Abstract
Mutations in two genes encoding the putative kinases LRRK2 and PINK1 have been associated with inherited variants of Parkinson disease. The physiological role of both proteins is not known at present, but studies in model organisms have linked their mutants to distinct aspects of mitochondrial dysfunction, increased vulnerability to oxidative and endoplasmic reticulum stress, and intracellular protein sorting. Here, we show that a mutation in the Caenorhabditits elegans homologue of the PTEN-induced kinase pink-1 gene resulted in reduced mitochondrial cristae length and increased paraquat sensitivity of the nematode. Moreover, the mutants also displayed defects in axonal outgrowth of a pair of canal-associated neurons. We demonstrate that in the absence of lrk-1, the C. elegans homologue of human LRRK2, all phenotypic aspects of pink-1 loss-of-function mutants were suppressed. Conversely, the hypersensitivity of lrk-1 mutant animals to the endoplasmic reticulum stressor tunicamycin was reduced in a pink-1 mutant background. These results provide the first evidence of an antagonistic role of PINK-1 and LRK-1. Due to the similarity of the C. elegans proteins to human LRRK2 and PINK1, we suggest a common role of both factors in cellular functions including stress response and regulation of neurite outgrowth. This study might help to link pink-1/PINK1 and lrk-1/LRRK2 function to the pathological processes resulting from Parkinson disease-related mutants in both genes, the first manifestations of which are cytoskeletal defects in affected neurons.
Mutations in the Parkinson disease (PD)2-related gene PINK1 have been associated with increased sensitivity to oxidative stress and mitochondrial dysfunction (1–4). Although a series of reports support a localization of PINK1 only in mitochondria (5–7), recent studies have shown that a portion of endogenous PINK1 is also distributed to the cytoplasm (8–11). Notably, the cytoplasmic kinase activity and not the mitochondrial targeting of PINK1 seems to be prerequisite of its protective effects against mitochondrial stress (12).
The GTPase-regulated kinase LRRK2, another gene associated with familial PD, has been linked to the biogenesis and regulation of vesicular transport (13, 14). In support of this notion, the C. elegans lrk-1, the LRRK2 homologue, has recently been shown to be involved in synaptobrevin-associated vesicular transport (15). Moreover, the Dictyostelium homologue of LRRK2/LRK-1 proteins, GbpC, a cGMP-binding protein is required for the normal phosphorylation and cytoskeletal assembly of myosin (16). Based on this data, it had been anticipated that PINK1/PINK-1 and LRRK2/LRK-1 might be involved in distinct cellular functions, whereas pathological mutations in both genes, leading either to loss-of-function (PINK1) or gain-of-function (LRRK2), result in a similar phenotype, the loss of dopaminergic neurons.
In the present study, we found a functional connection between their Caenorhabditits elegans homologues pink-1 and lrk-1. We demonstrate that loss of C. elegans pink-1 results in oxidative stress sensitivity and neurite outgrowth defects. All aspects of pink-1 deficiency were suppressed in the absence of lrk-1 that, as a single mutant, displayed a profound hypersensitivity to the ER stressor tunicamycin. Because the latter phenotype was in turn suppressed by deleting pink-1, our results support an antagonistic role of pink-1 and lrk-1 in stress response and neuronal activities.
EXPERIMENTAL PROCEDURES
Strains
All strains used in this study were maintained on NGM plates (different diameters) seeded with Escherichia coli OP50 and cultivated at 20 °C as previously described (17). The C. elegans strains were obtained from Dr. Shohei Mitani (National Bioresource Project, Japan) and provided by Dr. Erik Lundquist (University of Kansas) and Dr. Kunihiro Matsumoto (Nagoya University, Japan). The following mutations and integrated arrays were used in this study: LGI, lrk-1(tm1898, km41); LGII, pink-1(tm1779); LGX, lqIs4 [ceh-10::gfp, lin-15(n765)]. N2 (Bristol) was used as the C. elegans wild type strain.
Molecular Cloning
For the determination of the expression pattern of pink-1, the translational gfp fusion Ppink-1::pink-1::gfp (pBY2101), was generated by cloning the complete genomic fragment of pink-1 (3191 bp) fused to a 6-kb upstream promoter region and to GFP at the C terminus in pPD117.01 (gift of A. Fire, Stanford School of Medicine, Stanford, CA). 10 ng/μl of the resulting plasmid (pBY2101) was injected into pha-1(e2123) animals along with the pha-1 rescue plasmid (pBX1) to yield strain BR3645 (pha-1(e2123);byEx686 [pink-1]) and BR3646 (pha-1(e2123);byEx687 [pink-1]), respectively. For the pink-1 rescue experiment, 10 ng/μl pBY2101 was co-injected with Pmyo-2::mCherry (pBY2550) as injection maker (pBY2550) and herring sperm DNA into pink-1(tm1779) animals (BR4006: pink-1(tm1779);byEx655 [pink-1]; BR4007: pink-1(tm1779);byEx681 [pink-1]). For the lrk-1 expression and rescue experiments, the translational fusion construct, Plrk-1::lrk-1::gfp (pBY2254) was generated by cloning the complete genomic region of lrk-1 (14 kb) to a 4.5-kb upstream promoter region and to GFP at the C terminus in pPD117.01 (provided by Dr. A. Fire, Stanford School of Medicine, Stanford, CA). For generating byIs137 [lrk-1], Plrk-1::lrk-1::gfp (pBY2254) and myo-2::mCherry (pBY2109) were co-injected into N2 wild type and lrk-1(tm1898) animals, UV integrated, and backcrossed multiple times. Furthermore, 0.1 ng/μl of the cosmid T27C10 as well as 0.1 ng/μl of the plasmid Paex-3::lrk-1(G1876S) (pBY2617) were co-injected with pBY2550 and herring sperm DNA into lqIs4 [ceh-10::gfp, lin-15(n765)] or lrk-1(tm1898) animals to obtain three independent extrachromosomal lines [lqIs4[ceh-10::gfp, lin-15(n765)];byEx737–739[T27C10] (BR5303–5305), or BR5300–5302:lrk-1(tm1898);byEx734–736[lrk-1(G1876S)].
Phenotypic Analysis and Drug Treatment
To determine the brood size, individual L4 hermaphrodites were placed onto fresh NGM plates and incubated at 20 °C. To prevent overcrowding, worms were transferred daily onto new NGM plates for three consecutive days. The progeny was counted 2–3 days after removal of the P0 generation (18). For oxidative stress assays (paraquat), seeded NGM plates (Ø 3.5 cm) containing E. coli OP50 and 150 mm paraquat (Sigma) were used. Synchronized L4 animals were analyzed for their survival at 20 °C after 3 days (19). For the ER stress assay (tunicamycin), adult worms were allowed to lay eggs for 3–4 h at 20 °C on NGM agar plates (Ø 3.5 cm) containing 1.5 μg/ml tunicamycin (Sigma). Eggs were counted and the number of worms at the L4 or young adult stage was scored 3 days later (20). Tunicamycin, which is isolated from Streptomyces sp., consists of at least four homologous antibiotics that differ in their fatty acid composition. As a consequence, the strength of the inhibitory activity on N-linked glycosylation varies in each individual batch. Mean values of the number of progeny that reached the L4 or the young adult stage (tunicamycin assay) and survivors (paraquat assay) were calculated from different experimental groups of at least three independent assays each. For analyzing canal-associated neurons (CAN) axon pathfinding, L4 animals, harboring an integrated ceh-10::gfp transgene (lqIs4 kindly provided by Erik Lundquist), were scored for defects as described (21).
