual screening, we identified several such inhibitors.
Wnt signaling plays an important role in embryonic development and the regulation of cell growth, and its inappropriate activation has been implicated in cancer and other human diseases such as ocular vascular disease (36, 43–45). For this reason, different elements within the Wnt signaling pathways have emerged as potentially useful targets for the development of therapeutic reagents (40). The cytoplasmic protein Dvl relays the Wnt signal from the membrane-bound Wnt receptor to downstream partners through the interaction between its PDZ domain and Wnt receptor Fz. Therefore, the ligand binding site on the surface of the PDZ domain provides a perfect site for formulating small molecule inhibitors to block the Wnt signaling pathway, and such small molecules may be a powerful tool to study Wnt signaling. In addition, it may also be helpful in formulating rational approaches to the development of novel pharmaceutical agents that can interfere with specific Wnt signal events that contribute to human diseases.
The best Dvl PDZ domain inhibitor identified in our studies, compound 3289-8625, has a binding affinity to Dvl, which is similar to that of Fz, a native Dvl PDZ domain binding partner in the Wnt signaling pathway (3). Therefore, this compound may be useful for blocking the Wnt signaling pathway at the Fz-Dvl interaction point. Indeed, different lines of experiments in our studies demonstrated that this compound does have the capability of blocking Wnt signaling in vivo.
For a successful small molecule inhibitor that can be used to target an intracellular protein-protein interaction event, the compound also has to be able to penetrate through the cell membrane. The molecular mass of compound 3289-8625 is 374 daltons. In addition, the calculated logP coefficient of the compound is 3.15, and the calculated polar surface area of the molecules is about 116.3 Å. All these parameters indicate that the compound is membrane-permeable, although we have not experimentally assessed its membrane permeability. Indeed, our studies with Xenopus embryos and cell culture showed that the compound was able to penetrate through the membranes of Xenopus embryos, 293 cells, and PC-3 cells. Furthermore, in vivo studies also showed that the compound could penetrate the membrane of vascular endothelial cells.
In summary, by screening data bases of small organic molecules that were readily synthesized, we were able to identify several PDZ binding compounds without the need for in-house design and synthesis of the ligand molecules. In addition, we also showed that one of the best inhibitors could effectively inhibit the Wnt signals in vivo. Therefore, the identified small molecule inhibitors not only can be used as templates for further chemical optimization(s) but also provide us a tool for dissection of the Wnt signaling pathways.
Acknowledgments
We thank Dr. David Kimelman for the siamois reporter construct and Youming Shao, Dr. Ho-Jin Lee, and the Protein Production Facility staff at St. Jude Children's Research Hospital for producing and supplying proteins; Dr. Weixing Zhang for support with NMR experiments; Dr. Charles Ross for computer support; and Sharon Naron for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM081492. This work was also supported, in whole or in part, by the American Lebanese Syrian Associated Charities and grants from the Ligue National Contre le Cancer and the Association pour la Recherche surle Cancer.
- Fz
- Frizzled
- Dvl
- Dishevelled
- Dpr
- Dapper (Dpr)
- TMR
- 2-((5(6)-tetramethylrhodamine)carboxyamino)ethyl methanethiosulfonate
- DMSO
- dimethyl sulfoxide
- HSQC
- heteronuclear single quantum correlation
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- TES
- 2-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}ethanesulfonic acid
- TUNEL
- terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling.
