Abstract
Objectives
We sought to determine whether polymorphisms in the large-conductance calcium and voltage-dependent potassium (BK) channel β1 subunit gene, KCNMB1, are associated with blood pressure response to verapamil SR or adverse outcomes in the GENEtic substudy of the INternational VErapamil SR/trandolapril STudy (INVEST-GENES).
Background
KCNMB1 is involved in calcium sensitivity and hypertension. The association between variability in KCNMB1 and calcium antagonist response, however, has not been assessed.
Methods
Genetic samples were collected from 5979 patients in INVEST. Blood pressure response to verapamil SR and time to achieve blood pressure control was assessed in relation to Glu65Lys and Val110Leu genotypes. The primary outcome (all cause mortality, nonfatal myocardial infarction or nonfatal stroke) was compared between genotype groups, and interaction with verapamil SR therapy was assessed.
Results
Systolic blood pressure response to verapamil SR did not differ by KCNMB1 genotype. Lys65 variant carriers, however, achieved blood pressure control earlier than Glu65Glu individuals [1.47 (interquartile ratio 2.77) versus 2.83 (interquartile ratio 4.17) months, P = 0.01] and were less likely to require multiple drugs at the time of blood pressure control (adjusted odds ratio 0.43, 95% confidence interval 0.19–0.95). Leu110 variant carriers had a reduced risk of primary outcome (hazard ratio 0.68, 95% confidence interval 0.47–0.998). Subgroup analysis revealed this finding to be more pronounced in verapamil SR-assigned patients (hazard ratio 0.587, 95% confidence interval 0.33–1.04) compared with atenolol-assigned patients (hazard ratio 0.946, 95% confidence interval 0.56–1.59). No difference was seen in the occurrence of the primary outcome compared by codon 65 genotype.
Conclusions
Our findings suggest that KCNMB1 genotype influences responsiveness to verapamil SR and risk of adverse cardiovascular outcomes.
Keywords: KCNMB1, polymorphism, verapamil SR
Introduction
The large-conductance calcium and voltage-dependent potassium (BK) channel found in vascular smooth muscle is comprised of pore-forming-α and regulatory-β1 subunits [1]. The BK channel, particularly the β1 subunit, functions in a negative feedback mechanism to enhance calcium sensitivity, decrease cell excitability, and limit smooth muscle contraction [1]. The gene that encodes the β1 subunit of the BK channel is KCNMB1, located on chromosome 5q34. KCNMB1 knockout mice have decreased calcium sensitivity, elevated blood pressure (BP), and cardiac hypertrophy [2,3].
KCNMB1 has two commonly occurring, nonsynonymous, single nucleotide polymorphisms (SNPs), Glu65Lys and Val110Leu. The Lys65 variant of the Glu65Lys polymorphism was associated with decreased risk of diastolic hypertension in a Spanish population and has been found to have functional consequences [1]. The variant Lys65 resulted in a BK channel with enhanced calcium sensitivity, which would support more efficient negative feedback on the l-type calcium channels resulting in vascular smooth muscle relaxation [1]. In addition, the protective effect of the Glu65Lys polymorphism on diastolic hypertension appears to interact with age and sex, with the most significant effect being in women > 54 years of age [4]. Furthermore, 5-year risk of cardiovascular events appears to be significantly lower in the Lys65 carriers compared with the Glu65 homozygous individuals [4]. In a study of healthy twins, the Val110Leu polymorphism was one of four polymorphisms associated with heart rate variability and baroreflex activity [5].
Calcium channel blockers are commonly used in the treatment of hypertension and angina but the response is widely variable. For example, a study of individuals with hypertension found systolic BP (SBP) response to verapamil to range from a 33 mmHg decline to a 4 mmHg increase, with an average 12 mmHg decrease [6]. A genetic component to this variability likely exists, and identifying sources of variability in response to calcium channel blockers is important, as these agents are among the most widely prescribed drug classes.
Given its relevance to calcium channel blocker pharmacology and role in BP control and adverse outcomes, KCNMB1 is a high priority candidate for containing polymorphisms that could influence interpatient variability in response to calcium antagonists. Accordingly, we investigated whether Glu65Lys and Val110Leu are associated with variable BP response to verapamil SR. We studied patients in a cohort of a recent randomized control trial the INVEST-GENES, GENEtic Substudy of the INternational VErapamil SR-trandolapril STudy (INVEST) [7]. INVEST participants who were at selected study sites for which the University of Florida IRB served as the site IRB were invited to join the Genetics Substudy. All of these sites were in the United States and Puerto Rico. No unique design issues or additional data collection were seen in INVEST-GENES versus INVEST, other than the collection of a genomic DNA sample. Additionally, we sought to determine whether the recently reported association [4] between KCNMB1 genotype and adverse cardiovascular outcomes and the influence of drug treatment on this genetic association could be confirmed in a different laboratory and independent population.
Methods
INVEST clinical trial design overview
The INVEST evaluated BP and adverse outcomes occurring with either an atenolol-based or a verapamil SR-based hypertension treatment strategy in 22 576 patients with documented coronary artery disease (CAD) and hypertension [7]. The design, protocol, and primary outcome have been published in detail elsewhere [7,8]. Briefly, the protocol required patients to be seen at baseline, 6, 12, 18, and 24 weeks, and then every 6 months thereafter until 2 years after the last patient was enrolled. At each visit, patients had BP and heart rate measured, clinical assessment performed, and additional antihypertensive medications added as needed to meet JNC VI BP goals [7,9]. The primary outcome was the first occurrence of death (all cause), nonfatal myocardial infarction (MI), or nonfatal stroke. Clinical Trial Registration Identifier: NCT00133692, URL: http://clinicaltrials. gov/ct/gui/show/NCT00133692?order=5.
INVEST-GENES
Genetic samples were collected from 5979 INVEST patients residing in mainland United States and Puerto Rico. Genomic DNA was collected using buccal cells from mouthwash samples as previously described [10]. All patients provided written informed consent for participation in the genetic substudy and the study was approved by the University of Florida IRB.
As in most recent hypertension trials conducted in high-risk patients, the INVEST protocol permitted entry of patients receiving background antihypertensive therapy, as well as those receiving no drug treatment. Therefore, to study the genetic contribution to BP response to verapamil SR, we analyzed only patients in whom changes in BP could reliably be attributed to verapamil SR. This BP response cohort of INVEST-GENES was made up of (i) the verapamil SR monotherapy group (i.e. those patients who entered INVEST with untreated hypertension who were only prescribed verapamil SR monotherapy at the first study visit) and (ii) the stable background therapy group (i.e. patients receiving antihypertensive therapy at entry which was maintained, with addition of verapamil SR as the only change to their antihypertensive regimen at the first study visit). BP response analyses were conducted in the entire BP response cohort (verapamil SR monotherapy plus stable background therapy groups) and in the verapamil SR monotherapy patients separately.
Tissue samples
Given positive association studies with codon 110 and a lack of published functional data, we chose to undertake allelic expression imbalance (AEI) studies as an initial functional assessment. AEI is observed in target tissues in participants where the studied gene harbors a functional polymorphism that affects gene regulation and mRNA processing. Polymorphisms in coding regions (synonymous or nonsynomous) in a majority of cases affect mRNA folding, and hence have an inherent potential to affect mRNA processing and functions [11].
Alternatively, such polymorphisms could be in linkage disequilibrium (LD) with regulatory polymorphisms. Given the limited tissues of expression for this protein, and the difficulties in studying ion channel function in human target tissues, we chose this approach as a starting point of the potential functional effects of this polymorphism.
Sixty-five left ventricle heart tissues were obtained from patients undergoing heart transplantation at Ohio State University Medical Center, Ohio, USA. Genomic DNA and total RNA were prepared from these tissues as described previously [11,12]. Complementary DNA (cDNA) was generated from 0.5 µg total RNA using oligo-dT and gene-specific primers (5′-GATTGGACTGGAAGAGTGGG) as described in Refs. [11,12].