Electron Microscopy
About 20 young adult worms were transferred into a 100-μm deep aluminum platelet/chamber (Microscopy Services, Flintbek) that was filled with E. coli OP50 suspension and immediately frozen using a BalTec HPM 10. Freeze substitution was carried out in a Leica EM AFS at −90 °C for 100 h in 0.1% tannic acid and another 7 h in 2% OsO4 (each w/v in dry acetone) as described (22). For electron microscopy, 40-nm sections were cut using a Leica UC6 ultramicrotome. Ribbons of sections were transferred on Formvar-coated copper slot-grids. The grids were placed for 10 min on drops of 4% (w/v) uranyl acetate in 75% methanol and then washed in distilled water. After air drying the grids were placed on lead citrate (23) for 2 min in a CO2-free chamber, and rinsed in distilled water. Micrographs were taken with a 1024 × 1024 CCD detector (Proscan CCD HSS 512/1024; Proscan Electronic Systems, Scheuring, Germany) in a Zeiss EM 902A, operated in the bright field mode. Longitudinal sections of muscle mitochondria were analyzed as the following: a square of 1 × 0.2 μm was placed longitudinally to the inner face of the membrane surrounding the mitochondrium. The total length of the apparent cristae in the square was determined manually and summed. The value for each square was calculated as membrane length/area (μm/μm2) and 1–4 squares were used per mitochondrium. The total area of cross-sectioned neuronal mitochondria in axons in the ventral nerve cord and the length of all apparent cristae were determined manually and calculated as membrane length/area (μm/μm2).
Northern Blot Analysis
Total RNA was isolated from mixed staged worm plates (wild type and pink-1(tm1779) animals) and prepared with an RNAeasy kit according to the manufacturer's instructions (Qiagen). 10 μg of total RNA was loaded per lane on the gel. PCR fragments for the probes of interest were labeled radioactively with [α-32P]dCTP using random hexamer primers and the Klenow fragment of the DNA polymerase. For amplification of the pink-1-specific probe the following oligonucleotides were used: GTCTATGAAACGATTCGG and CATTCTTTCCAGGAACAGCCGTC. Hybridization with the probes was done in standard hybridization buffer (10% dextran sulfate, 10× Denhart's, 50 mm Tris/HCl, pH 7.5, 1% SDS, 0.1% sodium pyrophosphate, 1 m NaCl, 50% formamide (deionized), 0.2 mg/ml salmon sperm DNA) overnight at 65 °C. After three washing steps (30 min, 2× SSC, 1% SDS at 65 °C; 15 min, 0.2 SSC at 65 °C, 15 min; 0.2 × SSC at room temperature). The radioactive signal was detected using the Bio-Rad Molecular Imager FX (Bio-Rad). act-1-specific probes were used as controls.
Software and Microscopy
The amino acid alignment was generated using ClustalW. Pictures of GFP expression were taken using a Zeiss AxioImager Microscope, the AxioCam camera, and the AxioVision Rel.4.6 software. For statistical analyses GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego, CA) was used. Statistical methods were applied as used in previous studies (3, 24).
RESULTS
C. elegans pink-1 Is Localized in the Cytoplasm and Mitochondria of Multiple Tissues
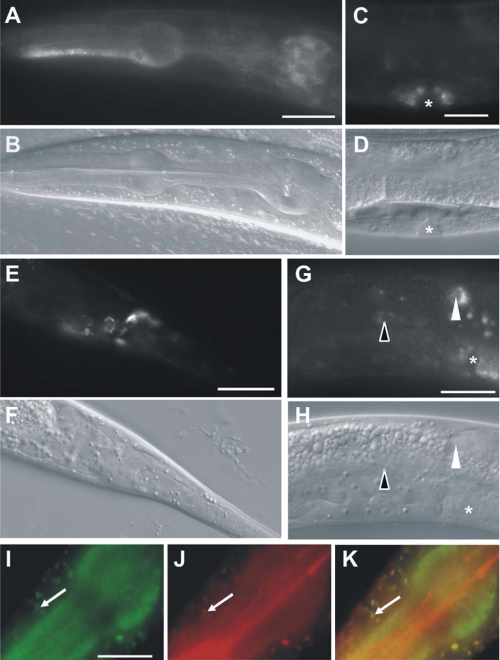
Sequence analysis of C. elegans, Drosophila melanogaster, and human PINK-1 revealed a single C. elegans homologue, which we termed PINK-1 (PTEN-induced kinase-1 homolog) (supplemental Fig. S1A). Like human PINK1, PINK-1 consists of two characteristic motifs, an N-terminal mitochondrial targeting sequence and a serine/threonine kinase domain that shares 36% identity and 54% similarity to human PINK1 (supplemental Fig. S1B) (1). Due to the absence of a functional antibody against C. elegans PINK-1, we used a translational GFP fusion that rescues the pink-1 mutant phenotype (see below) to determine the expression pattern of pink-1. PINK-1::GFP is detected in many cell types and is maintained throughout all stages of postembryonic development and adulthood (Fig. 1). GFP staining was observed in head and tail neurons (Fig. 1, A–B and E–F) and in neurons of the midbody region, such as the canal-associated neurons (CAN) (Fig. 1, G and H). Expression was also seen in many non-neuronal cells including the pharyngeal muscles (Fig. 1, A and B) and the vulva (Fig. 1, C and D). We found that a fraction of C. elegans PINK-1 co-localized with the mitochondrial marker Mitotracker Red (Fig. 1, I and K), indicating both mitochondrial and cytoplasmic distribution of the fusion protein. Similarly, endogenous PINK1 in human cells may also be localized both to mitochondria and the cytoplasm (8–11). The broad distribution of PINK-1 in many tissues suggests that its function is not limited to the nervous system.
FIGURE 1.
pink-1 is ubiquitously expressed in C. elegans. The expression pattern of pink-1 was investigated by transgenic expression of a rescuing Ppink-1::pink-1::gfp [pink-1] construct in two independent pha-1 lines (BR3645 and BR3646). pink-1 is ubiquitously, but weakly expressed in all body regions including head and tail neurons (A and E), the pharynx (A), the distal tip cells (arrowheads), the CAN (open arrowheads), and vulva epithelium (asterisk). A and B, head region (head neurons, pharyngeal muscles). C and D, mid-body region (vulva epithelium). E and F, tail region (tail neurons). G and H, mid-body region (distal tip cell, CAN). The corresponding DIC pictures are shown (B, D, F, and H). I–K, co-localization of PINK-1-GFP (I) with Mitotracker Red (J) in the mitochondria (arrows) is shown in a merged image (K). Scale bars represent 20 (A–H) and 10 μm (I–K).
