Abstract
Objective
Mutations in the GJB2, GJB6 and SLC26A4 genes are a frequent cause of hearing loss in a number of populations. However, little is known about the genetic causes of hearing loss in the Korean population.
Methods
We sequenced the GJB2 and GJB6 genes to examine the role of mutations in these genes in twenty-two hearing loss patients. We also sequenced the SLC26A4 gene in seven patients with inner ear malformations, including enlarged vestibular aqueduct (EVA) revealed by computer tomography.
Results
Coding sequence mutations in GJB2 were identified in 13.6% of the patients screened. Two different mutations, 235delC and T86R were found in three unrelated patients. The 235delC was the most prevalent mutation with an allele frequency of 6.9% in our patient group. No mutations, including 342 kb deletion, were found in GJB6 gene. Three different variants of SLC26A4 were identified in the EVA patients, including one novel mutation. Four EVA patients carried two mutant alleles of SLC26A4, and at least one allele in all patients was the H723R mutation, which accounted for 75% of all mutant alleles.
Conclusions
Our results suggest that GJB2 and SLC26A4 mutations together make up a major cause of congenital hearing loss in the Korean population. Further studies may be able to identify other common variants that account for a significant fraction of hearing loss in the Korean population.
Keywords: Connexin, Hearing loss, Koreans, Mutation, Pendred Syndrome
1. Introduction
Congenital hearing loss is one of the most common sensory impairments in humans, occurring in approximately one in 1000 live births. More than 50% of these cases are hereditary [1]. Of the hearing loss disorders attributable to genetic causes, approximately 70% are classified as nonsyndromic since hearing impairment is the only symptom while 30% are classified as syndromic and are associated with other clinical features. Hereditary hearing loss can also be divided by the mode of inheritance. The majority of cases, up to ~80%, shows autosomal recessive transmission, while 15–20% of the cases are autosomal dominant, and the remaining show X-linked or mitochondrial inheritance [2].
To date, approximately 120 genetic loci have been mapped as sites of genes causally related to nonsyndromic hearing loci, and 41 different causative genes, encoding proteins with a wide variety of functions, have been identified [3]. Among these genes, mutations in the GJB2 (connexin 26) (OMIM 121011) are responsible for approximately half of cases of autosomal recessive nonsyndromic hearing loss (ARNSHL) in many populations [3, 4]. So far, more than 100 different mutations in GJB2 have been reported to be associated with recessive hearing loss. However, a few GJB2 mutations have been described to be associated with autosomal dominant hearing loss [5]. Among the recessive mutations, 35delG is the most frequent in Caucasians, 167delT in Ashkenazi Jews, 235delC in East Asians, and R143W in Africans, suggesting the existence of founder effects in different ethnic groups [6–18]. In addition, some studies have shown the high prevalence of the IVS1 + 1G to A mutation in the non-coding part of the GJB2 gene [19, 20].
Recently, it has been shown that mutations in GJB6, the gene encoding connexin 30, are another common cause of hearing loss [21]. In particular, a 342 kb deletion disrupting the GJB6 gene (delGJB6-D13S1830) is common in patients from Spain, France, Germany, the United Kingdom, Brazil, and Ashkenazi Jews [22–26]. This mutation is found in both monogenic del (GJB6-D13S1830) and digenic GJB2/del (GJB6-D13S1830) patterns of inheritance.
Despite the high prevalence of GJB2 and GJB6 gene mutations in western populations, these mutations seem to account for a smaller percentage of hereditary hearing loss in Asian populations. It has been suggested that mutations in GJB2 account for 10% of hearing loss in Korea and 20–30% in Japan, but the frequency of GJB6 mutations in these populations is unknown [27, 28]. In contrast, mutations of SLC26A4 gene which are responsible for both Pendred syndrome (PS) and nonsyndromic hearing loss with enlarged vestibular aqueduct (EVA) (DFNB4, OMIM 600791) appear to be common in Asian populations. Such mutations account for approximately 5% of recessive hearing loss in Asians [29]. Moreover, Park et al. (2004) have shown that a high proportion of Korean hearing loss patients with EVA have mutations of this gene, and five mutations account for 80% of all mutant alleles [30].
Since no reports have yet provided a systematic study of the prevalence of GJB2, GJB6 and SLC26A4 gene mutations in Korean population, we decided to analyze the each gene in the Korean hearing loss patients in order to better characterize the prevalence of GJB2, GJB6, and SLC26A4 gene mutations in the Korean population.
2. Materials and Methods
2.1. Patients
A total of twenty-nine subjects with hearing impairment were recruited from the Department of Otorhinolaryngology-Head and Neck Surgery, Kyungpook National University Hospital, Daegu, Korea. There were 15 female and 14 male patients, with an age range of 2 to 51 years (mean age, 18.2 years). Nineteen of the cases had affected relatives and 10 were sporadic.
After a complete physical and otoscopic examination, audiological studies were carried out including pure tone audiometry, tympanometry, or auditory brainstem response in a sound treated room. Pure-tone average (PTA) was calculated as an average of the threshold measured at 0.5, 1.0, 2.0 and 3.0 KHz for comparing subgroups of patients. The level of hearing loss is described as follows depending on PTA: normal hearing, below 20 dB; mild hearing impairment; 21 to 40 dB; moderate hearing impairment, 41 to 70 dB; severe hearing impairment, 71 to 95 dB; and profound hearing impairment above 95 dB. In 20 of 29 patients with hearing loss it was possible to perform temporal bone computed tomography (CT) to search for inner ear malformation. All patients with unilateral hearing loss, past history of meningitis, head trauma, noise trauma, infectious disease associated with hearing loss, and other acquired hearing loss were excluded from the study. DNA samples from 50 unrelated normal Koreans were used as controls. All participants provided written informed consent according to the protocol approved by the Ethics Committee of Kyungpook National University Hospital prior to the study.