Nested epidemiological studies
We conducted three studies within the INVEST-GENES patient population. First, we conducted a cohort study where the outcome of interest was SBP response to verapamil SR at 6 weeks and the secondary outcomes were time to BP control and number of drugs required at the time of BP control, compared by KCNMB1 genotype.
Second, we conducted a nested case–control study among the 258 INVEST-GENES patients who experienced a primary outcome event (death, nonfatal MI, or nonfatal stroke) during study follow-up (cases). A total of 813 individuals who did not have an event during study follow-up were frequency matched in a ratio of approximately 3 : 1 to cases for age, sex, and race/ethnicity (controls).
Last, because codon 110 trended toward significance in our case–control study, we conducted a cohort study of all INVEST-GENES patients to evaluate the association of codon 110 genotype with the occurrence of the primary outcome. This cohort study was conducted to evaluate the appropriateness of our case–control sample and to estimate the probability of missing a significant relationship with codon 110, if indeed one existed, because of a lack of power.
Genotyping
Genomic DNA was extracted from buccal cells collected in mouthwash samples according to standard protocols [10]. The Glu65Lys and Val110Leu polymorphisms were genotyped by pyrosequencing (PSQ HS 96A) or Taqman (Applied Biosystem, Foster City, California, USA) methods. The PSQ HS 96 genotyping platform (Biotage AB, Uppsala, Sweden) was used for the pyrosequencing assay (primer sequences available upon request). The codon 65 and codon 110 PCR reactions were carried out using HotStar Taq mix (Qiagen, Valencia, California, USA), 10 pmmol each of forward and reverse primers, water, and 20 ng of genomic DNA. The annealing temperature was 58°C for codon 65 and 63°C for codon 110. The Applied Biosystems 7900 HT SNP genotyping platform was used for the Taqman assay. The PCR primers and probes for KCNMB1 codon 65 and 110 assays (IDs C_302618_10, and C_3026206_1) were purchased from Applied Biosystems (Applied Biosystems, Foster City, California, USA). 5 µl reactions in 384-well plate were prepared and the assays were performed and analyzed according to the manufacturer’s recommendations.
Ancestry informative markers (AIMs) were genotyped using either allele-specific PCR with universal energy transfer labeled primers [13] or competitive allele-specific PCR at Prevention Genetics (Marshfield, Wisconsin, USA).
Haplotypes were computationally derived and pairwise LD (D′) calculated separately for each racial/ethnic group using Polymorphism and Haplotype Analysis Suite (http://ilya.wustl.edu/~pgrn/programs.html).
Quantitative analysis of allelic ratios in genomic DNA and mRNA using SNaPshot
To determine whether Val110Leu affects mRNA level in the heart tissue, we used this SNP as a marker for measuring the amount of mRNA derived from one allele over the other (allelic RNA ratio) in heterozygous samples. Deviation of RNA ratios from DNA ratios demonstrates the presence of AEI, as an indicator of cis-acting functional polymorphisms affecting gene regulation and mRNA processing and turnover [11,14]. Details of the SNaPshot assay procedure have been published [11,12,14]. Briefly, a segment of DNA or cDNA ( ~ 100 bp) surrounding the marker SNP was amplified by PCR using forward primer (5′-TGCTCCTACATCCCAGGCA) and reverse primer (5′-AATTTGGCTCTGACCTTCTCC). Then the PCR product was subjected to a primer extension assay (SNaPshot, Applied Biosystems) using an extension primer (5′-GCCGTCTGGTAATTGTCCA) designed to anneal to the amplified DNA adjacent to the SNP site. Allelic DNA ratios, normalized to 1, served as internal control. Allelic mRNA ratios were normalized by DNA ratios.
Statistical analysis
Baseline characteristics were compared by genotype using χ2 test or analysis of variance, as appropriate. Owing to the low minor allele frequency for both the codon 65 and codon 110 SNPs, we decided a priori to divide patients assuming a dominant model of inheritance with heterozygote patients pooled with the homozygous variant patients for all analyses. Hardy–Weinberg equilibrium was calculated separately by race/ethnicity using χ2 test with one degree of freedom. All statistical analyses were conducted using SAS version 9.1 (Cary, North Carolina, USA) or SPSS version 11.5 (Chicago, Illinois, USA). A P < 0.05 (two-sided) was considered significant for all analyses. BP response after 6 weeks of verapamil SR therapy was compared by genotype using a general linear model with genotype included as a fixed effect adjusted for prespecified baseline covariates of age, sex, race/ethnicity, BP, and body mass index (BMI), all of which in univariable analysis had a P < 0.2 in either verapamil SR monotherapy or all BP response patients. Treatment BPs estimated using least square means (adjusted for above covariates) ± standard error are presented. Secondary BP analyses included time to BP control, defined as the time after receiving drug when BP control ( < 140/90mmHg) was achieved and maintained for at least 50% of subsequent visits, and number of drugs required at time of BP control. Kaplan–Meier analysis was used to estimate time to BP control and comparison between genotype groups were made using a log-rank test. Cox regression modeling using forward inclusion was also performed with the prespecified covariates. Number of drugs at time of BP control was assessed using a cumulative logit model with factors known to influence the number of antihypertensive medications required including the prespecified covariates plus history of renal insufficiency, heart failure, and diabetes. Percent change in SBP in response to verapamil SR was compared among haplotypes (0, 1, or 2 copies) using test for trend.
Unadjusted and adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for occurrence of the primary outcome were calculated using logistic regression for the case–control group. The model contained all covariates from the primary INVEST analysis that were found to influence prognosis (with the exception of US residency because all INVEST-GENES participants were US residents) [15]. These variables include age, sex, race/ethnicity, BMI, smoking, INVEST treatment strategy, previous MI, previous stroke, heart failure, diabetes, renal insufficiency, peripheral vascular disease, baseline SBP, and strategy assignment. Additionally, we included diuretic and angiotensin converting enzyme (ACE) inhibitor use, codon 65 and 110 genotype, and the interaction term between genotype.
A Cox proportional hazards model using forward inclusion with all of the above variables as prespecified covariates was used to evaluate the effect of codon 110 genotype on outcomes and Kaplan–Meier curves were estimated. As the INVEST protocol called for diuretic (HCTZ) and ACE inhibitor (trandolapril) therapy to be added to the primary study drugs for patients failing to achieve BP goals with primary agents or to those with heart failure, renal insufficiency, or diabetes (for trandolapril), these variables were treated as time-dependent covariates in the Cox proportional hazards model. The model was also conducted separately by study strategy (i.e. in the verapamil SR-based group and in the atenolol-based group) and in subgroups by age ( < 70 years and ≥70 years), sex, and race/ethnicity.
To control for the potential of population stratification in our racially and ethnically diverse population, we used a total panel of 87 AIMs, selected to show large allele frequency differences across three parental populations (West Africans, Indigenous Americans, and Europeans) selected from a large panel of over 10 000 SNPs [16]. Maximum likelihood was then used to estimate each patient’s individual genomic ancestry proportions on these three axes and these terms were included in statistical models instead of the race/ethnicity term.
Results
Study population and baseline characteristics
Baseline characteristics for all INVEST-GENES patients are shown in Table 1. As expected, owing to the entry criterion of ≥ 50 years of age, the patients were elderly with a mean age of 66 years. Over half of the patients were female, with a high percentage of Hispanic patients. Baseline characteristics were similar when compared by codon 110 genotypes, with the exception of the variant allele being slightly more frequent in Hispanic and Black patients than White patients. In addition, hypercholesterolemia was present in 55% of Val110Val patients compared with 51.3% of Leu110 carriers (P = 0.03). Baseline characteristics for 1071 patients selected as cases and controls are similar to the overall INVEST-GENES cohort (Table 1). A summary of the other antihypertensive medications patients from the stable background therapy group were receiving at baseline is presented in Table 2.