PINK-1 Functions in Mitochondrial Homeostasis and Oxidative Stress Response
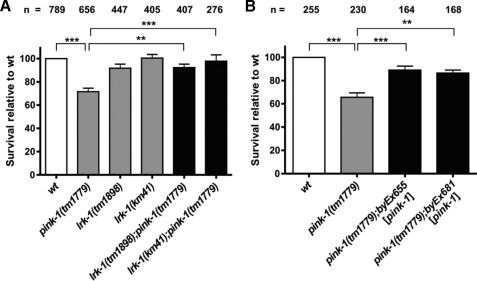
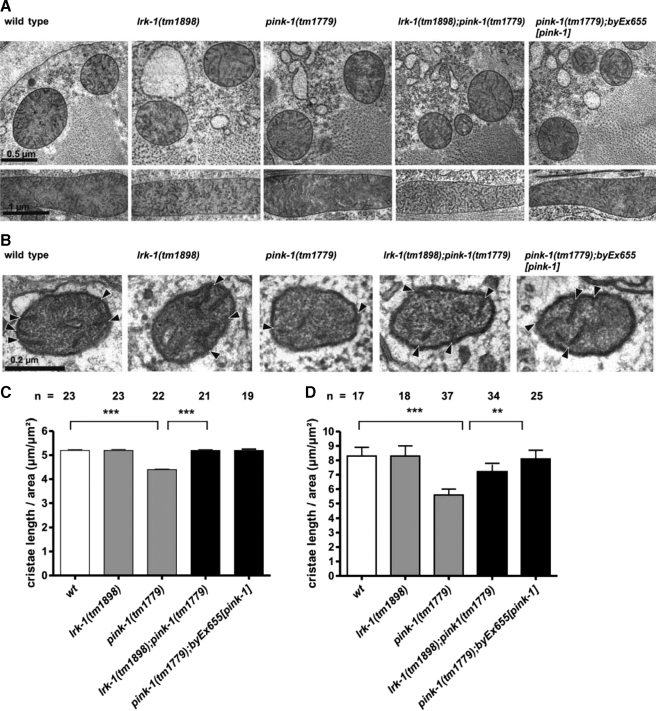
Previous experiments in cultivated mammalian cells, mouse, and D. melanogaster have shown that loss of PINK1 resulted in both an increased vulnerability to oxidative stress and morphological alterations of mitochondrial structures, indicated by reduced or disrupted mitochondrial cristae (2–4, 24). To compare the consequences of loss-of-function of pink-1 in C. elegans with findings from other organisms, we characterized a deletion mutant of pink-1 (kindly provided by S. Mitani). The pink-1(tm1779) allele harbors a 350-bp deletion that eliminates part of the promoter region and the first two exons including the proposed transcriptional and translational start sites. Northern blot analysis revealed that pink-1(tm1779) is indeed a null allele (supplemental Fig. S2). Thus, the deletion most likely results in a complete functional loss (supplemental Fig. S2). pink-1(tm1779) animals were viable, developed normally, and showed a slightly reduced brood size at 20 °C (179 ± 5; Table 1). To investigate the role of C. elegans pink-1 in oxidative stress response, we exposed pink-1(tm1779) animals to 150 mm paraquat and measured their survival rates as previously described (19). Interestingly, pink1(tm1779) animals displayed an increased sensitivity to a 3-day exposure to paraquat compared with wild type (71.5 ± 3.1% survivors; Fig. 2A). Expression of transgenic pink-1 in pink-1(tm1779) mutants was sufficient to partially rescue the animals against paraquat-induced toxicity (Fig. 2B) and other defects (see below). This indicates that the phenotype of the pink-1(tm1779) animals was indeed due to the deletion in the pink-1 locus. Because down-regulation of PINK1 in D. melanogaster was accompanied by mitochondrial defects especially of the wing musculature (2–3), we examined the morphological integrity of mitochondria in C. elegans pink-1 mutants using rapid freeze-electron microscopy (Fig. 3). In longitudinal sections of C. elegans we determined that the apparent length of mitochondrial cristae was reduced by 12% in muscle cells and by more than 30% in the neurons of pink-1(tm1779) animals compared with wild type (Fig. 3, C and D). Notably, transgenic expression of a wild type copy of pink-1 was able to rescue the mitochondrial defects due to loss of pink-1 function in both neuronal and muscle cells (Fig. 3, C and D). The coincidence of both oxidative stress sensitivity and mitochondrial phenotypes may support a conserved function of PINK1/PINK-1 in mitochondrial homeostasis. In summary, wild type levels of PINK-1 protect C. elegans against oxidative stress, which is in agreement with reports from other model organisms (2–4). This suggests a conserved role of PINK-1/PINK1 among several species.
TABLE 1.
Brood size at 20 °C
| Genotype | Progenya (mean ± S.E.) | nb |
|---|---|---|
| Wild type | 233 ± 8cd | 16 |
| pink-1(tm1779) | 179 ± 5ce | 18 |
| lrk-1(tm1898) | 208 ± 9df | 17 |
| lrk-1(tm1898);pink-1(tm1779) | 249 ± 8ef | 19 |
a Number of progeny is presented as mean ± S.E.
b Total number of animals scored.
c Significantly different int-test,p< 0.0001.
d Significantly different int-test,p = 0.0236.
e Significantly different int-test,p< 0.0001.
f Significantly different int-test,p = 0.0017.
FIGURE 2.
The pink-1(tm1779)-induced oxidative stress sensitivity is suppressed by lrk-1 alleles tm1898 and km41 (A) and rescued by a wild type copy of pink-1 (B). Synchronized L4 larval animals were grown at 20 °C on NGM plates containing 150 mm paraquat and the survivors were counted 3 days later. Shown are mean values ± S.E. The percentage of survivors was normalized to wild type (wt). The total numbers of animals analyzed from individual strains are listed above each column (n). p values were calculated by t test analysis. **, p < 0.0017; and ***, p < 0.0007.
FIGURE 3.
lrk-1(tm1898) suppresses mitochondrial cristae defects of pink-1(tm1779). A, cross (top) and longitudinal (below) sections of mitochondria in C. elegans muscle cells. B, cross-sections of mitochondria in C. elegans neurons. C, quantitative analysis of cristae membrane length in muscle cells with respect to total mitochondrial area (μm2). D, cristae membrane length in neurons/μm2 mitochondrium area. Shown are mean ± S.E. The number of analyzed mitochondrial profiles from at least 19 (in muscle cells) and 17 (in neurons), derived from at least two independent animals is listed above each column (n). p values were calculated by analysis of variance. **, p < 0.0050 and ***, p < 0.0001.