2.2. Molecular Genetic Analysis
We sequenced the GJB2 and GJB6 genes in twenty-two hearing loss patients. We also sequenced the SLC26A4 gene in seven patients with inner ear malformations revealed by computer tomography.
Genomic DNA was extracted from peripheral blood using FlexiGene DNA extraction kit (QIAGEN, Hilden, Germany).
For the analysis of the GJB2, the entire region including non-coding exon1 were amplified by polymerase chain reaction (PCR) using the appropriate intronic primer sets reported previously [19, 31]. For the study of GJB6 and SLC26A4 genes, all the exons including exon-intron bounderies of each gene were amplified by polymerase chain reaction (PCR) using the primer sets reported previously [24, 32] and designed by Primer 3.0 software. PCR was performed in a total of 25 µl reaction, containing 0.2 mM of each deoxynucleotide, 15 pmol of each forward and reverse primers, 1.0–1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.75 U of Taq DNA polymerase (Solgent, Korea), and 25 ng of genomic DNA. PCR conditions were as follows: 35 cycles of denaturation at 95°C for 30 sec; annealing at 55°C or 57°C, depending on the primers for 30 sec; and extension at 72°C for 1 min. The first denaturation step and the last extension step were at 95°C for 2 min and 72°C for 10 min, respectively. Five microliters of the PCR products were separated and visualized on a 2% agarose gel. Fifteen microliters of this PCR product were then treated with 0.3U of shrimp alkaline phosphatase (USB) and 3U of exonuclease I (USB) at 37°C for 1 hr, followed by incubation at 80°C for 15 min. This was diluted with an equal volume of dH2O, and 6 µl was used for the final sequencing reaction. Sequencing reactions were performed in both directions on the PCR products in reactions containing 5 pmol of primer, 0.25 µl of ABI Big Dye Terminator v3.1 Cycle Sequencing Kit, and 1 µl of a dilution buffer (400 mM Tris-HCl, pH 9 and 10 mM MgCl2). Cycling conditions were 95°C for 2 min followed by 35 cycles of 94°C for 20 sec, 55°C for 20 sec, and 60°C for 4 min. Sequencing reaction products were ethanol precipitated, and the pellets were resuspended in 10 µl of formamide loading dye. An ABI 3130XL DNA sequencer was used to resolve the products, and data was analyzed by using ABI sequencing Analysis (v.5.0) and LASERGENE-SeqMan software.
The 342 kb deletion of GJB6 gene (GJB6-D13S1830), was assayed by PCR using an internal primer that is located in the deleted segment of GJB6 as previously reported by del Castillo et al., 2002 [24]. Using these primers, two different PCR products were obtained, providing discrimination of homozygous wild-type (651bp product), homozygous deleted (405bp product), and heterozygous (both products) in a single reaction. The PCR products were electrophoresed on 2% agarose gel and visualized with ethidium bromide staining.
One novel missense mutation in SLC26A4 gene was evaluated for potential pathogenicity using SIFT (http://blocks.fhcrc.org/sift/SIFT.html) and SNPs3D (http://www.snps3d.org) and by multiple sequence alignment using ClustalX (www-igbmc.u-strasbg.fr/BioInfo/ClustalX/Top.html).
3. Results
3.1. Clinical features of patients with congenital hearing loss
Twenty-nine patients had congenital moderate to profound sensorineural hearing loss. The PTAs of these patients were over 40 dB in both right and left ears, and 6 patients (20.7%) had moderate hearing loss, 12 patients (41.4%) had severe hearing loss, and in 11 patients (37.9%), hearing loss was profound.
Twenty patients with hearing loss were evaluated by temporal bone CT. Seven of these patients were found to have inner ear malformations. Three of 7 patients had bilateral EVA which was defined as greater than 1.5 mm width of the vestibular aqueduct at the middle portion between the endolymphatic sac and the vestibule, as described by Valvassori and Clemis [33]. Incomplete partition of cochlea described as Mondini’s malformation [34] was found in 2 in 7 patients and 2 of 7 had Mondini’s malformation with bilateral EVA.