Table 1.
Baseline characteristics among those with complete Val110Leu genotypea
| Characteristic | Overall INVEST (n = 22,576) | INVEST-GENES (n = 5486) | Cases (n = 255) | Controls (n = 798) |
|---|---|---|---|---|
| Age, mean (SD), years | 66.2 (9.7) | 66.1 (9.7) | 71.6 (9.9) | 69.9 (9.3) |
| Women | 11770 (52.1) | 3047 (55.5) | 131 (51.4) | 400 (50.1) |
| BP, mean (SD), mmHg | ||||
| Systolic | 150.9 (19.5) | 148.0 (18.4) | 150.7 (19.0) | 147.4 (19.1) |
| Diastolic | 87.2 (11.9) | 85.4 (10.7) | 83.6 (11.1) | 83.5 (11.2) |
| Race/ethnicity | ||||
| White | 10925 (48.4) | 2076 (37.8) | 152 (59.6) | 468 (58.7) |
| Black | 3029 (13.4) | 588 (10.7) | 32 (12.6) | 93 (11.7) |
| Hispanic | 8045 (35.6) | 2598 (47.4) | 63 (24.7) | 219 (27.4) |
| Other/multiracial | 577 (2.6) | 224 (4.1) | 8 (3.1) | 18 (2.2) |
| BMI, mean (SD), kg/m2 | 29.2 (7.1) | 29.3 (5.5) | 27.5 (4.8) | 29.1 (5.6) |
| Lys65 allele frequency | n/a | n/a | 0.135 | 0.11 |
| Leu110 allele frequency | n/a | 0.09 | 0.06 | 0.09 |
| Past Medical History | ||||
| Myocardial infarction | 7218 (32.0) | 1281 (23.4) | 95 (37.3) | 236 (29.6) |
| Angina pectoris | 15045 (66.6) | 4070 (74.2) | 151 (59.2) | 503 (63.0) |
| Revascularization > 1 month ago | 6166 (27.3%) | 783 (14.3) | 62 (24.3) | 149 (18.7) |
| Stroke/TIA | 1629 (7.2) | 381 (6.9) | 36 (14.1) | 73 (9.2) |
| Left ventricular hypertrophy | 4948 (21.9) | 823 (15.0) | 45 (17.7) | 139 (17.4) |
| Heart failure (class I–III) | 1256 (5.6) | 179 (3.3) | 28 (10.3) | 32 (4.0) |
| Peripheral vascular disease | 2699 (12.0) | 607 (11.1) | 43 (16.9) | 95 (11.9) |
| Smoking | ||||
| Past | 10454 (46.3) | 2265 (41.3) | 130 (51.0) | 364 (45.6) |
| Within 30 days | 2809 (12.4) | 558 (10.2) | 32 (12.6) | 81 (10.2) |
| Never | 12122 (53.7) | 3221 (58.7) | 125 (49.0) | 434 (54.4) |
| Diabetesb | 6401 (28.4) | 1542 (28.1) | 107 (39.5) | 232 (29.1) |
| Hypercholesterolemiab | 12448 (55.1) | 2998 (54.7) | 159 (62.4) | 497 (62.3) |
| Renal impairmentc | 424 (1.9) | 85 (1.6) | 14 (5.2) | 19 (2.4) |
| Cancer | 760 (3.4) | 222 (4.1) | 20 (7.8) | 46 (5.8) |
| Medication | ||||
| Aspirin/other antiplatelet agent | 12795 (56.7) | 2493 (45.4) | 159 (62.4) | 457 (57.3) |
| Antidiabetic medication | 5428 (24.0) | 1351 (24.6) | 85 (33.3) | 195 (24.4) |
| Any lipid-lowering agent | 8294 (36.7) | 1985 (36.2) | 104 (40.8) | 338 (42.4) |
| Nitrates | 8128 (36.0) | 1540 (28.1) | 91 (35.7) | 243 (30.5) |
BP, blood pressure; BMI, body mass index; n/a, not applicable because entire group not genotyped; TIA, transient ischemic attack.
Values expressed as number (percentage) unless otherwise indicated. Percentages may not equal 100 owing to rounding.
History of or currently taking antidiabetic or lipid-lowering medications.
History of or currently have elevated serum creatinine level but less than 4 mg/dl (< 354 µmol/l).
Table 2.
Antihypertensive medications in stable background therapy group (n = 440)
| Percentage | |
|---|---|
| Verapamil added to monotherapy AH | |
| HCTZ/nonstudy diuretic | 3.4 |
| Trandolapril/other ACEI | 25.9 |
| α-Blocker/other vasodilator | 11.8 |
| Centrally acting AH | 5.5 |
| Other AH | 31.1 |
| Verapamil added to multiple AH ( ≥ 2) | |
| HCTZ + ACEI | 9.3 |
| Diuretic + other AH | 7.3 |
| ACEI + other AH | 3.0 |
| Other combination | 2.7 |
ACEI, angiotensin converting enzyme inhibitor; AH, antihypertensives; HCTZ, hydrochlorothiazide.
Genotyping was successful for codon 65 in 99% of the BP response cohort and 97% of cases (98% overall). For codon 110, genotyping was conducted in the entire INVEST-GENES cohort and was successful in 99% of the BP response cohort and 99% of cases (92% overall). For quality control purposes, 470 samples were genotyped in duplicate with a 99% concordance rate.
All but three of those patients missing genotypes for codon 110 were patients in the overall cohort who did not experience an event. Additional efforts to successfully genotype these samples was deemed unnecessary given the limited power gained with the addition of 490 controls to the existing controls with genotype data. Codon 65 genotype frequencies did not deviate from Hardy–Weinberg equilibrium for any of the racial/ethnic groups for codon 65 (P > 0.14 for all groups). Codon 110 genotypes did not deviate from Hardy–Weinberg equilibrium in Hispanic or Black patients. However, codon 110, deviated among White individuals (P < 0.01). Genotype frequencies are shown in Table 3. The two SNPs were in strong LD in White patients using D′ (D′ = 1), but weakly linked using r2 as a measure of LD (r2 = 0.009). The SNPs were weakly linked in Hispanic (D′ = 0.52, r2 = 0.007) and Black patients (D′ = 0.55, r2 = 0.004) using either measure of LD.
Table 3.
KCNMB1 genotype frequency by race/ethnicity and in INVEST-GENES
| INVEST-GENES | Verapamil monotherapy |
Stable background therapy |
White | Hispanic | Black | Other | |
|---|---|---|---|---|---|---|---|
| Glu65Glu | 1412 (77%) | 121 (75%) | 454 (75%) | 743 (81%) | 466 (73%) | 153 (76%) | 50 (79%) |
| Glu65Lys | 392 (22%) | 40 (24%) | 143 (24%) | 165 (18%) | 169 (26%) | 45 (23%) | 13 (21%) |
| Lys65Lys | 20 (1%) | 2 (1%) | 4 (1%) | 9 (1%) | 9 (1%) | 2 (1%) | 0 |
| Lys65 allele frequency | 0.12 | 0.135 | 0.13 | 0.10 | 0.14 | 0.13 | 0.10 |
| Val110Val | 4591 (84%) | 128 (79%) | 493 (82%) | 1782 (86%) | 2143 (83%) | 480 (82%) | 186 (83%) |
| Val110Leu | 843 (15%) | 32 (20%) | 103 (17%) | 273 (13%) | 432 (17%) | 103 (18%) | 35 (16%) |
| Leu110Leu | 52 (1%) | 2 (1%) | 2 ( < 1%) | 21 (1%) | 23 (1%) | 5 (1%) | 3 (1%) |
| Leu110 allele frequency | 0.09 | 0.105 | 0.09 | 0.08 | 0.09 | 0.10 | 0.09 |
Baseline demographic characteristics did not differ between the 5486 individuals from INVEST-GENES in whom codon 110 genotyping was successful and the 493 individuals in whom genotyping was not successful (data not shown).