lrk-1 Mutants Suppress the pink-1(tml779) Phenotype
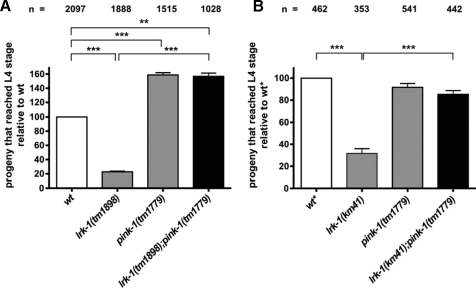
Human PINK1 has already been linked to DJ-1 and Parkin, the other two genes associated with autosomal-recessive PD (25–26). However, no functional connection between PINK1 and the GTPase-regulated kinase LRRK2 has been demonstrated so far. As it was shown before by a systematic study that disease-associated genes are functionally connected and genes causing similar diseases form subnetworks (27), we were encouraged to investigate whether C. elegans LRK-1 and PINK-1 may contribute to overlapping biological functions. The C. elegans lrk-1 shares 35% identity (57% similarity) within the GTPase domain, and 37% identity (57% similarity) within the kinase domain of human LRRK2, respectively. We confirmed the widespread expression pattern of lrk-1 that has been reported earlier (15). lrk-1 is co-expressed with pink-1 in several tissues including the cells of the nervous system and many non-neuronal tissues, such as the body wall musculature and the epithelial cells of the nematode vulva (supplemental Fig. S3). We examined two deletion alleles of lrk-1 and first confirmed the synaptobrevin sorting defect for lrk-1(tm1898) animals (data not shown) that was previously reported for the allele km41 (15). Similar to lrk-1(km41), the lrk-1(tm1898) allele harbors an out-of-frame deletion, resulting in loss of both the GTPase and kinase domains. Thus, both alleles probably represent severe loss-of-function alleles that express truncated proteins only consisting of the N-terminal ankyrin repeat. Although pink-1 mutant animals were sensitive to paraquat treatment, the survival rates of both lrk-1(tm1898) and lrk-1(km41) animals were not different from wild type (Fig. 2A). Surprisingly, both alleles of lrk-1 suppressed the paraquat sensitivity of pink-1(tm1779) mutants, resulting in survival levels comparable with wild type (Fig. 2A). Although lrk-1(tm1898) showed a mild reduction in the number of progeny at 20 °C (208 ± 9; Table 1), the double mutants lrk-1(tm1898);pink-1(tm1779) produced a wild type number of progeny (249 ± 7; Table 1), suggesting antagonistic activities of the two genes.
Because oxidative stress sensitivity is often the consequence of mitochondrial dysfunction, we next tested mitochondrial integrity in lrk-1 single and lrk-1(tm1898);pink-1(tm1779) double mutants. In agreement with a report of D. melanogaster LRRK mutants (28), the mitochondria of C. elegans lrk-1(tm1898) single mutants were indistinguishable from wild type (Fig. 3). Remarkably, lrk-1(tm1898) suppressed the cristae defects of pink-1(tm1779) animals to wild type levels (Fig. 3, C and D). Taken together, our results indicate that genetic deletion of lrk-1 could compensate for both the oxidative stress sensitivity and the morphological defects observed in a pink-1 loss-of-function allele.
pink-1(tml779) Suppresses the ER Stress Sensitivity of Mutant lrk-1
In addition to alterations in the oxidative stress response, defects in protein trafficking as well as impaired protein degradation have been reported to contribute to PD pathology (29). To investigate whether lrk-1 and pink-1 are involved in the ER stress response, we exposed pink-1 and lrk-1 single and double mutants to 1.5 μg/ml tunicamycin. This toxin functions as a specific inhibitor of N-linked glycosylation, thus leading to the accumulation of unfolded proteins in the ER (30). C. elegans lrk-1(tm1898) single mutants displayed an increased sensitivity to ER stress, indicated by the developmental arrest of the majority of animals (22.7 ± 1% survival relative to wild type; Fig. 4A). Similar levels of lethality were observed in lrk-1(km41) animals (31.7 ± 4.3% survival; Fig. 4B). To test whether the ER stress sensitivity observed in lrk-1 mutants was indeed caused by loss of lrk-1 function, we tried to rescue the phenotype by generating transgenic animals expressing a genomic copy of lrk-1 fused to GFP. Transgenic expression of wild type LRK-1 resulted in an early larval arrest at 20 °C (57.9 ± 3.4%; Table 2), the penetrance of which was further increased upon treatment with tunicamycin. Larval arrest could have been caused either by toxicity of the LRK-1 fusion protein or by a dose-dependent increase of LRK-1 activity expressed from the multicopy transgene. To distinguish both models, we analyzed transgenic animals expressing the G1876S mutant of LRK-1 that corresponds to the prominent G2019S mutation in human LRRK2 from a C. elegans pan-neuronal promoter. This variant of LRRK2 is supposed to have increased kinase activity (31). Because both wild type and also the G1876S variant caused embryonic lethality (data not shown), we conclude that expression levels of lrk-1 have to be tightly controlled, and that the phenotype is not caused by tagging LRK-1 with GFP. In contrast to the hypersensitivity of lrk-1 mutant animals, pink-1(tm1779) did not show an altered response to tunicamycin (Fig. 4). Notably, the ER stress sensitivity of both lrk-1 alleles tested (km41 and tm1898) was suppressed in a pink-1(tm1779) mutant background (Fig. 4, A and B). These results further substantiate our observation that lrk-1 and pink-1 have opposing activities, affecting both C. elegans ER and oxidative stress responses.
FIGURE 4.
Loss of pink-1 suppresses the tunicamycin sensitivity of lrk-1 mutants. The sensitivity of two lrk-1 alleles, tm1898 (A) and km41 (B) to treatment with 1.5 μg/ml tunicamycin resulted in a reduced survival rate of the animals that is suppressed by pink-1(tm1779). The total numbers of animals analyzed from individual strains are listed above each column (n). The percentage of survivors was normalized to wild type (wt). Shown are mean ± S.E. The distinct survival rate of pink-1(tm1779) animals between (A) and (B) is caused by the use of different tunicamycin batches for each set of experiments. Data of experiments obtained for B were carried out with strains harboring an additional integrated transgene ceh-10::gfp (indicated, for simplicity, as wild type (wt*)). **, p = 0.0030 and ***, p < 0.0003.
TABLE 2.
Lethality of animals expressinglrk-1
| Genotype | Lethality/arresta (mean ± S.E.) | nb |
|---|---|---|
| % | ||
| OP50 | ||
| Wild type | 0.4 ± 0.4c | 205 |
| lrk-1(tm1898) | 10.3 ± 5.9d | 234 |
| lrk-1(tm1898);byIs137P[lrk-1] | 57.9 ± 3.4cd | 530 |
| 1.5 μg/ml tunicamycin | ||
| Wild type | 62.5 ± 4.0ef | 527 |
| lrk-1(tm1898) | 82.0 ± 3.4e | 406 |
| lrk-1(tm1898);byIs137P[lrk-1] | 100.0 ± 0f | 317 |
a Number of lethal/arrested progeny is presented as mean ± S.E.
b Total number of animals scored.
c Significantly different int test,p< 0.0001.
d Significantly different int test,p< 0.0001.
e Significantly different int test,p = 0.0009.
f Significantly different int test,p< 0.0001.
