Abstract
The inhibitor peptide DT-2 (YGRKKRRQRRRPPLRKKKKKH) is the most potent and selective inhibitor of the cGMP-dependent protein kinase (PKG) known today. DT-2 is a construct of a PKG tight binding sequence (W45, LRKKKKKH, KI = 0.8 μm) and a membrane translocating sequence (DT-6, YGRKKRRQRRRPP, KI = 1.1 μm), that combined strongly inhibits PKG catalyzed phosphorylation (KI = 12.5 nm) with ∼1000-fold selectivity toward PKG over protein kinase A, the closest relative of PKG. However, the molecular mechanism behind this inhibition is not entirely understood. Using a combination of photoaffinity labeling, stable isotope labeling, and mass spectrometry, we have located the binding sites of PKG-specific substrate and inhibitor peptides. Covalent linkage of a PKG-specific substrate analogue was localized in the catalytic core on residues 356–372, also known as the glycine-rich loop, essential for ATP binding. By analogy, the individual inhibitor peptides W45 and DT-6 were also found to cross-link near the glycine-rich loop, suggesting these are both substrate competitive inhibitors. A bifunctional photoreactive analogue of DT-2 was found to generate dimers of PKG. This cross-linking induced covalent PKG dimerization was not observed for an N-terminal deletion mutant of PKG, which lacks the dimerization domain. In addition, non-covalent mass spectrometry was used to determine binding stoichiometry and binding order of the inhibitor peptides. Dimeric PKG binds two W45 and DT-6 peptides, whereas only one DT-2 molecule was observed to bind to the dimeric PKG. Taken together, these findings imply that (i) the two individual components making up DT-2 are both targeted against the substrate-binding site and (ii) binding of a single DT-2 molecule inactivates both PKG monomers simultaneously, which is an indication that (iii) in cGMP-activated PKG the catalytic centers of both subunits may be in each other's proximity.
Among the superfamily of protein kinases the two cyclic nucleotide-regulated protein kinases, cAMP-dependent protein kinase and cGMP-dependent protein kinase, form a closely related subfamily of serine/threonine protein kinases (1–4). Both proteins share several structural elements, such as the N-terminal dimerization domain, an autoinhibition site, two in-tandem cyclic nucleotide-binding sites, and a highly conserved catalytic core (Fig. 1, A and B). Despite these similarities, these two enzymes display differences, which account for their unique properties. Whereas PKA2 is nearly ubiquitous, PKG is primarily found in the lung, cerebellum, and smooth muscles (5, 6). From a structural point of view these cyclic nucleotide-dependent protein kinases differ as well. The holoenzyme of PKA is a tetramer composed of two regulatory and two catalytic subunits. The catalytic subunits are non-covalently attached to the regulatory subunit dimer. Upon interaction with cAMP, the catalytic subunits dissociate from the holoenzyme and are free to catalyze heterophosphorylation (Fig. 1C). The mammalian type I PKGs are homodimeric cytosolic proteins containing two identical polypeptides of ∼76 kDa. Alternative mRNA splicing produces type Iα and type Iβ PKG, which are identical proteins apart from their first ∼100 N-terminal residues (7). Each PKG subunit is composed of a regulatory and a catalytic domain on a single polypeptide chain. Consequently, when cGMP activates PKG, the catalytic and regulatory components remain physically attached (Fig. 1D). Within the catalytic domain PKA and PKG share a strong primary sequence homology (8). Not surprisingly, these enzymes also exhibit overlapping substrate specificities, a feature that often interferes with efforts to elucidate their distinct biological pathways. Peptide substrates with a primary amino acid sequence motif RRX(S/T)X are in general recognized by both PKA and PKG (9). Besides this strong overlapping substrate specificity, several studies report on subtle differences in determinants that discriminate for PKA and PKG substrate specificity (10–16). To specifically discriminate between PKG and PKA activity in biological assays a highly specific PKG peptide inhibitor was developed (Table 1) (17). This peptide, YGRKKRRQRRRPPLRKKKKKH (DT-2), is the most potent and selective PKG inhibitor known today. Recently, the validity of DT-2 as a superior inhibitor of PKG in terms of potency, selectivity, and membrane permeability has been demonstrated (18–24). The inhibitor is a construct of a substrate competitive sequence, LRKKKKKH (W45), derived from a library screen that selected for tight PKG binding sequences, with a significant specificity toward PKG over PKA, and a membrane translocating signal peptide, YGRKKRRQRRRPP (DT-6). DT-2 strongly inhibits PKG-catalyzed phosphorylation (Ki = 12.5 nm), however, the molecular nature of DT-2 inhibition is not entirely understood (25). Because high resolution structural data are not available for PKG, one of our goals is to elucidate binding sites for PKG-specific substrates and inhibitors in more detail using a combination of mass spectrometric techniques and photoaffinity labeling. To further delineate the nature of inhibition we have developed photoaffinity analogues of DT-2 and related inhibitory peptides, as well as a high affinity peptide substrate. The method of photoaffinity labeling enables the direct probing of target proteins through a covalent bond, which is photochemically introduced between a ligand and its specific receptor (26). In combination with modern mass spectrometric techniques this is a powerful approach for the characterization of peptide-protein interactions (27). Substrate and inhibitor peptides containing photoactivatable analogues of phenylalanine, 4-benzoyl-l-phenylalanine (Phe(Bz)) or 4′-(3-(trifluoromethyl)-3H-diazirin-3-yl)-l-phenylalanine (Phe(Tmd)) were synthesized and used to locate their substrate/inhibitor-binding sites on PKG. These measurements indicate that the substrate peptide resides near the glycine-rich loop within the catalytic domain and that the inhibitor peptides are directed similarly toward this substrate-binding site, thereby acting as competitive inhibitors. In addition, nanoflow electrospray ionization time of flight mass spectrometry (ESI-TOF-MS) was performed to study the interaction between DT-2 and PKG in more detail. ESI-MS has proven to be a useful tool to analyze the non-covalent interaction of proteins with ligands, oligonucleotides, peptides, or other proteins (28–31). Using this technique, important information on conformational changes (32–35), measurement of relative dissociation constants (36, 37), and sequential binding order and cooperativity (38, 39) can be obtained. ESI-MS confirms that PKG is primarily a homodimer and is able to bind four cGMP molecules. Binding of DT-2 was strongly enhanced in the presence of cGMP. Surprising is the observation that only one DT-2 molecule binds to dimeric PKG. The information derived from these measurements allows for molecular modeling and structural refinements of the next generation of PKG-selective inhibitors.
FIGURE 1.
Linear arrangement of the functional domains of the regulatory and catalytic subunit of PKA (A) and PKG (B) type I and schematic representation of the current working models of the activation process of PKA (C) and PKG (D) type 1. Binding of cAMP to the PKA induces a conformational change that results in the dissociation of the catalytic subunits. Binding of cGMP to PKG also induces a conformational change, which exposes the catalytic domains, but both catalytic domains remain near each other via the N-terminal dimerization domain. (Images adapted from Scholten et al. (4).)
TABLE 1.