As sample sizes for the various analyses were determined by patients’ participation in INVEST, statistical power was estimated post hoc for each subgroup of interest. For the verapamil SR monotherapy patients, we had 80% power with a two-sided α of 0.05 to detect an 8 mmHg difference in treatment SBP between variant carriers and homozygous wild-type individuals for codon 65 and a 9 mmHg difference for codon 110. For the entire BP response cohort of patients, we had 80% power to detect a 5 mmHg difference in SBP for both codon 65 and codon 110. For the case–control logistic regression, we had 80% power to detect an alternative hypothesis with an odds ratio of 1.58 for codon 65 and 1.6 for codon 110. For the entire INVEST-GENES cohort, we had 80% power to detect a hazard ratio of 1.55 for codon 110.
Blood pressure response to verapamil SR
Among INVEST-GENES patients, 163 met criteria for verapamil SR monotherapy patients and 603 met criteria for being considered in the overall BP response cohort. In general, the BP response cohort was similar to the entire INVEST-GENES cohort with a few exceptions. White patients made up 47% of verapamil SR monotherapy patients compared with 39% of the overall BP response cohort and 41% of overall INVEST-GENES. In addition, mean baseline BP was higher in verapamil SR monotherapy patients at 157/91 ± 16/8 mmHg compared with 149/86 ± 18/10 in the overall BP response group and 148/85 ± 18/11 in the entire INVEST-GENES cohort, consistent with the untreated status of the monotherapy groups at study entry.
Verapamil SR-mediated change in blood pressure
Among verapamil SR monotherapy patients, mean SBP response to verapamil SR treatment at 6 weeks adjusted for baseline SBP, age, sex, race, and BMI was 139 ± 1.3 mmHg in Glu65Glu patients, compared with 132 ± 4.0 mmHg in Lys65 carriers, P=0.077. Diastolic blood pressure (DBP) adjusted for the same variables was 80 ± 0.73 mmHg in Glu65Glu patients and 79 ± 1.2 mmHg in Lys65 carriers, P = 0.326 when compared by codon 110 genotype, treatment SBP was 139 ± 1.2 mmHg in Val110Val patients and 133 ± 4.0 mmHg in Leu110 carriers, P = 0.147. DBP was 80 ± 0.71 mmHg in Val110Val patients and 80 ± 1.4 mmHg in Leu110 carriers, P=0.669.
When patients with stable background therapy were considered in addition to the verapamil SR monotherapy patients, neither SBP nor DBP response differed by codon 65 or codon 110 genotype. SBP and DBP were 136 ± 0.9 and 79 ± 0.39 mmHg, respectively, in Glu65Glu patients and 135 ± 1.8 mmHg and 79 ± 0.68 in Lys65 carriers, P > 0.219 for SBP and DBP. Compared by codon 110 genotype, SBP and DBP were 135 ± 0.7 and 79 ± 0.37 mmHg, respectively, in Val110Val patients and 135 ± 1.9 and 79 ± 0.81 mmHg, respectively, in Leu110 carriers, P > 0.764 for SBP and DBP. We did not find a significant interaction between genotype and age or sex (data not shown).
When codon 65 and codon 110 were considered jointly as haplotypes, the analysis was less informative than either single SNP analysis.
Time to blood pressure control
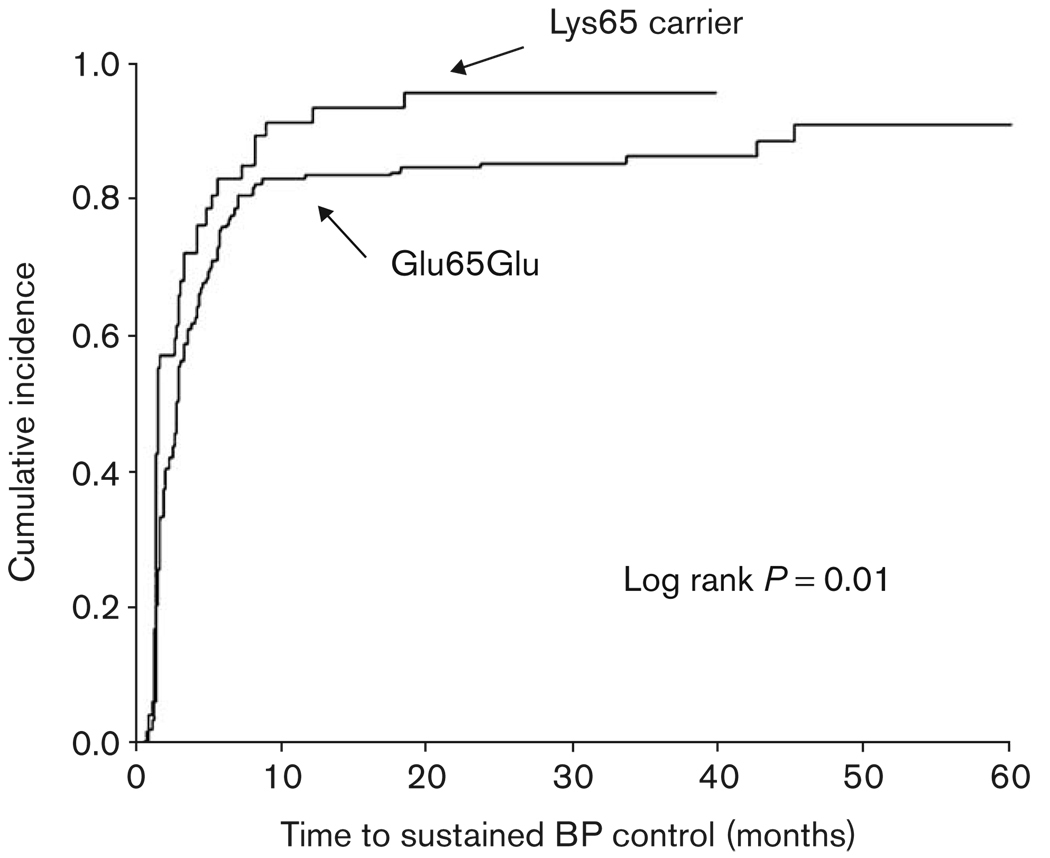
For the time to BP control analyses, 218 patients were available in the verapamil SR monotherapy and 791 were available in the entire BP response group. The median time to achieve BP control (first visit with SBP < 140 and DBP < 90 mmHg and 50% of visits thereafter) among verapamil SR monotherapy patients was 2.83 [interquartile range (IQR) 4.17] months in Glu65Glu patients and only 1.47 (IQR 2.77) months in Lys65 carriers, P = 0.01 (Fig. 1). Median time to BP control in the entire BP response cohort was 2.8 (IQR 1.33) months in Glu65Glu patients and 2.0 (IQR 1.40) months in Lys65 carriers, P = 0.06. Time to BP control did not differ by codon 110 genotype in verapamil SR monotherapy patients or among all BP response patients (data not shown).
Fig. 1.
Kaplan–Meier curves of time to sustained BP control by KCNMB1 codon 65 genotype in verapamil SR monotherapy patients. Sustained BP control was defined as the time point at which BP control ( < 140/90 mmHg) was achieved and maintained for at least 50% of subsequent visits. Log rank P = 0.01.