3.2. Molecular Analyses
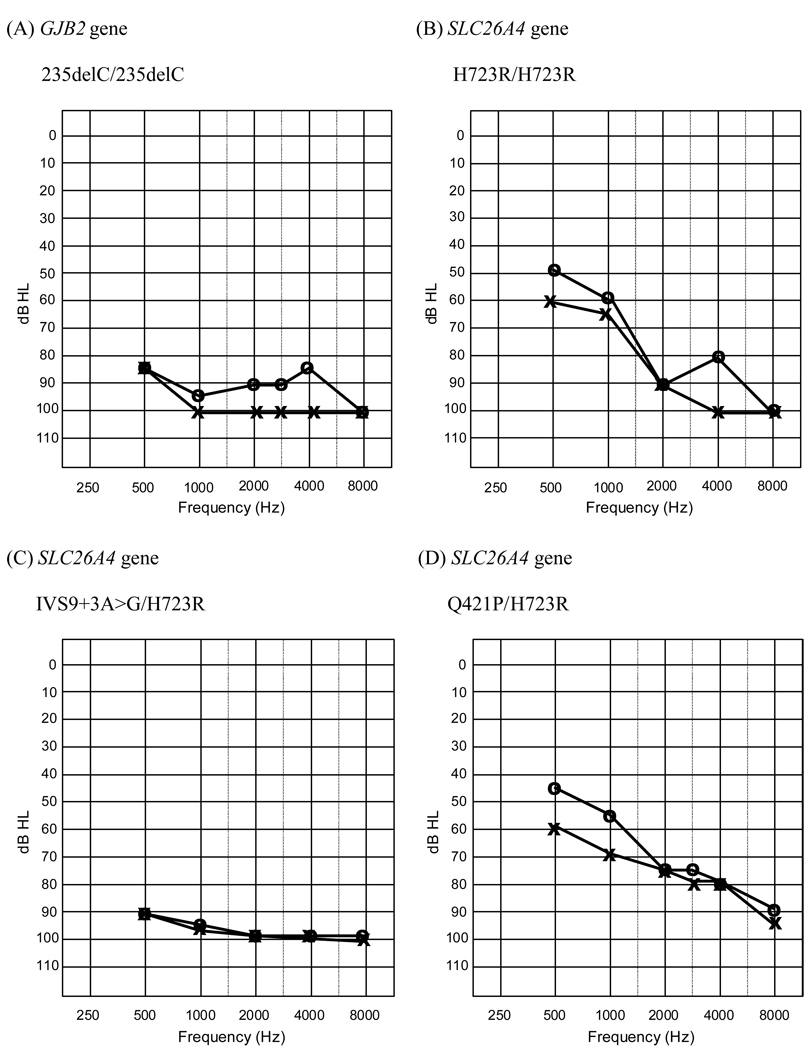
Table 1 shows the demographic features, audiometric features, radiological abnormalities, and genotypes of the patients with hearing impairments in the present study. Two patients homozygous for 235delC had profound hearing loss over 90 dB HEARING LOSS in pure tone audiometry respectively. The PTAs of 7 patients with inner ear malformations revealed moderate to profound hearing loss. Figure 1 shows the audiograms of the different genotypic classes of patients.
Table 1.
Demographic features, audiometric features, radiological abnormalities, familial history and genotypes of the patients
| Case | Sex | Age | Degree of hearing loss * |
Temporal bone CT |
Familial Hx |
Mutation |
||
|---|---|---|---|---|---|---|---|---|
| Rt | Lt | GJB2 | SLC26A4 | |||||
| 1 | M | 10 | 70 | 71.25 | normal | + | - | |
| 2 | F | 19 | 95 | 100 | - | + | - | |
| 3 | F | 5 | NR** | NR** | normal | − | - | |
| 4 | M | 6 | 96.25 | 96.25 | EVA | − | IVS9+3A>G/ H723R |
|
| 5 | M | 2 | 87.5 | 93.75 | mondini | + | - | |
| 6 | M | 5 | 97.5 | 95 | mondini | + | - | |
| 7 | M | 5 | NR | NR | normal | − | - | |
| 8 | F | 4 | NR | NR | EVA, mondini | − | - | |
| 9 | F | 14 | 48.75 | 48.75 | normal | + | - | |
| 10 | M | 7 | 65 | 53.75 | normal | + | - | |
| 11 | F | 38 | 42.5 | 48.75 | - | + | - | |
| 12 | F | 27 | 87.5 | 85 | normal | + | - | |
| 13 | M | 23 | 95 | 96.25 | normal | + | - | |
| 14 | M | 9 | NR | NR | normal | + | 235delC/235delC | |
| 15 | F | 3 | NR** | NR** | normal | − | - | |
| 16 | M | 10 | 77.5 | 82.5 | EVA | − | Q421P/H723R | |
| 17 | F | 2 | 80** | 80** | normal | + | T86R/T86R | |
| 18 | F | 11 | 90 | 96.25 | normal | − | 235delC/235delC | |
| 19 | F | 15 | 96.25 | 91.25 | - | + | - | |
| 20 | F | 9 | 71.25 | 77.5 | EVA | − | H723R/H723R | |
| 21 | M | 12 | 100 | 82.5 | normal | − | - | |
| 22 | F | 51 | 87.5 | 91.25 | - | + | - | |
| 23 | F | 49 | 100 | 100 | - | + | - | |
| 24 | M | 44 | 85 | 85 | - | + | - | |
| 25 | M | 47 | 98 | 98.75 | - | + | - | |
| 26 | M | 42 | 93.75 | 97 | - | + | - | |
| 27 | F | 2 | 90** | 80** | normal | + | - | |
| 28 | F | 6 | 80** | 50** | EVA, mondini | − | H723R/H723R | |
| 29 | F | 44 | 46.25 | 43.75 | - | + | - | |
Notes: pure tone average (dB HL) at 500, 1000, 2000, and 3000 Hz except the patients with ** marking
the result of auditory brainstem responses Rt; right ear, Lt; left ear, Hx; history, NR; no response, EVA; bilateral enlarged vestibular aqueduct, mondini; mondini’s malformation.
Figure 1.
The audiograms of the different genotypic classes of patientsshow a diversity of hearing threshold (no audiogram is available for T86R, only ABR data).