Inhibition contants (KI) of PKA- or PKG-specific peptide inhibitors and the PKA/PKG specificity index
| Peptide | Sequence | PKGKi | PKAKi | Specificity index (PKA/PKG) | Ref. |
|---|---|---|---|---|---|
| μm | μm | ||||
| PKI(5–24) | TTYDFIASGRTGRRNAIHD-NH2 | 150 | 0.003 | 0.0002 | (11) |
| WW21 | TQAKRKKALAMA-NH2 | 7.5 | 750 | 100 | (11) |
| W45 | LRKKKKKH | 0.82 ± 0.33 | 559 | 680 | (17) |
| DT-6 | YGRGGRRQRRRPP | 1.1 ± 0.22 | 26 ± 4 | 23.6 | (17) |
| DT-2 | YGRKKRRQRRRPPLRKKKKKH | 0.0125 ± 0.003 | 16.5 ± 3.8 | 1320 | (17) |
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
ArgoGelTM Rink-NH-Fmoc resin (Argonaut Technologies, Muttenz, Switzerland) functionalized with a 4-((2′,4′-dimethoxyphenyl)aminomethyl)phenoxyacetamido moiety (Rink amide linker) (40) was used to obtain C-terminal peptide amides. The coupling reagent 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) (41) was obtained from Richelieu Biotechnologies Inc. (Montreal, Canada). The coupling reagents N-(dimethylamino)-1H-1,2,3-triazolo[4,5-bipyridin-1-yl-methylene]-N-methylmethanaminium hexafluorophosphate/N-hydroxy-7-azabenzotriazole (HATU/HOAt) (42) were obtained from Applied Biosystems. N-Hydroxybenzotriazole (HOBt), Fmoc-hydroxylsuccinimide and Nα-9-fluorenylmethyloxycarbonyl (Fmoc) amino acids were obtained from Advanced ChemTech (Machelen, Belgium). The side-chain protecting groups were: Boc, tert-butyloxycarbonyl, for lysine; tBu, tert-butyl, for threonine, tyrosine, and serine; Pbf, 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl, for arginine; and Trt, trityl, for glutamine and histidine. Fmoc-Phe(Tmd)-OH (N-9-fluorenylmethyloxycarbonyl-4′-[3-(trifluoromethyl-3H-diazirin-3-yl)]phenylalanine) was obtained from photoprobes (Sins, Switzerland). U-13C6/15N-labeled leucine was obtained from Cambridge Isotope Laboratories (Andover, MA). This amino acid was converted into its corresponding Fmoc derivative according to the method of ten Kortenaar (43). U-13C6/15N4-labeled arginine was obtained from Sigma. Peptide grade dichloromethane, N-methylpyrrolidone (NMP), methyl-tert-butyl ether, trifluoroacetic acid and HPLC-grade acetonitrile were purchased from Biosolve (Valkenswaard, The Netherlands). Piperidine and N,N-diisopropylethylamine (DIPEA) were obtained from Acros Organics (‘s-Hertogenbosch, The Netherlands). HPLC grade trifluoroacetic acid and 1,2-ethanedithiol were obtained from Merck (Amsterdam, The Netherlands). AMPPNP, ATP, cGMP, dithiothreitol, iodoacetamide, and MgCl2 were obtained from Sigma. Sequencing grade trypsin and GluC were purchased from Roche Diagnostics GmbH (Mannheim, Germany). Bovine PKG type Iα was expressed in SF9 insect cells and purified to homogeneity essentially as described (17).
Peptide Synthesis
Peptide synthesis was performed automatically on an Applied Biosystems 433A Peptide Synthesizer (Foster City, CA). Analytical and preparative HPLC runs were performed on a Gilson HPLC workstation (Middleton, WI). As an example, the peptide acetyl-Thr-Gln-Ala-Lys-Arg-Lys-Lys- Ser-Leu(12C/14N,13C/15N)-Ala-Met-Phe(Tmd)-Leu-Arg-NH2 (W64-[Phe(Tmd14)]) was synthesized using the FastMoc protocol on a 0.25-mmol scale (44). Each synthetic cycle consisted of Nα-Fmoc removal by a 10-min treatment with 20% piperidine in NMP, a 6-min NMP wash, a 45-min coupling step with 1.0 mmol of preactivated Fmoc amino acid in the presence of 2 eq of DIPEA, and a 6-min NMP wash. Nα-Fmoc amino acids were activated in situ with 1.0 mmol of HBTU/HOBt (0.36 m in NMP) in the presence of DIPEA (2.0 mmol). After the final Fmoc removal the free amine was acetylated with an excess of acetic acid anhydride/DIPEA/HOBt in NMP. Fmoc-Phe(Tmd)-OH was coupled with HATU/HOAt in an excess of only 2 eq (with respect to the loading of the resin) for 90 min. Fmoc-Leu(13C/15N)-OH was coupled in the presence of 1 equimolar equivalent of Fmoc-Leu(12C/14N)-OH with HATU/HOAt in an excess of only 2 eq for 90 min. After these coupling steps any remaining free amine functionalities were acetylated with acetic acid anhydride/DIPEA/HOBt. The peptide was cleaved from the resin and deprotected by treatment with trifluoroacetic acid/H2O/1,2-ethanedithiol (95:2.5:2.5, v/v) at room temperature for 3 h. The peptide was precipitated with methyl-tert-butyl ether/hexane (1:1, v/v) at −20 °C. The precipitate was decanted and subsequently washed with cold methyl-tert-butyl ether/hexane (1:1, v/v, 3×) and finally lyophilized from tert-BuOH/H2O (1:1, v/v).
Peptide Purification
The crude lyophilized peptides were dissolved in a minimum amount of 0.1% trifluoroacetic acid in CH3CN/H2O (5:95, v/v) and loaded onto the HPLC column (Phenomenex Luna C8, 100-Å pore size, 10-μm particle size, 2.2 × 25 cm). The peptides were eluted with a flow rate of 10.0 ml/min using a linear gradient of buffer B (100% in 96 min) from 100% buffer A (buffer A: 0.1% trifluoroacetic acid in CH3CN/H2O (5:95, v/v), buffer B: 0.1% trifluoroacetic acid in CH3CN/H2O (60:40, v/v)).
Peptide Characterization
To determine purity, the synthesized peptides were subjected to liquid chromatography ESI-MS using a Shimadzu LCMS-QP8000 (Duisburg, Germany) single quadrupole bench-top mass spectrometer operating in a positive ionization mode. The purified peptides were loaded onto a Phenomenex Luna C8 column (100-Å pore size, 5-μm particle size, 0.46 × 25 cm) at a flow rate of 1 ml/min using a linear gradient of buffer B (100% in 48 min) from 100% buffer A (buffer A: 0.1% trifluoroacetic acid in CH3CN/H2O (5:95, v/v); buffer B: 0.1% trifluoroacetic acid in CH3CN/H2O (60:40, v/v)). The mass of purified peptides was finally confirmed by ESI-MS.
Photoaffinity Labeling and Proteolysis
Upon irradiation with UV-light (∼350 nm), 4′-(3-(trifluoromethyl)-3H-diazirin-3-yl)-l-phenylalanine irreversibly generates a highly reactive singlet carbene by a photodissociative process. This intermediate can enforce hydrogen abstraction on appropriately oriented target C–H bonds within a distance ∼3 Å or less (45, 46). Because the electrophilic nature of the carbene intermediate renders solvent O–H bonds as appropriate H-donors, the radius of the photochemical insertion reaction is limited. Specific labeling by Phe(Tmd) would therefore suggest a closer proximity between target protein and the photoprobe (45). Upon excitation (∼350 nm), the 4-benzoyl-l-phenylalanine irreversibly generates a highly reactive intermediate that rapidly enforces hydrogen abstraction on appropriately oriented target C–H bonds and has a lower reactivity toward water compared with Phe(Tmd). The protein, at a concentration of 0.2 mg/ml in a 25 mm potassium phosphate buffer (pH 7.4), containing 50 mm NaCl, 1.2 mm EDTA, 4 mm Mg(OAc)2, and 200 μm AMPPNP, was incubated with 200 μm photoaffinity label at 30 °C for 10 min in the dark. The protein photolabel mixture was illuminated at 366 nm for 15 min in a 96-well plate on ice using a Sylvania light source (F8T5/BTB). For intact protein mass measurements, the sample was dialyzed overnight against 200 mm NH4OAc and finally concentrated using an Ultrafree-0.5 centrifugal filter (Millipore, Bedford, MA). Protein masses were determined by nanoflow ESI-TOF-MS on a Micromass LC-T time-of-flight instrument using homemade borosilicate nano-electrospray needles. For optimal detection of the intact PKG source, interface pressures were optimized as previously described (47, 48). For the HPLC tandem MS analysis the PKG protein was digested. Prior to digestion all cysteines were reduced with dithiothreitol and alkylated using iodoacetamide. The protein sample was subsequently diluted 10 times with 50 mm NH4HCO3, pH 8.0, and spun in an Ultrafree-0.5 centrifugal filter. This step was repeated three times. Finally, either trypsin or GluC was added to a final protein:protease concentration of 20:1 by weight. The digestion mixture was incubated at 37 °C for 16 h.