Number of drugs at time of blood pressure control
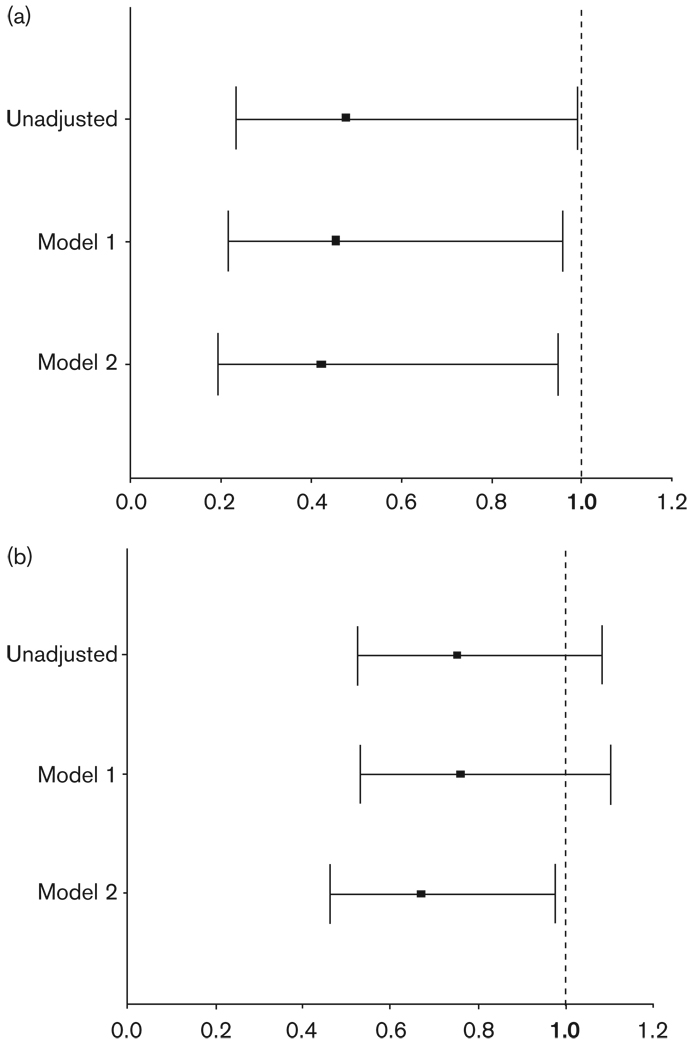
Consistent with the observation that less time was required to achieve BP control, Lys65 variant carrier status was significantly associated with the need for fewer drugs to achieve BP control (OR 0.48, 95% CI 0.23–0.99) in the verapamil SR monotherapy group (Fig. 2a). Additionally, as multiple covariates were added to the model including demographics and disease states, the effect of Lys65 variant carrier status remained significant. The effect of Lys65 had similar trends in the entire BP response cohort, but was less pronounced than in the verapamil SR monotherapy group (Fig. 2b).
Fig. 2.
Odds ratios for Lys65 variant on probability of requiring multiple drugs to achieve BP control. Odds ratios < 1 indicate lower likelihood of Lys65 variant carriers requiring as many drugs as Glu65Glu individuals to achieve BP control. Odds ratios > 1 indicate higher likelihood of requiring more drugs than Glu65Glu individuals. Model 1 is adjusted for age, race, and sex. Model 2 is adjusted for all model 1 variables plus baseline SBP and DBP, BMI, and history of heart failure, diabetes, renal insufficiency, and left ventricular hypertrophy. Panel A is verapamil SR monotherapy patients and Panel B is stable background therapy patients. BP, blood pressure; DBP, diastolic blood pressure; SBP, systolic blood pressure.
Effect of KCNMB1 genotype on primary outcome Case–control group
In the case–control population, the Leu110 variant was associated with a 33% reduced risk of primary outcome (OR 0.66, 95% CI 0.43–1.01). Adjustment for all prespecified covariates did not affect this association (OR 0.64, 95% CI 0.41–1.01). When the AIMs were included in the model, we found a similar association (OR 0.59, 95% CI 0.34–0.99), suggesting that our findings are not spurious owing to population stratification. No significant association between codon 65 and occurrence of primary outcomes was observed (OR 1.15, 95% CI 0.83–1.61).
Entire INVEST-GENES cohort
To ensure that our case–control study design was representative of the entire INVEST-GENES cohort and that we were not missing important associations because of lack of power, we genotyped all the INVEST-GENES patients for the Val110Leu polymorphism.
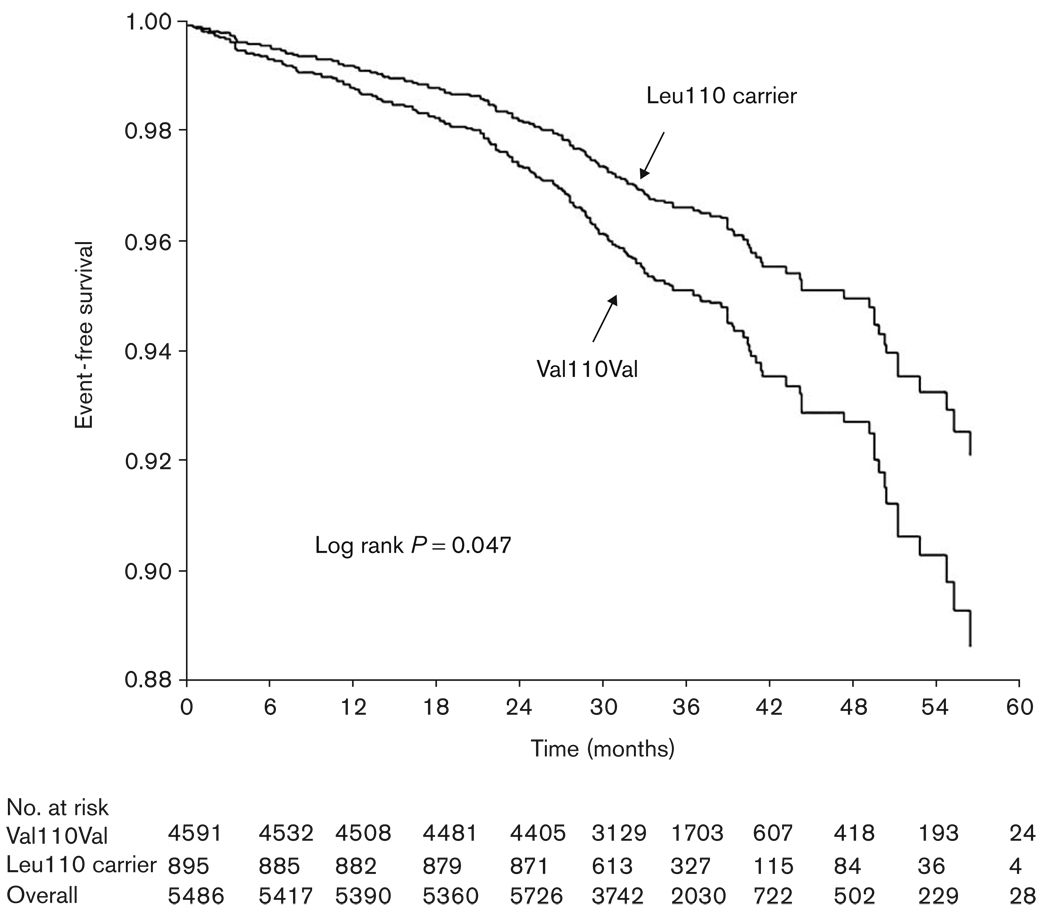
Unadjusted for differences in baseline characteristics, codon 110 variant carrier state was associated with a 32% lower risk of primary outcome compared to noncarriers (HR 0.68, 95% CI 0.47–0.99) (Fig. 3). After full adjustment for prespecified covariates, this risk still trended toward significance (HR 0.72, 95% CI 0.49–1.05). Results were consistent when the AIMs were included in the modeling instead of the race/ethnicity term (HR 0.66, 95% CI 0.42–1.03). Haplotype analysis did not provide additional information over consideration of genotype alone (data not shown). As a secondary analysis, we tested the components of the primary outcome individually and found a significant reduction in the risk of nonfatal MI in Leu110 variant carriers in the unadjusted model that trended toward significance in the fully adjusted model (Table 4).