GJB2 gene
Molecular analysis of the entire GJB2 gene including non-coding exon 1 was performed in twenty-two patients. Two different mutations were identified in the coding region (Table 1). Two patients were homozygous for 235delC, which has been described as the most frequent mutation in the Korean population [27]. We also detected a coding sequence variant which was homozygous for threonine to arginine change at the position 86 (T86R) in one patient. This variant was not observed in 100 normal Korean chromosomes and encoded changes in amino acids that are conserved across humans, mice, and rats, further supporting our hypothesis that it is causative for hearing loss. Three previously described polymorphisms, V27I, E114G, and I203T, were also found in both patients and in controls. The minor allele frequencies of V27I and E114G variants were 22.9 and 6.3% respectively, but I203T was found only in two heterozygotes, suggesting it is rare in Koreans.
GJB6 gene
The 342 kb deletion in GJB6 was not detected in the patients and 50 normal Korean controls examined. We also sequenced the coding region of GJB6, but did not find any variants in these patients.
SLC26A4 gene
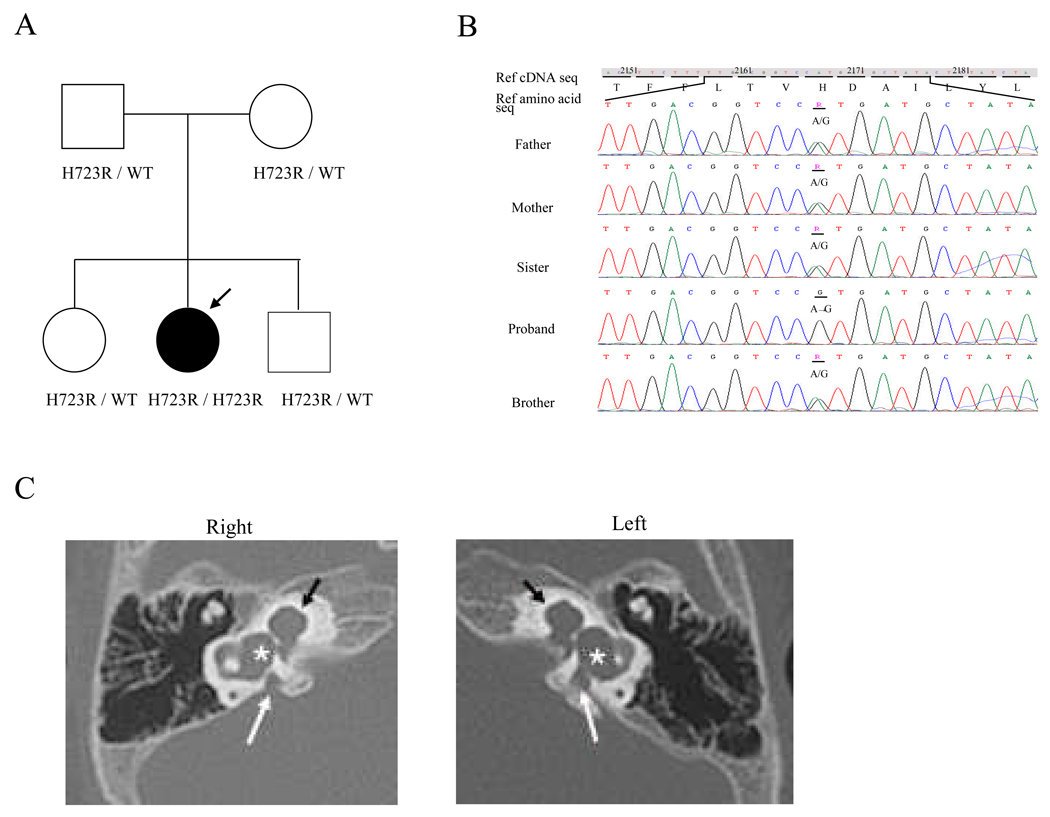
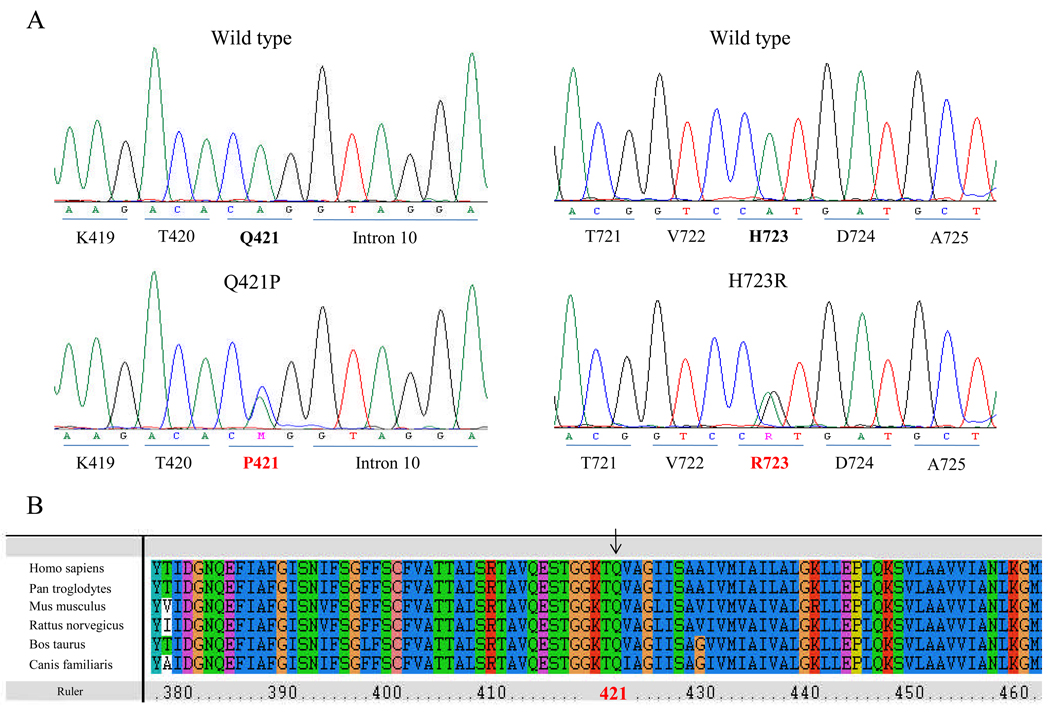
We observed two known pathogenic variants in the SLC26A4 gene in unrelated Korean patients with EVA. One was an adenine to guanine change at position +3 transition donor splice site of intron 9 (IVS9+3A>G), while the second was a histidine to arginine change at position 723 (H723R). H723R was the most common mutation we observed, accounting for 75% of all the mutant alleles. H723R was identified in the homozygous state in two patients. Figure 2 shows the sequence of this gene in one of the patients and his family members and images showing bilateral EVA with Mondini’s malformation from temporal bone CT scans. H723R was observed in the compound heterozygous state in other two patients. In one of the heterozygote cases, the SLC26A4 mutation at IVS9+3A>G was observed on the other chromosome. In the other case, a novel mutation, Q421P, was observed in the other copy of the SLC26A4 gene (Fig. 3A). This variant was not observed in 100 normal Korean control chromosomes, and has not been reported in other populations. To evaluate the evolutionary conservation of the amino acid affected by this mutation, we made an alignment of pendrin amino acid sequences of human with other species. As shown in Figure 3B, glutamine at this position is evolutionarily conserved. In three patients with inner ear malformation , no SLC26A4 mutation was detected.
Figure 2.