LC-ESI-MS and MS/MS Peptide Analysis
Proteolytic peptide mixtures were analyzed using an Ultimate nano-HPLC system (LC Packings, Amsterdam, The Netherlands) operating essentially as described by Meiring et al. (49). Samples were loaded onto a Phenomenex Aqua C18 pre-column (200-Å pore size, 5-μm particle size) at 5 μl/min in 100% solvent A (0.6% acetic acid). Subsequently, peptides were separated on an Aqua C18 nano-reversed phase column (50 μm inner diameter, 20 cm/liter) with a flow rate of 100–150 nl/min. Elution was performed using a gradient of 0–60% solvent B (80% CH3CN and 0.6% acetic acid) at 1.2% min−1. The column effluent of the nano-LC was directly infused into a Qtof-1 quadrupole time-of flight instrument (Micromass Ltd., Manchester, UK) using in-house prepared nanospray emitters. Mass spectra were acquired in positive ion mode, and argon was used as collision gas. During LC-MS runs, analysis time of cross-linked products was increased by performing peak parking essentially as described by Meiring et al. (49). In short, prior to elution of an analyte of interest the column flow rate was lowered to ∼20–40 nl/min by instantaneously dropping the column pressure using an extra flow-splitter. Hereby, the elution time was broadened to more than 2 min, and this allowed sufficient time to perform a low energy collision-induced dissociation analysis. For the fragmentation of labeled peptides the quadruple mass resolution parameters were set to a relatively large mass window (∼5 m/z units) to mass select both light and heavy leucine- or arginine-labeled precursor ions.
RESULTS
Strategy for Determining Peptide-binding Sites Using Photoactivatable Peptide Analogues
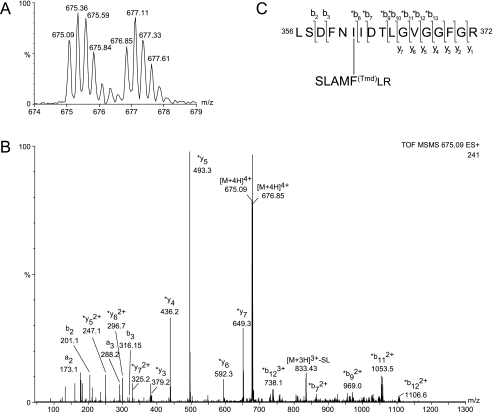
Substrate and inhibitor peptide analogues containing either 4′-(3-(trifluoromethyl)-3H-diazirin-3-yl)-l-phenylalanine or 4-benzoyl-l-phenylalanine were synthesized using standard solid phase peptide synthesis (Table 2). These two phenylalanine derivatives are commonly used as photoactivatable residues that allow for the covalent cross-linking between peptide ligands and the target protein (50, 51). The substrate peptide TQAKRKKSLAMFLR (W64), in which the serine is the phosphate acceptor site (P0), was derived from a library screen directed to identify highly PKG-specific substrates (11, 52). This peptide with a Km of 260 nm and a PKG/PKA specificity index of 19.2 is the most selective and specific synthetic substrate for PKG known today (Table 3). A photoactivatable analogue of this substrate was designed by incorporation of either Phe(Bz) or Phe(Tmd) at the p + 4 position, replacing the original phenylalanine. This position for the photoactivatable residue was chosen for two reasons. Firstly, in this manner the bulky side chains of Phe(Bz) and Phe(Tmd) are positioned on a site where a bulky amino acid is tolerated, so negative effects due to steric hindrance are minimized. Secondly, previously it was shown that a phenylalanine at the p + 4 position has a positive effect on selectivity toward PKG and increases PKG/PKA specificity index (10, 11) (Table 3). The photoactivatable substrate peptide was cross-linked to PKG in the presence and absence of cGMP. Fig. 2 shows the ESI-TOF-MS spectra of PKG after cross-linking with W64-[Phe(Bz)-12)] both in absence (Fig. 2A) and presence of cGMP (Fig. 2B). In Fig. 2A the abundant ion signals between m/z 5500 and 7500 correspond to a protein of 153,010, matching to dimeric apo-PKG with a calculated mass of 152,875 Da. A second less-abundant ion series corresponding to a total mass of 154,850 Da was also observed, and this matches apo-PKG with 1 W64-[Phe(Bz)-12] peptide (mass 1840 Da), indicating a small percentage of protein is labeled. When the same sample was not subjected to UV light, this product between PKG and W64-[Phe(Bz)-12] was not observed (data not shown), suggesting the product in Fig. 2A is covalent and specifically formed after irradiation. More importantly, Fig. 2B shows multiple abundant ion series, matching to cGMP-saturated PKG with an apparent mass of 154,421 (four molecules of cGMP cause a net mass increase of 1380 Da). Next to that cGMP-saturated PKG with 1 and 2 W64-[Phe(Bz)-12] peptide labels are observed. Hence, for cGMP-activated PKG the incorporation level of W64-[Phe(Bz)-12] is much higher and up to two peptide substrates can be attached, which was, again, only specific for the irradiated sample. To identify directly the sites of proximity between PKG and peptide substrate, the cross-linked complex was digested with trypsin or GluC, and the proteolytic digests were subjected to nanoflow HPLC and ESI quadrupole TOF-MS. To facilitate the retrieval of cross-linked peptides, the photoaffinity labeled peptides were additionally labeled isotopically during synthesis, by incorporating for 50% 13C6,15N1-leucine. This will give the substrate peptide and subsequently all cross-linked products a characteristic isotopic doublet with a 7-Da difference that will distinguish them from the majority of non-cross-linked peptides, and this will enhance the identification of cross-linked products tremendously. To facilitate the visualization of cross-linked products even further, a two-dimensional display of the LC-MS run was constructed in which the retention time is orthogonally plotted against the m/z ratio, and the ion intensity is the variable parameter (Fig. 3B). Manual inspection of this peptide display identified an ion-doublet around 41 min at m/z 675.09 and m/z 676.85 (Fig. 3, B inset, and C). Subsequently, the sample was injected once more, and the ion-doublet was selected for tandem mass spectrometric analysis. Fig. 4A displays the isotopic pattern of one doublet that was isolated and subsequently fragmented. This doublet corresponds to the [M+4H]4+ of a cross-linked product with an apparent monoisotopic mass of 2696.4 Da. Tandem mass spectrometry was performed on the [M+4H]4+ ions (Fig. 4B) to identify the binding site on PKG. The fragmentation spectrum displays a series of y″ ions, which match to the PKG sequence 366GVGGFGR372. Further manual interpretation revealed the composition of the cross-linked product (Fig. 4C) as the tryptic PKG-peptide (K)-356LSDFNIIDTLGVGGFGR372-(V) (monoisotopic mass of 1779.92 Da) and the substrate peptide SLAMFLR after cleavage by trypsin, with monoisotopic mass 916.44 Da, taking into account the loss of 28 Da (N2, specific for the Phe(Tmd) coupling) and conversion of the C-terminal amide of the arginine into a carboxyl group during digestion. The cross-link between PKG and the substrate was determined to be Ile361. To demonstrate that this is the truly functional binding site of the substrate peptide, ATP and Mg2+ were directly added after cross-linking. After proteolytic digestion and MS analysis, a phosphorylated cross-linked product could be identified (supplemental Fig. S1), which validates that the cross-linked peptide is properly orientated to become phosphorylated.
TABLE 2.