Fig. 3.
Kaplan–Meier curve of time to primary outcome event by Val110Leu variant carrier status in all INVEST-GENES cohort. Black line represents Leu110 carriers, grey line represents Val110Val; Log rank P = 0.047.
Table 4.
Primary and secondary outcomes by codon 110 genotype
| Leu110 carrier (n = 895) | Val110Val (n = 4591) | Unadjusted HR (95% CI) for variant carrier status |
Adjusteda HR (95% CI) for variant carrier status |
|
|---|---|---|---|---|
| Primary outcome event | 30 (0.55) | 225 (4.1) | 0.682 (0.466, 0.998) | 0.719 (0.491, 1.053) |
| Secondary outcomes | ||||
| Death (all cause) | 13 (0.24) | 85 (1.6) | 0.792 (0.442, 1.419) | 0.814 (0.453, 1.463) |
| Nonfatal MI | 6 (0.11) | 74 (1.4) | 0.415 (0.181, 0.954) | 0.448 (0.195, 1.031) |
| Nonfatal stroke | 13 (0.24) | 72 (1.3) | 0.925 (0.513, 1.670) | 0.984 (0.544, 1.779) |
ACE, angiotensin converting enzyme; BMI, body mass index; CI, confidence interval; HR, hazard ratio; MI, myocardial infarction; SBP, systolic blood pressure.
Adjusted for: age, sex, race/ethnicity, BMI, smoking, INVEST treatment strategy, previous myocardial infarction, previous stroke, heart failure, diabetes, renal insufficiency, baseline SBP, diuretic use, and ACE inhibitor use.
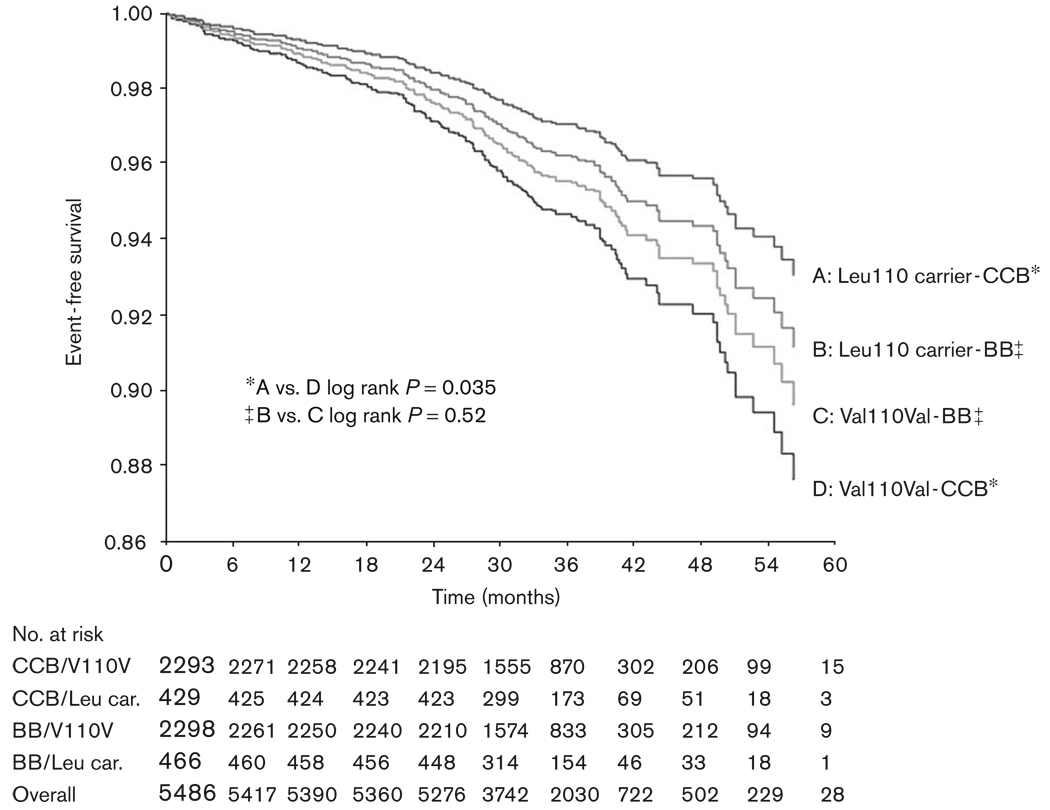
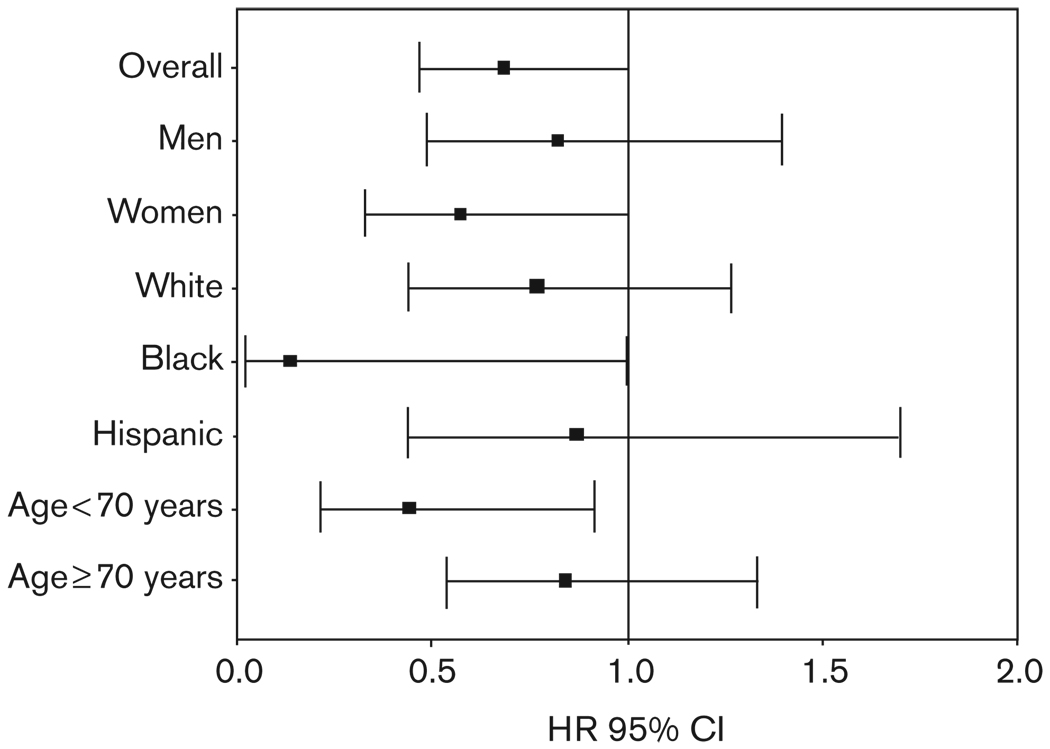
Although the interaction term was not significant between treatment strategy and codon 110 genotype, when the analyses were conducted separately by treatment strategy, the protective effect of the KCNMB1 variant allele appeared to be confined to those randomized to verapamil SR (HR 0.55, 95% CI 0.31–0.97), with no benefit in the atenolol-treated group (HR 0.84, 95% CI 0.51–1.41) (FIG. 4). The association between codon 110 genotype and adverse outcomes was more prominent in women (HR 0.57, 95% CI 0.33–0.99), individuals < 70 years of age (HR 0.45, 95% CI 0.22–0.92), and Black individuals (HR 0.14, 95% CI 0.02–1.00) (Fig. 5).