(A) Pedigree of family with the H723R mutation. Filled symbol indicates affected person. The proband is marked with an arrow. (B) Direct sequencing chromatographs of the H723R mutation. Father, mother, sister, brother panel: the heterozygote of the H723R mutation. Proband panel: the homozygote of the H723R mutation. Missense mutation at nucleotide 2168 in exon 19 substituted arginine for histidine at amino acide 723. (C) Temporal bone CT images of the inner ear in the proband with bilateral EVA and Mondini’s malformation. The vestibular aqueducts in both ears are enlarged (white arrow). And the lateral semicircular canal and vestibule of both ears are dilated (asterisk). There are cochleae with incomplete partition (black arrow). Interscalar septum is absent between the each cochlear turns.
Figure 3.
(A) Sequencing traces showing the Q421P/H723R compound heterozygote. (B) Multiple-sequence alignment of selected proteins with significant sequence homology to human pendrin. The amino-acid sequence of human pendrin is aligned relative to the sequences of other species. The arrow denotes the missnese mutation detected in our study.
4. Discussion
We investigated the DNA sequence of the GJB2, GJB6, and SLC26A4 genes in Korean hearing loss patients. Mutations in the GJB2 gene are a major contributor to autosomal recessive hearing loss and also constitute a small percentage of autosomal dominant hearing loss in many populations [35]. In this study, we screened the entire GJB2 gene including the non-coding region for the first time in Korean hearing loss patients. GJB2 mutations were identified in three unrelated patients. All of these mutations occur in the coding region, and they occurred at two different sites, 235delC and T86R. The 235delC, which we observed 4 times, was the most prevalent mutation. The T86R mutation has been reported previously in a Japanese group but was found in Koreans for the first time in this study [7]. The T86R is located in the second transmembrane domain (M2) which is essential for oligomerization to form the connexon hemichannel [36]. The substitution of threonine for arginine results in a change from polar uncharged amino acid to positively charged amino acid, which may affect the tertiary structure and function of the connexin protein. Absence of this mutation in the normal population and a previously observed missense mutation at the same position suggest that this mutation is causative for hearing loss. Additional proof of this hypothesis could come from a functional assay for this mutation in vitro. Our study further supports the view that mutations in GJB2 gene are a less common cause of hearing loss in East Asian populations compared to Caucasians. We did not find GJB6 coding region variants or del(GJB6-D13S1830) in any of the patients. Our result is consistent with the results of Liu et al. (2002), who did not detect this mutation in a sample from the Chinese population [37]. Since a number of other studies also found a low frequency or absence of this mutation in different populations, including Austrian, Russian, and Moroccan [23, 38–41], this deletion may be restricted to certain populations [23].