List of synthesized PKG-specific substrate and inhibitor photoaffinity labels
| Synthesis |
Sequencea,b | Photolabel | Cross-link | |
|---|---|---|---|---|
| Peptide no. | Name | |||
| a-0 | W64-[Phe(Bz)-12] | TQAKRKKSLAMF*LR | Ph(Bz) | yes |
| b-1 | W64-[Phe(Tmd)-12] | TQAKRKKSLAMF*LR | Ph(Tmd) | yes |
| b-2a | W45-[Phe(Tmd)-8] | LRKKKKKF* | Ph(Tmd) | no |
| b-2b | DT-2-[Phe(Tmd)-21] | YGRKKRRQRRRPPLRKKKKKF* | Ph(Tmd) | no |
| b-2c | DT-2-[Phe(Tmd)-1] | F*GRKKRRQRRRPPLRKKKKKF* | Ph(Tmd) | no |
| b-3 | DT-2-[Phe(Tmd)-1,21] | F*GRKKRRQRRRPPLRKKKKKH | Ph(Tmd) | no |
| b-4a | W45-[Phe(Tmd)-8] | LRKKKKKF* | Ph(Tmd) | no |
| b-4b | DT-2-[Phe(Tmd)-21] | YGRKKRRQRRRPPLRKKKKKF* | Ph(Tmd) | no |
| b-4c | DT-2-[Phe(Tmd)-1] | F*GRKKRRQRRRPPLRKKKKKF* | Ph(Tmd) | no |
| b-5 | DT-2-[Phe(Tmd)-1,21] | F*GRKKRRQRRRPPLRKKKKKH | Ph(Tmd) | no |
| c-1a | W45-[Phe(Bz)-1] | F*RKKKKKY | Ph(Bz) | yes |
| c-1b | W45-[Phe(Bz)-2] | LF*RKKKKKY | Ph(Bz) | yes |
| c-2 | W45-[Phe(Bz)-6] | LRKKKF*KY | Ph(Bz) | yes |
| d-3a | DT-6-[Phe(Bz)-6] | YGRKKF*RQRRRPP | Ph(Bz) | yes |
| d-3b | DT-2-[Phe(Bz)-6,14] | YGRKKF*RQRRRPPF*RKKKKKY | Ph(Bz) | yes |
a Bold letter indicates the position of the isotopically labeled Leucine (L) or Arginine (R) or the position of the photoactivatable phenylalanine derivative (F*).
b Peptides are N-terminally acetylated and C-terminally amidated.
TABLE 3.
Km and kcatvalues for the phos pho ryl a tion by PKA and PKG of synthetic peptides derived from previous studies
| Peptidea | PKG |
PKA |
Index |
||||
|---|---|---|---|---|---|---|---|
| Km | kcat | kcat/Km | Km | kcat | kcat/Km | PKG/PKA | |
| μm | μmol/min/mg | μm | μmol/min/mg | ||||
| PKI(5–24): | |||||||
| TYADFIASGRTGRRNSIb | 2.4 | 1.0 | 0.42 | 0.12 | 1.0 | 8.33 | 0.05 |
| IASGRTGRRNSIb | 2.7 | 0.74 | 0.27 | 0.18 | 1.4 | 7.78 | 0.03 |
| IASGRTGRRNSIb | 7.0 | 0.79 | 0.11 | 0.32 | 0.56 | 1.75 | 0.63 |
| W64: | |||||||
| TQAKRKKSLAMFLRc | 0.26 | 11.5 | 44.23 | 1.7 | 3.9 | 2.29 | 19.2 |
| TQAKRKKSLAMFc | 0.5 | 11.0 | 22 | 3.3 | 5.7 | 1.72 | 12.9 |
| TQAKRKKSLAc | 1.74 | 11.1 | 6.37 | 2.68 | 7.9 | 5.22 | 2.16 |
| RKISASEFDRPLRd | 68 | 11 | 0.162 | 320 | 3.2 | 0.010 | 16.2 |
| RKISASEFd | 120 | 5.2 | 0.043 | 480 | 2.6 | 0.006 | 8.1 |
| RKISASEd | 80 | 5.6 | 0.070 | 93 | 3.7 | 0.040 | 1.8 |
FIGURE 2.
Nanoflow ESI-TOF mass spectra of PKG that was irradiated in the presence of the photolabel W64-[Phe(Tmd)-12] for 10 min at 366 nm in the absence of cGMP (A) and the presence of 100 μm cGMP (B). Shown are on the left are the m/z spectra containing the charge state envelope of dimeric PKG, and shown are on the right are the corresponding deconvoluted mass spectra. These spectra clearly indicate that cGMP activation is a prerequisite for the binding of peptides substrates.
FIGURE 3.
Analysis of a tryptic digest of the PKG/W64-[Phe(Bz)-12] cross-link product. A, base peak intensity ion chromatogram of the trypsin digest. B, peptide display of the LC-MS chromatogram. Inset, enlarged view of the peptide display around 41 min shows the presence of an ion doublet. C, three-dimensional image of the [M+4H]4+ confirms the presence of ion doublet with 7 Da mass difference due to the 50% 13C6,15N1-leucine in the photoaffinity substrate.
FIGURE 4.
Tandem mass spectrometry (MS/MS) analysis of the tryptic cross-linked peptide derived from PKG/W64-[Phe(Bz)-12]. A, the [M+4H]4+ precursor ion that was selected by the quadrupole using a broad selection window (∼5 m/z units) to allow transmission and subsequent simultaneous fragmentation of the light and heavy leucine labeled cross-linked products. B, MS/MS spectrum of the [M+4H]4+ at m/z 675–676. C, primary sequence of the cross-linked product and identified fragment ions. The b and y″ ions corresponding the primary sequence of PKG, without covalently attached substrate are annotated by bn or yn, whereas b or y″ ions of the primary sequence of PKG that carries the covalent attached substrate are labeled *bn or *yn.
Location of the Binding Site of the Peptide Inhibitor W45 on PKG
Using the above mentioned strategy, several photoaffinity-labeled analogues of W45, DT-6, and DT-2 were synthesized (see Table 2). On basis of information from the peptide library screen that led to the discovery of W45 (17), two different photolabeled peptides were designed. In the first peptide, W45-[Phe(Bz)-2], the photoreactive amino acid was positioned between the N-terminal leucine and the adjacent arginine. At this position, bulky side chains positively contribute to binding and hence the photoaffinity label is expected not to lead to a decreased affinity. For the second peptide, W45-[Phe(Bz)-6], the fourth lysine was replaced with the photoreactive residue, because this particular lysine showed to be an important determinant in binding. After irradiation the obtained cross-linked products were proteolytically digested by either trypsin or GluC. From the GluC digest of PKG cross-linked with W45-[Phe(Bz)-2], a peptide was identified with an apparent mass of 2947.9 Da (Fig. 5A). The tandem mass spectrum of the [M+5H]5+ peak, shown in Fig. 5B, displays several multiply charged ions from which a partial sequence of FNIID can be sequenced. Again, manual interpretation determines the composition of the product as the GluC fragment of PKG (D)-359FNIIDTLGVGGFGRVE374-(L), with a monoisotopic mass of 1692.8 and the intact photolabel W45-[Phe(Bz)-2], with a monoisotopic mass of 1254.7 Da (Fig. 5C). Unfortunately, the exact location of the covalent linkage could not be determined from this spectrum, however, it could be pinpointed to the residues 364TLGVGGFGRVE374 (Fig. 5C). From the tryptic digest of PKG cross-linked with W45-[Phe(Bz)-6]), a cross-linked product of 3170.2 Da was detected. The low energy collision-induced dissociation spectrum of this product (supplemental Fig. S2) was identified as the tryptic fragment of PKG (K)-356LSDFNIIDTLGVGGFGRVELVQLK379-(S), with monoisotopic mass 2589.4 Da and the trypsin-cleaved inhibitor peptide acetyl-LF(Bz)R, with monoisotopic mass 580.3 Da. Also for this cross-linked product no exact site of cross-linkage could be determined, but the fragmentation spectrum restricted the site of linkage to the part 366GVGGFGRVELVQLK379. In addition, a second cross-linked product was identified from this digest. Fig. 6 displays the fragmentation spectrum of a cross-linked product with a monoisotopic mass of 1946.2 Da. From this a partial protein sequence of Q-(312 amu)-FQTI could be derived. This partial sequence tag matches to the PKG primary sequence 194QCFQTIMMR202. The exact composition of this cross-linked product is composed of acetyl-LF*R (monoisotopic mass 580.3), and the trypsin cleavage fragment QCFQTIMMR, in which the cysteine is actually carrying an additional dithiothreitol adduct that is carbamidomethylated. This is clear from y″-ions series in the tandem mass spectrum in which a “gap” of 312 Da is seen, which matched to cysteine (103), a carbamidomethyl group (+58), and dithiothreitol (+154) and the loss of three protons (−3 Da) for the cysteine (−1) and the oxidized dithiothreitol-adduct (−2).
FIGURE 5.
MS/MS analysis of a cross-linked product derived from a GluC-digest of PKG, which was cross-linked with W45-[Phe(Bz)-6]. A, the [M+5H]5+ precursor ion at m/z 590–593 that was selected by the quadrupole using a broad selection window (∼5 m/z units). The characteristic doublet is indicative for the presence of the isotopically labeled leucine from the photoaffinity labeled peptide. B, MS/MS spectrum of the [M+5H]5+ at m/z 590–593. C, primary sequence of the cross-linked product and identified fragment ions. The b and y″ ions corresponding the primary sequence of PKG, without covalently attached inhibitor are annotated by bn or yn, whereas b or y″ ions of the primary sequence of PKG that carries the covalent attached photolabel are labeled *bn or *yn.