Down-regulation of lrk-1 Suppresses pink-1(tml779)-mediated Axon Guidance Defects
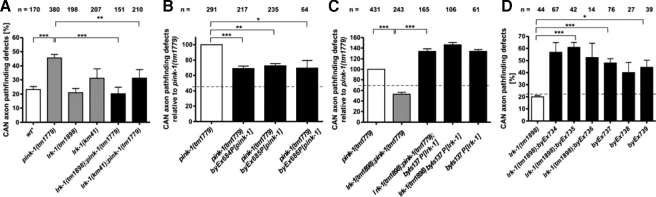
PD-associated dominant mutations in LRRK2 result in a progressive reduction in neurite length and branching in mammalian cell culture (31). A function for C. elegans lrk-1 in neurite outgrowth has not been described so far. Because lrk-1 and pink-1 were co-expressed in many neurons, we investigated their role in the positioning and morphology of neurons in vivo. Several previous reports have provided mechanistic insights into the control of cell migration and neurite integrity in C. elegans, involving the CAN (32). Hence, we used a previously described integrated GFP reporter strain, carrying the lqIs4[ceh-10::gfp] transgene to monitor the morphological integrity of these neurons (21). The somata of the bilateral CAN neurons are generated in the anterior body region during embryogenesis and migrate posteriorly to the midbody to localize closely to the vulva. Their unbranched, linear processes extend anteriorly to the head and posteriorly to the tail (33). Wild type animals exhibited occasional defects in axon pathfinding (Fig. 5A), which had already been observed.3 Although the frequency of defects in CAN axon pathfinding in lrk-1(tm1898) animals (21.0 ± 2.9%; Fig. 5A) was comparable with that of wild type (23.3 ± 2.1%), defective CAN processes were observed twice as often in pink-1(tm1779) animals (45.7 ± 2.3%), resulting in either premature neurite termination, misguidance, or branching (supplemental Fig. S4). Transgenic expression of pink-1 was sufficient to rescue a considerable portion of these guidance defects (Fig. 5B). The incomplete rescue by the pink-1 transgenic lines may be due to a mosaic expression and/or the generally low expression levels of the transgene. On the other hand, deletion of lrk-1 in tm1898 fully suppressed the pink-1(tm1779) axon outgrowth defects at 20 °C (20.2 ± 4.7% in the double mutant; Fig. 5A). Similar to tm1898, lrk-1(km41) suppressed CAN neurite defects in pink-1(tm1779) animals to a level comparable with that of the lrk-1(km41) single mutant (31.2 ± 6.1% in the double mutant; Fig. 5A). These results are consistent with the opposing activity of LRK-1 to PINK-1 in oxidative stress response (Fig. 2).
FIGURE 5.
The CAN axon pathfinding defects of pink-1 mutants are suppressed by loss of lrk-1 function. CAN axon pathfinding was visualized using the integrated transgene lqIs4[ceh-10::gfp] (20). Shown is the percentage (mean ± S.E.) of posterior CAN axons that terminated prematurely, were misguided, or branched inappropriately. A, both lrk-1 alleles tm1898 and km41 suppress CAN axon pathfinding defects of pink-1(tm1779). B, the CAN axon pathfinding defects of pink-1(tm1779) is rescued by transgenic expression of extrachromosomal pink-1::gfp. C, a genomic copy of lrk-1::gfp restores CAN axon pathfinding defects in lrk-1(tm1898);pink-1(tm1779) double mutants and, thus, suppresses the lrk-1(tm1898) phenotype. D, a genomic copy of lrk-1 (cosmid T27C10; byEx737–739) as well as a dominant version of LRK-1(G1876S) (lrk-1(tm1898);byEx734–736) leads to CAN axon pathfinding defects in both lrk-1(tm1898) mutant and in a wild type (wt) background. Dotted line in B–D, the mean value of pathfinding defects in wild type (wt) animals carrying the lqIs4-integrated ceh-10::gfp transgene is shown. The total numbers of animals analyzed from individual strains are listed above each column (n). *, p = 0.0295; **, p < 0.0064; and ***, p < 0.0007.
Rescue of the suppressor activity of lrk-1 by transgenic copies of lrk-1 further corroborate this finding. As described above, transgenic expression of lrk-1::gfp resulted in larval arrest of more than half of the transgenic animals, but the animals escaping lethality showed restoration of the pink-1 mutant phenotype (Fig. 5C). In addition, transgenic animals expressing a genomic copy of lrk-1 (cosmid T27C10) or the G1876S variant (see “Experimental Procedures”) resulted in significant CAN axon guidance defects in a wild type or lrk-1(tm1898) mutant background (Fig. 5D). We conclude that both reduced and enhanced lrk-1 expression and activity cause a phenotype, and may either result in suppression of pink-1(tm1779) neurite outgrowth defects or may induce pathfinding defects on its own. This is in accordance with other studies showing that slight changes in the expression level or activity of human LRRK2 could either result in extension (small interfering RNA against LRRK2) or reduction (point mutations in the kinase domain, e.g. I2020T or G2019S) of neurite processes (31). Therefore, correct levels of activity of LRK-1/LRRK2 are critical for the cell.
Axon Guidance Control by pink-1 and lrk-1 Is Not Affected by Oxidative Stress
We wondered whether the neurite defects of pink-1(tm1779) mutants were simply the result of their increased oxidative stress sensitivity or whether they might resemble a phenotypic aspect not related to stress sensitivity. Therefore, we tested neurite outgrowth of CAN neurons after pre-exposure to a variety of stressors in both wild type and mutant backgrounds. An increase of the growth temperature from 20 to 23 °C resulted in mild CAN axon pathfinding defect of lrk-1(tm1898) animals (33.3 ± 6.3%; Table 3) that were inconspicuous at the low temperature (Fig. 5A). Moreover, at 23 °C lrk-1(tm1898) showed a less pronounced suppression of the pink-1 mutant phenotype (53.6 ± 4.0% compared with 41.5 ± 3.6% in the lrk-1(tm1898);pink-1(tm1779); Table 3). This indicates that lrk-1(tm1898) suppression of pink-1(tm1779) is affected by increased temperature. To investigate whether sublethal doses of paraquat are sufficient to induce or enhance neurite outgrowth defects, we tested mutant combinations of lrk-1 and pink-1 in the background of low doses of this toxin (2 mm paraquat). Paraquat pre-treatment did not affect the CAN axon guidance defects in pink-1(tm1779) mutant animals (32.5 ± 4.4%) or the suppressive effect of lrk-1(tm1898) (20.5 ± 5.4%; Table 3). This result suggests that the increased oxidative stress sensitivity of pink-1(tm1779) is not the cause of neurite defects and that both phenotypic aspects are independently suppressed by loss of lrk-1 function.
TABLE 3.
Summary of CAN axon pathfinding defects at different stress conditions
| Genotype | Posterior CAN axon outgrowth defectsa ± S.E. | nb |
|---|---|---|
| % | ||
| Temperature stress: 23 °C | ||
| +c | 24.7 ± 0.3de | 144 |
| pink-1(tm1779) | 53.6 ± 4.0df | 251 |
| lrk-1(tm1898) | 33.3 ± 6.3 | 228 |
| lrk-1(tm1898);pink-1(tm1779) | 41.5 ± 3.6ef | 163 |
| Oxidative stress: 2 mmparaquat | ||
| +c | 18.0 ± 4.2g | 122 |
| pink-1(tm1779) | 32.5 ± 4.4gh | 173 |
| lrk-1(tm1898) | 16.4 ± 2.9 | 112 |
| lrk-1(tm1898);pink-1(tm1779) | 20.5 ± 5.4h | 108 |
| ER stress: 1.5 μg/ml tunicamycin | ||
| +c | 28.1 ± 9.1 | 140 |
| pink-1(tm1779) | 41.8 ± 2.2 | 167 |
| lrk-1(tm1898) | 42.0 ± 4.6 | 149 |
| lrk-1(tm1898);pink-1(tm1779) | 38.8 ± 3.1 | 115 |
a The percentage of posterior CAN neurites that terminated prematurely, branched, or were misguided (20).
b Total number of animals scored.
c CAN axon pathfinding was determined using the integrated transgene lqIs4[ceh-10::gfp] (20).
d Significantly different int test,p = 0.0009.
e Significantly different int test,p = 0.0047.
f Significantly different int test,p = 0.0421.
g Significantly different int test,p = 0.0381.
h Significantly different int test,p = 0.0441.