We performed nucleotide sequence analysis of SLC26A4 gene from seven unrelated Korean patients affected with inner ear malformations. Four patients carried three mutant alleles, and the H723R mutation accounted for the majority of all mutant alleles. Previous studies demonstrated that different ethnic populations can have their own distinctive, diverse series of SLC26A4 mutant alleles, which can include one or a few prevalent founder alleles [31, 42]. The mutations, IVS8+1G>A, L236P, and T416P, account for nearly half of all SLC26A4 mutation alleles in Caucasians. In contrast, H723R, IVS7-2A>G, and IVS9+3A>G account for the majority of SLC26A4 mutations in East Asian populations. Combined with the results of Park et al. (2004) and Cho et al. (2006), our data indicate the H723R mutation accounts for approximately 40% of SLC26A4 mutations in Korea, which is similar to the 53% observed in Japanese patients [30, 43]. Our observations confirm that H723R is the most frequently detected mutation in both Korean and Japanese populations, perhaps as a result of a common founder effect. In addition to two common mutations, the novel Q421P mutation was identified in this study. The Q421R mutation at the same position was reported in the study of Prasad et al. (2004) [44] and the functional studies of the similar location such as T416P mutation suggest that the change of amino acid at this position occurred the cause of disease [45, 46]. We demonstrated severe to profound hearing impairment in 7 patients with inner ear malformations, consistent with the results of previous studies [30, 42]. The phenotypic features of SLC26A4 mutations are variable, ranging from typical PS to nonsyndromic recessive hearing loss associated with EVA (DFNB4). In this study, none of 7 patients with bilateral EVA had a goiter. Although these young patients do not have a goiter, the clinical diagnosis is difficult to differentiate from PS because the goiter usually is not developed until adolescence [47]. Thus, it will be of interest to follow up the clinical features of these patients as they progress to adolescence.
In conclusion, GJB2 and SLC26A4 mutations appear to be responsible for major cause of congenital hearing loss in the Korean population. Further studies may be able to identify other common variants that account for a significant fraction of hearing loss in the Korean population.
Acknowledgements
We thank all the subjects who participated in the present study. We also thank Young-Jun Choi and Jung Ree Lee for collecting patients and family members. This work was supported by the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korean government (MOST) (No. R01-2008-000-10431-0). This work was supported by BioMedical Research Institute grant, Kyungpook National University Hospital (2007). SY Choi and JW Bae were supported by a the Korean Ministry of Education through the Brain Korea 21 project.
References
- 1.Marazita ML, Ploughman LM, Rawlings B, Remington E, Arnos KS, Nance WE. Genetic epidemiological studies of early-onset deafness in the U.S. school-age population. Am. J. Med. Genet. 1993;46(5):486–491. doi: 10.1002/ajmg.1320460504. [DOI] [PubMed] [Google Scholar]
- 2.Morton NE. Genetic epidemiology of hearing impairment. Ann. N. Y. Acad. Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- 3.Vam Camp G, Smith RJH. Hereditary Hearing Loss Homepage. http://webh01.ua.ac.be/hhh.
- 4.Zelante L, Gasparini P, Estivill X, Melchionda S, D'Agruma L, Govea N, et al. Connexin26 mutations associated with the most common form of non-syndromic neurosensory autosomal recessive deafness (DFNB1) in Mediterraneans. Hum. Mol. Genet. 1997;6(9):1605–1609. doi: 10.1093/hmg/6.9.1605. [DOI] [PubMed] [Google Scholar]
- 5.Connexin-Deafness Homepage. http://davinci.crg.es/deafness/
- 6.Van Laer L, Coucke P, Mueller RF, Caethoven G, Flothmann K, Prasad SD, et al. A common founder for the 35delG GJB2 gene mutation in connexin 26 hearing impairment. J. Med. Genet. 2001;38(8):515–518. doi: 10.1136/jmg.38.8.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohtsuka A, Yuge I, Kimura S, Namba A, Abe S, Van Laer L, et al. GJB2 deafness gene shows a specific spectrum of mutations in Japan, including a frequent founder mutation. Hum. Genet. 2003;112(4):329–333. doi: 10.1007/s00439-002-0889-x. [DOI] [PubMed] [Google Scholar]
- 8.Brobby GW, Muller-Myhsok B, Horstmann RD. Connexin 26 R143W mutation associated with recessive nonsyndromic sensorineural deafness in Africa. N. Engl. J. Med. 1998;338(8):548–550. doi: 10.1056/NEJM199802193380813. [DOI] [PubMed] [Google Scholar]
- 9.Green GE, Scott DA, McDonald JM, Woodworth GG, Sheffield VC, Smith RJ. Carrier rates in the midwestern United States for GJB2 mutations causing inherited deafness. JAMA. 1999;281(23):2211–2216. doi: 10.1001/jama.281.23.2211. [DOI] [PubMed] [Google Scholar]