REFERENCES
- 1.Bhanot P., Brink M., Samos C. H., Hsieh J. C., Wang Y., Macke J. P., Andrew D., Nathans J., Nusse R. ( 1996) Nature 382, 225– 230 [DOI] [PubMed] [Google Scholar]
- 2.Dann C. E., Hsieh J. C., Rattner A., Sharma D., Nathans J., Leahy D. J. ( 2001) Nature 412, 86– 90 [DOI] [PubMed] [Google Scholar]
- 3.Wong H. C., Bourdelas A., Krauss A., Lee H. J., Shao Y., Wu D., Mlodzik M., Shi D. L., Zheng J. ( 2003) Mol. Cell 12, 1251– 1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallingford J. B., Habas R. ( 2005) Development 132, 4421– 4436 [DOI] [PubMed] [Google Scholar]
- 5.Uematsu K., He B., You L., Xu Z., McCormick F., Jablons D. M. ( 2003) Oncogene 22, 7218– 7221 [DOI] [PubMed] [Google Scholar]
- 6.Uematsu K., Kanazawa S., You L., He B., Xu Z., Li K., Peterlin B. M., McCormick F., Jablons D. M. ( 2003) Cancer Res. 63, 4547– 4551 [PubMed] [Google Scholar]
- 7.Bui T. D., Beier D. R., Jonssen M., Smith K., Dorrington S. M., Kaklamanis L., Kearney L., Regan R., Sussman D. J., Harris A. L. ( 1997) Biochem. Biophys. Res. Commun. 239, 510– 516 [DOI] [PubMed] [Google Scholar]
- 8.Mizutani K., Miyamoto S., Nagahata T., Konishi N., Emi M., Onda M. ( 2005) Tumori 91, 546– 551 [DOI] [PubMed] [Google Scholar]
- 9.Wong H. C., Mao J., Nguyen J. T., Srinivas S., Zhang W., Liu B., Li L., Wu D., Zheng J. ( 2000) Nat. Struct. Biol. 7, 1178– 1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheyette B. N., Waxman J. S., Miller J. R., Takemaru K., Sheldahl L. C., Khlebtsova N., Fox E. P., Earnest T., Moon R. T. ( 2002) Dev. Cell 2, 449– 461 [DOI] [PubMed] [Google Scholar]
- 11.Axelrod J. D., Miller J. R., Shulman J. M., Moon R. T., Perrimon N. ( 1998) Genes Dev. 12, 2610– 2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Axelrod J. D. ( 2001) Genes Dev. 15, 1182– 1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boutros M., Paricio N., Strutt D. I., Mlodzik M. ( 1998) Cell 94, 109– 118 [DOI] [PubMed] [Google Scholar]
- 14.Rothbächer U., Laurent M. N., Deardorff M. A., Klein P. S., Cho K. W., Fraser S. E. ( 2000) EMBO J. 19, 1010– 1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shan J., Shi D. L., Wang J., Zheng J. ( 2005) Biochemistry 44, 15495– 15503 [DOI] [PubMed] [Google Scholar]
- 16.London T. B., Lee H. J., Shao Y., Zheng J. ( 2004) Biochem. Biophys. Res. Commun. 322, 326– 332 [DOI] [PubMed] [Google Scholar]
- 17.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. ( 1995) J. Biomol. NMR 6, 277– 293 [DOI] [PubMed] [Google Scholar]
- 18.Goddard T. D., Kneller D. G. ( 1998) SPARKY 3.0, University of California, San Francisco, CA [Google Scholar]
- 19.Umbhauer M., Djiane A., Goisset C., Penzo-Méndez A., Riou J. F., Boucaut J. C., Shi D. L. ( 2000) EMBO J. 19, 4944– 4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zi X., Guo Y., Simoneau A. R., Hope C., Xie J., Holcombe R. F., Hoang B. H. ( 2005) Cancer Res. 65, 9762– 9770 [DOI] [PubMed] [Google Scholar]
- 21.Shimizu H., Julius M. A., Giarré M., Zheng Z., Brown A. M., Kitajewski J. ( 1997) Cell Growth Differ. 8, 1349– 1358 [PubMed] [Google Scholar]