ER Stress Prevents Antagonistic Function of lrk-1
We have shown that elimination of lrk-1 results in increased ER stress sensitivity, we wanted to know whether the suppressive effect of loss of lrk-1 function on the CAN pathfinding defect caused by deletion of pink-1 is modulated after tunicamycin treatment. Exposure of mutant combinations to this toxin resulted in elevated levels of CAN axon pathfinding defects of lrk-1(tm1898) mutants (42.0 ± 4.6%; Table 3). Strikingly, tunicamycin treatment abolished the lrk-1-mediated suppression of pink-1(tm1779), yet it did not affect axonal outgrowth of pink-1 single mutants (38.8 ± 3.1% in the lrk-1(tm1898);pink-1(tm1779) double mutant; Table 3). We conclude that lrk-1 mutants are hypersensitive to ER stress and that, in turn, ER stress prevents lrk-1 mutants from antagonizing pink-1 function.
In summary, the putative C. elegans kinases LRK-1 and PINK-1 have opposing roles both in the regulation of axon guidance and stress response and are, thus, mechanistically connected. This is to our knowledge the first report of such a functional relationship between both proteins.
DISCUSSION
Our results presented here suggest a functional linkage between lrk-1 and pink-1, whose human homologues LRRK2 and PINK1 segregate with familial cases of PD. We show that in C. elegans lrk-1 and pink-1 have opposing activities, and mutations in both genes suppress the phenotypic aspects observed in the respective single mutants.
First of all, we demonstrate that loss of C. elegans pink-1 function results in an increased paraquat sensitivity of the animals. This suggests that the function of PINK-1 helps to protect C. elegans from reactive oxygen species, as reported earlier for its homologues in D. melanogaster and vertebrate models (2–4). The mitochondria are the initial site of action of the herbicide and toxin paraquat, indicating that the increased oxidative stress sensitivity mediated by loss of pink-1 function could be linked to mitochondrial dysfunction also in the nematode. The source of paraquat-induced reactive oxygen species has not been determined in detail, however, recent findings suggest that they require the presence of respiratory substrates, in particular complexes of the electron transport chain III. Consistent with an increased paraquat sensitivity of C. elegans pink-1(tm1779) animals, recent studies have shown that PINK1 knock-out mice have defects in the electron transport chain complexes I and II–IV of the inner membrane (34). Thus, our data support a role of PINK-1 for the functional integrity of the inner mitochondrial membrane that harbors the respiratory chain complexes. Indeed, rapid freeze-electron microscopy studies on C. elegans muscular and neuronal mitochondria revealed that the cristae of the inner membrane show low, but significant alterations of length. PINK1 has recently been found to encode a protein inserted into the mitochondrial outer membrane, with its C-terminal kinase tail facing into the cytosol (35). The details of its localization have not been figured out so far, and it is not known at present how a protein inserted in the outer membrane can affect the respiratory chain situated in the inner membrane. Similar to mouse models, the defects linked to putative mitochondrial functions of pink-1(tm1779) are rather subtle, although the deletion represents a null allele. We therefore suggest that PINK-1 may have additional roles in other subcellular compartments. This notion is supported by the broad intracellular distribution pattern of PINK-1::GFP.
Both putative kinases LRK-1 and PINK-1 are expressed in many tissues of C. elegans (and other organisms) from embryo to adulthood, and the lack of additional, profound phenotypic aspects of both mutants was, therefore, somewhat surprising. A possible reason for this could be the existence of related proteins with overlapping functions to both kinases, or could indicate very specific functions of both proteins. The former notion is not very likely, given the profound level of suppression of both paraquat and tunicamycin sensitivity, general fecundity, and axonal outgrowth defects observed in the lrk-1(tm1898);pink-1(tm1779) double mutant.
The pink-1(tm1779) mutation resulted in defective outgrowth of the CAN neurons, a pair of migratory neurons in the midbody region of the worm that are frequently used as an indicator for axonal guidance and cell migration defects (21). We demonstrate for the first time that PINK-1 may contribute to the control of neurite outgrowth. Previous studies suggested a role of human LRKK2 in the regulation of actin cytoskeleton-related processes, such as neurite outgrowth and synaptic plasticity (36, 37). However, in C. elegans, we did not detect a respective defect in the two lrk-1 loss-of-function alleles that we used in this study. However, transgenic expression of Plrk-1::lrk-1::gfp [lrk-1] as well as a dominant-active version or LRK-1 [Paex-3::lrk-1(G1876S)] in a wild type or lrk-1(tm1898) mutant background was sufficient to cause aberrant CAN axon pathfinding in those animals that escaped early larval arrest/lethality, a hallmark observed in all lrk-1 variants expressed in multiple copies. This data implicates that the expression and activity of lrk-1 needs to be tightly regulated in C. elegans, as it has been suggested in other studies (31).
Strikingly, both loss-of-function alleles of lrk-1 suppressed the CAN axon outgrowth defects of pink-1 mutant animals, indicating that, in addition to an antagonistic activity of PINK-1 and LRK-1 upon oxidative and ER stress, lrk-1 deficiency can also suppress the lack of pink-1 in these neurons. This suppression is temperature-sensitive, because it is reduced already at slightly elevated temperatures (23 °C), at which lrk-1(tm1898) animals on their own displayed mild defects in CAN morphology. This could indicate that either lrk-1 single mutants are sensitive to the elevated temperature or that the suppression mechanism by loss of lrk-1 in the pink-1(tm1779) is affected by additional stress. A general temperature sensitivity of lrk-1(tm1898) animals is unlikely, because we observed no difference in the phenotype of lrk-1 single mutants at altered temperature or after heat shock induction (data not shown).
Are the CAN defects observed in pink-1(tm1779) simply the cause of increased oxidative stress sensitivity and mitochondrial dysfunction, or do they represent the involvement of pink-1 in functions distinct from cellular stress control? If the former were the case, then preincubation of pink-1(tm1779) animals with lower doses of paraquat should increase the axonal defects observed in this mutant. This was not the case. Moreover, paraquat treatment also did not affect lrk-1 nor the lrk-1-mediated suppression of pink-1(tm1779) CAN axon guidance defects. We currently have no evidence that axonal guidance control mediated by antagonistic activities of PINK-1 and LRK-1 is linked to the response of oxidative stress. Our data would also be consistent with an up-regulation of lrk-1 activity as a consequence of loss of pink-1 function, the former, as we have shown, being indeed sufficient to result in defective CAN axon guidance.
Is the lrk-1 sensitivity to tunicamycin related to its role in suppressing pink-1(tm1779) neurite defects? A toxic effect of tunicamycin on axon outgrowth has been suggested previously (38). We indeed found that exposure of lrk-1 mutant animals to this toxin resulted in an observable CAN axon guidance defect compared with wild type and a loss of pink-1 suppression. This data supports the hypothesis that axon guidance in the lrk-1(tm1898) mutant is modulated by heat and ER stress, but not by oxidative stress. Given that a localization of LRRK2/LRK-1 at or in the ER has not been shown convincingly, it is currently unclear how the mutant could affect ER stress sensitivity.