- 10.Morell RJ, Kim HJ, Hood LJ, Goforth L, Friderici K, Fisher R, et al. Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N. Engl. J. Med. 1998;339(21):1500–1505. doi: 10.1056/NEJM199811193392103. [DOI] [PubMed] [Google Scholar]
- 11.Gasparini P, Rabionet R, Barbujani G, Melchionda S, Petersen M, Brondum-Nielsen K, et al. High carrier frequency of the 35delG deafness mutation in European populations. Genetic Analysis Consortium of GJB2 35delG. Eur. J. Hum. Genet. 2000;8(1):19–23. doi: 10.1038/sj.ejhg.5200406. [DOI] [PubMed] [Google Scholar]
- 12.Kudo T, Ikeda K, Kure S, Matsubara Y, Oshima T, Watanabe K, et al. Novel mutations in the connexin 26 gene (GJB2) responsible for childhood deafness in the Japanese population. Am. J. Med. Genet . 2000;90(2):141–145. doi: 10.1002/(sici)1096-8628(20000117)90:2<141::aid-ajmg10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 13.Lerer I, Sagi M, Malamud E, Levi H, Raas-Rothschild A, Abeliovich D. Contribution of connexin 26 mutations to nonsyndromic deafness in Ashkenazi patients and the variable phenotypic effect of the mutation 167delT. Am. J. Med. Genet. 2000;95(1):53–56. doi: 10.1002/1096-8628(20001106)95:1<53::aid-ajmg11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 14.Sobe T, Erlich P, Berry A, Korostichevsky M, Vreugde S, Avraham KB, et al. High frequency of the deafness-associated 167delT mutation in the connexin 26 (GJB2) gene in Israeli Ashkenazim. Am. J. Med. Genet. 1999;86(5):499–500. [PubMed] [Google Scholar]
- 15.Sobe T, Vreugde S, Shahin H, Berlin M, Davis N, Kanaan M, et al. The prevalence and expression of inherited connexin 26 mutations associated with nonsyndromic hearing loss in the Israeli population. Hum. Genet. 2000;106(1):50–57. doi: 10.1007/s004390051009. [DOI] [PubMed] [Google Scholar]
- 16.Kudo T, Ikeda K, Oshima T, Kure S, Tammasaeng M, Prasansuk S, Matsubara Y. GJB2 (connexin 26) mutations and childhood deafness in Thailand. Otol. Neurotol. 2001;22(6):858–861. doi: 10.1097/00129492-200111000-00025. [DOI] [PubMed] [Google Scholar]
- 17.Wang YC, Kung CY, Su MC, Su CC, Hsu HM, Tsai CC, et al. Mutations of Cx26 gene (GJB2) for prelingual deafness in Taiwan. Eur. J. Hum. Genet. 2002;10(8):495–498. doi: 10.1038/sj.ejhg.5200838. [DOI] [PubMed] [Google Scholar]
- 18.Yan D, Park HJ, Ouyang XM, Pandya A, Doi K, Erdenetungalag R, et al. Evidence of a founder effect for the 235delC mutation of GJB2 (connexin 26) in east Asians. Hum. Genet. 2003;114(1):44–50. doi: 10.1007/s00439-003-1018-1. [DOI] [PubMed] [Google Scholar]
- 19.Santos RL, Aulchenko YS, Huygen PL, van der Donk KP, de Wijs IJ, Kemperman MH, et al. Hearing impairment in Dutch patients with connexin 26 (GJB2) and connexin 30 (GJB6) mutations. Int. J.Pediatr. Otorhinolaryngol. 2005;69(2):165–174. doi: 10.1016/j.ijporl.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 20.Seeman P, Sakmaryova I. High prevalence of the IVS 1 + 1 G to A/GJB2 mutation among Czech hearing impaired patients with monoallelic mutation in the coding region of GJB2. Clin. Genet. 2006;69(5):410–413. doi: 10.1111/j.1399-0004.2006.00602.x. [DOI] [PubMed] [Google Scholar]
- 21.Grifa A, Wagner CA, D'Ambrosio L, Melchionda S, Bernardi F, Lopez-Bigas N, et al. Mutations in GJB6 cause nonsyndromic autosomal dominant deafness at DFNA3 locus. Nat. Genet. 1999;23(1):16–18. doi: 10.1038/12612. [DOI] [PubMed] [Google Scholar]
- 22.Bolz H, Schade G, Ehmer S, Kothe C, Hess M, Gal A. Phenotypic variability of non-syndromic hearing loss in patients heterozygous for both c.35delG of GJB2 and the 342-kb deletion involving GJB6. Hear. Res. 2004;188(1–2):42–46. doi: 10.1016/S0378-5955(03)00346-0. [DOI] [PubMed] [Google Scholar]
- 23.del Castillo I, Moreno-Pelayo MA, Del Castillo FJ, Brownstein Z, Marlin S, Adina Q, et al. Prevalence and evolutionary origins of the del(GJB6-D13S1830) mutation in the DFNB1 locus in hearing-impaired subjects: a multicenter study. Am. J. Hum .Genet. 2003;73(6):1452–1458. doi: 10.1086/380205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.del Castillo I, Villamar M, Moreno-Pelayo MA, del Castillo FJ, Alvarez A, Telleria D, et al. A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N. Engl. J. Med. 2002;346(4):243–249. doi: 10.1056/NEJMoa012052. [DOI] [PubMed] [Google Scholar]
- 25.Lerer I, Sagi M, Ben-Neriah Z, Wang T, Levi H, Abeliovich D. A deletion mutation in GJB6 cooperating with a GJB2 mutation in trans in non-syndromic deafness: A novel founder mutation in Ashkenazi Jews. Hum. Mutat. 2001;18(5):460. doi: 10.1002/humu.1222. [DOI] [PubMed] [Google Scholar]
- 26.Pallares-Ruiz N, Blanchet P, Mondain M, Claustres M, Roux AF. A large deletion including most of GJB6 in recessive non syndromic deafness: a digenic effect? Eur. J. Hum. Genet. 2002;10(1):72–76. doi: 10.1038/sj.ejhg.5200762. [DOI] [PubMed] [Google Scholar]
- 27.Park HJ, Hahn SH, Chun YM, Park K, Kim HN. Connexin26 mutations associated with nonsyndromic hearing loss. Laryngoscope. 2000;110(9):1535–1538. doi: 10.1097/00005537-200009000-00023. [DOI] [PubMed] [Google Scholar]