- 22.Zhang X. A., Bontrager A. L., Stipp C. S., Kraeft S. K., Bazzoni G., Chen L. B., Hemler M. E. ( 2001) Mol. Biol. Cell 12, 351– 365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shan J., Zheng J. J. ( 2009) J. Comput. Aided Mol. Des 23, 37– 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H. J., Wang N. X., Shao Y., Zheng J. J. ( 2009) Bioorg. Med. Chem. 17, 1701– 1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramer B., Rarey M., Lengauer T. ( 1999) Proteins 37, 228– 241 [DOI] [PubMed] [Google Scholar]
- 26.Böhm H. J. ( 1992) J Comput. Aided Mol. Des 6, 61– 78 [DOI] [PubMed] [Google Scholar]
- 27.Böhm H. J. ( 1994) J Comput. Aided Mol. Des 8, 243– 256 [DOI] [PubMed] [Google Scholar]
- 28.Klebe G., Mietzner T. ( 1994) J Comput. Aided Mol. Des 8, 583– 606 [DOI] [PubMed] [Google Scholar]
- 29.Eldridge M. D., Murray C. W., Auton T. R., Paolini G. V., Mee R. P. ( 1997) J Comput. Aided Mol. Des 11, 425– 445 [DOI] [PubMed] [Google Scholar]
- 30.Muegge I., Martin Y. C. ( 1999) J Med. Chem. 42, 791– 804 [DOI] [PubMed] [Google Scholar]
- 31.Jones G., Willett P., Glen R. C., Leach A. R., Taylor R. ( 1997) J. Mol. Biol. 267, 727– 748 [DOI] [PubMed] [Google Scholar]
- 32.Kuntz I. D., Blaney J. M., Oatley S. J., Langridge R., Ferrin T. E. ( 1982) J. Mol. Biol. 161, 269– 288 [DOI] [PubMed] [Google Scholar]
- 33.Wüthrich K. ( 2000) Nat. Struct. Biol. 7, 188– 189 [DOI] [PubMed] [Google Scholar]
- 34.Xu Q., Wang Y., Dabdoub A., Smallwood P. M., Williams J., Woods C., Kelley M. W., Jiang L., Tasman W., Zhang K., Nathans J. ( 2004) Cell 116, 883– 895 [DOI] [PubMed] [Google Scholar]
- 35.Brannon M., Gomperts M., Sumoy L., Moon R. T., Kimelman D. ( 1997) Genes Dev. 11, 2359– 2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lobov I. B., Rao S., Carroll T. J., Vallance J. E., Ito M., Ondr J. K., Kurup S., Glass D. A., Patel M. S., Shu W., Morrisey E. E., McMahon A. P., Karsenty G., Lang R. A. ( 2005) Nature 437, 417– 421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gavrieli Y., Sherman Y., Ben-Sasson S. A. ( 1992) J. Cell Biol. 119, 493– 501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagahata T., Shimada T., Harada A., Nagai H., Onda M., Yokoyama S., Shiba T., Jin E., Kawanami O., Emi M. ( 2003) Cancer Sci. 94, 515– 518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wissmann C., Wild P. J., Kaiser S., Roepcke S., Stoehr R., Woenckhaus M., Kristiansen G., Hsieh J. C., Hofstaedter F., Hartmann A., Knuechel R., Rosenthal A., Pilarsky C. ( 2003) J. Pathol. 201, 204– 212 [DOI] [PubMed] [Google Scholar]
- 40.Barker N., Clevers H. ( 2006) Nat. Rev. Drug Discov. 5, 997– 1014 [DOI] [PubMed] [Google Scholar]
- 41.Shoichet B. K. ( 2004) Nature 432, 862– 865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leach A. R., Shoichet B. K., Peishoff C. E. ( 2006) J. Med. Chem. 49, 5851– 5855 [DOI] [PubMed] [Google Scholar]
- 43.Giles R. H., van Es J. H., Clevers H. ( 2003) Biochim. Biophys. Acta 1653, 1– 24 [DOI] [PubMed] [Google Scholar]
- 44.Logan C. Y., Nusse R. ( 2004) Annu. Rev. Cell Dev. Biol. 20, 781– 810 [DOI] [PubMed] [Google Scholar]
- 45.Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. ( 2004) Nat. Rev. Genet. 5, 691– 701 [DOI] [PubMed] [Google Scholar]