Fig. 4.
Kaplan–Meier curve of time to primary outcome event by Val110Leu and treatment strategy in all INVEST-GENES cohort. CCB indicates verapamil SR-based strategy; BB indicates atenolol-based strategy; Leu car. indicates Leu 110 variant carrier.
Fig. 5.
Association between Val110Leu variant carrier status and primary outcome in subgroups.
Val110Leu functional assessment
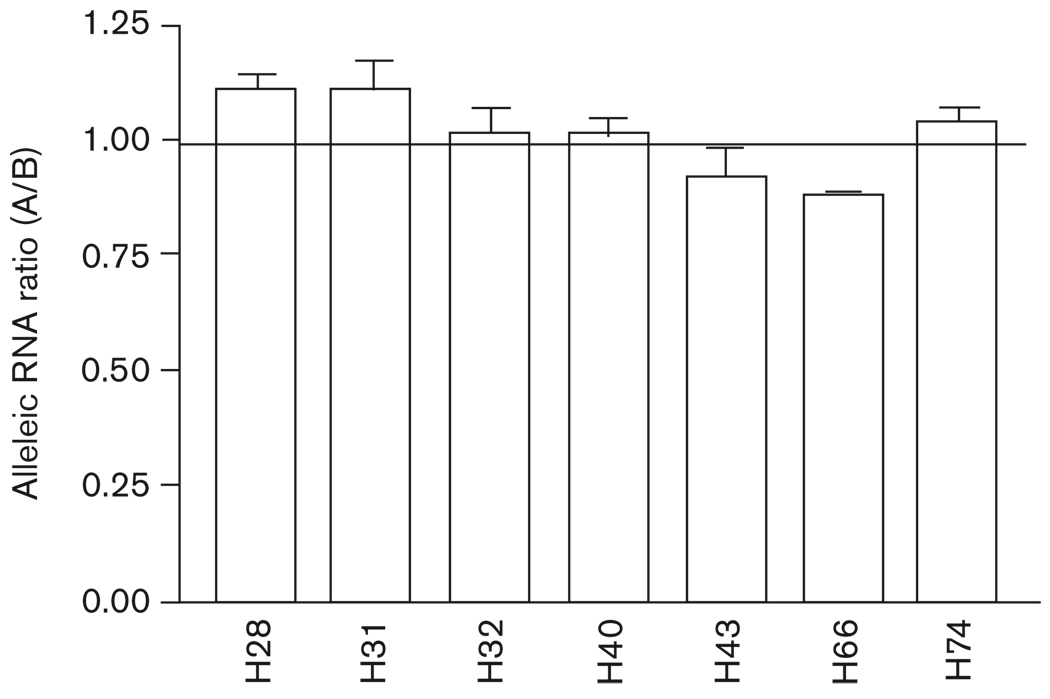
Of 65 heart tissue samples, seven samples were heterozygous for Val110Leu. We measured allelic expression in these heterozygous DNA and RNA (converted to cDNA) samples. The presence of any polymorphism that affects transcription or mRNA processing should result in discrepant DNA and cDNA allelic ratios (AEI). Shown in Fig. 6, RNA ratios from all seven samples did not differ from DNA ratios when using Val110Leu as a marker, indicating that this nonsynonymous SNP was not associated with different mRNA levels of KCNMB1 in cardiac tissues. Using another two SNPs in 3′-UTR (rs2656842, rs2656841) as marker SNPs, we confirmed this result in three samples also heterozygous for these two SNPs (data not shown).
Fig. 6.
Allelic RNA ratios for KCNMB1 measured with marker SNP Val110Leu in heterozygous human heart tissues. Allelic expression ratios of DNA and RNA were measured with SNaPshot assay. RNA ratios were normalized by that of DNA, which were set to 1. Allelic DNA ratio is 1 ± 0.13 (SD). A, major allele; B, minor allele.
Discussion
Our findings suggest that in hypertensive patients with CAD, two nonsynonymous polymorphisms in the KCNMB1 gene, Glu65Lys and Val110Leu, may account for a component of the interpatient variability in BP response to verapamil SR. Additionally, our findings suggest that the Val110Leu polymorphism may be associated with adverse outcomes. These conclusions are based on results of several different types of analyses. First, we found that Lys65 variant carriers achieved BP goals more rapidly than Glu65 homozygote individuals in the verapamil SR monotherapy group and required fewer drugs to achieve BP control. These results are consistent with functional studies of this polymorphism indicating that the variant BK channel has enhanced calcium sensitivity [1] and would be predicted to have enhanced responsiveness to calcium channel blockers. All of our BP response analyses (i.e. change in SBP at 6 weeks, time to BP control, and number of drugs needed for BP control) consistently identified Lys65 variant carriers to have the most favorable BP response to verapamil SR in patients on verapamil SR monotherapy. This effect was also consistently weaker when individuals being treated with other antihypertensive agents on stable background therapy were added to the analyses. This may be because the effect of KCNMB1 on antihypertensive response to verapamil SR is small and may be overcome by the addition of other antihypertensive medications. For example, individuals on multidrug regimens had lower BP at study entry and may be less dependent on vascular smooth muscle relaxation from the BK channel pathway because their other antihypertensive agents are decreasing vascular resistance through other mechanisms. On the other hand, patients on verapamil SR monotherapy might be more dependent on the BK channel pathway to decrease vascular tone, allowing us to observe the apparent pharmacogenetic effect of the Glu65Lys SNP. The second potential reason our findings with antihypertensive response to verapamil SR were only identified in the smaller group of patients is because of a type I error. We do not anticipate this to be the case given that our findings are in line with Fernandez–Fernandez and Senti and colleagues [1,4].
The second major finding in our study is that the Leu110 variant allele is associated with a decreased risk of adverse outcome among patients with hypertension and CAD. Interestingly, this finding was significant only in the patients randomized to a verapamil SR-based treatment strategy, suggesting a pharmacogenetic effect, with the majority of the protective effect of the variant occurring only when the patients are treated with a calcium channel blocker. If these findings were replicated in an independent population, it would suggest that hypertensive CAD patients who are Leu110 carriers should receive a calcium channel blocker.
Our secondary analyses suggest that the protective effect of the Leu110 allele on the occurrence of adverse outcomes may be driven by a reduction in nonfatal MI. The mechanism for this reduction in risk, however, remains unclear. The subgroup analyses, suggested that greater protective effective effect associated with the variant allele was found in women, individuals less than 70 years, and Black individuals. Although these findings are hypothesis generating, they could be viewed as consistent with a previous study that found the association of codon 65 with hypertension to be strongest among older women [4]. Thus, it is possible that the effects of this gene’s variants are greater in women and with increased age.