FIGURE 6.
MS/MS analysis of a cross-linked product derived from a trypsin-digest of PKG, which was cross-linked with W45-[Phe(Bz)-2]. A, the [M+3H]3+ precursor ion at m/z 649–652 that was selected by the quadrupole using a broad selection window (∼5 m/z units). The characteristic doublet is indicative for the presence of the isotopically labeled leucine from the photoaffinity-labeled peptide. B, MS/MS spectrum of the [M+3H]3+ at m/z 649–652. C, primary sequence of the cross-linked product and identified fragment ions. The b and y″ ions corresponding the primary sequence of PKG, without covalently attached inhibitor are annotated by bn or yn, whereas b or y″ ions of the primary sequence of PKG that carries the covalent attached photolabel are labeled *bn or *yn. The cysteine was found to carry a dithiothreitol adduct that was subsequently modified by iodoacetamide.
Location of the Binding Site of the Peptide Inhibitor DT-6 on PKG
The DT-6 photoaffinity label acetyl-YGRKKRRQRF(Bz)RPP-amide (DT-6-[Phe(Bz)2]) was synthesized to determine the DT-6-binding site on PKG. The arginine nearest to the C terminus of the peptide was chosen for isotopic labeling. However, in contrast to the leucine labeling, it was observed that during peptide synthesis incorporation of the heavy arginine was less efficient than normal arginine. It appeared that only 10% heavy arginine (13C6,15N4-arginine) was incorporated instead of the desired/expected 50% (supplemental Fig. S3). Albeit, cross-linking of DT-6-[Phe(Bz)6] to PKG resulted in the detection of a quadruply charged cross-linked product at m/z 616.3 that displayed the same isotopic pattern as the unreacted label (supplemental Fig. S4). The fragmentation spectrum of this cross-link product showed an abundant series of y″-ions, matching to PKG sequence; (K)-356LSDFNIIDTLGVGGFGR372-(V). This series of y″-ions still contains part of the photoaffinity label KKF(Bz)R (mass 681.3), which is also observed as a singly charged fragment ion.
Cross-linking Behavior of a DT-2-based Dual Photoaffinity Label to PKG
Now that both W45 and DT-6 were individually cross-linked to the catalytic domain of PKG, a dual DT-2 photoaffinity label that was synthesized containing two photochemical residues and two isotopically labeled arginine residues. After cross-linking to PKG and subsequent proteolytic digestion using trypsin of GluC no cross-linked products could be detected. Either the cross-linked product was too large and/or too basic too be seen using the LC-MS method, or the level of incorporation was not high enough. In an attempt to determine labeling efficiency, the cross-link product was subjected to SDS-PAGE analysis. This revealed an interesting result, namely the dual DT-2 label initiated a cross-link between the two individual PKG monomeric subunits, resulting in a high molecular weight cross-link products, presumably dimeric in nature (Fig. 7A). In-gel digestion of these high molecular weight bands unfortunately also did not yield any cross-linked product. Although the cross-link product (with an expected mass of 155 kDa) migrates at a relatively larger mass (above 200 kDa), it is not unlikely that both the positive character of the DT-2 label and the non-linear nature of the formed product will cause this effect. To confirm that this DT-2 induced specific cross-linking of the two individual PKG monomers, we performed a similar cross-linking, using the dual DT-2 label and a deletion mutant of PKG, Δ77, in which the first 77 residues at the N terminus have been deleted. This mutant is a constitutively active monomer of PKG, which, due to the lack of the dimerization and autoinhibition domain, cannot form dimers (52). No DT-2-induced dimerization was observed for this deletion mutant, demonstrating that the observed high molecular weight bands are specific for full-length and functional PKG dimers. In addition it was tested whether regular DT-2 could cause the PKG dimerization, and it was also tested whether the individual W45-[Phe(Bz)-2], W45-[Phe(Bz)-6], and DT-6-[Phe(Bz)-6] photolabels could be responsible for this dimerization (Fig. 7B), but all these tests show that the dimerization is due to the dual DT-2-[Phe(Bz)-6,14] photoaffinity label.
FIGURE 7.
SDS-PAGE analysis of reduced PKG DT-2-[Phe(Bz)-6,14] cross-link product shows dimerization of monomer. A, PKG wild type was cross-linked in the presence of 100 μm cGMP at three different DT-2-[Phe(Bz)-6,14] concentrations (1, 10, and 100 μm). At 100 μm DT-2-[Phe(Bz)-6,14] a high molecular weight band is clearly visible. The DT-2-photoaffinity label is unable to form covalent dimers of the constitutively active monomer PKG Δ77. B, the formation of covalent PKG dimers is specifically seen with the dual DT-2-photolabel and not with regular DT-2 or with the W45 or DT-6-based photoaffinity labels.
Binding Stoichiometry and Binding Order of the Interaction between PKG and Various Inhibitor Peptides
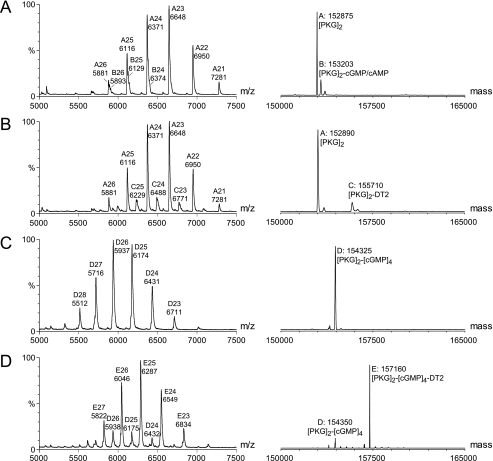
Now that the binding sites of the individual components of DT-2 were determined, the binding of DT-2 to PKG was studied in more detail using non-denaturing ESI-MS. Previously, we have shown that, when non-covalent complexes between PKG and ligands are conserved during the ionization process, important information, such as stoichiometry and/or binding order can be deduced (4, 47). Fig. 8A displays the positive electrospray ionization mass spectrum of a solution of PKG. The abundant ion signals between m/z 6000 and 7500 belong to dimeric protein, PKG2, with a calculated mass of 152,875 Da. When the inhibitor peptide DT-2 was added to the electrospray solution, a very similar mass spectrum was observed (Fig. 8B), albeit a small amount of the PKG2-DT-2 complex was visualized. In the presence of cGMP, a homogeneous complex of PKG2 with four cGMP molecules was obtained (Fig. 8C). Strikingly, when DT-2 was added to this solution a highly abundant complex of dimeric PKG, four cGMP molecules, and only one DT-2 molecule was observed (Fig. 8D). When the concentration of DT-2 was increased no binding of a second molecule of DT-2 to the PKG2-cGMP4-DT-2 complex was observed (supplemental Fig. S5), which would suggest that there is a single binding site for DT-2 on dimeric PKG. The binding of W45 and DT-6 were also studied and from these spectra (supplemental Fig. S6) it became clear that both W45 and DT-6 bind the strongest to cGMP-saturated PKG and more importantly up to two inhibitor peptides per PKG-dimer were observed.
FIGURE 8.
Nanoflow positive ESI-MS of ∼1.5 μm PKG (A), ∼1.5 μm PKG in the presence of 5 μm DT-2 (B), ∼1.5 μm PKG in the presence of 20 μm cGMP (C), ∼1.5 μm PKG and in the presence of 5 μm DT-2 and 20 μm cGMP (D). On the left, the raw m/z spectra are shown, and each multiply charge ion is labeled with a single letter, followed by the charge state and the centroid m/z value. On the right, the corresponding deconvoluted mass spectra are shown. Each peak is labeled with a single-letter code as in the raw spectra, followed by the calculated mass and determined complex composition.