Noteworthy, a not thoroughly investigated role of tunicamycin to induce growth cone collapse has been suggested (39). Moreover, Hess et al. (40) provided evidence that functional Golgi contributes to axon growth, because inhibition of ADP-ribosylation factor, a protein involved in ER to Golgi and intra-Golgi transport, caused rapid retraction of growth cones. Based on its sequence homology, LRRK2 has been associated with the family of Rab-GTPases that are involved in ER to Golgi trafficking and the docking of ER-derived transport vesicles at the Golgi membrane (41). Tunicamycin has been shown to disturb the ER to Golgi transport, resulting in increased Golgi fragmentation (42). Furthermore, defects in Golgi function and trafficking have been shown to affect the ER system by causing a traffic jam in the early secretory system. Strikingly, C. elegans lrk-1 mutants displayed abnormalities in the secretory apparatus, namely Golgi fragmentation and aberrant distribution of the endosomes and other vesicular structures.4 In agreement with this, LRK-1 has been shown to localize to the Golgi system and affect sorting of synaptic vesicle proteins at the Golgi level (15). This phenotypic aspect of loss of lrk-1 was also observed in the lrk-1(tm1898) mutant allele used in this study (data not shown). Consistent with a role of LRK-1 in modulating synaptic function, a physical interaction between human LRRK2 and Rab5, a key regulator of endocytotic vesicular transport was demonstrated recently (43). This is supported by the localization of LRRK2 in lipid rafts that have been shown to be important for axon guidance during development (13). Future studies will be required to understand the biochemical consequences of the pink-1 and lrk-1 interaction and the guidance of axons under activated stress response.
Three models might help to describe the antagonistic activities of LRK-1 and PINK-1: (i) LRK-1 could act as a negative regulator of PINK-1. However, because we have shown that pink-1(tm1779) is a null allele, it is difficult to imagine how in such case lrk-1 loss-of-function could directly suppress pink-1(tm1779). (ii) LRK-1 could be a downstream factor that is negatively regulated by PINK-1. Loss of pink-1 should consequently lead to an activation of lrk-1 manifested by increased oxidative stress sensitivity. Additional deletion of lrk-1 would then restore the basal level of the downstream signal and thus suppress the pink-1(tm1779) phenotype. This should be phenocopied by lrk-1 overexpression in a wild type background. This is indeed the case for neurite outgrowth, but not for the ER stress response, because transgenic expression of lrk-1 resulted in increased lethality upon exposure to tunicamycin (Table 2). (iii) Both genes act in parallel on antagonistic pathways controlling neurite outgrowth and stress sensitivity. Further investigations are required to distinguish between the latter two models.
In summary, the genetic data we present here strongly indicate antagonistic roles of PINK-1 and LRK-1 in C. elegans. Based on mutant analyses and pharmacological studies, we suggest that the response to reactive oxygen species and the control of neurite outgrowth are two independent phenotypic aspects controlled by opposing activities of pink-1 and lrk-1. The functional connection of both genes in a genetic network provides the basis for further studies to help uncover the detailed functions of both kinases. In particular, it will be interesting to determine how PINK-1 and LRK-1 link mitochondrial and cytoplasmic functions and what the substrates of their respective enzymatic activities are. Our data support a model that perturbations of the intricate cross-talk between membrane-associated, cytoplasmic functions, and mitochondrial activities may contribute to the onset of PD. Mechanistic links between mitochondrial (dys-) function and cytoplasmic signaling pathways have also been considered to account for cellular aging. Analyzing these intracellular cross-talks may, therefore, provide the basis for understanding the age dependence of PD.
Supplementary Material
Acknowledgments
We thank Mark Seifert, Ursula Schäffer, Jörg Weiß, Xu Huang, and Pamela A. Padilla for helpful discussions and for critically reading the manuscript. We thank Shohei Mitani for providing lrk-1(tm1898) and pink-1(tm1779), Erik Lundquist for the lqIs4[ceh-10::gfp] transgene, Kunihiro Matsumoto for providing lrk-1(km41), and Andy Fire for the pPD117.01 plasmid.
This work was supported by FORSYS Grant 03139219, the Freiburg Initiative in Systems Biology (a program of the German Federal Ministry in Research and Education), Deutsche Forsch ungs ge mein schaft Grant SFB780, European Community 6th Framework Network of Excellence LIFESPAN Grant LSHG-CT-2007-036894, the Deutsche Forsch ungs ge mein schaft German Excellence Initiative (FRIAS LIFENET, EXC 294 BIOSS), and the European Community 7th Framework MEMOSAD Grant HEALTH-F2-2007-200611.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
E. Lundquist, personal communication.
J. Sämann, J. Hegermann, E. von Gromoff, S. Eimer, R. Baumeister, and E. Schmidt, unpublished results.
- PD
- Parkinson disease
- ER
- endoplasmic reticulum
- GFP
- green fluorescent protein
- LRRK2
- leucine-rich repeat kinase 2
- PINK1/PINK-1
- PTEN-induced kinase 1
- CAN
- canal-associated neurons.