To our knowledge, this is the first report to date to evaluate the association of the Val110Leu polymorphism with either the response to verapamil SR or adverse outcomes. Although evidence for the functional role of the gene and the codon 65 SNP in hypertension exist (as discussed in the introduction), the functional significance of Val110Leu remains unclear. The codon 110 SNP represents a conservative amino acid substitution, but functional changes at the protein level cannot be ruled out. Alternatively, any nucleotide change in the transcribed region has the potential to affect mRNA folding, processing, turnover, and translation [11]. Regulatory polymorphisms, including those affecting mRNA processing, are prevalent in determining human phenotypic variations [17–19]. Therefore, we measured allelic KCNMB1 mRNA expression in human heart tissues from transplant patients, using the codon 110 SNP as the marker. Differential expression from the two alleles, or AEI, is a definitive indicator of the presence of a cis-acting factor. This approach did not detect the presence of significant AEI, indicating that the codon 110 SNP does not affect mRNA expression levels in the myocardium. The use of human failing heart tissues obtained before transplantation, rather than vascular smooth muscle, however, could confound the interpretation of the results because mRNA expression (including allelic expression) may be tissue-dependent. Therefore, measuring myocardial allelic expression represents but a first step in clarifying the potential function of the Val110Leu polymorphism. Whether Val110Leu is functional owing to altered expression in vascular tissue, altered protein function owing to amino acid change or another mechanism, or is merely a tag for the true functional SNP, however, remains unknown. One other group has reported a genetic association with Val110Leu, specifically that Val110Leu heterozygote individuals had higher heart rate variability and baroreflex sensitivity in normotensive individuals [5]. These results could be considered consistent with our findings given increased heart rate variability is associated with improved outcomes in epidemiological studies and Leu110 carriers had a better prognosis than Val110Val individuals in our study. Furthermore, because the r2 measures of LD between codons 65 and 110 were so low, it seems unlikely that the differences in genetic associations between these SNPs (i.e. codon 65 with BP response and codon 110 with adverse outcomes) are accounted for by LD between the two SNPs. Additionally, haplotype analysis considering the joint effects of these SNPs afforded no further improvement in defining the genetic association. In fact, the consideration of both SNPs only weakened the associations. Therefore, these data suggest that the mechanisms of association for the Glu65Lys and BP response and the Val110Leu and outcomes response to verapamil are different. As discussed above additional research is needed to define the functional basis for the codon 110 associations.
The association of the Glu65Lys polymorphism with the BP response to verapamil SR was not linked to a reduction in clinical outcomes. Given that the BP effect was only evident in those individuals on verapamil SR monotherapy (n = 163), it, however, is likely that this effect was not present in a large enough subset to be seen in the overall INVEST-GENES cohort.
Conclusion
Our data suggest that variability in KCNMB1 is associated with the antihypertensive response to verapamil SR, and also with adverse cardiovascular outcomes in patients having hypertension with CAD. Of particular interest regarding the latter finding is that the protective effect of the variant allele was largely confined to who received verapamil SR monotherapy. To our knowledge, this is the first study evaluating the influence of KCNMB1 on the antihypertensive response to verapamil SR and highlights the importance of considering pharmacodynamic pathway genes.
Acknowledgements
The authors would like to gratefully acknowledge Ms Lynda Stauffer and Ms Lauren Burt and Dr James Moss for their laboratory assistance and Dr Issam Zineh for his insightful comments regarding the manuscript. This study was supported by NIH grants HL 74730, HL 69758, RR017568, and by a grant from Abbott Laboratories.
References
- 1.Fernandez-Fernandez JM, Tomas M, Vazquez E, Orio P, Latorre R, Senti M, et al. Gain-of-function mutation in the KCNMB1 potassium channel subunit is associated with low prevalence of diastolic hypertension. J Clin Invest. 2004;113:1032–1039. doi: 10.1172/JCI20347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner R, Perez GJ, Bonev AD, Eckman DM, Kosek JC, Wiler SW, et al. Vasoregulation by the beta1 subunit of the calcium-activated potassium channel. Nature. 2000;407:870–876. doi: 10.1038/35038011. [DOI] [PubMed] [Google Scholar]
- 3.Pluger S, Faulhaber J, Furstenau M, Lohn M, Waldschutz R, Gollasch M, et al. Mice with disrupted BK channel beta 1 subunit gene feature abnormal Ca(2 + ) spark/STOC coupling and elevated blood pressure. Circ Res. 2000;87:E53–E60. doi: 10.1161/01.res.87.11.e53. [DOI] [PubMed] [Google Scholar]
- 4.Senti M, Fernandez-Fernandez JM, Tomas M, Vazquez E, Elosua R, Marrugat J, et al. Protective effect of the KCNMB1 E65K genetic polymorphism against diastolic hypertension in aging women and its relevance to cardiovascular risk. Circ Res. 2005;97:1360–1365. doi: 10.1161/01.RES.0000196557.93717.95. [DOI] [PubMed] [Google Scholar]
- 5.Gollasch M, Tank J, Luft FC, Jordan J, Maass P, Krasko C, et al. The BK channel beta 1 subunit gene is associated with human baroreflex and blood pressure regulation. J Hypertens. 2002;20:927–933. doi: 10.1097/00004872-200205000-00028. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen BN, Parker RB, Noujedehi M, Sullivan JM, Johnson JA. Effects of COER-verapamil on circadian pattern of forearm vascular resistance and blood pressure. J Clin Pharmacol. 2000;40(12 Pt 2):1480–1487. [PubMed] [Google Scholar]
- 7.Pepine CJ, Handberg EM, Cooper-DeHoff RM, Marks RG, Kowey P, Messerli FH, et al. A calcium antagonist vs. a non-calcium antagonist hypertension treatment strategy for patients with coronary artery disease. The International Verapamil-Trandolapril Study (INVEST): a randomized controlled trial. JAMA. 2003;290:2805–2816. doi: 10.1001/jama.290.21.2805. [DOI] [PubMed] [Google Scholar]
- 8.Pepine CJ, Handberg-Thurmond E, Marks RG, Conlon M, Cooper-DeHoff R, Volkers P, et al. Rationale and design of the International Verapamil SR/Trandolapril Study (INVEST): an Internet-based randomized trial in coronary artery disease patients with hypertension. J Am Coll Cardiol. 1998;32:1228–1237. doi: 10.1016/s0735-1097(98)00423-9. [DOI] [PubMed] [Google Scholar]
- 9.The sixth report of the Joint National Committee on prevention, detection, evaluation, and treatment of high blood pressure. Arch Intern Med. 1997;157:2413–2446. doi: 10.1001/archinte.157.21.2413. [DOI] [PubMed]
- 10.Andrisin TE, Humma LM, Johnson JA. Collection of genomic DNA by the noninvasive mouthwash method for use in pharmacogenetic studies. Pharmacotherapy. 2002;22:954–960. doi: 10.1592/phco.22.12.954.33598. [DOI] [PubMed] [Google Scholar]
- 11.Johnson AD, Wang D, Sadee W. Polymorphisms affecting gene regulation and mRNA processing: broad implications for pharmacogenetics. Pharmacol Ther. 2005;106:19–38. doi: 10.1016/j.pharmthera.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Pinsonneault J, Nielsen CU, Sadee W. Genetic variants of the human H+ /dipeptide transporter PEPT2: analysis of haplotype functions. J Pharmacol Exp Ther. 2004;311:1088–1096. doi: 10.1124/jpet.104.073098. [DOI] [PubMed] [Google Scholar]
- 13.Myakishev MV, Khripin Y, Hu S, Hamer DH. High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome Res. 2001;11:163–169. doi: 10.1101/gr.157901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Wang D, Johnson AD, Papp AC, Sadee W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280:32618–32624. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]
- 15.Pepine CJ, Kowey PR, Kupfer S, Kolloch RE, Benetos A, Mancia G, et al. Predictors of adverse outcome among patients with hypertension and coronary artery disease. J Am Coll Cardiol. 2006;47:547–551. doi: 10.1016/j.jacc.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 16.Shriver MD, Mei R, Parra EJ, Sonpar V, Halder I, Tishkoff SA, et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genomics. 2005;2:81–89. doi: 10.1186/1479-7364-2-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rockman MV, Wray GA. Abundant raw material for cis-regulatory evolution in humans. Mol Biol Evol. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- 18.Buckland PR. Allele-specific gene expression differences in humans. Hum Mol Genet. 2004;13(Spec No 2):R255–R260. doi: 10.1093/hmg/ddh227. [DOI] [PubMed] [Google Scholar]
- 19.Bray NJ, Buckland PR, Owen MJ, O’Donovan MC. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum Genet. 2003;113:149–153. doi: 10.1007/s00439-003-0956-y. [DOI] [PubMed] [Google Scholar]