DISCUSSION
DT-2 is the most potent inhibitor of PKG Iα and has the highest specificity for PKG over PKA. The inhibitor is a construct of the substrate competitive sequence, LRKKKKKH (W45) and a membrane translocating signal peptide YGRKKRRQRRRPP (DT-6). The combination of W45 and DT-6 resulted in a 65-fold increase in inhibition potency. DT-2 displayed a linear mixed competitive/non-competitive inhibition behavior. From this it has been suggested that the membrane-translocating sequence DT-6 binds and inhibits PKG at a different site from the catalytic cleft, and fusion of DT-6 to W45 accounts for the observed synergistic inhibition effect (17). To further unravel the unusual inhibition pattern of PKG by DT-2, several photoaffinity inhibitor peptides were synthesized. These were cross-linked to PKG in the presence of cGMP, subsequently proteolytically digested, and screened for cross-link products. The success of such an approach is largely dependent on the position of the photoreactive amino acid within the peptide. Phenylalanine-based photoaffinity probes, such as Phe(Tmd) and Phe(Bz), are rather bulky. To avoid a decrease in affinity, it is important that the photoreactive residue is positioned on a place where a bulky residue is tolerated. Also, it should be considered that the position of the photoactivatable amino acid is important in the recognition between peptide and protein. This will enhance the possibilities that a covalent cross-link between protein and peptide is achieved. Several W45-, DT-2-, or DT-6-based photoaffinity labels were synthesized in which the photoactivatable amino acid was positioned on either the N or C terminus to avoid negative effects in binding (Table 2). However, this strategy was not successful, because none of these photoaffinity labels cross-linked to PKG, although they were observed to bind in ESI-TOF-MS analyses. Most likely the termini of the inhibitor peptides are not near enough the surface of PKG to form a cross-link with it. Additionally, these labels were synthesized with Phe(Tmd), which is more reactive to water than Phe(Bz). Overall, this reiterates that the positioning of the photoactivatable amino acid with the peptide is important. To test the potential of cross-linking peptides to PKG via photoaffinity labeling we initially synthesized a photolabeled substrate peptide, based on the sequence TQAKRKKSLAMFLR, which is a high specificity peptide substrate of PKG (11). The phenylalanine was replaced by a photoactivable amino acid Phe(Tmd) at the p + 4 position (P0 is the phosphate accepting serine). The phenylalanine at this position is important for substrate recognition and is a critical determinant in PKG/PKA specificity (see Table 3) (10). Kinetic analysis showed that both Km and kcat for phosphorylation of W64-[Phe(Bz)-12] by PKG are within the same order as the original substrate (data not shown), indicating that the Phe(Bz) at this position does not influence binding affinity. The fragmentation spectra of the cross-link product indicated that cross-linking occurred on Ile361, which is a part of the small lobe of the catalytic core. Also, 10 residues upstream of this cross-link is the glycine-rich loop. This loop contains a GXGXXG motif, which is the highest conserved motif among protein kinases and is essential for ATP binding. To further determine the specific nature of this cross-link, the protein-photolabel product was incubated with Mg2+ and ATP, and indeed it was found that after cross-linking the substrate could still be phosphorylated by PKG (supplemental Fig. S1). Recently, hydrogen/deuterium exchange analysis of PKG in absence and presence of cGMP also revealed this particular part, residues 358–372, becomes more solvent-exposed upon cGMP activation (35). This is in agreement with the observation that the degree of cross-linking was significantly higher in the presence of cGMP. In a previous study, PKA-specific substrates were cross-linked via the P0 position to the catalytic subunit of PKA (53). Two sites of cross-linkage, namely Gly125 and Met128, were identified as close sites of proximity between PKA and the substrate. These residues reside in the large lobe of the catalytic subunit. This is in contrast to the PKG substrate, which was cross-linked via the p + 4 position to the small lobe. For comparison the sites of cross-linkage in both these studies are represented in Fig. 9, using the crystal structure of PKA in complex with Mn2+/ATP and the inhibitor peptide PKI. In this structure PKI docks preferentially on the large lobe of the catalytic domain, the p + 0 position is very near to the glycine-rich loop. The sites of the P0 cross-link for the PKA substrate (53) are highlighted in yellow, the equivalents of the p + 4 cross-link site for PKG are highlighted in pink. Previously, a refined molecular model for the catalytic domain of PKG was presented (11). The electrostatic surface representation of this model displayed a different interaction site for peptide substrate when compared with the crystal structure of PKA in complex with PKI. In addition, molecular modeling of the docking of the TQAKRKKALA-amide inhibitor peptide onto this PKG model demonstrated that the peptide primarily binds to the small lobe (11). This in contrast to the PKI/PKA systems, in which the N-terminal part of PKI is largely associated with the large lobe of the catalytic domain and the C-terminal part of PKI is more outside the catalytic cleft. This further fits with the general observation that C-terminal residues determine specificity for PKG, whereas N-terminal residues contribute to the specificity of PKA (also see Table 3), suggesting both proteins utilize different peptide conformations to achieve their substrate specificity.
FIGURE 9.
Ribbon diagrams of the catalytic subunit of PKA co-crystallized with MnATP and the inhibitor peptide PKI 5–24 (TTYADFIASGRTGRRNAIHD) (PDB entry code 1APM). The glycine-rich loop (shown in blue) participates in ATP binding and/or catalysis. The inhibitor peptide (shown in green) largely docks on the large lobe of the catalytic subunit. The P0 site, representing the phosphate-accepting residues in the substrate, is shown in cyan. The two arginines at the P3 and P2 positions, important for substrate recognition, are shown in red. Site of cross-linking of PKA-specific substrate are shown in yellow, and the sites of cross-linking of PKG are shown in pink. The ribbon diagram on the right (II) is a 90-degree clockwise rotation of the diagram on the left (I).
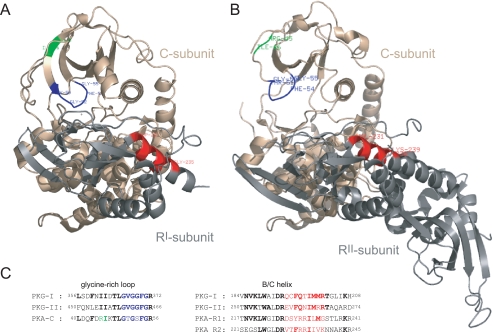
Two inhibitory peptide analogues of W45, W45-[Phe(Bz)-2] and W45-[Phe(Bz)-6], were cross-linked to PKG near the glycine-rich loop in the small lobe of the catalytic domain of PKG, very near the same area as the substrate peptides. For W45-[Phe(Bz)-6] a second cross-linkage to PKG on residues 199–210 was observed. This part of PKG lies within the high affinity cGMP-binding site A of the regulatory domain. This somewhat unexpected observation could indicate that W45 has either two different binding sites on PKG or more likely, that both PKG sequences (residues 360–370 and residues 194–202) are very near each other in space and form the same binding pocket. This latter assumption is strengthened by examining the crystal structure of the PKA holoenzyme. In two separate structures of the catalytic subunit in complex with (i) the regulatory RIα subunit (54, 55) and with (ii) the regulatory RIIα subunit (56), is it seen that the catalytic core and the regulatory core are attached by three different contact points. One of these is the b/c-helix, which is shown in red in Fig. 10 in both structures. This helix of the regulatory domain is in very close distance to the glycine-rich loop of the catalytic domain, although it should be noted that is in an inactive state for PKA. The b/c-helix of both RI and RII regulatory subunits of PKA aligns with 184VNVKLWAIDRQCFQTIMMRTGLIKH208 of PKG (Fig. 10C), to which W45-[Phe(Bz)-6] was cross-linked. In addition, several residues within this part are conserved within cGMP-dependent protein kinase type I and type II (Fig. 10C) Taken all these observations together this implies that W45 has one binding site on PKG. This binding site of W45 is almost identical to the substrate-binding site, i.e. very near the glycine-rich loop in the small lobe of the catalytic core. In analogy to W45, DT-6 also binds PKG in this area. This latter observation supports the idea that DT-6, like W45, acts as a competitive inhibitor, blocking the entrance for substrate peptides/proteins.
FIGURE 10.