REFERENCES
- 1.Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del T. D., Bentivoglio A. R., Healy D. G., Albanese A., Nussbaum R., Gonzalez-Maldonado R., Deller T., Salvi S., Cortelli P., Gilks W. P., Latchman D. S., Harvey R. J., Dallapiccola B., Auburger G., Wood N. W. ( 2004) Science 304, 1158– 1160 [DOI] [PubMed] [Google Scholar]
- 2.Park J., Lee S. B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J. M., Chung J. ( 2006) Nature 441, 1157– 1161 [DOI] [PubMed] [Google Scholar]
- 3.Clark I. E., Dodson M. W., Jiang C., Cao J. H., Huh J. R., Seol J. H., Yoo S. J., Hay B. A., Guo M. ( 2006) Nature 441, 1162– 1166 [DOI] [PubMed] [Google Scholar]
- 4.Exner N., Treske B., Paquet D., Holmstrom K., Schiesling C., Gispert S., Carballo-Carbajal I., Berg D., Hoepken H. H., Gasser T., Kruger R., Winklhofer K. F., Vogel F., Reichert A. S., Auburger G., Kahle P. J., Schmid B., Haass C. ( 2007) J. Neurosci. 27, 12413– 12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silvestri L., Caputo V., Bellacchio E., Atorino L., Dallapiccola B., Valente E. M., Casari G. ( 2005) Hum. Mol. Genet 14, 3477– 3492 [DOI] [PubMed] [Google Scholar]
- 6.Muqit M. M., bou-Sleiman P. M., Saurin A. T., Harvey K., Gandhi S., Deas E., Eaton S., Payne S., Venner K., Matilla A., Healy D. G., Gilks W. P., Lees A. J., Holton J., Revesz T., Parker P. J., Harvey R. J., Wood N. W., Latchman D. S. ( 2006) J. Neurochem. 98, 156– 169 [DOI] [PubMed] [Google Scholar]
- 7.Pridgeon J. W., Olzmann J. A., Chin L. S., Li L. ( 2007) PLoS Biol. 5, e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weihofen A., Ostaszewski B., Minami Y., Selkoe D. J. ( 2008) Hum. Mol. Genet. 17, 602– 616 [DOI] [PubMed] [Google Scholar]
- 9.Takatori S., Ito G., Iwatsubo T. ( 2008) Neurosci. Lett. 430, 13– 17 [DOI] [PubMed] [Google Scholar]
- 10.Gandhi S., Muqit M. M., Stanyer L., Healy D. G., Abou-Sleiman P. M., Hargreaves I., Heales S., Ganguly M., Parsons L., Lees A. J., Latchman D. S., Holton J. L., Wood N. W., Revesz T. ( 2006) Brain 129, 1720– 1731 [DOI] [PubMed] [Google Scholar]
- 11.Beilina A., Van Der, Brug M., Ahmad R., Kesavapany S., Miller D. W., Petsko G. A., Cookson M. R. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 5703– 5708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haque M. E., Thomas K. J., D'Souza C., Callaghan S., Kitada T., Slack R. S., Fraser P., Cookson M. R., Tandon A., Park D. S. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 1716– 1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatano T., Kubo S., Imai S., Maeda M., Ishikawa K., Mizuno Y., Hattori N. ( 2007) Hum. Mol. Genet 16, 678– 690 [DOI] [PubMed] [Google Scholar]
- 14.Biskup S., Moore D. J., Celsi F., Higashi S., West A. B., Andrabi S. A., Kurkinen K., Yu S. W., Savitt J. M., Waldvogel H. J., Faull R. L., Emson P. C., Torp R., Ottersen O. P., Dawson T. M., Dawson V. L. ( 2006) Ann. Neurol. 60, 557– 569 [DOI] [PubMed] [Google Scholar]
- 15.Sakaguchi-Nakashima A., Meir J. Y., Jin Y., Matsumoto K., Hisamoto N. ( 2007) Curr. Biol. 17, 592– 598 [DOI] [PubMed] [Google Scholar]
- 16.Bosgraaf L., Russcher H., Smith J. L., Wessels D., Soll D. R., Van Haastert P. J. ( 2002) EMBO J. 21, 4560– 4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenner S. ( 1974) Genetics 77, 71– 94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong A., Boutis P., Hekimi S. ( 1995) Genetics 139, 1247– 1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hertweck M., Gobel C., Baumeister R. ( 2004) Dev. Cell 6, 577– 588 [DOI] [PubMed] [Google Scholar]
- 20.Springer W., Hoppe T., Schmidt E., Baumeister R. ( 2005) Hum. Mol. Genet. 14, 3407– 3423 [DOI] [PubMed] [Google Scholar]
- 21.Lundquist E. A., Reddien P. W., Hartwieg E., Horvitz H. R., Bargmann C. I. ( 2001) Development 128, 4475– 4488 [DOI] [PubMed] [Google Scholar]
- 22.Rostaing P., Real E., Siksou L., Lechaire J. P., Boudier T., Boeckers T. M., Gertler F., Gundelfinger E. D., Triller A., Marty S. ( 2006) Eur. J. Neurosci. 24, 3463– 3474 [DOI] [PubMed] [Google Scholar]
- 23.Reynolds E. S. ( 1963) J. Cell Biol. 17, 208– 212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J. W., Yang L., Beal M. F., Vogel H., Lu B. ( 2006) Proc. Natl. Acad. Sci. U. S. A. 103, 10793– 10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith W. W., Pei Z., Jiang H., Moore D. J., Liang Y., West A. B., Dawson V. L., Dawson T. M., Ross C. A. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 18676– 18681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore D. J., Zhang L., Troncoso J., Lee M. K., Hattori N., Mizuno Y., Dawson T. M., Dawson V. L. ( 2005) Hum. Mol. Genet. 14, 71– 84 [DOI] [PubMed] [Google Scholar]
- 27.Gandhi T. K., Zhong J., Mathivanan S., Karthick L., Chandrika K. N., Mohan S. S., Sharma S., Pinkert S., Nagaraju S., Periaswamy B., Mishra G., Nandakumar K., Shen B., Deshpande N., Nayak R., Sarker M., Boeke J. D., Parmigiani G., Schultz J., Bader J. S., Pandey A. ( 2006) Nat. Genet. 38, 285– 293 [DOI] [PubMed] [Google Scholar]
- 28.Lee S. B., Kim W., Lee S., Chung J. ( 2007) Biochem. Biophys. Res. Commun. 358, 534– 539 [DOI] [PubMed] [Google Scholar]
- 29.Ryu E. J., Harding H. P., Angelastro J. M., Vitolo O. V., Ron D., Greene L. A. ( 2002) J. Neurosci. 22, 10690– 10698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patterson S. I., Skene J. H. ( 1994) J. Cell Biol. 124, 521– 536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacLeod D., Dowman J., Hammond R., Leete T., Inoue K., Abeliovich A. ( 2006) Neuron 52, 587– 593 [DOI] [PubMed] [Google Scholar]
- 32.Wu Y. C., Cheng T. W., Lee M. C., Weng N. Y. ( 2002) Dev. Biol. 250, 145– 155 [DOI] [PubMed] [Google Scholar]
- 33.Forrester W. C., Perens E., Zallen J. A., Garriga G. ( 1998) Genetics 148, 151– 165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gautier C. A., Kitada T., Shen J. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 11364– 11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou C., Huang Y., Shao Y., May J., Prou D., Perier C., Dauer W., Schon E. A., Przedborski S. ( 2008) Proc. Natl. Acad. Sci. U. S. A. 105, 12022– 12027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaleel M., Nichols R. J., Deak M., Campbell D. G., Gillardon F., Knebel A., Alessi D. R. ( 2007) Biochem. J. 405, 307– 317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Habig K., Walter M., Poths S., Riess O., Bonin M. ( 2008) Neurogenetics. 9, 83– 94 [DOI] [PubMed] [Google Scholar]
- 38.Finnie J. W., Manavis J., Blumbergs P. C., Kuchel T. R. ( 2000) Vet. Pathol. 37, 677– 680 [DOI] [PubMed] [Google Scholar]
- 39.Patterson S. I., Skene J. H. ( 1995) Methods Enzymol. 250, 284– 300 [DOI] [PubMed] [Google Scholar]
- 40.Hess D. T., Smith D. S., Patterson S. I., Kahn R. A., Skene J. H., Norden J. J. ( 1999) J. Neurobiol. 38, 105– 115 [DOI] [PubMed] [Google Scholar]
- 41.Zerial M., McBride H. ( 2001) Nat. Rev. Mol. Cell Biol. 2, 107– 117 [DOI] [PubMed] [Google Scholar]
- 42.Lin T. Y., Wang S. M., Fu W. M., Chen Y. H., Yin H. S. ( 1999) J. Cell. Biochem. 74, 638– 647 [DOI] [PubMed] [Google Scholar]
- 43.Shin N., Jeong H., Kwon J., Heo H. Y., Kwon J. J., Yun H. J., Kim C. H., Han B. S., Tong Y., Shen J., Hatano T., Hattori N., Kim K. S., Chang S., Seol W. ( 2008) Exp. Cell Res. 314, 2055– 2065 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.