- 28.Abe S, Usami S, Shinkawa H, Kelley PM, Kimberling WJ. Prevalent connexin 26 gene (GJB2) mutations in Japanese. J. Med. Genet. 2000;37(1):41–43. doi: 10.1136/jmg.37.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, et al. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J. Med. Genet. 2003;40(4):242–248. doi: 10.1136/jmg.40.4.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park HJ, Lee SJ, Jin HS, Lee JO, Go SH, Jang HS, et al. Genetic basis of hearing loss associated with enlarged vestibular aqueducts in Koreans. Clin. Genet. 2005;67(2):160–165. doi: 10.1111/j.1399-0004.2004.00386.x. [DOI] [PubMed] [Google Scholar]
- 31.Scott DA, Kraft ML, Carmi R, Ramesh A, Elbedour K, Yairi Y, et al. Identification of mutations in the connexin 26 gene that cause autosomal recessive nonsyndromic hearing loss. Hum. Mutat. 1998;11(5):387–394. doi: 10.1002/(SICI)1098-1004(1998)11:5<387::AID-HUMU6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 32.Everett LA, Glaser B, Beck JC, Idol JR, Buchs A, Heyman M, et al. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS) Nat. Genet. 1997;17(4):411–422. doi: 10.1038/ng1297-411. [DOI] [PubMed] [Google Scholar]
- 33.Valvassori GE, Clemis JD. The large vestibular aqueduct syndrome. Laryngoscope. 1978;88(5):723–728. doi: 10.1002/lary.1978.88.5.723. [DOI] [PubMed] [Google Scholar]
- 34.Mondini C. Minor works of Carlo Mondini: the anatomical section of a boy born deaf. Am. J. Otol. 1997;18(3):288–293. [PubMed] [Google Scholar]
- 35.Kelley PM, Harris DJ, Comer BC, Askew JW, Fowler T, Smith SD, Kimberling WJ. Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. Am. J. Hum. Genet. 1998;62(4):792–799. doi: 10.1086/301807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar NM, Gilula NB. The gap junction communication channel. Cell. 1996;84(3):381–388. doi: 10.1016/s0092-8674(00)81282-9. [DOI] [PubMed] [Google Scholar]
- 37.Liu XZ, Xia XJ, Ke XM, Ouyang XM, Du LL, Liu YH, et al. The prevalence of connexin 26 ( GJB2) mutations in the Chinese population. Hum. Genet. 2002;111(4–5):394–397. doi: 10.1007/s00439-002-0811-6. [DOI] [PubMed] [Google Scholar]
- 38.Gunther B, Steiner A, Nekahm-Heis D, Albegger K, Zorowka P, Utermann G, Janecke A. The 342-kb deletion in GJB6 is not present in patients with non-syndromic hearing loss from Austria. Hum. Mutat. 2003;22(2):180. doi: 10.1002/humu.9167. [DOI] [PubMed] [Google Scholar]
- 39.Erbe CB, Harris KC, Runge-Samuelson CL, Flanary VA, Wackym PA. Connexin 26 and connexin 30 mutations in children with nonsyndromic hearing loss. Laryngoscope. 2004;114(4):607–611. doi: 10.1097/00005537-200404000-00003. [DOI] [PubMed] [Google Scholar]
- 40.Gazzaz B, Weil D, Rais L, Akhyat O, Azeddoug H, Nadifi S. Autosomal recessive and sporadic deafness in Morocco: high frequency of the 35delG GJB2 mutation and absence of the 342-kb GJB6 variant. Hear. Res. 2005;210(1–2):80–84. doi: 10.1016/j.heares.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 41.Posukh O, Pallares-Ruiz N, Tadinova V, Osipova L, Claustres M, Roux AF. First molecular screening of deafness in the Altai Republic population. BMC Med. Genet. 2005;6:12. doi: 10.1186/1471-2350-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang JJ, Tsai CC, Hsu HM, Shiao JY, Su CC, Li SY. Hearing loss associated with enlarged vestibular aqueduct and Mondini dysplasia is caused by splice-site mutation in the PDS gene. Hear. Res. 2005;199(1–2):22–30. doi: 10.1016/j.heares.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Cho MA, Jeong SJ, Eom SM, Park HY, Lee YJ, Park SE, et al. The H723R mutation in the PDS/SLC26A4 gene is associated with typical Pendred syndrome in Korean patients. Endocrine. 2006;30(2):237–243. doi: 10.1385/ENDO:30:2:237. [DOI] [PubMed] [Google Scholar]
- 44.Prasad S, Kolln KA, Cucci RA, Trembath RC, Van Camp G, Smith RJ. Pendred syndrome and DFNB4-mutation screening of SLC26A4 by denaturing high-performance liquid chromatography and the identification of eleven novel mutations. Am. J. Med. Genet. A. 2004;124(1):1–9. doi: 10.1002/ajmg.a.20272. [DOI] [PubMed] [Google Scholar]
- 45.Scott DA, Wang R, Kreman TM, Andrews M, McDonald JM, Bishop JR, et al. Functional differences of the PDS gene product are associated with phenotypic variation in patients with Pendred syndrome and non-syndromic hearing loss (DFNB4) Hum. Mol. Genet. 2000;9(11):1709–1715. doi: 10.1093/hmg/9.11.1709. [DOI] [PubMed] [Google Scholar]
- 46.Yoon JS, Park HJ, Yoo SY, Namkung W, Jo MJ, Koo SK, et al. Heterogeneity in the processing defect of SLC26A4 mutants. J. Med. Genet. 2008 doi: 10.1136/jmg.2007.054635. [DOI] [PubMed] [Google Scholar]
- 47.Fraser GR. Association of Congenital Deafness with Goitre (Pendred's Syndrome) a Study of 207 Families. Ann. Hum. Genet. 1965;28:201–249. doi: 10.1111/j.1469-1809.1964.tb00479.x. [DOI] [PubMed] [Google Scholar]