Ribbon diagrams of the PKA Type I holoenzyme (PDB entry code 3FHI) (A) and PKA Type IIα (PDB entry code 2QVS) (B). The catalytic subunit is shown in brown, and the glycine-rich loop is highlighted in blue. The site of cross-linkage between PKG and the substrate and W45 and DT-6 analogues is highlighted in green. The regulatory subunit in type I (A) and in type IIα (B) are both shown in gray. The homologous part in both regulatory subunits represents the second site of cross-linking of W45 in the regulatory subunit of PKG, and is colored in red. C, sequence alignment of the glycine-rich loop in the catalytic domain of PKA and PKG type I and type II and sequence alignment of the b/c helix in the regulatory subunit of PKA (PKA-RI and PKA-RII) and the corresponding primary sequence in PKG type I and type II.
Although a dual photolabel derivative of DT-2 was successively coupled to PKG, unfortunately no cross-link products could be detected from its proteolytic digest, and hence the true binding sites of DT-2 remain unknown. However, SDS-PAGE analysis of the intact DT-2-[Phe(Bz)-6,14]/PKG cross-link product showed that this dual photolabel yields a high molecular weight species that migrate much slower than the monomeric PKG. This result implies that the dual photolabel bound to dimeric PKG, cross-links to each monomer upon irradiation, forming a covalent dimer. To strengthen this latter assumption, we have used a deletion mutant of PKG, Δ77. This mutant lacks the N-terminal dimerization and autoinhibition domain and is a constitutively active monomer. Although DT-2 is able to block the activity of the deletion mutant, the dual DT-2-[Phe(Bz)-6,14] photolabel does not form high molecular weight bands. Furthermore, neither regular DT-2 nor the photoaffinity labels of W45 and DT-6 produced these high molecular weight complexes suggesting that this would be an artifact.
Next to these cross-linking experiments, native mass spectrometry was used to further study the interactions between PKG and the various inhibitor peptides. The native mass spectrometry data clearly indicated that binding of cGMP to PKG is a prerequisite for the binding of all three inhibitor peptides (Fig. 8 and supplemental Fig. S6). This is in full agreement with the currently working model of PKG, in which the N-terminal autoinhibition domain blocks entrance to the active site of PKG in absence of cGMP (57). Upon cGMP activation, the autoinhibition domain is released from the catalytic domain, which is then accessible for target peptides/proteins. This is exemplified from the analysis of the intact PKG/W64-[Phe(Bz)-12] complexes (Fig. 2) in which peptide incorporation was substantially higher in the presence of cGMP. The observation that W45 and DT-6 also require cGMP activation is in perfect alignment with the cross-link data that showed that these peptides are targeted near the substrate-binding site.
In summary, the ultimate goal of these measurements was to determine the exact binding site of DT-2 in an attempt to understand its peculiar inhibition behavior, and even more to understand its high selectivity toward PKG. Although the exact binding of site of DT-2 remains unresolved, the present data contain a series of novel and significant clues about the molecular mechanisms of DT-2 inhibition. It is now clear that both individual components of DT-2, namely W45 and DT-6, are targeted to the same pocket in the catalytic domain as is the substrate peptide W64. Also, native mass spectrometry confirmed that cGMP activation is required to bind inhibitor/substrate peptides, which suggest all inhibitor/substrate peptides compete with the N-terminal autoinhibition domain for binding to the catalytic core. Taken together, the peculiar observations, that one DT-2 molecule is preferentially bound to dimeric PKG, combined with the results from the dual DT-2-photoprobe, which specifically produces high molecular weight dimers/multimers, lead us to put forth the model that DT-2 is able to inactivate two active sites simultaneously. This potentially far-reaching model suggests that, within dimeric PKG, both active sites are in close proximity of each other. In addition our model may explain the remarkable potency and specificity of DT-2 toward PKG. As mentioned above, in PKG both regulatory and catalytic domain lie within one single polypeptide chain. Hence, the two catalytic subunits remain always in close proximity with each other via the dimerization of the N termini. This is in contrast to the PKA system, in which the catalytic and regulatory domains reside on individual subunits that dissociate from each other once the regulatory subunits bind cAMP (Fig. 1). In summary, DT-2 acts as a bidentate inhibitor, targeting the catalytic domains of dimeric PKG. The resulting inhibition indicates that these two catalytic domains are spatially proximal, implying a unique architecture for PKG holoenzyme.
Supplementary Material
This work was supported in part by National Institutes of Health Grant HL68891 (to W. R. D.). This work was also supported by the Netherlands Proteomics Centre and the Totman Trust for Medical Research.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- PKA
- 3′,5′-cAMP-dependent protein kinase
- PKG
- 3′,5′-cGMP-dependent protein kinase
- PKI
- cAMP-dependent protein kinase inhibitor
- ESI-TOF-MS
- electrospray ionization time-of-flight mass spectrometry
- Phe(Bz)
- 4-benzoyl-l-phenylalanine
- Phe(Tmd)
- 4′-(3-(trifluoromethyl)-3H-diazirin-3-yl)-l-phenylalanine
- DIPEA
- N,N-diisopropylethylamine
- Fmoc
- Nα-9-fluorenylmethyloxycarbonyl
- HPLC
- high-performance liquid chromatography
- AMPPNP
- adenosine 5′-(β,γ-imino)triphosphate
- LC
- liquid chromatography
- HATU
- N-(dimethylamino)-1H-1,2,3-triazolo[4,5-bipyridin-1-yl-methylene]-N-methylmethanaminium hexafluorophosphate
- HOAt
- N-hydroxy-7-azabenzotriazole
- HOBt
- N-hydroxybenzotriazole
- HBTU
- 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- NMP
- N-methylpyrrolidone.
REFERENCES
- 1.Adams J. A. ( 2001) Chem. Rev. 101, 2271– 2290 [DOI] [PubMed] [Google Scholar]
- 2.Francis S. H., Corbin J. D. ( 1994) Annu. Rev. Physiol. 56, 237– 272 [DOI] [PubMed] [Google Scholar]
- 3.Taylor S. S., Buechler J. A., Yonemoto W. ( 1990) Annu. Rev. Biochem. 59, 971– 1005 [DOI] [PubMed] [Google Scholar]
- 4.Scholten A., Aye T. T., Heck A. J. ( 2008) Mass Spectrom. Rev. 27, 331– 353 [DOI] [PubMed] [Google Scholar]
- 5.Hofmann F., Dostmann W., Keilbach A., Landgraf W., Ruth P. ( 1992) Biochim. Biophys. Acta 1135, 51– 60 [DOI] [PubMed] [Google Scholar]
- 6.Lohmann S. M., Vaandrager A. B., Smolenski A., Walter U., De Jonge H. R. ( 1997) Trends Biochem. Sci. 22, 307– 312 [DOI] [PubMed] [Google Scholar]
- 7.Francis S. H., Woodford T. A., Wolfe L., Corbin J. D. ( 1988) Second Messengers Phosphoproteins 12, 301– 310 [PubMed] [Google Scholar]
- 8.Takio K., Wade R. D., Smith S. B., Krebs E. G., Walsh K. A., Titani K. ( 1984) Biochemistry 23, 4207– 4218 [DOI] [PubMed] [Google Scholar]
- 9.Glass D. B., Krebs E. G. ( 1979) J. Biol. Chem. 254, 9728– 9738 [PubMed] [Google Scholar]
- 10.Colbran J. L., Francis S. H., Leach A. B., Thomas M. K., Jiang H., McAllister L. M., Corbin J. D. ( 1992) J. Biol. Chem. 267, 9589– 9594 [PubMed] [Google Scholar]
- 11.Dostmann W. R., Nickl C., Thiel S., Tsigelny I., Frank R., Tegge W. J. ( 1999) Pharmacol. Ther. 82, 373– 387 [DOI] [PubMed] [Google Scholar]
- 12.Glass D. B., el-Maghrabi M. R., Pilkis S. J. ( 1986) J. Biol. Chem. 261, 2987– 2993 [PubMed] [Google Scholar]
- 13.Mitchell R. D., Glass D. B., Wong C. W., Angelos K. L., Walsh D. A. ( 1995) Biochemistry 34, 528– 534 [DOI] [PubMed] [Google Scholar]
- 14.Thomas N. E., Bramson H. N., Nairn A. C., Greengard P., Fry D. C., Mildvan A. S., Kaiser E. T. ( 1987) Biochemistry 26, 4471– 4474 [DOI] [PubMed] [Google Scholar]
- 15.Wood J. S., Yan X., Mendelow M., Corbin J. D., Francis S. H., Lawrence D. S. ( 1996) J. Biol. Chem. 271, 174– 179 [DOI] [PubMed] [Google Scholar]
- 16.Yan X., Corbin J. D., Francis S. H., Lawrence D. S. ( 1996) J. Biol. Chem. 271, 1845– 1848 [DOI] [PubMed] [Google Scholar]
- 17.Dostmann W. R., Taylor M. S., Nickl C. K., Brayden J. E., Frank R., Tegge W. J. ( 2000) Proc. Natl. Acad. Sci. U. S. A. 97, 14772– 14777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bove P. F., Wesley U. V., Greul A. K., Hristova M., Dostmann W. R., van der Vliet A. ( 2007) Am. J. Respir. Cell Mol. Biol. 36, 138– 146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dey N. B., Foley K. F., Lincoln T. M., Dostmann W. R. ( 2005) J. Cardiovasc. Pharmacol. 45, 404– 413 [DOI] [PubMed] [Google Scholar]
- 20.Foley K. F., De Frutos S., Laskovski K. E., Tegge W., Dostmann W. R. ( 2005) Front. Biosci. 10, 1302– 1312 [DOI] [PubMed] [Google Scholar]
- 21.Hou Y., Wong E., Martin J., Schoenlein P. V., Dostmann W. R., Browning D. D. ( 2006) Cell Signal. 18, 882– 888 [DOI] [PubMed] [Google Scholar]
- 22.Krieg T., Philipp S., Cui L., Dostmann W. R., Downey J. M., Cohen M. V. ( 2005) Am. J. Physiol. Heart. Circ. Physiol. 288, H1976– 1981 [DOI] [PubMed] [Google Scholar]
- 23.Taylor M. S., Okwuchukwuasanya C., Nickl C. K., Tegge W., Brayden J. E., Dostmann W. R. G. ( 2004) Mol. Pharmacol. 65, 1111– 1119 [DOI] [PubMed] [Google Scholar]
- 24.Zhu C. B., Carneiro A. M., Dostmann W. R., Hewlett W. A., Blakely R. D. ( 2005) J. Biol. Chem. 280, 15649– 15658 [DOI] [PubMed] [Google Scholar]
- 25.Dostmann W. R., Tegge W., Frank R., Nickl C. K., Taylor M. S., Brayden J. E. ( 2002) Pharmacol. Ther. 93, 203– 215 [DOI] [PubMed] [Google Scholar]
- 26.Brunner J. ( 1996) Trends Cell Biol. 6, 154– 157 [DOI] [PubMed] [Google Scholar]
- 27.Jahn O., Eckart K., Tezval H., Spiess J. ( 2004) Anal. Bioanal. Chem. 378, 1031– 1036 [DOI] [PubMed] [Google Scholar]
- 28.Heck A. J., Van Den Heuvel R. H. ( 2004) Mass Spectrom. Rev. 23, 368– 389 [DOI] [PubMed] [Google Scholar]
- 29.Loo J. A. ( 1997) Mass Spectrom. Rev. 16, 1– 23 [DOI] [PubMed] [Google Scholar]
- 30.Przybylski M., Glocker M. O. ( 1996) Angew. Chem., Int. Ed. Engl. 35, 807– 826 [Google Scholar]
- 31.Sharon M., Robinson C. V. ( 2007) Annu. Rev. Biochem. 76, 167– 193 [DOI] [PubMed] [Google Scholar]
- 32.Samalikova M., Matecko I., Müller N., Grandori R. ( 2004) Anal. Bioanal. Chem. 378, 1112– 1123 [DOI] [PubMed] [Google Scholar]
- 33.Simmons D. A., Konermann L. ( 2002) Biochemistry 41, 1906– 1914 [DOI] [PubMed] [Google Scholar]
- 34.Veenstra T. D., Johnson K. L., Tomlinson A. J., Craig T. A., Kumar R., Naylor S. ( 1998) J. Am. Soc. Mass Spectrom. 9, 8– 14 [DOI] [PubMed] [Google Scholar]
- 35.Alverdi V., Mazon H., Versluis C., Hemrika W., Esposito G., van den Heuvel R., Scholten A., Heck A. J. ( 2008) J. Mol. Biol. 375, 1380– 1393 [DOI] [PubMed] [Google Scholar]
- 36.Bligh S. W., Haley T., Lowe P. N. ( 2003) J. Mol. Recognit. 16, 139– 148 [DOI] [PubMed] [Google Scholar]
- 37.Tjernberg A., Carnö S., Oliv F., Benkestock K., Edlund P. O., Griffiths W. J., Hallén D. ( 2004) Anal. Chem. 76, 4325– 4331 [DOI] [PubMed] [Google Scholar]
- 38.Rogniaux H., Sanglier S., Strupat K., Azza S., Roitel O., Ball V., Tritsch D., Branlant G., Van Dorsselaer A. ( 2001) Anal. Biochem. 291, 48– 61 [DOI] [PubMed] [Google Scholar]
- 39.van Duijn E., Bakkes P. J., Heeren R. M., van den Heuvel R. H., van Heerikhuizen H., van der Vies S. M., Heck A. J. ( 2005) Nat. Methods 2, 371– 376 [DOI] [PubMed] [Google Scholar]
- 40.Rink H. ( 1987) Tetrahedron Lett. 28, 3787– 3790 [Google Scholar]
- 41.Knorr R., Trzeciak A., Bannwarth W., Gillessen D. ( 1989) Tetrahedron Lett. 30, 1927– 1930 [Google Scholar]
- 42.Carpino L. A., Elfaham A., Minor C. A., Albericio F. ( 1994) J. Chem. Soc. Chem. Commun. 2, 201– 203 [Google Scholar]
- 43.Ten Kortenaar P. B., Van Dijk B. G., Peeters J. M., Raaben B. J., Adams P. J., Tesser G. I. ( 1986) Int. J. Pept. Protein Res. 27, 398– 400 [Google Scholar]
- 44.Fields C. G., Lloyd D. H., Macdonald R. L., Otteson K. M., Noble R. L. ( 1991) Pept. Res. 4, 95– 101 [PubMed] [Google Scholar]
- 45.Ploug M., Ostergaard S., Hansen L. B., Holm A., Danø K. ( 1998) Biochemistry 37, 3612– 3622 [DOI] [PubMed] [Google Scholar]
- 46.Weber P. J., Beck-Sickinger A. G. ( 1997) J. Pept. Res. 49, 375– 383 [DOI] [PubMed] [Google Scholar]
- 47.Pinkse M. W., Heck A. J., Rumpel K., Pullen F. ( 2004) J. Am. Soc. Mass. Spectrom. 15, 1392– 1399 [DOI] [PubMed] [Google Scholar]
- 48.Tahallah N., Pinkse M., Maier C. S., Heck A. J. ( 2001) Rapid Commun. Mass Spectrom. 15, 596– 601 [DOI] [PubMed] [Google Scholar]
- 49.Meiring H. D., van der Heeft E., ten Hove G. J., de Jong A. P. ( 2002) J. Sep. Sci. 25, 557– 568 [Google Scholar]
- 50.Kauer J. C., Erickson-Viitanen S., Wolfe H. R., Jr., DeGrado W. F. ( 1986) J. Biol. Chem. 261, 10695– 10700 [PubMed] [Google Scholar]
- 51.Nassal M. ( 1984) J. Am. Chem. Soc. 106, 7540– 7545 [Google Scholar]
- 52.Tegge W., Frank R., Hofmann F., Dostmann W. R. ( 1995) Biochemistry 34, 10569– 10577 [DOI] [PubMed] [Google Scholar]
- 53.Miller W. T., Kaiser E. T. ( 1988) Proc. Natl. Acad. Sci. U. S. A. 85, 5429– 5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim C., Cheng C. Y., Saldanha S. A., Taylor S. S. ( 2007) Cell 130, 1032– 1043 [DOI] [PubMed] [Google Scholar]
- 55.Kim C., Xuong N. H., Taylor S. S. ( 2005) Science 307, 690– 696 [DOI] [PubMed] [Google Scholar]
- 56.Wu J., Brown S. H., von Daake S., Taylor S. S. ( 2007) Science 318, 274– 279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Francis S. H., Poteet-Smith C., Busch J. L., Richie-Jannetta R., Corbin J. D. ( 2002) Front. Biosci. 7, d580– 592 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.